- 1USDA, Agricultural Research Service, Beltsville Agricultural Research Center, Animal Parasite Disease Laboratory, Beltsville, MD, United States
- 2USDA, Agricultural Research Service, National Animal Disease Center, Virus and Prion Research Unit, Ames, IA, United States
- 3Department of Agricultural and Environmental Sciences, College of Agriculture, Tennessee State University, Nashville, TN, United States
- 4USDA, National Animal Disease Center, Center for Veterinary Biologics: Policy, Evaluation and Licensing, Ames, IA, United States
One of the largest impediments for commercial swine production is the presence of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV), a devastating RNA viral infection that is responsible for over $1 billion in loss in the U.S. annually. The challenge with combating PRRSV is a combination of the effect of an extraordinary rate of mutation, the ability to infect macrophages, and subversion of host immune response through a series of actions leading to both immunomodulation and immune evasion. Currently there are a handful of commercial vaccines on the market that have been shown to be effective against homologous infections, but struggle against heterologous or mixed strain infections. However, vaccination is the current best strategy for combating PRRSV, making research into new vaccine technology key. To address these issues with PRRSV and host antiviral functions a novel modified-live vaccine (MLV) able to stimulate known antiviral interferons was created and examined for its ability to potentiate effective immunity and better protection. Here, we examine gene expression in the liver of pigs vaccinated with our novel vaccine, given the liver's large role in antiviral responses and vaccine metabolism. Our study indicated that pigs administered the novel vaccine experience homeostatic gene expression consistent with less inflammation and T-cell depletion risk than pigs administered the commercial vaccine.
Introduction
Growing populations can only be sustained by the availability of healthy, abundant, and affordable food. Safeguarding these supplies requires maintaining herds in the face of established and emerging pathogens. One of the largest impediments to this for commercial swine production is Porcine Reproductive and Respiratory Syndrome Virus (PRRSV), a devastating RNA viral infection that is responsible for over $1 billion in loss in the U.S. annually (1, 2). The virus exists as two separate types, PRRSV-1 and PRRSV-2, originally named for the geographic regions in which they originated (3). The challenge with combating PRRSV is a combination of the effect of an extraordinary rate of mutation, the ability to infect macrophages, and subversion of host immune response through a series of actions leading to both immunomodulation and immune evasion (4, 5). PRRSV's ability to suppress interferon signaling compromises the antiviral response. Available commercial vaccines protect animals against homologous infections but provide less protection than heterologous or mixed strain infections (6, 7). Nonetheless, vaccination remains the best available strategy for combating PRRSV. Research into new vaccine technology may offer tools to offset anticipated viral adaptation to evade the host immune system, given the selective pressures arising from widespread vaccine use throughout the swine industry. To address these issues with PRRSV and host antiviral functions, we evaluated a novel modified-live vaccine (MLV) able to stimulate known antiviral interferons for its ability to potentiate effective immunity and better protection. Clinical studies of the vaccine prototype showed efficacy comparable to a commercially available PRRSV vaccine. Here, we examine gene expression in the liver of unvaccinated and vaccinated pigs treated with our novel or commercial MLV vaccine. The porcine liver plays a role in the ability to mount an immune response against pathogens, is affected by the vaccine's metabolism, and is large source of monocytic cells at various stages of the porcine life cycle. Monocytic cells in the form of macrophages are the cells which PRRSV infect, and though the liver is a substantial producer of monocytic cells, very little is known if the organ acts as a reservoir of the virus (8–10).
Materials and Methods
Vaccine Design
Our work was governed by the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). The pCMV-P129 infectious cDNA clone was constructed from the virulent PRRSV-2 (Linage 8) field virus P129, isolated in Indiana in 1995. The infectious clone pKermit (Zoetis) (4) contains the GFP gene within an additional dedicated sgRNA expression cassette. The GFP gene was replaced with genes encoding a cohort of optimized antiviral interferons (IFNs) of different sub-types. The IFN cohort-expressing virus (PRRSV-P129-IFNmix) was examined with and without an adjuvant whilst being compared to the commercial Ingelvac PRRS® ATP (MLV-ATP) MLV vaccine. The original vaccine challenge study used outbred pigs (5-week-old, n = 10/group).
