- 1Program of Veterinary Clinical Studies, Graduate School, Kasetsart University, Nakorn Pathom, Thailand
- 2Department of Cardio-Thoracic Technology, Faculty of Allied Health Sciences, Naresuan University, Phitsanulok, Thailand
- 3Department of Large Animal and Wildlife Clinical Sciences, Faculty of Veterinary Medicine, Kasetsart University, Nakorn Pathom, Thailand
Background: Myosin-binding protein C3 A31P (MYBPC3-A31P) missense mutation is a genetic deviation associated with the development of hypertrophic cardiomyopathy (HCM) in Maine Coon cats. The standard detection of the MYBPC3-A31P mutation is complicated, time-consuming, and expensive. Currently, there has been a focus on the speed and reliability of diagnostic tools. Therefore, this study aimed to develop a loop-mediated isothermal amplification assay (LAMP) coupled with a lateral flow dipstick (LFD) test to detect MYBPC3-A31P mutations in Maine Coon cats.
Materials and Methods: Fifty-five Maine Coon cats were enrolled in this study, and blood samples were collected. MYBPC3-A31P was genotyped by DNA sequencing. Primers for LAMP with a LFD test were designed. The optimal conditions were determined, including temperature and time to completion for the reaction. The sensitivity of A31P-LAMP detection was compared between agarose gel electrophoresis (the standard method) and the LFD test. The A31P-LAMP-LFD test was randomly performed on seven cats (four with the A31P mutation and three wild-type cats).
Results: The A31P-LAMP procedure was able to distinguish between cats with MYBPC3-A31P wild-type cats and MYBPC3-A31P mutant cats. The LAMP reactions were able to be completed in 60 min at a single temperature of 64◦C. Moreover, this study demonstrated that A31P-LAMP coupled with the LFD test allowed for A31P genotype detection at a lower DNA concentration than agarose gel electrophoresis.
Discussions: This new A31P-LAMP with a LFD test is a successful and reliable assay with a rapid method, cost-effectiveness, and low requirements for sophisticated equipment for the detection of MYBPC3-A31P mutations. Thus, this assay has excellent potential and can be recognized as a novel screening test for hypertrophic cardiomyopathy associated with MYBPC3-A31P mutations in felines.
Introduction
Hypertrophic cardiomyopathy (HCM) is the most common hereditary cardiovascular disease in cats (1–4). The HCM phenotype has a prevalence of approximately 15% in cats (5). Moreover, the HCM phenotype in cats is related to an increase in age. Cats with clinical HCM commonly demonstrated left-sided congestive heart failure due to an impairment of diastolic function such as respiratory distress, cardiogenic pulmonary edema, pleural effusion, syncope, hypothermia, and atrial thromboembolism (ATE) (6–9). Specific breeds are highly associated with HCM development, especially the Maine Coon (10) and Ragdoll (11).
Previous research investigated the cause of HCM in cats. Myosin binding protein C3 A31P (MYBPC3-A31P), a single nucleotide polymorphism (SNP), was possibly associated with HCM development in cats, especially Maine Coon cats (12). MYBPC3, the sarcomeric protein in the myocardium, plays a crucial role in heart muscle contraction. Hence, MYBPC3-A31P mutation disturbs normal heart function and leads to cardiac concentric hypertrophy (12, 13).
The diagnosis of the HCM phenotype routinely relies on echocardiographic measurements. While detection of the SNP missense mutation MYBPC3-A31P is required for genetic screening in Maine Coon cats with familial HCM. However, cats without MYBPC3-A31P mutation may develop HCM. For the detection of SNP mutations, DNA sequencing is widely recognized as the gold standard technique (14). However, due to the inconvenience of DNA sequencing, requiring labor-intensive skills, prohibitive cost, and a long time, numerous researchers have developed different methods for SNP detection, such as allele-specific loop-mediated isothermal amplification (AS-LAMP) (15), microarray genotyping (16), molecular beacon genotyping (17), allele-specific polymerase chain reaction (AS-PCR) (18), and high-resolution melting (HRM) (19). Based on advanced genetic research, LAMP is one of the accepted powerful methods for SNP detection. Applying a constant single temperature, LAMP can be used for point-of-care (POC) diagnostic tests owing to the utilization of simple instruments such as thermocyclers, heated blocks, or water baths. The advantages of LAMP are rapid detection with high accuracy because of four to six specific primers used for LAMP and isothermal amplification as a one-step process (20, 21).
In contrast, one disadvantage of LAMP is the challenge of designing a specific primer set. The detection of LAMP products is also usually performed by agarose gel electrophoresis. However, this method uses complex procedures and requires specialized equipment. The other commonly used methods for LAMP product verification are the visualization of colorimetric and lateral flow dipstick (LFD) tests, which are simple and straightforward for carrying out POC tests in a small animal hospital or small laboratory unit (22). Nonetheless, the application of LAMP combined with LFD for the detection of MYBPC3-A31P mutations has not yet been reported. Therefore, this study aimed to develop a specific and rapid diagnostic tool for detecting MYBPC3-A31P mutations in Maine Coon cats using a LAMP assay coupled with an LFD.
Materials and Methods
Animals and Sample Collection
Fifty-five Maine Coon cats were recruited into this study. The procedures of animal use were permitted by Kasetsart University Institutional Animal Care and Use Committee, Kasetsart University, Bangkok, Thailand (Approval number: ACKU 62-VET-059). The blood samples were collected from venous vessels for 2–3 ml per cat and stored in an ethylenediaminetetraacetic acid (EDTA) tube. The stored blood was aliquoted and kept at −20°C until DNA extraction. As described previously, the process of DNA extraction was conducted via Blood Genomic DNA Extraction Mini Kit (Favorgen, Taiwan) regarding manufacturer's recommendations (23).
