- 1Dipartimento di Medicina Veterinaria e Produzioni Animali, Università degli Studi di Napoli “Federico II”, Napoli, Italy
- 2Dipartimento di Medicina Veterinaria, Università degli Studi di Bari “Aldo Moro”, Bari, Italy
- 3Department of Pathobiological Sciences and Louisiana Animal Disease Diagnostic Laboratory-LADDL, School of Veterinary Medicine, Louisiana State University, Baton Rouge, LA, United States
- 4Pathology Department, Faculty of Veterinary Medicine, University of Agricultural Sciences and Veterinary Medicine, Cluj-Napoca, Romania
ERas is a new gene of the Ras family found in murine embryonic stem (ES) cells. Its human ortholog is not expressed in human ES cells. So far ERas gene has only been found to be expressed in the tissues of adult cynomolgus monkeys and cattle; however, information about ERAS expression or its potential functions in equine tissues is lacking. This study was performed to investigate whether Eras is an equine functional gene and whether ERAS is expressed in the tissues of adult horses and determine its potential physiological role. Expression of the ERas gene was detected in all examined adult tissues, and the RT-PCR assay revealed ERAS transcripts. Protein expression was also detected by Western blot analysis. Quantitative real time RT-qPCR analysis revealed that different expression levels of ERAS transcripts were most highly expressed in the testis. Immunohistochemically, ERAS was found to be localized prevalently in the plasmatic membrane as well as cytoplasm of the cells. ERAS was a physical partner of activated PDGFβR leading to the AKT signaling. ERAS was found to interact with a network of proteins (BAG3, CHIP, Hsc70/Hsp70, HspB8, Synpo2, and p62) known to play a role in the chaperone-assisted selective autophagy (CASA), which is also known as BAG3-mediated selective macroautophagy, an adaptive mechanism to maintain cellular homeostasis. Furthermore, ERAS was found to interact with parkin. PINK1, BNIP3, laforin. All these proteins are known to play a role in parkin-dependent and -independent mitophagy. This is the first study demonstrating that Eras is a functional gene, and that ERAS is constitutively expressed in the tissues of adult horses. ERAS appears to play a physiological role in cellular proteostasis maintenance, thus mitigating the proteotoxicity of accumulated misfolded proteins and contributing to protection against disease. Finally, it is conceivable that activation of AKT pathway by PDGFRs promotes actin reorganization, directed cell movements, stimulation of cell growth.
Introduction
Embryonic Ras (ERas) is a novel member of the Ras family, that was first identified in murine embryonic stem (ES) cells. It is localized on the X chromosome and encodes a small GTPase protein composed of 227 amino acids that shared 43, 46, and 47% identity to the conventional Ras oncogenes H-ras, K-ras, and N-ras, respectively (1). Unlike other proteins of the Ras family, ERAS is constitutively active without any mutations (1, 2). Expressed sequence tag (EST) databases indicate that orthologs of this gene are expressed in other mammals (3). A truncated noncoding ERas transcript has been identified in human ES cells resulting from a premature polyadenylation signal upstream of its coding sequence (3). By in vivo studies, ERAS was found to be expressed both in ES cells and in tissues of adult cynomolgus monkeys, Asian long-tailed macaques (Macaca fascicularis) (4), and in tissues of adult cattle (5, 6). It has been shown that ERAS modulates the Akt signaling pathway and, thus appears to be involved in promoting cell proliferation and tumorigenicity. It has been suggested that ERAS coordinates cell proliferation and cell differentiation, plays a central role in the stimulation of somatic cell reprogramming, which is similar to the cancer initiation process (7, 8). Eras regulates epithelial-mesenchymal transition in pancreatic cancer cells via the ERK/AKT signaling pathway (9). ERAS expression has been found in certain human cancers, including colorectal, pancreatic, neuroblastoma, and breast carcinoma cell lines, as well as in mouse mammary tumors (10–13). ERAS is also expressed in human gastric cancer, where it may play a crucial role in gastric cancer cell survival and metastasis to the liver via downregulation of E-cadherin (14, 15). Recently, ERAS overexpression appeared to have strong oncogenic ability in triggering human colorectal cancer due to its ability to activate AKT signaling (16).
ERAS was found to be constitutively expressed in bovine placental tissue (6) and ERAS overexpression was detected in naturally occurring bladder cancer of cattle associated with bovine papillomavirus (BPV) infection (5). ERAS was found to interact physically with the activated platelet-derived growth factor β receptor (PDGFβR), thus forming a ternary complex with BPV E5 oncoprotein. This complex appeared to play a crucial role in the phosphorylation of AKT, a downstream effector of both ERAS and PDGFβR, in these tumors (5). Recently, ERAS was found to interact with a protein network involved in macroautophagy, including mitophagy induced by BPVs (17, 18).
Our goal is to provide evidence about ERAS expression in tissues of adult horses and its interaction with a network of proteins, some of them known to be interactors with the cellular autophagy machinery.
Materials and Methods
Tissue Samples
Tissue samples such as the medulla oblongata, pons, cerebellum, heart, lung, liver, kidneys, spleen, intestine, uterus, ovary, and testis, were collected from fifteen 3 to 5-year-old horses. All animals were from eastern Europe. Furthermore, at term placenta samples were obtained from ten 3 to 5-year-old Italian standardbred pregnant mares after placenta expulsion during physiological parturition. Tissue samples were immediately divided into several parts with some fixed in 10% buffered formalin for microscopic investigations and others immediately stored at −80 °C for subsequent molecular analysis. The remaining parts were submerged in RNA later storage reagent to stabilize and protect RNA.
