- 1Hunan Provincial Key Laboratory of Animal Intestinal Function and Regulation, Hunan International Joint Laboratory of Animal Intestinal Ecology and Health, Laboratory of Animal Nutrition and Human Health, College of Life Sciences, Hunan Normal University, Changsha, China
- 2Hunan Provincial Key Laboratory of Animal Nutritional Physiology and Metabolic Process, Key Laboratory of Agro-Ecological Processes in Subtropical Region, Hunan Provincial Engineering Research Center of Healthy Livestock, Scientific Observing and Experimental Station of Animal Nutrition and Feed Science in South-Central, Ministry of Agriculture, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China
- 3Key Lab of Non-wood Forest Nurturing and Protection of National Ministry of Education, Hunan Provincial Key Laboratory for Forestry Biotechnology, Central South University of Forestry and Technology, Changsha, China
- 4Key Laboratory of Tea Science of Ministry of Education, National Research Center of Engineering Technology for Utilization of Functional Ingredients From Botanicals, College of Horticulture, Hunan Agricultural University, Changsha, China
The objective of the present study was to assess the effects of replacing corn silage with Phragmites australis shoot remainder (PSR) silage on intake, growth performance, serum biochemical parameters, and rumen microbial diversity of growing-finishing beef. Fifteen Angus beef cattle with an average body weight of 253 ± 2.94 kg were randomly divided into three groups (five replicas vs. each group vs. Angus beef cattle). The three treatments were group A fed 60% PSR silage + 40% concentrate, group B fed 30% PSR silage + 30% corn silage + 40% concentrate, and group C fed 60% corn silage + 40% concentrate. The adaptation period was 15 days, and the trial period lasted for 45 days. Results showed that the ADG was significantly higher, and FCR was significantly lower both in groups A and B compared with group C. The results of serum biochemical parameters showed that the concentration of GLU was significantly lower in group B than both groups A and C. Microbial diversity results showed that the OTUs, Shannon, Chao1, and ACE indices were significantly lower in group A compared with groups B and C. At the phyla level, the relative abundances of Tenericutes and Melainabacteria had significant differences among the three groups, and the relative abundances of Papillibacter, Anaeroplasma, and Anaerovorax had significant differences among the three groups at the genus level. Additionally, Rikenellaceae was the unique biomarker among the three groups. Furthermore, the results of function prediction showed that the gene families associated with metabolism of cofactors and vitamins, cellular processes and signaling, metabolism, biosynthesis of other secondary metabolites, infectious diseases, signaling molecules and interaction, nervous system, and digestive system were significantly decreased, while lipid metabolism was dramatically increased from groups A to C at KEGG level 2. At KEGG level 3, 11 metabolic pathways were significantly influenced among the three groups. In summary, these findings indicated that PSR silage substituted the corn silage totally or partially improved the growth performance, and altered the rumen microbial composition and diversity and the corresponding change in prediction function of rumen bacteria in Angus beef cattle.
Introduction
Phragmites australis is a kind of native perennial grass, which is a very good non-competitive feed resource. P. australis has excellent nutritional value and a broad ecological distribution and adaptation in the world, and other characteristics (1). Therefore, rational utilization of P. australis resources is one of the effective methods to enlarge the feed source and relieve the shortage of roughage when ensuring that P. australis has rich nutrient content and high yield. According to statistics, China is rich in P. australis resources, and the distribution area is about 800,000 ha, among which Hunan has about 80,000 ha and with an annual output of up to 400,000 tons, mainly distributed in Dongting Lake and along the Yangtze River (2). P. australis feed has good palatability, which contains high crude protein and comprehensive mineral nutrition, and also contains a variety of amino acids and vitamins. In particular, the organic matter of starch, protein, and cellulose in P. australis feed degrades into monosaccharide, disaccharide, amino acid, and trace elements after fermentation, which makes the feed become soft, fragrant, and more palatable (1). Therefore, P. australis has the potential to be an important roughage for livestock. According to the determination (data from the American Feed Regulation Society NRC2-01-113), the dry matter of the stem and leaves of the young P. australis contained metabolizable energy 9.20 MJ/kg, crude protein 12.2%, crude fiber 26.8%, calcium 0.4%, and phosphorus 0.3%, which was higher than that of the common forage (3, 4). Existing studies have found that adding a certain amount of dried reed to the diet can accelerate the growth of livestock and improve the feed utilization rate (4).