Experimental Design, Animals, and Sample Collection
This study design employed four of the eight treatment groups used in the clinical study of the vaccines effectiveness to protect pigs infected with the NADC-34 strain of PRRSV. The four treatments were: (a) commercial PRRSV vaccine Ingelvac PRRS® ATP (MLV-ATP), (b) ARS-novel vaccine (MLV-129), Sham-challenge (positive control), and Sham-No challenge (Negative control). Control groups were given a sham vaccination of cell culture medium at the beginning of the clinical study in concert with the vaccine treatment groups (-42 days pre-infection); all groups were challenged with a PRRSV dose of 2 ml/pig IM at 1 ×104 TCID50/ml (except the negative control group which was given a sham challenge of cell culture medium). A total of 5 pigs (n = 5/group) were randomly selected from the four treatment groups at 14 days post infection (dpi) and had liver tissue harvested for transcriptomic analysis. This timepoint coincided with the end of the vaccine trial. Vaccinated animals were treated with 2 ml/pig of either the commercial or novel vaccine, which was administered through intramuscular shot. Pigs in all treatment groups were monitored for symptoms of illness pre and post inoculation. All animals were monitored for temperature daily and was compiled weekly. Animal weight was also observed and recorded weekly. Additional evaluations of the treatment of all treatment groups were examined and recorded daily for temperature (°C) using a subdermal chip. The threshold for determining if an animal was experiencing a febrile response was set at 40°C (104 F°). Animals were examined weekly for individual weight gain. Weight was measured in pounds and values averaged within treatment group over the time course of the study. Serology was performed on serum samples by ELISA to measure the presence of host antibodies in response to the PRRS viral challenge. Detection was based on mean S/P ratio within treatment groups for days−42 (vaccination), 0 (challenge), and 14 (necropsy). Positive antibody response was based on S/P ratios ≥0.4 as the cut-off.
Sequencing, Quality Control, and Mapping