Antibodies
Antibodies against HspB8 (SC-22393), Hsc70/Hsp70 (SC-33575), carboxyl terminus of Hsc70-interacting protein (CHIP) (SC-133083), total PDGFβR (SC-432), phosphorylated PDGFβR (pPDGFβR) (SC-12909), β-actin (SC-47778), p62 (SC-28359), PTEN-induced putative kinase1 (PINK1) (SC-33796) and BNIP3 (SC-56167) obtained from Santa Cruz Biotechnology (TX, USA) were diluted 1:500 in BSA. AKT (#9272) antibody obtained from Cell Signaling (LID, NL) was diluted 1:1000 in BSA. Rabbit polyclonal anti-Bag3 (27 F 189C3) obtained from Biouniversa s.r.l. (AV, Italy) was diluted 1:2000 in BSA. Rabbit polyclonal anti-Synaptopodin 2 (Synpo2) (it) (orb453548) obtained from Biorbyt (CA, USA) was diluted 1:1000 in BSA. Antibody against laforin obtained from Novus Biologicals (NBP2-24474) (CO, USA) was diluted 1:500 in BSA. Rabbit antibody against human embryonic stem cell-expressed Ras (ERAS) (TA324562), diluted 1:1000 in BSA, with ERAS HEK 293T cell transient overexpression lysate and WB positive control (COD. LC405694) was obtained from OriGene Technologies, Inc. (MD, USA). This specific antibody was produced against a human ERAS epitope composed of 31 amino acid residues at position 51–81. The human epitope revealed a 100% identity with the same equine ERAS epitope.
Immunohistochemistry
Tissues fixed in 10% neutral buffered formalin were progressively alcohol dehydrated and paraffin embedded. Five-micron sections from several tissues were immunolabelled using the avidin-biotin-peroxidase complex (ABC) method with the Vectastain ABC kit (Vector Laboratories, Inc., CA, USA). Tissue sections were deparaffinized in decreasing alcohol solutions and endogenous peroxidase activity was blocked by incubation in 0.3% H2O2 in methanol for 20 min. Antigen retrieval was performed by pre-treatment with microwave heating (twice for 5 min each at 700 W) in EDTA, pH 8.0. The slides were washed three times with phosphate buffered saline (PBS, pH 7.4, 0.01 M), and then incubated for 1 h at room temperature with normal goat serum (Santa Cruz Biotechnology, TX, USA) diluted at 20% in PBS. The excess serum was gently drained and a polyclonal rabbit anti-ERAS primary antibody (OriGene Technologies, Inc. MD, USA) diluted at 1:100 in PBS was applied overnight at +4°C in a humid chamber. The slides were washed three times with PBS, and then incubated for 30 min with a goat anti-rabbit biotinylated secondary antibody (Vector Laboratories Inc., CA, USA) diluted at 1:200 in PBS. Sections were washed three times with PBS and then incubated with Vectastain ABC reagent (Vector Laboratories Inc., CA, USA) for 30 min in a humid chamber at room temperature. Color development was obtained by treatment with 3, 3'-diaminobenzidine (Vector Laboratories Inc., CA, USA) for 2–10 min. Sections were counterstained with Mayer's hematoxylin. Species- and isotype-matched immunoglobulin (IgG) replaced the primary antibody in the negative controls at the same concentration.
RNA Extraction and Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from 15 equine tissues (brain, cerebellum, pons, intestine, lung, spleen, heart, testis, uterus, ovary, kidney, liver and placenta) using an RNeasy Mini Kit (Qiagen TM, DE), in accordance with the manufacturer's instructions. Genomic DNA was removed from the RNA samples using RNase-free DNase I from Fermentas Life Sciences (Thermo Fisher Scientific, MA, USA). One microgram of the total RNA was used to generate a single strand of cDNA using the QuantiTect Reverse Transcription Kit (Qiagen TM, DE), according to the manufacturer's instructions. PCR was performed both on samples in which reverse transcriptase was added to the reaction mix and in those without adding RT with specific primer sets designed using Primer 3, an online tool, for the equine ERAS: forward 5'-GTAGAAGACCACGACCCCAC-3', reverse 5'- GAAGGGTCATCGAGGGCAAA-3'; (179 bp) and ß-actin: forward 5′- TCCCTGGAGAAGAGCTACGA-3′, reverse 5′-GGATTCCATGCCCAGGAAGG-3′ (111 bp). Primers for both ERAS and ß-actin were evaluated showing a 100% efficiency. The conditions used for PCR were: 94 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s.
Sequence Analysis
PCR products from cDNA, were purified using a Qiaquick PCR purification Kit (Qiagen TM, ME, DE) and bidirectionally sequenced using a BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, CA, USA) following the manufacturer's recommendations. Sequences were removed with a DyeEx_2.0 spin kit (Qiagen TM, DE) and run on a 3,500 Genetic Analyzer (Applied Biosystems, CA, USA). Electropherograms were analyzed using Sequencing analysis v5.2 and sequence scanner v1.0 software (Applied Biosystems, CA, USA). The sequences obtained were compared to others in GenBank using the BLAST program.
Real Time RT-QPCR
To perform real time RT-qPCR analysis, total RNA and cDNA from 15 equine tissues were generated as reported above. Real time PCR was carried out on a BioRad CFX Connect™ Real Time PCR Detection System (Bio Rad Hercules, CA, USA) using iTAq Universal SYBR® Green Supermix (Bio Rad CA, USA). Each reaction was performed in triplicate, and the primers used for ERas and ß-actin were the same as those for RT-PCR assay. The thermal profile for the PCR was: 95°C for 10 min, 40 cycles of 94°C for 15 s, and 58°C for 30 s, followed by a melting curve. The relative quantification (RQ) was carried out using CFX Manager™ software, based on the equation RQ=2−ΔCq, where Cq is the quantification cycle to detect fluorescence. Cq data were normalized to the equine β-actin gene (forward: 5′-TCCCTGGAGAAGAGCTACGA-3′, reverse 5′-GGATTCCATGCCCAGGAAGG-3′).