Genotyping of MYBPC3-A31P Mutation
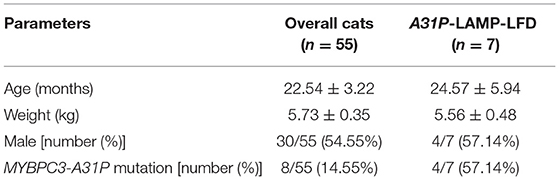
The protocol for genotyping of MYBPC3-A31P mutation was obtained from a previous study by Godiksen et al. (24). The details of forwarding and reverse primers were represented in Table 1. In brief, the steps of PCR included heat activation at 95°C for 15 min, 35 cycles of 3 steps of (i) denaturation at 95°C for 30 s, (ii) annealing at 58°C for 30 s, and (iii) extension at 72°C for 1 min and final extension at 72°C for 10 min. The size of the PCR product was 242 bp. PCR purification was then conducted following manufacturer's protocol (Favorgen, Taiwan). The Sanger sequencing was applied to determine the nucleotide of the MYBPC3 gene. The mutation of MYBPC-A31P was evaluated by using the Bioedit program. After MYBPC3-A31P genotyping, the enrolled cats were divided into two groups: wild-type genotype and mutant genotype. Three cats from the wild-type group and four cats from the mutant group were randomly selected, owing to the concentration and purity of DNA extraction for the detection of the A31P-LAMP method coupled with LFD test.
A31P-LAMP Primer Design
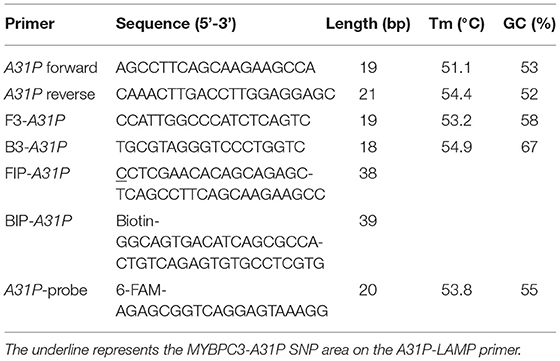
The A31P-LAMP primer set was designed by using Primer Explorer version 5 software (http://primerexplorer.jp/lampv5e/index.html). Based on our experience, the high guanine and cytosine (G/C) nucleotide in the sequence of the MYBPC3 required a manual design of the primer set. The DNA sequence of MYBPC3 protein was obtained from the National Center for Biotechnology Information (NCBI) database. In this research study, we designed the specific primer set based on the efficiency of recognition of SNP at position A31P for specifically selective amplification. The four designed primers can be used to detect mutant allele, cytosine (C), at the A31P missense mutation point from the MYBPC3. Four specific primers of A31P-LAMP, including forwarding inner primer (A31P-FIP), backward inner primer (A31P-BIP), A31P-F3 primer, and A31P-B3 primer were designed under the consideration of correlative comparable in the number of base pairs, percentage of G-C components, and melting temperatures. The position of MYBPC3-A31P was located at the 5' end of the designed forward inner primer (A31P-FIP). The designed primers set in this study were shown in Table 1. The illustration of the MYBPC3, location of A31P-LAMP primer, and steps of the A31P-LAMP process are represented in Figure 1.
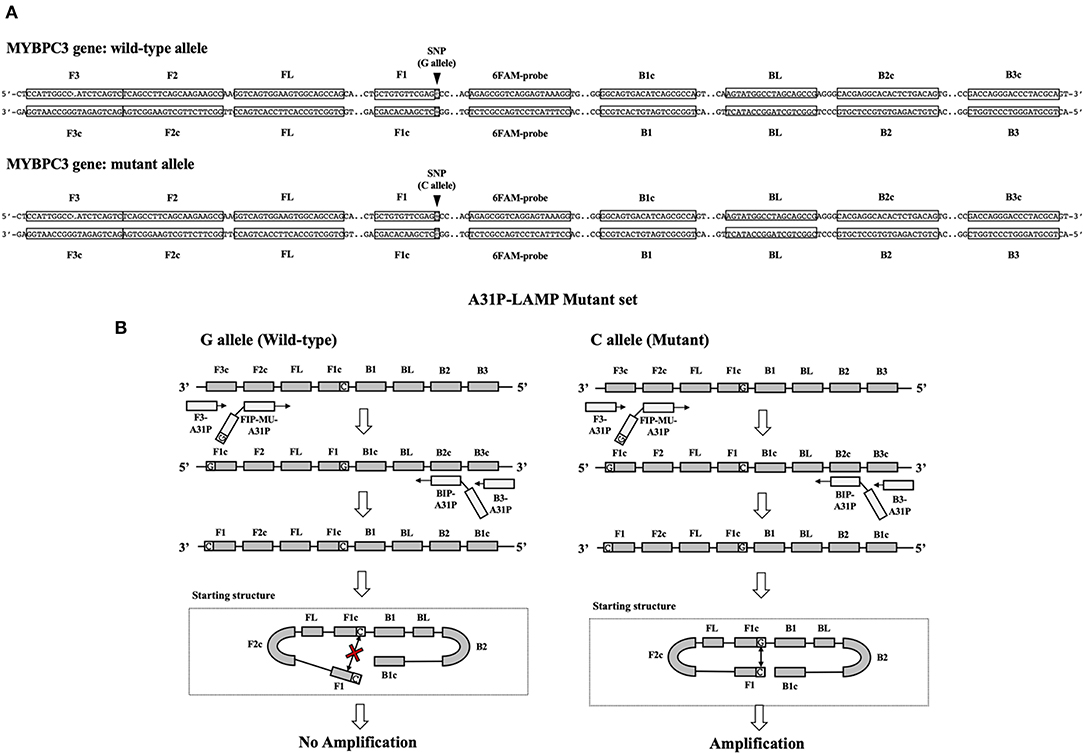
Figure 1. The schematic of target sequence A31P-LAMP for detecting the MYBPC3-A31P and A31P LAMP process. (A) The homozygous wild-type and homozygous mutation of the MYBPC3-A31P were represented. The target of the A31P-LAMP primer regions including outer primer (F3 and B3 primer) and inner primer (FIP and BIP) was demonstrated. The dark triangle symbols pointed to MYBPC3-A31P SNP. (B) The schematic displayed the A31P-LAMP process. The guanine (G) allele from the homozygous wild type was not amplified while the cytosine (C) allele from homozygous mutation was amplified.