Representative samples of liver tissue were extracted from several locations within the liver and combined at 14 dpi. Per the method described in Fleming et al. (5) total RNA was extracted, and quality was measured using an Agilent bioanalyzer (RIN 3.4-9). Library preparation and sequencing were performed by the Iowa State University Genomics center using the 3′ Quantseq fwd kit and the Illumina Hiseq 4000 to produce twenty 100 bp single-end reads. We analyzed sequences using the following tools present at usegalaxy.org (11) and following the method used in Fleming et al. (5) with the following changes and software updates. Quality control was performed using fastp Galaxy Version 0.19.5+galaxy1 and FastQC (12) to examine raw read data, remove adapters, and reads with a phred score below 30. Alignment to the Sscrofa 11.1 reference genome was carried out using Hisat2 version 2.1.0+galaxy5 (13) set to default parameters for forward strand alignment. Raw counts were generated using FeatureCounts (14) and the Ensembl Sscrofa11.1.104 GTF file. The FeatureCounts tool was set to default parameters. Differential gene expression (DEG) was identified using DeSeq2 version 2.11.40.6 (15). The parameters were set to use poscounts for the estimate SizeFactors to account for genes with zero counts and the fit type was set to local with all other parameters tuned to default. We sought evidence for differentially expressed genes (DEGs) with a false discovery rate (FDR) ≤ 0.1 and with no minimum threshold for log2fc up or down regulation. Gene ontology (GO) and Pathway analysis were conducted using software from gProfiler (16) and used an FDR of ≤ 0.05 to establish significance. We also used annotation and pathway information from homologous human genes in Ensembl, NCBI, Uniprot, and KEGG (17–20).
Results
MLV-129p-IFNmix vs. Sham-PRRSV (Vaccinated and Challenged Groups vs. Non-vaccinated and Challenged)
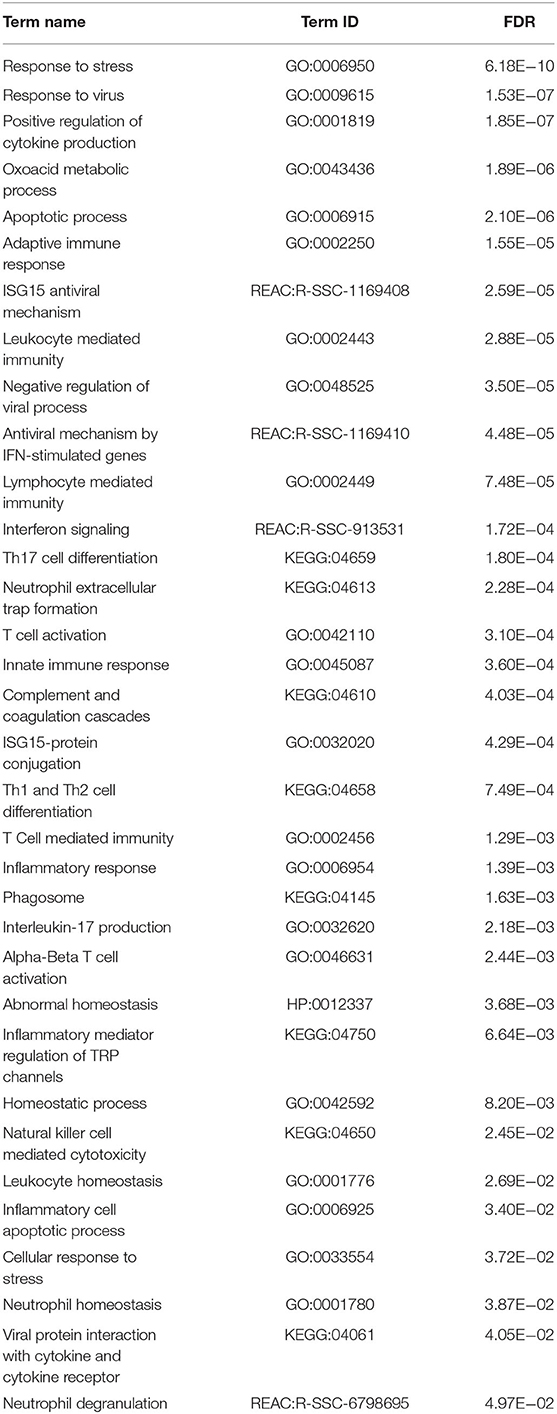
Analysis of the MLV-129p-IFNmix vaccinated pigs vs. non-vaccinated pigs (Sham-PRRSV) at 14 dpi yielded a total of 197 differentially expressed genes (122 upregulated in the vaccinated pigs, 75 downregulated) in the liver after statistical cut-off was (FDR ≤ 0.1) applied (Supplementary Table 1). Transcriptomic analysis of the commercial vaccine (MLV-ATP) vs. the Sham-PRRSV group returned a total of 363 DEGs (187 upregulated, 176 downregulated) (Supplementary Table 2). Venn diagram analysis of the DEGs list for both vaccine groups displayed only a moderate number of overlapping genes (n = 96), however the number of uniquely differential expressed differed greatly between the vaccinated animals; giving the first indication that the vaccines diverged in molecular impact on the host liver (Figure 1).