Kadi et al. (5) reported that P. australis feed contained high N content, neutral detergent fiber (NDF), potassium, and magnesium. Tanaka et al. (1) investigated the timing of harvest and nutritive value of P. australis for ruminants in Lake Dianchi of China, which found that P. australis harvested in the early growing stage had relatively high concentrations of total digestible nutrients and demonstrated that P. australis can use a high-quality roughage for ruminants. Generally, there are three feed types of P. australis feed used in livestock: fresh, sun-dried, and ensiled. P. australis shoot remainder (PSR) is a by-product of processing P. australis shoots. By analyzing the nutrient composition of PSR, we detected that the crude protein and crude fiber contents reached 14.93 and 19.27%, respectively, which have high nutritional value, but barely have been utilized (unpublished data). While fresh PSR cannot be preserved for a long time, ensiled PSR is considered to be an effective long-term preservation method for beef cattle breeding. Different roughage may influence production performance and rumen microbial structure and function in ruminants (6–8). However, little research has indicated whether PSR can replace traditional feed ingredients in ruminants, especially affecting the rumen microbiota of beef cattle. Furthermore, effects of PSR silage on growth performance, blood biochemical indices, and rumen microbiota of beef cattle have no reports. Thus, the aim of this study was to explore the effect of PSR silage substitution for corn silage, totally or partially, on growth, serum biochemical indices, rumen microbial diversity, and predicted function in beef cattle.
Materials and Methods
Animals, Treatments, and Experimental Procedures
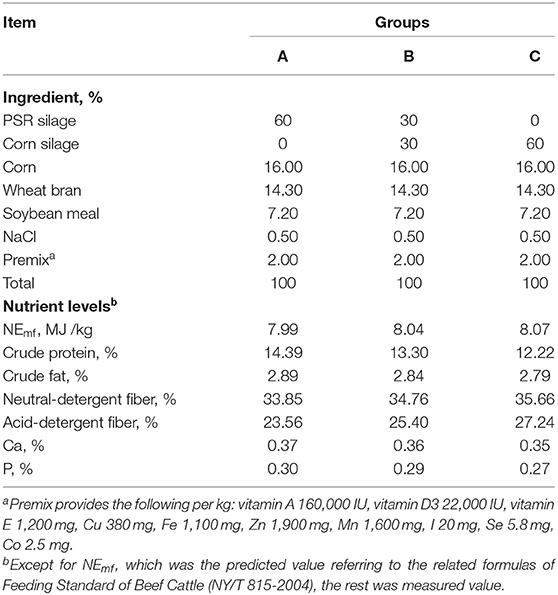
Fifteen Angus beef cattle with an average initial body weight (IBW) of 253 ± 2.94 kg were chosen and randomly allotted to three experimental treatments consisting of three dietary levels of PSR silage (DM basis): 60% (group A), 30% (group B), and 0 (group C) as a substitute of corn silage, respectively. Experimental diets were composed of 60% of silage and 40% of concentrate (DM basis) and were formulated to meet nutritional requirements (9) of beef cattle, and feed ingredients, and the nutritional composition are shown in Table 1. Each bull was fed in individual pens with automatic drinking and free feeding intake, five pens per group. Before the trial, all bulls were weighed, dewormed, and vaccinated (foot and mouth disease vaccine and anthrax vaccine). Cattle were adapted to the diets for 15 days, and the experimental period lasted for 45 days.
Bulls were fed three times daily at 07:00, 12:00, and 17:00 h with total mixed diets. Residual feed was evaluated at 06:00 h each day to quantify and adjust daily feed allowance to a maximum of 5% residues. Feed samples were collected from each pen every 15 days and then composited, and were frozen at −20°C for nutritional ingredients analysis.
Growth Performance and Blood Biochemical Parameters
Each bull was weighed individually in the morning on an empty stomach at the beginning and end of the experiment. The ADG (average daily gain) was calculated by the weight gain per bull divided by the trial days. The ADFI (average daily feed intake) was calculated by the amount of diet offered minus the residues per pen and then divided by the total trial days. The FCR (feed conversion ratio) was calculated as ADFI per ADG (10). At the end of the experiment, blood samples were collected in a 5-ml vacuum tube without anticoagulant (Changsha Yiqun Medical Equipment) from the caudal vein of each bull in the morning on an empty stomach. After standing for 2–3 h, blood samples were centrifuged by the model TG16-WS H1650 centrifuges (Hunan Xiangyi Laboratory Instrument Development Co. Ltd.) at 3,000 r/min for 10 min, and then the supernatant was separated and stored at −20°C for further analysis. A TBA-120FR automatic biochemistry analyzer (Toshiba Corporation) was used to measure the concentrations of serum biochemical parameters (11–13).
DNA Extraction and Amplification of 16S rRNA Genes
The total microbial genomic DNA was extracted using the CTAB/SDS method. The V3–V4 regions of 16S rRNA genes were amplified with forward primer V515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and reverse primer V806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR reactions were performed in 30-μl systems. For specific PCR amplified procedures, refer to Wang et al. (14, 15). The sequencing libraries were constructed by Ion Plus Fragment Library Kit 48 rxns (Thermo Scientific). The Ion S5TM XL platform to sequence was further used, and 407- to 412-bp single-end reads were generated (14, 15).