A31P-LAMP Condition
A31P-LAMP solution was performed in a total volume of 25 μl per reaction. The mixture of reaction consisted of the following contents: 1X isothermal amplification buffer, containing 20 mM Tris-HCl, 10 mM (NH4)2SO4, 50 mM of KCl, 2 mM MgSO4, 0.1% Tween (New England Biolabs, United States), 6 mM of MgSO4 (8 mM in final concentration), 1.4 mM of dNTPs (Thermo Scientific, United States), 0.8 M of betaine (Sigma-Aldrich, United States), 7.5% of DMSO, 0.2 μM of A31P-F3 primer, 0.2 μM of A31P-B3 primer, 1.6 μM of A31P-FIP primer, 1.6 μM of A31P-BIP primer, 8 unit of Bst 2.0 DNA polymerase (New England Biolabs, United States), 9.4 μl of sterile water, and 2 μl of DNA template. RNase-free water was used as a negative control in each experiment. To optimize the reaction time of A31P-LAMP, the initial condition was modified from the manufacturer (New England Biolabs, United States). The optimal temperature of the A31P-LAMP reaction was evaluated at the gradient temperature, ranging from 60.8 to 68.0°C. Moreover, the mixtures of LAMP reaction were experimented with to incubate with different time points for 30, 45, 60, and 75 min.
Lateral Flow Dipstick Test
After the A31P-LAMP protocol was optimized, an LFD test was conducted for validating the test efficacy. Regarding this study, the designed A31P-LAMP products were labeled with biotin-tagged BIP. In our experiment, an LFD test was performed according to commercial manufacture protocol (Melinia Genline HybriDetect, GieBen, Germany). In short, the amplificated A31P-LAMP product was added with 1 μM of A31P-probe which was labeled with a 6-carboxyfluorescein (6-FAM) probe and allowed to hybridize in the same temperature of A31P-LAMP amplification for 5 min. The sequence of designed A31P-probe is represented in Table 1. Then, the 10 μl of hybridized LAMP products were directly dropped on lateral flow test strips. After that, lateral flow test strips were dipped into 80 μl of assay buffer that has been rewarmed at room temperature. Subsequently, the assay solution was emigrated by chromatography method along the test strips. The results of the LFD were observed by the naked eye within 5–15 min (Figure 2). A positive result for the MYBPC3-A31P mutation was represented by two bands, the control line and the test line, appearing on the strip of the LFD test in a purple-red color. One band, the control line, appeared for homozygous wild-type cats. If no band appeared for the control line, the results of the LFD test were considered invalid.
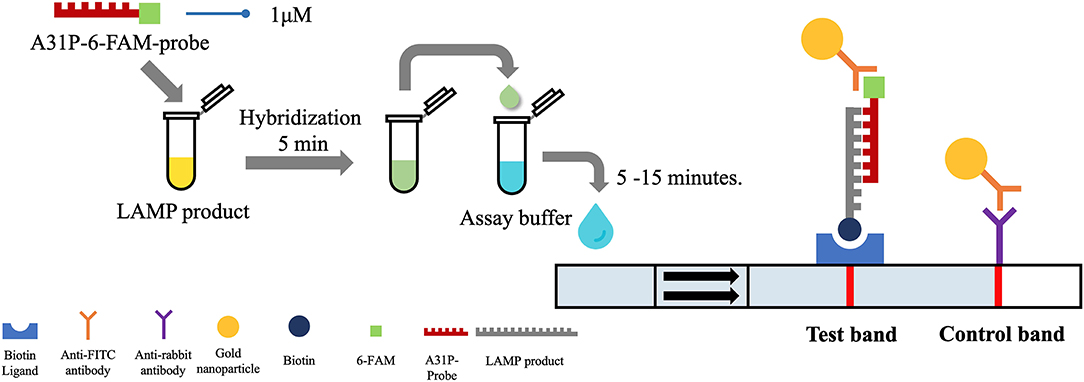
Figure 2. The illustration of the principle of LFD test. The A31P-LAMP assay coupled with the LFD test showed the positive purple-red color band in the control line. In homozygous wild-type cats, the band was not represented in the test band while the positive test band appeared in cats with the MYBPC3-A31P mutation.
Verity of the Diagnostic Test for the A31P-LAMP Test
The analytical sensitivity of the A31P-LAMP test was determined in this study. The DNA of enrolled cats with MYBPC3-A31P mutation was diluted in various concentrations, including 10, 1, 0.1, 0.01, 0.001, 0.0001, and 0.00001 ng/μl. A31P-LAMP reaction was performed according to the protocol as described above. Then, the A31P-LAMP product was detected via (LFD test in comparison with a standard 1.5% agarose gel electrophoresis technique (25).
Results
Descriptive Demographics of the Animals
In this study, 55 Maine Coon cats were recruited to the experiment. The average age of the enrolled cats was 22.54 ± 3.22 months (mean ± SEM). Their average body weight was 5.73 ± 0.35 kg (mean ± SEM). There were 30/55 (54.55%) male and 25/55 (45.45%) female Maine Coon cats. The DNA sequencing results revealed that 8/55 cats (14.55%) had the MYBPC3-A31P mutation. Among these, 1/55 (1.82%) was a homozygous mutation and 7/55 (12.73%) were heterozygous mutations. The demographics of the seven randomly selected Maine Coon cats are presented in Table 2. Figure 3 represents the DNA sequencing examination of the MYBPC3-A31P mutation.