Western Blot
Western blot analysis was performed on 15 equine tissues. The tissue samples were lysed in RIPA assay-morpholinepropanesulfonic acid (RIPA-MOPS) buffer (20 mM MOPS, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% deoxycholate, and 0.1% SDS) containing protease and phosphatase inhibitors, and extracted proteins were quantified by Bradford assay. Sixty micrograms of extracted proteins was boiled and electrophoresed for 1.5 h at 150 V on a 12% (wt/vol) polyacrylamide/SDS gel. Samples were transferred for 1.5 h at 100 V to PVDF membranes in transfer buffer (25 mM Tris base, 192 mM glycine, and 20% (vol/vol) methanol). Membranes were blocked in 5% (wt/vol) nonfat dry milk in TBST (10 mM Tris HCl (pH 7.4), 167 mM NaCl, 1% Tween-20) for 1 h and incubated overnight at 4 °C with anti-ERAS rabbit polyclonal antibody or anti-β-actin mouse monoclonal antibody. Blots were washed three times in TBST and subsequently incubated for 1 h at room temperature with horseradish peroxidase (HRP)-conjugated donkey anti-rabbit or donkey anti-mouse secondary antibody diluted 1:3000, in 5% (wt/vol) milk/TBST. The blots were then washed and visualized by enhanced chemiluminescence.
Immunoprecipitation
Total protein extracts from equine tissues such as the placenta, ovary, testis, kidney, spleen, and liver, obtained as previously described, were immunoprecipitated. Protein samples (500 μg) were incubated with anti-ERAS primary antibodies or with rabbit IgG for 1 h at 4 °C with gentle shaking. Following incubation, centrifugation (1,000 × g, 5 min at 4°C) was carried out, and the samples were collected and incubated overnight with 30 μl of Protein A/G-Plus Agarose (sc-2003) (Santa Cruz Biotechnology, TX, USA) at 4°C. Immunoprecipitates were washed four times in complete lysis RIPA buffer (as described above) and separated on polyacrylamide gels. Following the transfer of proteins, the membranes were blocked for 1 h at room temperature in 5% bovine serum albumin and incubated overnight at 4°C with primary antibodies. After three washes in Tris-buffered saline, the membranes were incubated with the respective secondary antibodies for 1 h at room temperature. Chemiluminescent signals were then developed with Western Blotting Luminol Reagent (Santa Cruz Biotechnology, TX, USA), and detected by the ChemiDoc XRS Plus gel documentation system (Bio-Rad, CA, USA).
Statistical Analysis
The results are presented as the means ± SE. GraphPad PRISM software version 8 (GraphPad Software, San Diego, CA) was used to assess the expression levels by one-way ANOVA, which was followed by Tukey's test for multiple comparisons of the means. P-values ≤ 0.05 were considered statistically significant.
Results
RT-PCR showed amplicons of ERAS cDNA in organs from all examined horses. The presence of ERAS mRNA was confirmed by sequencing the amplicons that showed 100% identity with the RNA sequences of Equus caballus ES cell expressed Ras (ERAS) (mRNA Sequence ID: XM_001493930.3) (Figure 1). The highest ERAS mRNA expression levels were detected in the testicular samples. Significant lower expression levels of ERAS mRNA (* p < 0.05 for liver; **p < 0.01 for spleen and kidneys; ***p < 0.001 for remaining ones) were detected in all examined organs compared to testicular samples (Figure 2). In particular, the exact p-values were reported in Supplemental Table 1. The brain appeared to have limited ERAS mRNA expression. Western blot analysis revealed the presence of protein expression in all examined organs. The testis, liver, kidneys, spleen, placenta showed the highest protein expression levels. Significant lower expression levels were detected in the organs such as the placenta and spleen (p-value *p < 0.05), lung, intestine, ovary, uterus (p-value **p < 0.01), medulla oblongata, pons and cerebellum (p-value *** p < 0.001) (Figure 3). The exact p-values were reported in Supplemental Table 2. Immunohistochemically, ERAS was found to be localized prevalently in the plasma membrane as well as cytoplasm of the cells (Figure 4).
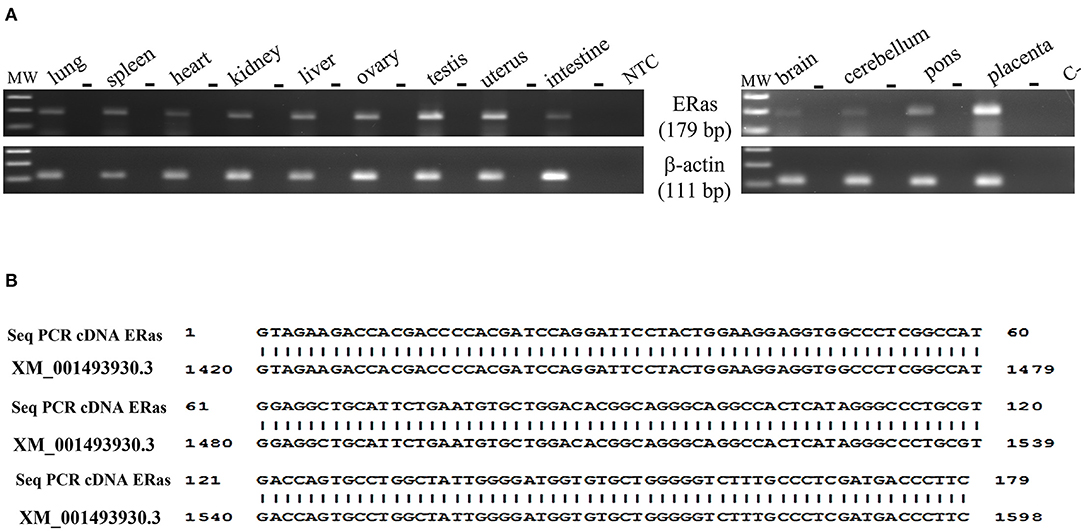
Figure 1. (A) Electrophoresis of RT-PCR products for evaluating ERAS mRNA expression in equine tissues. MW: DNA molecular weight marker (100 bp). The PCR was performed both on samples in which reverse transcriptase (RT) was added to the reaction mix and in those without RT (-), using the same amount of RNA. NTC: no template control, omitted RNA template from the reaction mix; C-: PCR negative control. (B) 100% identity between the sequence of the amplicons (Seq PCR cDNA ERas) and the sequences of Equus caballus ES cell expressed Ras (ERAS), mRNA Sequence ID: XM_001493930.3.