Sequencing and Bioinformatics Analysis
The raw reads were cleaned by the Cutadapt quality control process (16). The UCHIME algorithm (17) was used to detect and remove the chimera sequences and finally to obtain the clean reads. Sequence analysis was performed by Uparse software (Uparse v7.0.1001) (18) to cluster the operational taxonomic units (OTUs) with ≥97% similarity. The Silva Database (19) was used to annotate taxonomic information and normalize the OTU abundant information. The alpha diversity and beta diversity were analyzed subsequently by QIIME (Version 1.7.0) and displayed by R Software (Version 2.15.3). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was utilized to predict the metabolic function of the microbiota. The raw sequencing data of this study were submitted to the Sequence Read Archive (SRA) with accession numbers SRR15662882–SRR15662896.
Statistical Analysis
The experimental data were analyzed on SPSS 22.0 software packages (SPSS, Chicago, IL, USA). Using the one-way ANOVA and t-tests to test the significance of growth performances and serum biochemical parameters, and the non-parameter test was performed to analyze the rumen microbial diversities, relative abundance, and function prediction. Final results were presented with meaning values. Differences were considered to have a tendency at 0.05 < p < 0.10 and statistically significant at p ≤ 0.05.
Results
Growth Performance
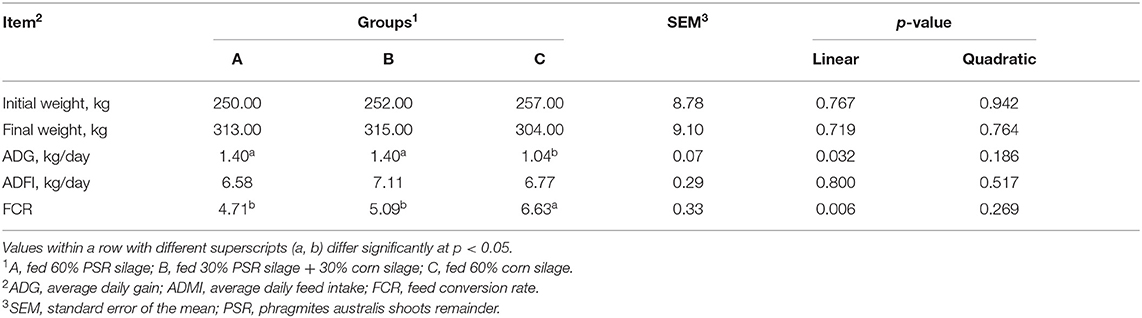
The results of growth performance are shown in Table 2. There was no significant difference in IBW (initial body weight) and ADFI among treatments, although the FBW (final body weight) did not differ among treatments and was higher in groups A and B than in group C. The ADG was significantly greater (linear, p = 0.032) in groups A and B than in group C. Similarly, the FCR was significantly lower (linear, p = 0.006) in groups A and B than in group C. Notably, the ADG and FCR were not different between A and B treatments.
Serum Biochemical Parameters
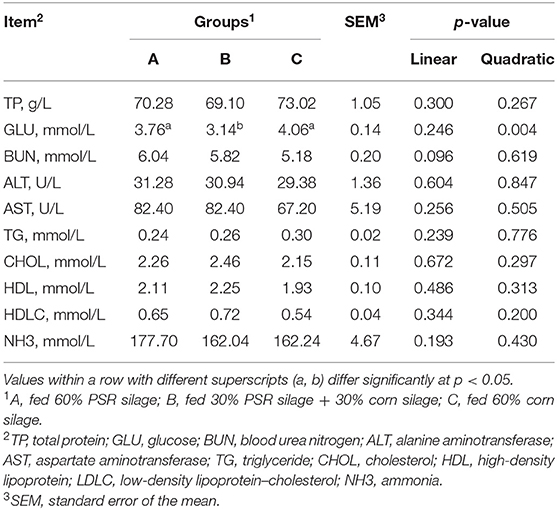
The concentrations of serum TP, ALT, AST, TG, CHOL, HDL, LDLC, and NH3 did not differ among treatments. The concentration of serum GLU was quadratically affected (quadratic, p = 0.004) among treatments, and the concentration of BUN showed a linearly downward trend (linear, p = 0.096) among treatments (Table 3).