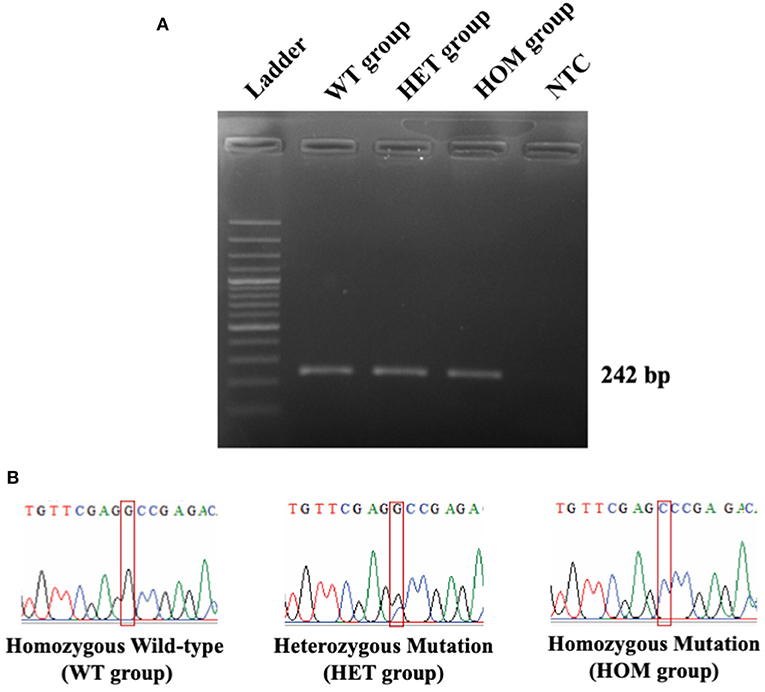
Figure 3. The DNA sequencing results of MYBPC3-A31P mutation. (A) The PCR result from MYBPC3 amplification. Lane 1: DNA ladder; Lane 2: homozygous wild-type group; Lane 3: heterozygous mutation group; Lane 4: homozygous mutation group; Lane 5: negative control (NTC). The product size relied on 242 bp. (B) The chromatograms from the Sanger sequencing method were demonstrated in three different types of MYBPC3-A31P mutation. The red box represents the location of the MYBPC3-A31P region.
Design of the A31P-LAMP Primer for Detecting the MYBPC3-A31P Mutation
In this study, we successfully designed a specific A31P-LAMP primer for detecting the mutant allele cytosine (C) of the MYBPC3-A31P mutation. MYBPC3-A31P is located at the 5' end of the forward inner primer (FIP) to distinguish between the wild-type genotype and mutant genotype in Maine Coon cats. The A31P-LAMP reaction on the MYBPC3-A31P mutation in cats is demonstrated in Figure 4. The mutated allele (C), including the heterozygous mutation (G/C) (Lane 3) and the homozygous mutation (C/C) (Lane 4), was amplified and evaluated via agarose gel electrophoresis as the standard method. However, homozygous wild-type (G/G) was not able to be observed by A31P-LAMP detection. Therefore, the designed A31P-LAMP primer set is an excellent LAMP primer set for identifying the MYBPC3-A31P mutation.
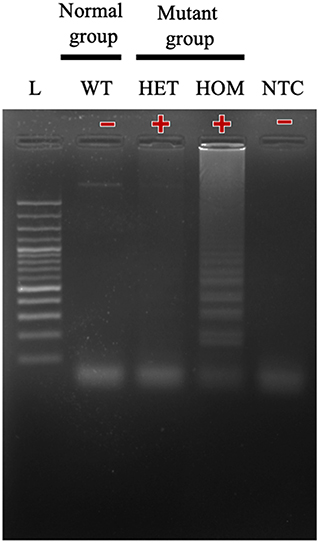
Figure 4. The results of the A31P-LAMP reaction for identifying MYBPC3-A31P mutation in Maine Coon cats by agarose gel electrophoresis. The results showed a positive amplified band in Lane 3 and Lane 4 from both heterozygous mutation and homozygous mutation. In Lane 1, the cats from the homozygous wild type did not amplify the A31P-LAMP.
Optimizations of the Designed A31P-LAMP Amplification
After designing an effective A31P-LAMP primer set, we sequentially optimized the conditions of the temperature and time to perform the A31P-LAMP reaction. The optimal temperatures were tested from 60.8 to 68.0°C. We observed that A31P-LAMP was successful at 62.4 to 66.2°C (Figure 5A). Thus, we selected the temperature of 64°C as optimal for the A31P-LAMP reaction. Moreover, the time required for the A31P-LAMP reaction was optimized. We discovered that A31P-LAMP had positive results from 60 to 75 min (Figure 5B). Therefore, the A31P-LAMP reaction at 60 min provided a better experimental outcome based on the higher intensity signal on gel electrophoresis.
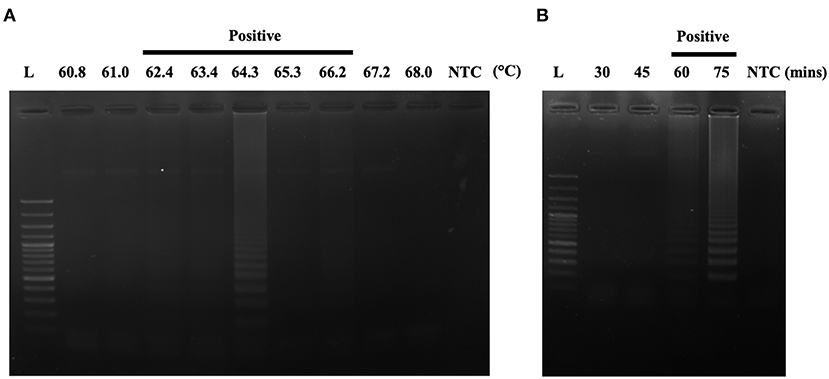
Figure 5. The optimization of the A31P-LAMP protocol. (A) The optimal temperature was examined from 60.8 to 68◦C. (B) The optimal temperature was evaluated from 30, 45, 60, to 75 min. The A31P-LAMP product was demonstrated within 60 min.