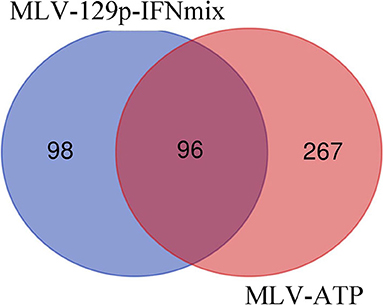
Figure 1. Venn diagram of statistically significant gene lists for MLV-129p-IFNmix and MLV-ATP vs. Sham-PRRSV. Genes were considered significant with FDR of ≤ 0.1.
Differential Gene Expression Involved in Biological Pathways of Vaccine Treatment Groups
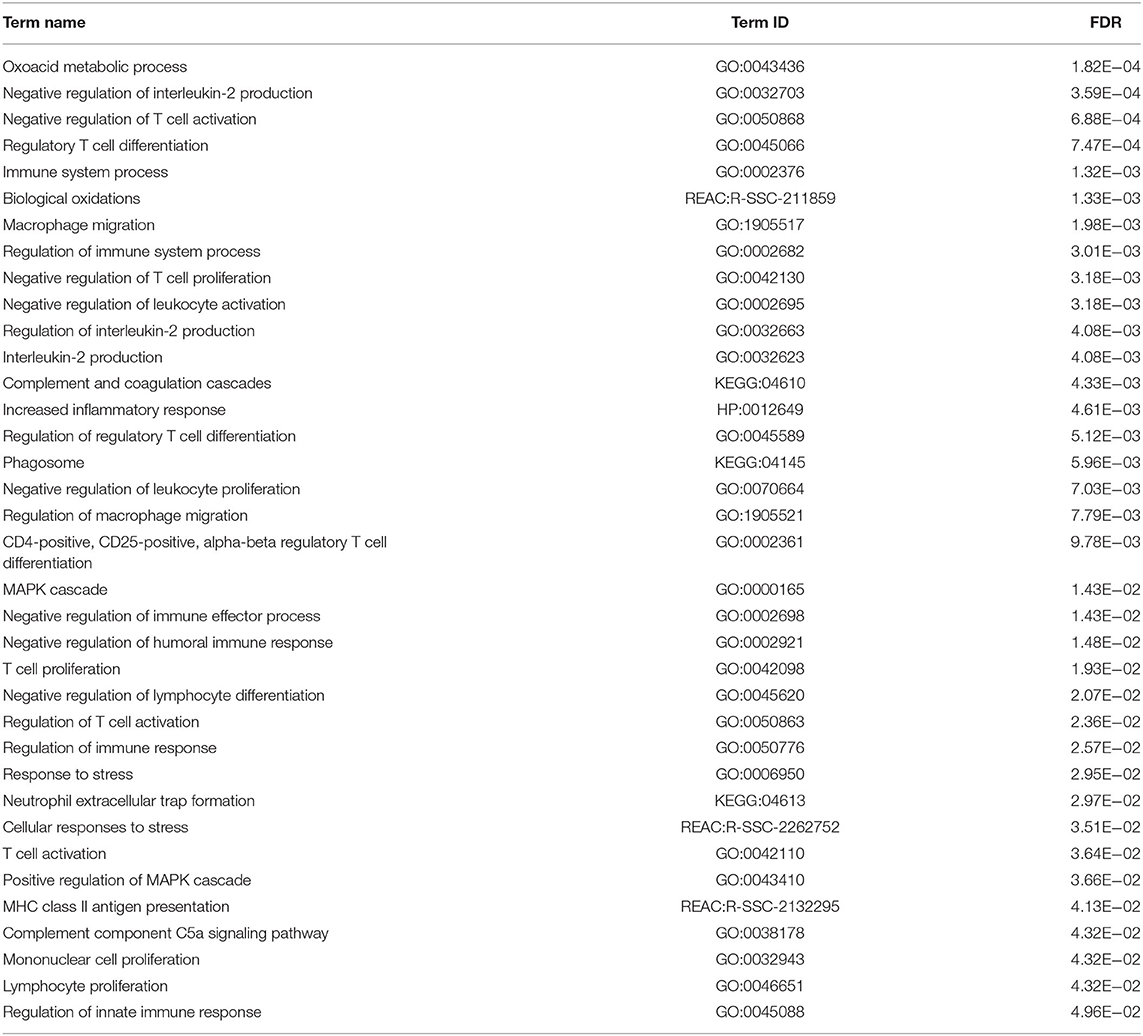
Examination of the list of genes being differentially expressed in the commercial MLV-ATP revealed observations related to the host response to PRRSV. The over-represented genes showed that the commercial vaccine stimulated acute inflammatory response (GO:0006954): nine of fourteen such genes were upregulated. Additionally, many upregulated genes were ascribed to the cellular response to stress (n = 24) (GO:0033554), homeostatic process (n = 26) (GO:0042592), and a human disease phenotype term Abnormal homeostasis (HP:0012337) (n = 31) (Table 1). These results were unique to MLV-ATP among the statistically significant terms present between both vaccinated groups. Analysis of the genes list of being differentially expressed in the MLV-129p-IFNmix also elucidated this vaccine's impact on viral immune response to PRRSV in the liver. The MLV-129p-IFNmix group included fewer differentially expressed genes related to inflammation and stress: 6 such genes are related to the human disease phenotype term increased inflammatory response (HP:0012649). The MLV-129p-IFNmix displayed a total of n = 8 genes involved in the cellular response to stress (REAC:R-SSC-2262752), however, three of these genes were unannotated loci. The MLV-129p-IFNmix treated group also displayed more GO terms related to negative regulation of immune response pathways, while also displaying differential expression of multiple T-cell and macrophage related pathways. The GO terms for MLV-129p-IFNmix included differential expression across innate, humoral, and adaptive immunity which was not observed with the group treated with the commercial vaccine (Table 2).