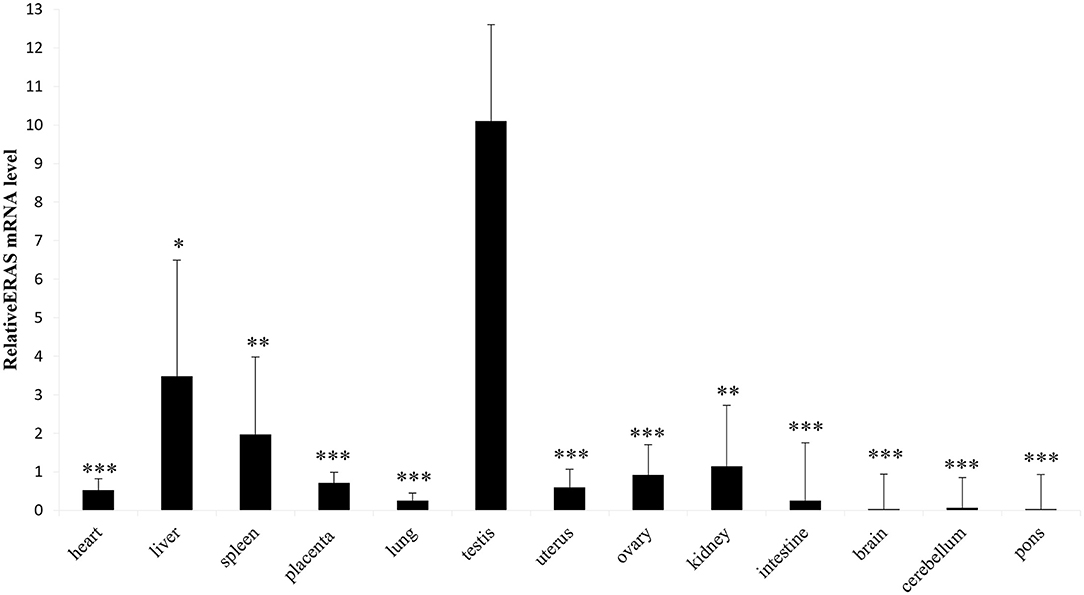
Figure 2. Representative RT-qPCR shows equine tissues ERAS mRNA levels. Data are expressed as mean and standard error mean (S.E.M) of three separate experiments from several animals, each performed in triplicate, related to ERAS levels in the testis (*p < 0.05; **p < 0.01; ***p < 0.001).
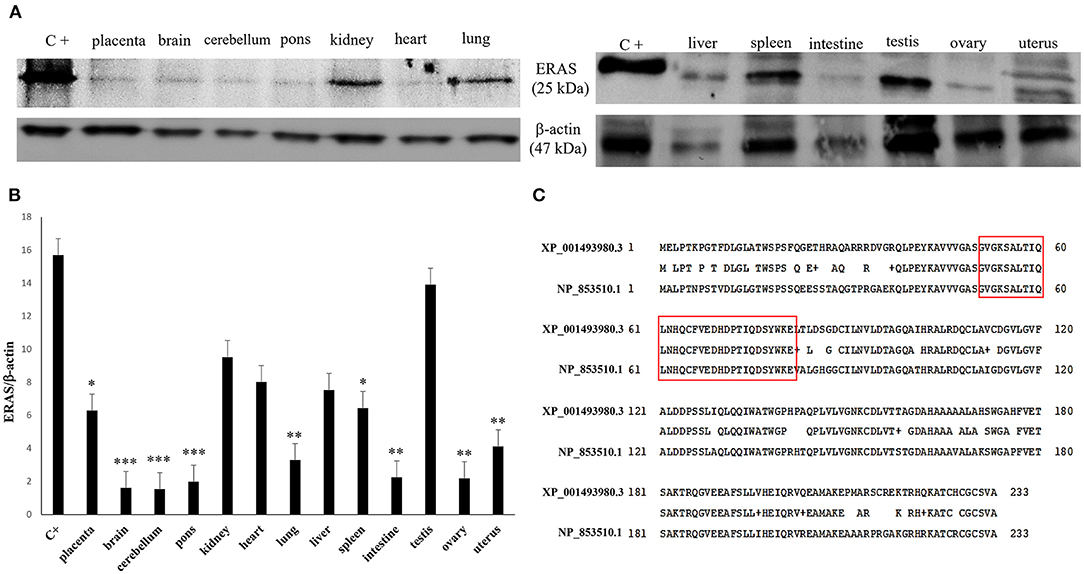
Figure 3. (A) Western blot (WB) analysis of ERAS protein in equine tissues. C+: WB positive control, ERAS HEK 293T cell transient overexpression lysate. An expected ~25kDa band was shown in all samples. Actin was used as loading control. (B) The values of the densitometric analysis were related to the ERAS protein levels in the positive control (*p < 0.05; **p < 0.01; ***p < 0.001). (C) Alignment between the sequences of the human (NP_853510.1) and equine (XP_001493980.3) ERAS protein showed an 81% identity. The amino acid residues of the epitope against the specific antibody used in the current study have underlined in red. This immunogenic portion of ERAS is composed of 31 amino acid residues (51–81 position) showing a 100% identity between the human and horse epitope.
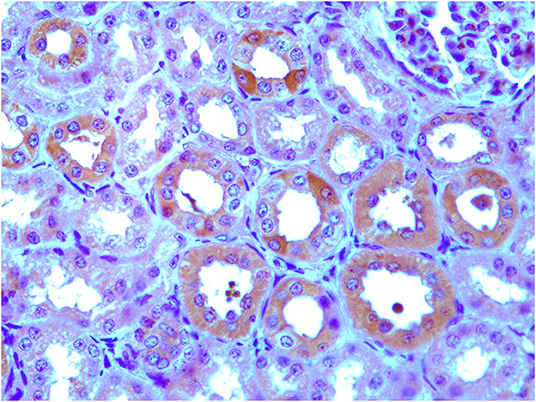
Figure 4. Immunohistochemical evidence of ERAS showing plasmatic and intracytoplasmic localization in epithelial cells of the kidneys and placenta. Inset: No immunoreactivity was seen in renal and placental epithelial cells where the primary anti-ERAS antibody was replaced by appropriate species- and isotype-matched immunoglobulins.