Evaluation of A31P-LAMP Coupled With Lateral Flow Dipstick Test
A31P-LAMP coupled with the LFD test was operated to detect the designed LAMP products. Biotin was tagged at the backward inner primer (A31P-BIP) and 6-carboxyfluorescein (6-FAM) was labeled in the A31P-probe. In this experiment, two purple-red color bands for the test line and control line appeared for mutant cats with either a heterozygous or homozygous mutation. However, only one band appeared at the control line in homozygous wild-type cats (Figure 6). Overall observations suggested that the A31P-LAMP coupled with the LFD test was potentially suitable for detecting MYBPC3-A31P.
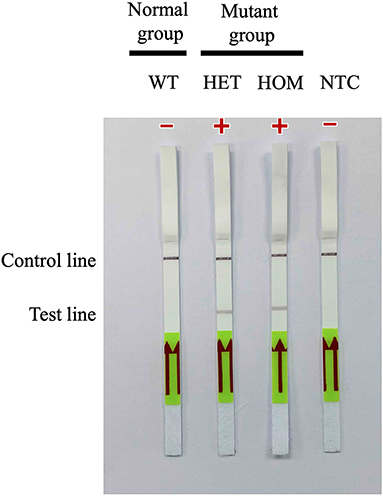
Figure 6. Detection of the DNA (10 ng per μL) of MYBPC3-A31P mutation using the recombinase polymerase amplification-LFD test. WT, wild-type cats; HET, heterozygous mutation; HOM, homozygous mutation; NTC, negative control.
Verity of a Diagnostic Test for A31P-LAMP Test
The sensitivity test of the designed A31P-LAMP reaction for detecting A31P-LAMP product was investigated using 10-fold serial dilutions of DNA concentration in comparison between agarose gel electrophoresis (standard method) and LFD test. In agarose gel electrophoresis, we found that the lowest DNA concentration to detect LAMP-A31P was 0.0001 ng/μl.
In this study, seven out of 55 cats were diagnosed using the A31P-LAMP assay coupled with the LFD test, all seven cats were used in various DNA concentrations (from 0.0001 to 0.00001 ng/μl) to detect A31P mutation. The results showed that 0.00001 ng/μl of DNA concentration is the minimal amount that was able to detect A31P mutation with the LFD test in all seven cats (Figure 7). The accumulated finding indicated that the LFD test provided more sensitivity to identify A31P-LAMP product than agarose gel electrophoresis.
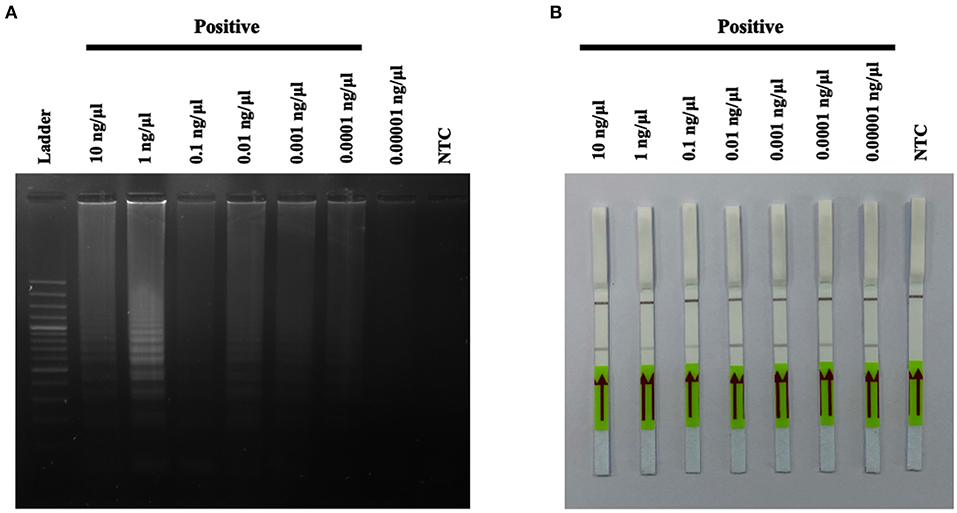
Figure 7. The results of A31P-LAMP reaction from various DNA concentrations (10–0.00001 ng per μL) for identifying MYBPC3-A31P mutation in Maine Coon cats by agarose gel electrophoresis (A). Detection of the DNA (10–0.00001 ng per μL) of MYBPC3-A31P mutation using the recombinase polymerase amplification-LFD test (B). NTC, negative control.
Discussion
The MYBPC3-A31P mutation is of great concern in Maine Coon cats due to the severity of the pathogenesis and the HCM disease progression (26). According to our findings in the recruited study animals, the MYBPC3-A31P mutation rate was 14.55%, which is low. In concordance with our recent prior publication, the mutation rates of MYBPC3-A31P and A74T in Maine Coon cats accounted for 21.43% in Thailand (23). The prevalence of MYBPC3-A31P mutations worldwide (not included in Thailand) is around 34% (27). Moreover, a study in a large European region showed that the prevalence of Maine Coon cats with the MYBPC3-A31P mutation was 41.5% (28).