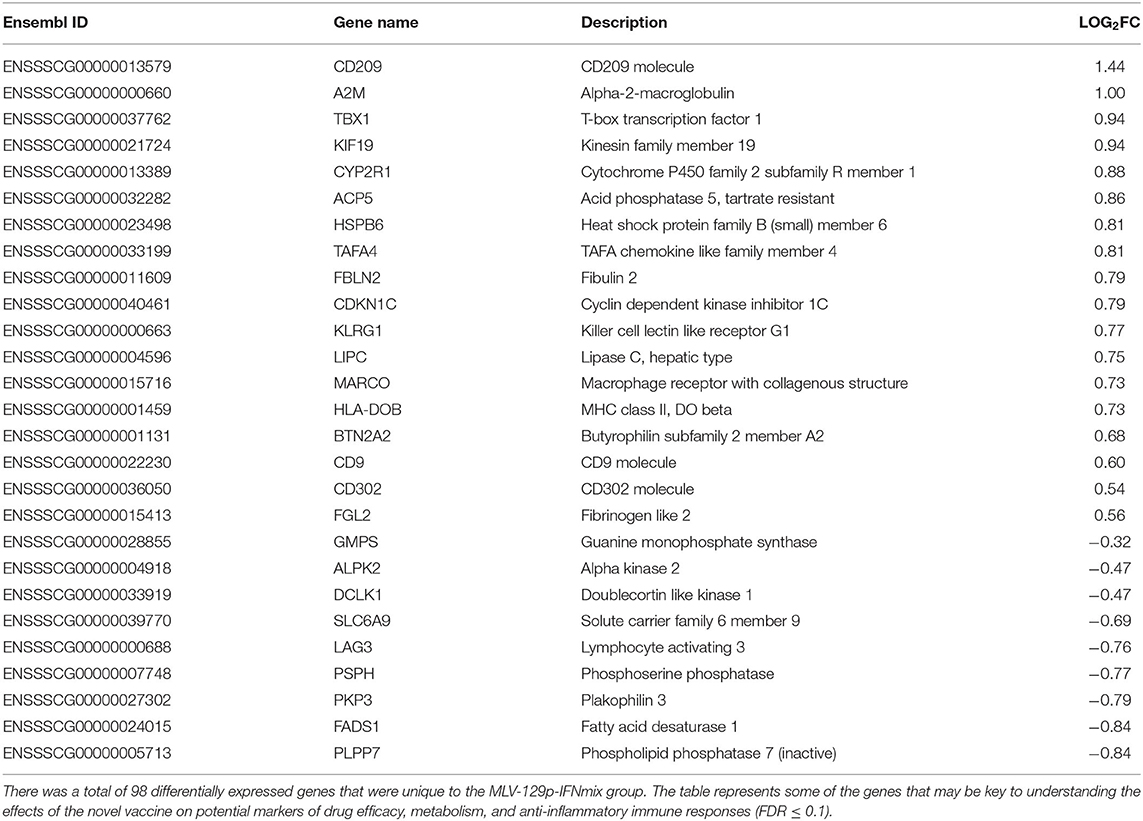
Unique Genes and Pathways of Interest Expressed by MLV-129p-IFNmix
We endeavored to characterize the immune response in the livers of challenged pigs who had been vaccinated with a novel PRRSV vaccine. The Venn diagram depicted that there were 98 genes uniquely differentially expressed in pigs administered MLV-129p-IFNmix. We explored gene function using porcine and human annotations to determine key up and down regulated genes of biological interest (Table 3). Many of the genes in the list are natively expressed in uninfected porcine liver and lung at detectable levels (18). Some key upregulated genes of interest included the most overexpressed gene in the MLV-129p-IFNmix analysis, CD209 molecule (CD209), a known receptor for PRRSV (9, 21). The gene is an innate and adaptive immunity C-type lectin receptor involved in recognizing various viral pathogens, regulating virus life cycles, and promoting T-cell proliferation. In addition to CD209, a small group of other CD markers, CD9 molecule (CD9) and CD302 molecule (CD302) were also upregulated as a cluster unique to MLV-129p-IFNmix (18, 22). Other genes of biological interest overexpressed in the liver included cytochrome P450 family 2 subfamily R member 1 (CYPR2R1) a gene with ubiquitous porcine liver and lung expression that in humans is known to act as a catalyst for drug metabolism and human Vitamin D metabolism (18, 22). Some of the genes of interest interact with the host immune system through anti-inflammatory, macrophage, and immune effector actions. Genes falling into this group included TAFA chemokine like family member 4 (TAFA4), which has immunoregulatory activity involved in macrophage chemotaxis and increased ROS release; acid phosphatase 5, tartrate resistant (ACP5) a negative regulator of inflammatory responses, IL-1B, and IL-12; killer cell lectin like receptor G1 (KLRG1) a killer and T cell inhibitor involved in innate and adaptive immune system immunoregulatory action; and MHC class II, DO beta (HLA-DOB) an antigen presenting negative regulator and positive T-cell regulator in humans (18, 22). The study also produced downregulated genes of interest; however, they were fewer in number. Some of the downregulated genes have a lower natural expression in uninfected pigs due to liver tissue specificity, however even native genes involved in normal host functioning were observed to be down regulated such as fatty acid desaturase 1 (FADS1) involved in the metabolism of inflammatory lipids and T-cell cytokine production (18, 22) which may possibly link to liver metabolism effects. Other genes of interest included: lymphocyte activating 3 (LAG3) an inhibitory immune receptor that is part of the MHC class II adaptive immune system responses. The effect of this gene is far-reaching and may prove to be of great interest due to its involvement in negative regulatory actions. Other genes of interest included doublecortin like kinase 1 (DCLK1) involved in viral responses and is a negative regulator of protein localization to the nucleus in humans and alpha kinase 2 (ALPK2) involved apoptotic regulation and mostly cardiac related functions, but also a negative regulator of the WNT signaling pathway (18, 22).