ERas is a functional gene that contributes to the homeostasis of bovine cells and may be involved in cellular signaling mechanisms under physiopathological conditions (5, 6). Therefore, to investigate the role, if any, of ERAS in normal equine cells, we sought to determine whether ERAS could be associated with factors that modulate cellular signaling. ERAS was found to physically interact with PDGFβR, as shown by immunoprecipitation studies. PDGFβR interacting with ERAS was shown to be highly phosphorylated, which suggested that the receptor was functionally activated. Furthermore, in all tissue samples, we found that ERAS interacted with AKT (Figure 5), an important downstream effector of activated PDGFβR (1, 5).
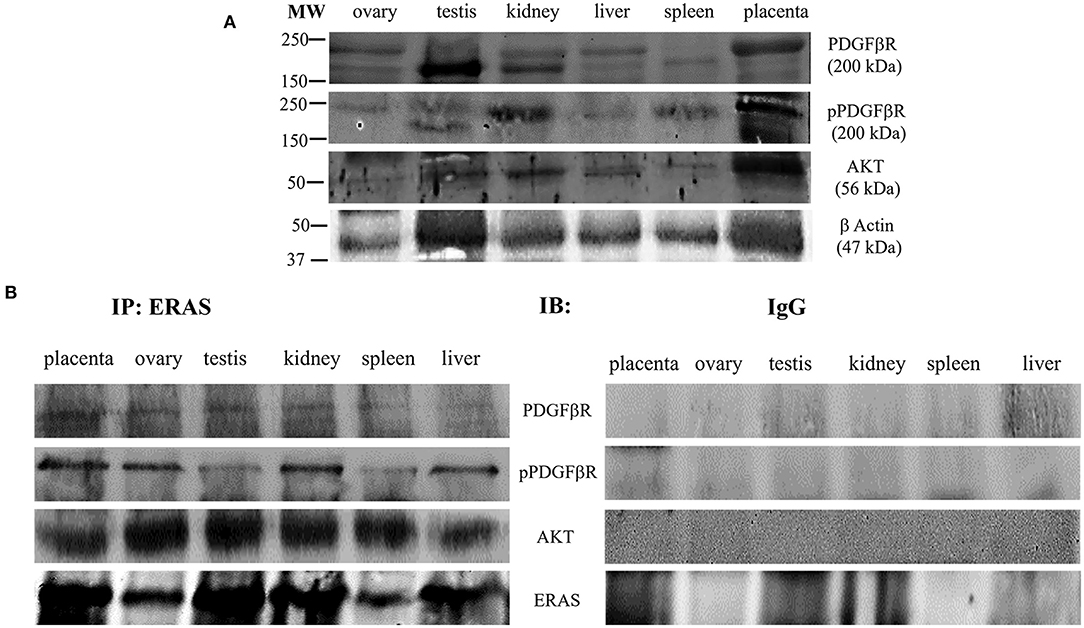
Figure 5. (A) Representative WB analysis and (B) ERAS immunoprecipitation (IP) of pPDGFβR, total PDGFβR and AKT in equine tissues from several animals. WB analysis performed on IP revealed that ERAS interacted with pPDGFβ, PDGFRβ and AKT.
It has been suggested that ERAS plays a crucial role in selective macroautophagy (19). Therefore, we investigated whether ERAS might be involved in cellular proteostasis via autophagy. ERAS was found to be a physical partner of the cochaperones BAG3, a Bcl-2-associated athanogene protein, and CHIP, a chaperone-associated ubiquitin ligase. ERAS also interacted with the chaperones Hsc70/Hsp70, and HspB8, as well as with Synpo2, a protein essential for autophagosome formation, and the autophagic ubiquitin adaptor p62 (Figure 6). Furthermore, ERAS may be an important factor of both parkin-dependent and parkin-independent activated mitophagy in viral infections (17, 18). Therefore, we sought to investigate the relationship, if any, between ERAS and mitophagy proteins in physiology. We found that ERAS interacted with PINK1, parkin, a ubiquitin E3 ligase, and laforin, which are proteins that play a crucial role in parkin-mediated mitophagy. Furthermore, ERAS was found to be a partner of BNIP3, a multifunctional mitochondrial outer membrane protein known to be an important mitophagy-mediating receptor (Figure 7).
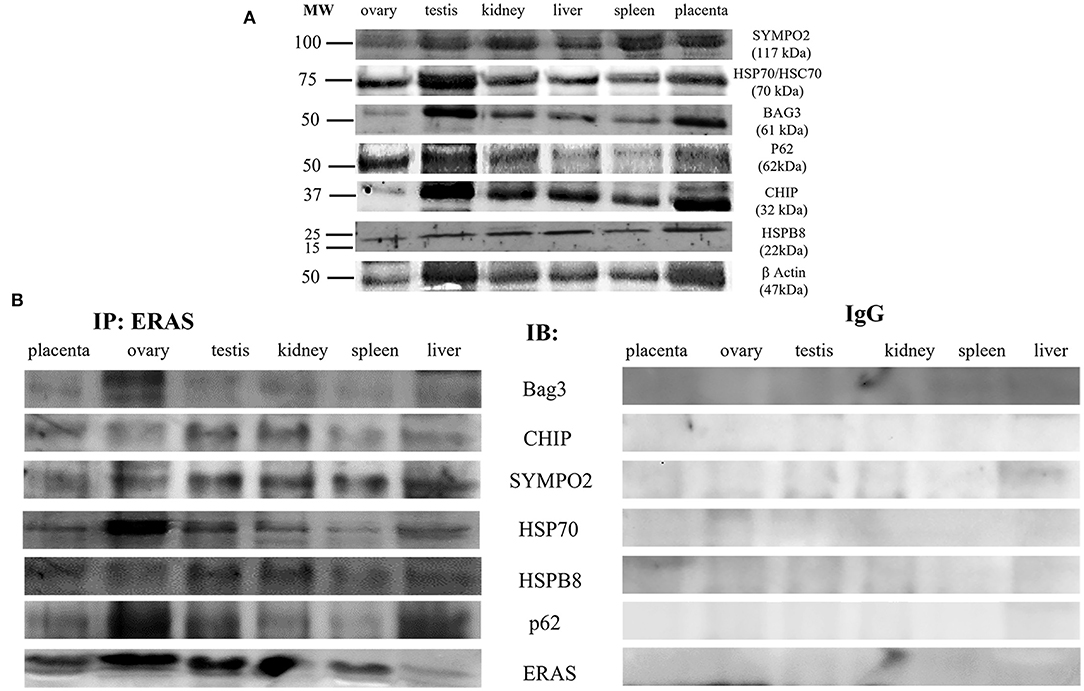
Figure 6. (A) Representative WB analysis and (B) ERAS immunoprecipitation (IP) in equine tissues from several animals. WB analysis carried out on IP revealed that ERAS interacted with some of proteins involved in autophagy.