To date, the gold standard for the detection of MYBPC3-A31P mutations is DNA sequencing. However, this method has various limitations, such as complicated processes, time-consuming, a prohibitive cost, and requiring special equipment. Over the past few decades, many researchers have attempted to develop better techniques for detecting SNP gene mutations. There is currently no perfect screening test that can be applied as a simple diagnostic tool for SNP gene mutations, especially MYBPC3-A31P mutations in cats.
This is the first study applying a LAMP coupled with an LFD test for screening MYBPC3-A31P mutations. Our findings demonstrated that A31P-LAMP associated with the LFD test can distinguish between wild-type cats and mutant cats with the MYBPC3-A31P mutation (Figure 6). Nevertheless, this technique has some limitations due to its inability to differentiate between homozygous and heterozygous mutation carriers.
After successfully developing a specific design of A31P-LAMP, the A31P-LAMP amplification was optimized for temperature and time (Figure 5). We found that the designed A31P-LAMP can be amplified in wide ranges of temperatures and with various equipment, such as thermocyclers, heated blocks, and water baths. This is consistent with a previous study, where the researchers developed a LAMP for detecting the Mycobacterium leprae pathogen that causes Hansen's disease. The researchers were able to conduct LAMP amplification in both a heated block and a water bath (29). Moreover, in this study, the average time to perform the MYBPC3-A31P test with LAMP was ~110 min, including the process of DNA extraction, DNA amplification, and SNP detection. These data indicated that the designed A31P-LAMP required less time than other methods for SNP detection, such as PCR with DNA sequencing. PCR with DNA sequencing takes a long time due to the numerous steps, including DNA extraction, PCR, purification of PCR products, and DNA sequencing methods (30).
The designed A31P-LAMP can be clinically utilized as a rapid assay for MYBPC3-A31P mutation detection. To reduce the SNP detection time, in future studies, DNA from various samples, such as blood, dry blood spots (DBSs), hair follicles, or buccal swaps, will be extracted with sodium hydroxide (NaOH). This method can save time for DNA extraction owing to its simple DNA extraction procedure (31).
The LFD test is a widely accepted technique for detecting LAMP products. Following numerous lines of previous evidence, a LAMP assay combined with an LFD test was initially used for detecting several diseases not only in animals but also in humans. For instance, LAMP with the LFD test has been reported for use in the detection of the hepatitis B virus in human fields (32). In this publication, the LAMP products were detected by agarose gel electrophoresis, LFD tests, and gold nanoparticles (AuNPs). The three methods had the same accuracy for hepatitis B virus detection. In humans infected with Plasmodium falciparum, LAMP with an LFD for specific SNP detection (Pf SNP-LAMP-LFD) was applied to investigate malarial disease. It has been reported that malarial disease is associated with antifolate molecular resistance to manage antimalarial drug strategies (33).
LAMP combined with LFD has been applied to detect hepatopancreatic parvovirus in shrimp (34). They documented that this method is more sensitive than one-step PCR with agarose gel electrophoresis due to the lower DNA concentration. In terms of the objective of this study, the combination of LAMP with the LFD test had superior sensitivity for the detection of the designed LAMP product compared with agarose gel electrophoresis at 0.00001 ng/μl (Figure 7). Based on these promising results, the designed A31P-LAMP coupled with the LFD test is a reliable assay for MYBPC3-A31P mutation screening. Similar to the previous study (35), our results showed that the LAMP assay only took 110 min, this method provides on-site detection of a pathogen without requiring complex equipment. The LAMP assay is a rapid, simple, and sensitive test. Therefore, the A31P-LAMP assay could be more suitable as a screening point-of-care test for detecting the MYBPC3-A31P mutation in Maine Coon cats than the conventional PCR methods. In our pilot study, only 7/55 cats (12.73%) were diagnosed using the A31P-LAMP assay coupled with the LFD test, which was a limitation. For future work, it will be useful to increase the number of cats tested; however, we still gained valuable results by using the A31P-LAMP-LFD test in this study. Another limitation was that we did not investigate the efficacy of diagnostic examinations, such as the sensitivity and specificity, for MYBPC3-A31P diagnosis with the A31P-LAMP assay coupled with the LFD test. This should be further investigated in a larger population of Maine Coon cats.
Novel methods for genetic testing of the MYBPC3-A31P mutation play a crucial role in reducing the effects of familial HCM in Maine Coon cats (26, 36). According to the American College of Veterinary Internal Medicine (ACVIM) consensus statement guidelines for the classification, diagnosis, and management of cardiomyopathies in cats, MYBPC3-A31P screening is suggested to decrease the abnormal gene pool and reduce HCM development in Maine Coon cats (26, 36). In this study, the A31P-LAMP method coupled with the LFD test is a rapid diagnostic tool for detecting MYBPC3-A31P mutations.
Conclusion
Loop-mediated isothermal amplification for the MYBPC3-A31P mutation (A31P-LAMP) coupled with an LFD test has enormous potential for screening the crucial SNP mutation of the sarcomeric protein in Maine Coon cats with HCM. Due to its high sensitivity and simple evaluation, this technique can be readily applied for on-site routine examinations in pedigree breeding in cats.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by Kasetsart University Institutional Animal Care and Use Committee, Kasetsart University, Bangkok, Thailand (Approval Number: ACKU 62-VET-059). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
SP and PS designed the study and wrote the original manuscript and prepared figures. SP, KD, and PS collected all samples and analyzed and interpreted the data. PS performed the experiments of genotyping and LAMP with LFD. SP finalized the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This research project was supported by the National Research Council of Thailand (NRCT): NRCT5-RGJ63002-035.
Conflict of Interest
The research was conducted with a patent application (ID: 2103003416).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to express our sincere thanks to Asst. Prof. Dr. Janjira Phavaphutanon from the Department of Companion Animals Clinical Sciences, Kasetsart University for her advice and suggestions. Also, thanks to Kasetsart University Veterinary Teaching Hospital, Faculty of Veterinary Medicine, Kasetsart University and Biotechnology Laboratory, Kamphaeng Saen Veterinary Diagnostic Center, Faculty of Veterinary Medicine, Kasetsart University for providing the facilities for this research.