Discussion
MLV-129p-IFNmix May Hasten Complement and Adaptive Immune Activation to PRRSV Infection Through Humoral-Mediated Expression
The results indicated interplay between MLV-129p-IFNmix and unique DEGs CD209, A2M, HLA-DOB, and other upregulated genes shared by both treatment groups involved in the complement cascade (Supplementary Tables 1, 2). The gene CD209 acts as a C-type lectin receptor (CLR) capable of pattern-recognition of PRRSV (23). It is possible that the observed overexpression of CD209 stimulates antigen presentation to T-cells, helping induce adaptive immune reactions. Additionally, as a lectin receptor CD209 plays a role in the complement cascade which supports the transition to adaptive immune functions. Multiple genes involved in both innate and adaptive immune function suggest that the novel vaccine engender quicker adaptive immune system recognition, reducing the cellular and physical stress of the host-pathogen interaction (24, 25).
MLV-129p-IFNmix Promotes Negative Regulation of Processes That May Bolster Immune Response to PRRSV
Pigs administered the novel vaccine exhibited changes in several negative regulatory processes and molecular functions (Table 2). The range of these pathways were centered around activation and proliferation of T-cells and leukocytes. The genes involved in these responses present in our study are mostly upregulated, pointing to a relaxing of the immune response characteristic of the innate arm of the porcine immune response. The preponderance of negative regulation at 14 dpi in the novel vaccine group was observable at both the pathway and gene level involving differential expression spanning both directions. The action of MLV-129p-IFNmix shows promise related to immune/T-cell exhaustion. Although T-cell exhaustion more commonly results from prolonged chronic infections, the abbreviated maturation experienced by commercial pigs could make a PRRSV infection essentially a lifelong infection. T-cell exhaustion risks depleting memory T-cells, severely handicapping the immune response. One of the first indicators of this is thought to be negative regulation of interleukin 2 (IL-2) (26). Here, of four genes involved in the negative regulation of interleukin-2 production (GO:0032703), three were downregulated in pigs receiving the novel vaccine (Table 2). One of these (LAG3) was unique to the novel vaccine's DEG list. Therefore, the novel vaccine may stimulate a better genomic-mediated immune response in the liver, reducing viral loads at 14 dpi. The gene LAG3 is an example of this. Its downregulation may affect negative regulatory processes that include IL-2 production and Treg cell differentiation. Recent studies of human LAG3 gene action suggest a possible role in immunomodulation and viral immune evasion (27). The downregulation around LAG3 is intriguing when viewed from the perspective that the difference in the liver response between vaccines may involve T-cell exhaustion. The phenomenon of T-cell exhaustion is thought to result from persistent signaling during chronic viral illness and cancers, which seems to be partly driven by LAG3 upregulation. It is possible that LAG3 downregulation may reduce inhibition of a particular type of T-cell response to PRRSV infection in pigs vaccinated with MLV-129p-IFNmix. It may be that the downregulation of the LAG3 inhibitory signaling keeps T-cell proliferation and activation at a metabolically sustainable level. In humans, overexpression of LAG3 is correlated with disease progression in viral infections. Downregulation may allow for less suppression surrounding regulatory T-cells, but limit CD8 or CD4 T-cells. Another gene uniquely expressed by the novel vaccine is, fibrinogen like 2 (FGL2) (Table 3); it is in the same gene family as FGL1, a major ligand and co-activator of LAG3's inhibitory action (18, 22). Porcine FGL1 may not downregulate LAG3 action, but rather upregulate FGL2, thereby negatively regulating dendritic cell, memory T-cells, and macrophage antigen processing. However, FGL2 does support T-cell activation on MHC molecules (18, 22, 28, 29). There was also unique macrophage (M2) gene expression. Action of the gene FGL2 on macrophages in concert with effector actions on regulatory T-cell expression has been observed to promote repair among macrophages, possibly in response to chronic illness or inflammation. The novel vaccine might improve macrophage, T-cell, and pro-inflammatory balance or may sort infected macrophages (needing opsonization or phagocytosis) from uninfected. Reduced expression may keep T-cell proliferation and activation at a metabolically sustainable level, improving the efficiency of established immune checkpoints against PRRSV.