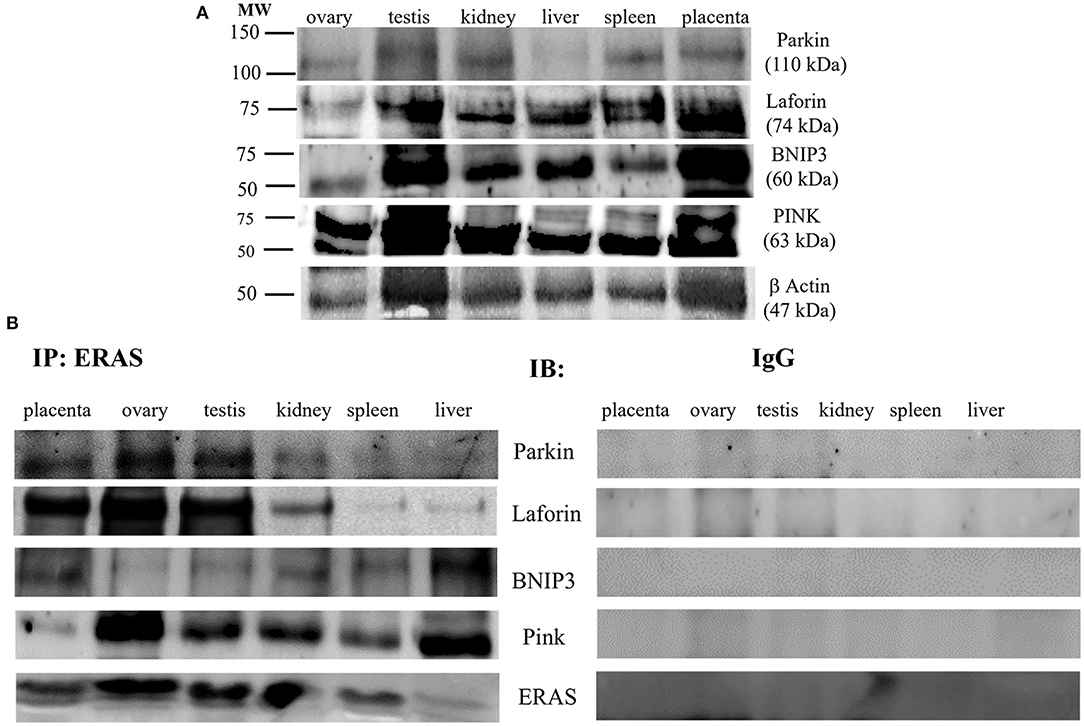
Figure 7. (A) Representative WB analysis and (B) ERAS immunoprecipitation (IP) in equine tissues from several animals. WB performed on IP revealed that ERAS interacted with some of proteins involved in mitophagy.
Discussion
Our study indicates that ERas, a new member of the Ras family, is a functional, constitutively expressed gene in the tissues of adult horses (Equus caballus). Usually, ERAS is localized to the plasma membrane (2, 4). The results presented here and those of other studies (6) reveal that ERAS expression can have both membranous and intracytoplasmic patterns in all tissues. It is conceivable that differential posttranslational modifications, including palmitoylation, could be responsible for promoting subcellular compartmentalization similar to other Ras GTPases, which can influence signaling differences (2, 20). It is worth noting that spatial location generates a distinct response, with broader outputs from Golgi-Ras and mitocondrion-Ras (21).
This study demonstrates that ERAS is a partner of numerous interactors that mediate many important signaling pathways crucial for metabolism and cell homeostasis. Therefore, ERAS appears to represent a signaling hub of the protein network known to play a role in fundamental cell processes.
Immunoprecipitation demonstrated that in the tissues of adult horses a ternary complex composed of ERAS, the activated form of PDGFβR and AKT, was present. These findings are consistent with those from experimental studies [(1); 22] and strengthen the molecular findings obtained in tissues from adult cattle under both physiological and pathological conditions (5, 6, 23). It is conceivable that ERAS also plays a crucial role in AKT signaling activation via PDGFβR in normal cells of adult horses. It is worth noting that PDGFβR signaling is of physiological importance in adult animals, as AKT signaling is important for many physiological activities of cells in adult tissues. In particular, it has been shown that activation of AKT pathway by PDGFRs promotes actin reorganization, directed cell movements, stimulation of cell growth, and inhibition of apoptosis (24, 25). As pPDGFβR is involved in blood vessel formation, ERAS may play an important role in tissue repair via angiogenesis. In addition, ERAS and its downstream AKT signaling pathway are important facilitators of the somatic reprogramming process, which is essential in development and tissue homeostasis (7, 26) and in maintaining quiescence in hepatic stellate cells (HSCs), which have been identified as liver-resident mesenchymal stem cells involved in liver development, immunoregulation, regeneration, and fibrogenesis (22).