References
1. Fox PR, Liu SK, Maron BJ. Echocardiographic assessment of spontaneously occurring feline hypertrophic cardiomyopathy. An animal model of human disease. Circulation. (1995) 92:2645–51. doi: 10.1161/01.CIR.92.9.2645
2. Ferasin L, Sturgess C, Cannon M, Caney S, Gruffydd-Jones T, Wotton P. Feline idiopathic cardiomyopathy: a retrospective study of 106 cats (1994–2001). J Feline Med Surg. (2003) 5:151–9. doi: 10.1016/S1098-612X(02)00133-X
3. Fox PR. Hypertrophic cardiomyopathy. Clinical and pathologic correlates. J Vet Cardiol. (2003) 5:39–45. doi: 10.1016/S1760-2734(06)70051-0
4. Granström S, Godiksen M, Christiansen M, Pipper CB, Martinussen T, Møgelvang R, et al. Genotype-phenotype correlation between the cardiac myosin binding protein C mutation A31P and hypertrophic cardiomyopathy in a cohort of Maine Coon cats: a longitudinal study. J Vet Cardiol. (2015) 17:S268–S81. doi: 10.1016/j.jvc.2015.10.005
5. Payne J, Brodbelt DC, Fuentes VL. Cardiomyopathy prevalence in 780 apparently healthy cats in rehoming centres (the CatScan study). J Vet Cardiol. (2015) 17:S244–57. doi: 10.1016/j.jvc.2015.03.008
6. Payne J, Borgeat K, Connolly D, Boswood A, Dennis S, Wagner T, et al. Prognostic indicators in cats with hypertrophic cardiomyopathy. J Vet Intern Med. (2013) 27:1427–36. doi: 10.1111/jvim.12215
7. Abbott JA. Feline hypertrophic cardiomyopathy: an update. Vet Clin North Am Small Anim Pract. (2010) 40:685–700. doi: 10.1016/j.cvsm.2010.04.004
8. Payne J, Fuentes VL, Boswood A, Connolly D, Koffas H, Brodbelt D. Population characteristics and survival in 127 referred cats with hypertrophic cardiomyopathy (1997 to 2005). J Small Anim Pract. (2010) 51:540–7. doi: 10.1111/j.1748-5827.2010.00989.x
9. Baty CJ, Malarkey DE, Atkins CE, DeFrancesco TC, Sidley J, Keene BW. Natural history of hypertrophic cardiomyopathy and aortic thromboembolism in a family of domestic shorthair cats. J Vet Intern Med. (2001) 15:595–9. doi: 10.1111/j.1939-1676.2001.tb01598.x
10. Kittleson MD, Meurs KM, Munro MJ, Kittleson JA, Liu S-K, Pion PD, et al. Familial hypertrophic cardiomyopathy in Maine coon cats: an animal model of human disease. Circulation. (1999) 99:3172–80. doi: 10.1161/01.CIR.99.24.3172
11. Meurs KM, Norgard MM, Ederer MM, Hendrix KP, Kittleson MD. A substitution mutation in the myosin-binding protein C gene in ragdoll hypertrophic cardiomyopathy. Genomics. (2007) 90:261–4. doi: 10.1016/j.ygeno.2007.04.007
12. Meurs KM, Sanchez X, David RM, Bowles NE, Towbin JA, Reiser PJ, et al. A cardiac myosin binding protein C mutation in the Maine Coon cat with familial hypertrophic cardiomyopathy. Hum Mol Genet. (2005) 14:3587–93. doi: 10.1093/hmg/ddi386
13. Coulton AT, Stelzer JE. Cardiac myosin binding protein C and its phosphorylation regulate multiple steps in the cross-bridge cycle of muscle contraction. Biochemistry. (2012) 51:3292–301. doi: 10.1021/bi300085x
14. Hawkins GA. Analysis of Human Genetic Variations Using DNA Sequencing. Basic Science Methods for Clinical Researchers. (2017). p. 77–98. doi: 10.1016/B978-0-12-803077-6.00005-9
15. Ding S, Chen R, Chen G, Li M, Wang J, Zou J, et al. One-step colorimetric genotyping of single nucleotide polymorphism using probe-enhanced loop-mediated isothermal amplification (PE-LAMP). Theranostics. (2019) 9:3723. doi: 10.7150/thno.33980
16. Lindroos K, Sigurdsson S, Johansson K, R¨nnblom L, Syvn AC. Multiplex SNP genotyping in pooled DNA samples by a four-color microarray system. Nucleic Acids Res. (2002) 30:e70. doi: 10.1093/nar/gnf069
17. Marras SA, Kramer FR, Tyagi S. Genotyping SNPs with molecular beacons. Methods Mol Biol. (2003) 212:111–28. doi: 10.1385/1-59259-327-5:111
18. Zhang W, Gianibelli M, Ma W, Rampling L, Gale K. Identification of SNPs and development of allele-specific PCR markers for γ-gliadin alleles in Triticum aestivum. Theor Appl Genet. (2003) 107:130–8. doi: 10.1007/s00122-003-1223-2
19. Garritano S, Gemignani F, Voegele C, Nguyen-Dumont T, Le Calvez-Kelm F, De Silva D, et al. Determining the effectiveness of high resolution melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet. (2009) 10:1–12. doi: 10.1186/1471-2156-10-5
20. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. (2000) 28:e63. doi: 10.1093/nar/28.12.e63
21. Yongkiettrakul S, Kampeera J, Chareanchim W, Rattanajak R, Pornthanakasem W, Kiatpathomchai W, et al. Simple detection of single nucleotide polymorphism in Plasmodium falciparum by SNP-LAMP assay combined with lateral flow dipstick. Parasitol Int. (2017) 66:964–71. doi: 10.1016/j.parint.2016.10.024