Conclusion
Each vaccine elicited distinct patterns of gene transcription in the liver of infected pigs. As compared to pigs administered the commercial vaccine, those administered the novel MLV-129p-IFNmix vaccine had fewer genes in flux at 14 dpi. They may therefore have been experiencing less viral-induced stress on the organ/immune system. Compared to the MLV-129p-IFNmix, pigs receiving the commercial vaccine experienced overexpression of more genes related acute inflammatory responses. Thus, pigs vaccinated with the commercial vaccine may experience a longer innate immune response to the virus, more pathology, and poorer growth. Pigs administered the commercial vaccine also showed differential expression of interferon stimulating genes (ISGs) not present in the MLV-129p-IFNmix. It is possible that by supplying interferons as part of the vaccine matrix, the experimental MLV-129p-IFNmix introduces interferon stimulation earlier than the commercial vaccine. However, more time points will need to be explored to determine when such stimulation occurs. From a transcriptomic standpoint, MLV-129p-IFNmix appears to show better cellular homeostasis in the liver. By contrast, animals administered the commercial vaccine experience less homeostasis and more metabolic and cellular stress.
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) (30) and are accessible through GEO Series accession number GSE197014 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197014).
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee at National Animal Disease Center Ames, IA.
Author Contributions
DF: methodology, computation software, transcriptomic data validation, formal analysis, review and editing, and writing and original draft preparation. DF, LM, and AV: animal tests and sample preparations. DF and LM: investigation, resources, data curation, and supervision. DF, LM, JL, and YS: writing and original draft preparation. DF, LM, and YS: project administration. YS and LM: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by USDA NIFA AFRI Grant 2013-67015-21236, and in part by USDA NIFA AFRI 2015-67015-23216 and USDA NIFA 2018-67016-28313. Post-doctoral funding for DF provided by the Oak Ridge Institute for Science and Education (ORISE).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jay G. Calvert (Pfizer Animal Health, Kalamazoo, MI, USA) for the gift of the PRRSV cDNA clone (pCMV-P129).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.791034/full#supplementary-material
Supplementary Table 1. MLV-129p-IFNmix differentially expressed gene list.
Supplementary Table 2. MLV-ATP differentially expressed gene list.
References
1. Holtkamp DJ, Kliebenstein JB, Neumann EJ, Zimmerman JJ, Rotto HF, Yoder TK. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J Swine Health Prod. (2013) 21:385–92. doi: 10.31274/ans_air-180814-28
2. Renken C, Nathues C, Swam H, Fiebig K, Weiss C, Eddicks M. Application of an economic calculator to determine the cost of porcine reproductive and respiratory syndrome at farm-level in 21 pig herds in Germany. Porcine Health Manag. (2021) 7:1–12. doi: 10.1186/s40813-020-00183-x
3. Rowland RRR, Lunney JK. Alternative strategies for the control and elimination of PRRS. Vet Microbiol. (2017) 209:1–4. doi: 10.1016/j.vetmic.2017.09.006
4. Miller LC, Fleming DS, Bayles LiX, Blecha DO, Sang F. Comparative analysis of signature genes in PRRSV-infected porcine monocyte-derived cells to different stimuli. PLoS ONE. (2017) 12:e0181256. doi: 10.1371/journal.pone.0181256
5. Fleming DS, Miller LC, Tian Y, Li Y, Ma W, Sang Y. Impact of porcine arterivirus, influenza B, and their coinfection on antiviral response in the porcine lung. Pathogens. (2020) 9:934. doi: 10.3390/pathogens9110934
6. Choi K, Park C, Jeong J, Kang I, Park SJ, Chae C. Comparison of commercial type 1 and type 2 PRRSV vaccines against heterologous dual challenge. Vet Rec. (2016) 178:291. doi: 10.1136/vr.103529
7. Oh T, Park SJ, Cho H, Yang S, Ham HJ, Chae C. Comparative evaluation of 4 commercial modified-live porcine reproductive and respiratory syndrome virus (PRRSV) vaccines against heterologous dual Korean PRRSV-1 and PRRSV-2 Challenge. Vet Med Sci. (2020) 6:846–53. doi: 10.1002/vms3.282
8. Duan X, Nauwynck HJ, Pensaert MB. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch Virol. (1997) 142:2483–97. doi: 10.1007/s007050050256