The immunoprecipitation findings of the current study showed that ERAS in somatic cells of adult horses is a physical partner of a large network of proteins known to play a role in cellular proteostasis. Indeed, molecular findings showed that ERAS interacted with the molecular chaperones Hsc70/Hsp70 and HSPB8 and the cochaperones BAG3 and CHIP. All these proteins are known to be part of so-called chaperone-assisted selective autophagy (CASA) machinery, also known as BAG3-mediated selective macroautophagy (27), a molecular complex believed to be an adaptive mechanism to maintain cellular homeostasis. Furthermore, p62, and Synpo2 were readily detectable in immunoprecipitated ERAS. The stress-inducible cellular protein p62, functions as a selective autophagy receptor for the degradation of ubiquitinated substrates (28). Synpo2, a cytoskeleton adaptor protein, has been shown to be responsible for autophagosome formation during CASA (28, 29). It has been shown that BAG3 facilitates the functional interplay between Hsc70/Hsp70 and HSPB8 and autophagic degradation of CASA-bound clients via p62. Molecular findings of this study suggest that ERAS could be a player in basal BAG3-mediated selective autophagy, which represents a pivotal adaptive safeguarding and emergency system of protein quality control (PQC), that operates physiologically to ensure cellular proteostasis (30). Our suggestions appeared to be corroborated by previous studies that demonstrated that ERAS plays a crucial role, through BAG3, in selective autophagy induced by BPVs in urothelial cells of cattle (31).
Furthermore, ERAS was shown to interact with a mitophagic network composed of the Parkin/PINK1/BNIP3/Laforin proteins. Damaged mitochondria accumulate PINK1, which then recruits parkin, resulting in ubiquitination of mitochondrial proteins, that are then bound by the autophagic protein p62, thus leading to degradation of mitochondria by mitophagy. In addition, BNIP3, a multifunctional mitochondrial outer membrane protein, harbors an LC3-interacting region (LIR), which strengthens the interaction with light chain3 (LC3) protein, which is involved in mitophagosome formation. Finally, laforin was shown to be a component of a novel pathway of mitophagy mediated by prohibitin 2, an inner mitochondrial membrane (IMM) (18). All these protein interactors found in immunoprecipitated ERAS indicate that ERAS may play an important role in the quality control of mitochondria, which is essential for maintaining cellular homeostasis. It is worth remembering that ERAS has been shown to be involved both in mitophagy mediated by parkin and receptors in naturally occurring papillomavirus-associated bladder tumors in cattle (17, 18).
In conclusion, ERAS is constitutively expressed in tissues of adult horses and its role seems to be critical in the modulation of various biological processes including cell growth, proliferation and immune response. ERAS appears to play a crucial role in the molecular mechanisms mediated by activated PDGFβR and in the BAG3-mediated selective autophagy, which is essential to maintain proteostasis via the dual function of protein transcription and degradation. Therefore, it will be of particular interest to improve our knowledge on the role of ERAS in the homeostasis of cells and tissues of adult horses, particularly those subjected to mechanical tension. The CASA complex appears to be essential for preservation of the cellular structure as well as the cardiovascular, respiratory and urogenital systems (32). Further studies are needed to address ERAS networking in an in vivo animal model.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
FDe and SR contributed to conception and design of the study. FDe organized the database. AP, NZ, FDel, CD, and IM performed the statistical analysis. SR wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank Dr. G. Salvatore of the Regione Basilicata, Dr. S. Morace of the University of Catanzaro “Magna Graecia”, Dr. N. Ercolino from ASL of Foggia, Drs. F. Di Domenico, E. Grieco, G. Pizzolante, G. Marino, S. Milone, F. Di Santi from Azienda Sanitaria Locale (ASL) of Salerno, Dr. R.N. La Rizza from ASL of Vibo Valentia, Dr. Giuseppe Rofrano of the Istituto Zooprofilattico Sperimentale del Mezzogiorno, and Dr. Domenico Rufrano, scholarship holder of Naples University for their technical help.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.818294/full#supplementary-material
References
1. Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. (2003) 423:541–5. doi: 10.1038/nature01646
2. Takahashi K, Nakagawa M, Young SG, Yamanaka S. Differential membrane localization of Eras and Rheb, two Ras-related proteins involved in the phosphatidylinositol 3-kinase/mTOR pathway. J Biol Chem. (2005) 280:32768–74. doi: 10.1074/jbc.M506280200
3. Kameda T, Thomson JA. Human ERas gene has an upstream premature polyadenylation signal that results in a truncated, noncoding transcript. Stem Cells. (2005) 23:1535–40. doi: 10.1634/stemcells.2005-0054
4. Tanaka Y, Ikeda T, Kishi Y, Masuda S, Shibata H, Takeuchi K, et al. ERas is expressed in primate embryonic stem cells but not related to tumorigenesis. Cell Transplant. (2009) 18:381–9. doi: 10.3727/096368909788809794
5. Russo V, Roperto F, Esposito I, Ceccarelli DM, Zizzo N, Leonardi L, et al. Eras protein is overexpressed and binds to the activated platelet-derived growth factor β receptor in bovine urothelial tumour cells associated with papillomavirus infection. Vet J. (2016) 212:44–7. doi: 10.1016/j.tvjl.2016.01.004
6. Roperto S, Russo V, Urraro C, Restucci B, Corrado F, De Falco F, et al. ERas is constitutively expressed in full term placenta of pregnant cows. Theriogenology. (2017) 103:162–8. doi: 10.1016/j.theriogenology.2017.07.047
7. Yu Y, Lang D, Tian Q, Chen X, Jiang B, Chou BK, et al. Stimulation of somatic cell reprogramming by Eras-Akt-Fox01 signaling axis. Stem Cells. (2014) 32:349–63. doi: 10.1002/stem.1447
8. Zhao ZA Yu Y, Ma HX, Wang XX, Lu X, Zhai Y, et al. The roles of ERAS during cell lineage specification of mouse early embryonic development. Open Biol. (2015) 5:150092. doi: 10.1098/rsob.150092
9. Liu Y, Qin P, Du L, Li F. ERas regulates cell proliferation and epithelial mesenchymal transition by affecting Erk/Akt signaling pathway in pancreatic cells. Hum Cell. (2020) 33:1186–96. doi: 10.1007/s13577-020-00401-2
10. Yasuda K, Yashiro M, Sawada T, Ohira M, Hirakawa K. ERas oncogene expression and epigenetic regulation by histone acetylation in human cancer cells. Anticancer Res. (2007) 27:4071–6.