22. Saechue B, Kamiyama N, Wang Y, Fukuda C, Watanabe K, Soga Y, et al. Development of a portable reverse transcription loop-mediated isothermal amplification system to detect the E1 region of Chikungunya virus in a cost-effective manner. Genes Cells. (2020) 25:615–25. doi: 10.1111/gtc.12797
23. Sukumolanan P, Phanakrop N, Thaisakun S, Roytrakul S, Petchdee S. Analysis of the serum peptidomics profile for cats with sarcomeric gene mutation and hypertrophic cardiomyopathy. Front Vet Sci. (2021) 8:771408. doi: 10.3389/fvets.2021.771408
24. Godiksen MT, Granström S, Koch J, Christiansen M. Hypertrophic cardiomyopathy in young Maine Coon cats caused by the p. A31P cMyBP-C mutation-the clinical significance of having the mutation. Acta Vet Scand. (2011) 53:7. doi: 10.1186/1751-0147-53-7
25. Günther S, Felten S, Wess G, Hartmann K, Weber K. Detection of feline Coronavirus in effusions of cats with and without feline infectious peritonitis using loop-mediated isothermal amplification. J Virol Methods. (2018) 256:32–6. doi: 10.1016/j.jviromet.2018.03.003
26. Fuentes VL, Abbott J, Chetboul V, Côté E, Fox PR, Häggström J, et al. ACVIM consensus statement guidelines for the classification, diagnosis, and management of cardiomyopathies in cats. J Vet Intern Med. (2020) 34:1062–77. doi: 10.1111/jvim.15745
27. Fries R, Heaney AM, Meurs KM. Prevalence of the myosin-binding protein C mutation in Maine Coon cats. J Vet Intern Med. (2008) 22:893–6. doi: 10.1111/j.1939-1676.2008.0113.x
28. Mary J, Chetboul V, Sampedrano CC, Abitbol M, Gouni V, Trehiou-Sechi E, et al. Prevalence of the MYBPC3-A31P mutation in a large European feline population and association with hypertrophic cardiomyopathy in the Maine Coon breed. J Vet Cardiol. (2010) 12:155–61. doi: 10.1016/j.jvc.2010.06.004
29. Garg N, Sahu U, Kar S, Ahmad FJ. Development of a Loop-mediated isothermal amplification (LAMP) technique for specific and early detection of Mycobacterium leprae in clinical samples. Sci Rep. (2021) 11:1–12. doi: 10.1038/s41598-021-89304-2
30. Gharahkhani P, O'Leary CA, Kyaw-Tanner M, Sturm RA, Duffy DL. A non-synonymous mutation in the canine Pkd1 gene is associated with autosomal dominant polycystic kidney disease in Bull Terriers. PLoS ONE. (2011) 6:e22455. doi: 10.1371/journal.pone.0022455
31. Liu X, Zhang C, Zhao M, Liu K, Li H, Li N, et al. A direct isothermal amplification system adapted for rapid SNP genotyping of multifarious sample types. Biosens Bioelectron. (2018) 115:70–6. doi: 10.1016/j.bios.2018.05.021
32. Augkarawaritsawong S, Srisurapanon S, Wachiralurpan S, Areekit S, Chansiri K. Comparatively rapid screening tests for diagnosis of hepatitis b virus infection using loop-mediated isothermal amplification (LAMP) paired with lateral flow dipstick (LFD), gold nanoparticles (AuNPs), and real-time turbidimetry. Sci Technol Asia. (2019) 45−57. doi: 10.14456/scitechasia.2019.13
33. Yongkiettrakul S, Kolié FR, Kongkasuriyachai D, Sattabongkot J, Nguitragool W, Nawattanapaibool N, et al. Validation of PfSNP-LAMP-lateral flow dipstick for detection of single nucleotide polymorphism associated with pyrimethamine resistance in Plasmodium falciparum. Diagnostics. (2020) 10:948. doi: 10.3390/diagnostics10110948
34. Nimitphak T, Kiatpathomchai W, Flegel T. Shrimp hepatopancreatic parvovirus detection by combining loop-mediated isothermal amplification with a lateral flow dipstick. J Virol Methods. (2008) 154:56–60. doi: 10.1016/j.jviromet.2008.09.003
35. Khan M, Wang R, Li B, Liu P, Weng Q, Chen Q. Comparative evaluation of the LAMP assay and PCR-Based assays for the rapid detection of Alternaria solani. Front Microbiol. (2018) 9:2089. doi: 10.3389/fmicb.2018.02089
Keywords: loop-mediated isothermal amplification, lateral flow dipstick test, myosin binding protein C3 A31P, mutation, Maine Coon cats
Citation: Sukumolanan P, Demeekul K and Petchdee S (2022) Development of a Loop-Mediated Isothermal Amplification Assay Coupled With a Lateral Flow Dipstick Test for Detection of Myosin Binding Protein C3 A31P Mutation in Maine Coon Cats. Front. Vet. Sci. 9:819694. doi: 10.3389/fvets.2022.819694
Received: 22 November 2021; Accepted: 07 January 2022;
Published: 07 March 2022.
Edited by:
Zeki Yilmaz, Faculty of Veterinary Medicine, TurkeyReviewed by:
Sena Ardicli, Uludaǧ University, TurkeyYu Ueda, North Carolina State University, United States
Copyright © 2022 Sukumolanan, Demeekul and Petchdee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soontaree Petchdee, ZnZldHN0ckBrdS5hYy50aA==
 Pratch Sukumolanan
Pratch Sukumolanan Kanokwan Demeekul
Kanokwan Demeekul Soontaree Petchdee
Soontaree Petchdee