9. Zhang Q, Yoo D. PRRS virus receptors and their role for pathogenesis. Vet Microbiol. (2015) 177:229–41. doi: 10.1016/j.vetmic.2015.04.002
10. An TQ, Li JN, Su CM, Yoo D. Molecular and cellular mechanisms for PRRSV pathogenesis and host response to infection. Virus Res. (2020) 286:197980. doi: 10.1016/j.virusres.2020.197980
11. Jalili V, Afgan E, Gu Q, Clements D, Blankenberg D, Goecks J. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. (2020) 48:W395–402. doi: 10.1093/nar/gkaa554
12. Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics. (2018) 34:i884–i90. doi: 10.1093/bioinformatics/bty560
13. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with Hisat2 and Hisat-Genotype. Nat Biotechnol. (2019) 37:907–15. doi: 10.1038/s41587-019-0201-4
14. Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
15. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for Rna-Seq Data with Deseq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
16. Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H. G:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. (2019) 47:W191–W8. doi: 10.1093/nar/gkz369
17. Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. (2008) 36(Database issue):D13–21. doi: 10.1093/nar/gkm1000
18. Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at Ncbi. Nucleic Acids Res. (2011) 39:D52-7. doi: 10.1093/nar/gkq1237
19. Howe KL, Achuthan P, Allen J, Alvarez-Jarreta J, Amode MR. Ensembl 2021. Nucleic Acids Res. (2021) 49:D884–D91. doi: 10.1093/nar/gkaa942
20. Bateman A, Martin MJ, O'Donovan C, Magrane M, Alpi E, Antunes R, et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. (2017) 45:gkw1099. doi: 10.1093/nar/gkw1099
21. Su CM, Rowland RRR, Yoo D. Recent advances in PRRS virus receptors and the targeting of receptor-ligand for control. Vaccines. (2021) 9:354. doi: 10.3390/vaccines9040354
22. Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. (2021) 49:gkaa1100. doi: 10.1093/nar/gkaa1100
23. Huang YW, Dryman BA, Li W, Meng XJ. Porcine Dc-Sign: molecular cloning, gene structure, tissue distribution and binding characteristics. Dev Comp Immunol. (2009) 33:464–80. doi: 10.1016/j.dci.2008.09.010
24. Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. (1998) 392:245–52. doi: 10.1038/32588
25. Engering A, Geijtenbeek TB, van Vliet SJ, Wijers M, van Liempt E, Demaurex N. The dendritic cell-specific adhesion receptor dc-sign internalizes antigen for presentation to T Cells. J Immunol. (2002) 168:2118–26. doi: 10.4049/jimmunol.168.5.2118
26. Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters Cd8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. (2003) 77:4911–27. doi: 10.1128/JVI.77.8.4911-4927.2003
27. Lucas F, Pennell M, Huang Y, Benson DM, Efebera YA, Chaudhry M. T cell transcriptional profiling and immunophenotyping uncover Lag3 as a potential significant target of immune modulation in multiple myeloma. Biol Blood Marrow Transplant. (2020) 26:7–15. doi: 10.1016/j.bbmt.2019.08.009
28. Shalev I, Liu H, Koscik C, Bartczak A, Javadi M, Wong KM. Targeted deletion of Fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. J Immunol. (2008) 180:249–60. doi: 10.4049/jimmunol.180.1.249
29. Hou XX, Wang XQ, Zhou WJ, Li DJ. Regulatory T cells induce polarization of pro-repair macrophages by secreting Sfgl2 into the endometriotic milieu. Commun Biol. (2021) 4:499. doi: 10.1038/s42003-021-02018-z
Keywords: Porcine Reproductive and Respiratory Syndrome Virus, liver, vaccine, transcriptome, immunity
Citation: Fleming DS, Miller LC, Li J, Van Geelen A and Sang Y (2022) Transcriptomic Analysis of Liver Indicates Novel Vaccine to Porcine Reproductive and Respiratory Virus Promotes Homeostasis in T-Cell and Inflammatory Immune Responses Compared to a Commercial Vaccine in Pigs. Front. Vet. Sci. 9:791034. doi: 10.3389/fvets.2022.791034
Received: 07 October 2021; Accepted: 14 February 2022;
Published: 24 March 2022.
Edited by:
Jiuzhou Song, University of Maryland, College Park, United StatesReviewed by:
Tao Lin, UCONN Health, United StatesXiuqing Wang, South Dakota State University, United States
Copyright © 2022 Fleming, Miller, Li, Van Geelen and Sang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Damarius S. Fleming, ZGFtYXJpdXMuZmxlbWluZ0B1c2RhLmdvdg==; Laura C. Miller, bGF1cmEubWlsbGVyQHVzZGEuZ292; Yongming Sang, eXNhbmdAdG5zdGF0ZS5lZHU=
 Damarius S. Fleming
Damarius S. Fleming Laura C. Miller
Laura C. Miller Jiuyi Li
Jiuyi Li Albert Van Geelen4
Albert Van Geelen4 Yongming Sang
Yongming Sang