11. Aoyama M, Kataoka H, Kubota E, Tada T, Asai K. Resistance to chemotherapeutic agents and promotion of transforming activity mediated by embryonic stem cell-expressed Ras (ERas) signal in neuroblastoma cells. Int J Oncol. (2010) 37:1011–6. doi: 10.3892/ijo_00000752
12. Wey M, Lee J, Kim HS, Jeong SS, Kim HS, Heo J. Kinetic mechanism of formation of hyperactive embryonic Ras in cells. Biochemistry. (2016) 55:543–59. doi: 10.1021/acs.biochem.5b00902
13. Suárez-Cabrera C, de la Peña B, González LL, Page A, Martinez-Fernández M, Casanova ML, et al. The Ras-related gene ERAS is involved in human and murine breast cancer. Sci Rep. (2018) 8:13038. doi: 10.1038/s41598-018-31326-4
14. Kubota E, Kataoka H, Aoyama M, Mizoshita T, Mori Y, Shimura T, et al. Role of ES cell-expressed Ras (ERas) in tumorigenicity of gastric cancer. Am J Pathol. (2010) 177:955–63. doi: 10.2353/ajpath.2010.091056
15. Liu Y, Wang Z, Li H, Wu Z, Wei F, Wang H. Role of the ERas gene in gastric cancer cells. Oncol Rep. (2013) 30:50–6. doi: 10.3892/or.2013.2417
16. Li X, Song N, Liu L, Liu X, Ding X, Song X, et al. USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat Commun. (2017) 8:14866. doi: 10.1038/ncomms14866
17. Roperto S, De Falco F, Perillo A, Catoi C, Roperto F. Mitophagy mediated by BNIP3 and BNIP3/NIX in urothelial cells of the urinary bladder cattle harbouring bovine papillomavirus infection. Vet Microbiol. (2018) 236:108396. doi: 10.1016/j.vetmic.2019.108396
18. De Falco F, Gentile I, Cerino P, Cutarelli A, Catoi C, Roperto S. Prohibitin 2 is involved in parkin-mediated mitophagy in urothelial cell of cattle infected with bovine papillomavirus. Pathogens. (2020) 9:621. doi: 10.3390/pathogens9080621
19. Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdelfatah M, Abdoli A, Abel S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy. (2021) 17:1–321. doi: 10.1080/15548627.2020.1797280
20. Lynch SJ, Snitkin H, Gumper I, Philips MR, Sabatini D, Pellicer A. The differential palmitoylation states of N-Ras and H-Ras determine their distinct Golgi sub-compartment localizations. J Cell Physiol. (2015) 230:610–9. doi: 10.1002/jcp.24779
21. Hernandez-Valladares M, Prior IA. Comparative proteomic analysis of compartmentalised Ras signalling. Sci Rep. (2015) 5:17307. doi: 10.1038/srep17307
22. Nakhaei-Rad S, Nakhaeizadeh H, Götze S, Kordes C, Sawitza I, Hoffmann MJ, et al. The role of embryonic stem cell-expressed RAS (ERAS) in the maintenance of quiescent hepatic stellate cells. J Biol Chem. (2016) 291:8399–413. doi: 10.1074/jbc.M115.700088
23. Roperto S. Role of BAG3 in bovine deltapapillomavirus-mediated autophagy. J Cell Biochem. (2022) 123:59–64. doi: 10.1002/jcb.30193
24. Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. (1999) 79:1283–316. doi: 10.1152/physrev.1999.79.4.1283
25. Andrae J, Gallini R, Betsholtz C. Role of the platelet-derived growth factors in physiology and medicine. Genes Dev. (2008) 22:1276–312. doi: 10.1101/gad.1653708
26. Miura K, Oiwa Y, Kawamura Y. Induced pluripotent stem cells from cancer-resistant naked mole-rats. Adv Exp Med Biol. (2021) 1319:329–39. doi: 10.1007/978-3-030-65943-1_13
27. Behl C. Breaking BAG: the co-chaperone BAG3 in health and disease. Trends Pharmacol Sci. (2016) 37:672–88. doi: 10.1016/j.tips.2016.04.007
28. Ulbricht A, Arndt V, Höhfeld J. Chaperone-assisted proteostasis is essential for mechanotransduction in mammalian cells. Commun Integr Biol. (2013) 6:e24925. doi: 10.4161/cib.24925
29. Ulbricht A, Höhfeld J. Tension-induced autophagy. Autophagy. (2013) 9:920–2. doi: 10.4161/auto.24213
30. Stürner E, Behl C. The role of the multifunctional BAG3 protein in cellular protein quality control and in disease. Front Mol Neurosci. (2017) 10:177. doi: 10.3389/fnmol.2017.00177
31. De Falco F, Urraro C, Cutarelli A, Roperto S. Bovine papillomavirus E5 oncoprotein upregulates parkin-dependent mitophagy in urothelial cells of cattle with spontaneous papillomavirus infection: a mechanistic study. Comp Immunol Microbiol Infect Dis. (2020) 70:101463. doi: 10.1016/j.cimid.2020.101463
Keywords: adult horse, basal autophagy, constitutive expression, ERas, mitophagy
Citation: De Falco F, Perillo A, Del Piero F, Del Prete C, Zizzo N, Marcus I and Roperto S (2022) ERAS Is Constitutively Expressed in the Tissues of Adult Horses and May Be a Key Player in Basal Autophagy. Front. Vet. Sci. 9:818294. doi: 10.3389/fvets.2022.818294
Received: 25 November 2021; Accepted: 03 May 2022;
Published: 24 May 2022.
Edited by:
Joanna Szyda, Wroclaw University of Environmental and Life Sciences, PolandReviewed by:
Alessandra Rosati, University of Salerno, ItalyEman Aly ELGhareeb Hamza, Cairo University, Egypt
Copyright © 2022 De Falco, Perillo, Del Piero, Del Prete, Zizzo, Marcus and Roperto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sante Roperto, c2FudGUucm9wZXJ0b0B1bmluYS5pdA==; orcid.org/0000-0001-6210-5519
 Francesca De Falco
Francesca De Falco Antonella Perillo2
Antonella Perillo2 Chiara Del Prete
Chiara Del Prete Nicola Zizzo
Nicola Zizzo Sante Roperto
Sante Roperto