Rumen Bacterial Communities
The results of sequencing analysis are presented in Figure 1. The difference in bacterial composition among the three groups was analyzed by the PCoA, and the PCoA plots showed that the group A data had a tendency to be separated from both the B and C groups (Figure 1A). Similarly, there were no significant differences in diversity and uniformity by the level of species richness existing between B and C based on the observed species (OTUs), Shannon, Chao1, and Ace index analyses (Figure 1B). Compared with both groups B and C, the A group had less OTUs (p < 0.001), Chao1 (p < 0.001), ACE (p < 0.001), and Shannon (p < 0.01) unexpectedly (Figure 1B).
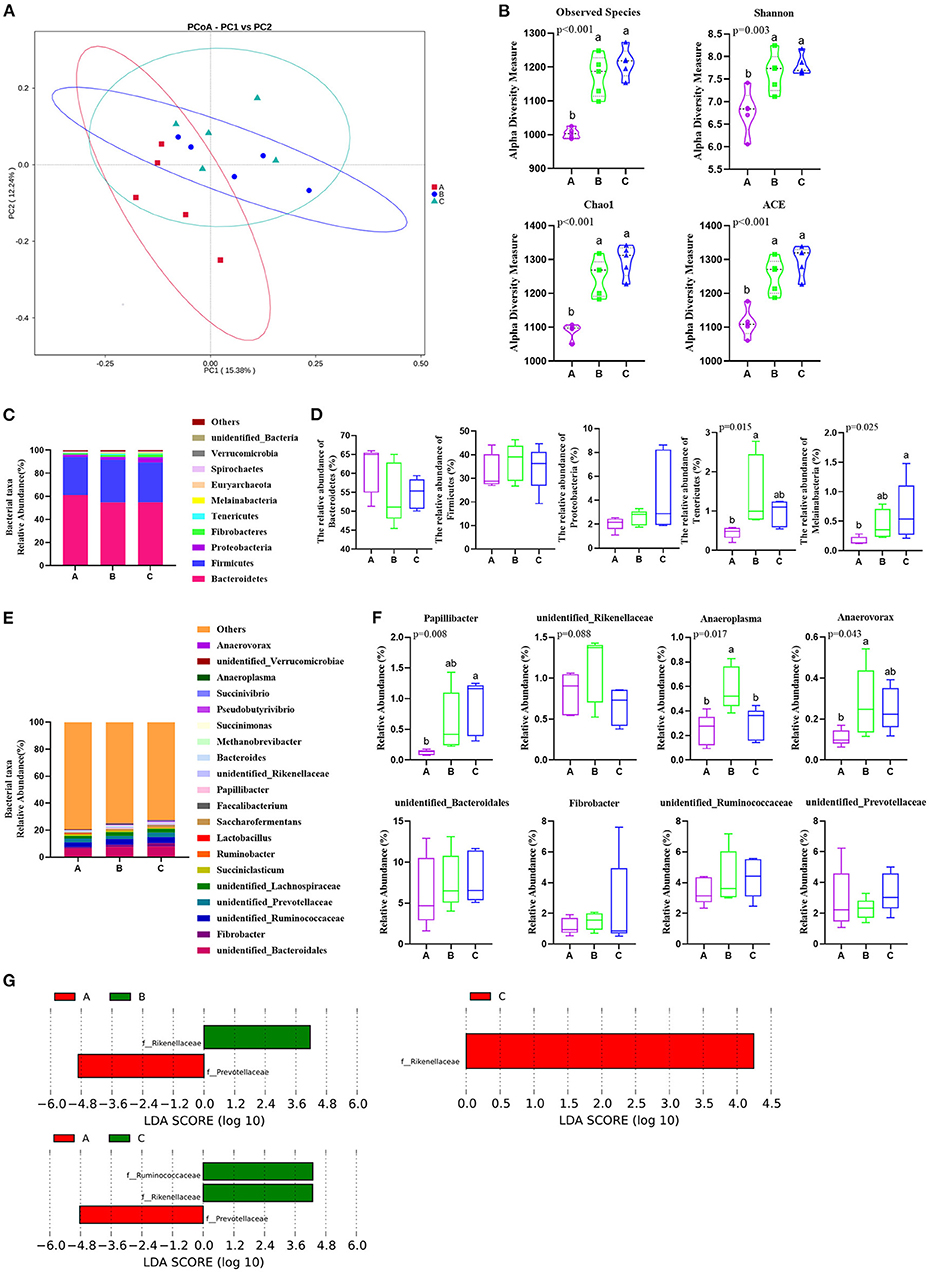
Figure 1. Phragmites australis shoot remainder (PSR) silage totally or partially substituting the corn silage alters rumen microbiota composition in Angus beef cattle (N = 5). Group A, fed 60% PSR silage; Group B, fed 30% PSR silage + 30% corn silage; and Group C, fed 60% corn silage. (A) PCoA analysis of rumen microbiota based on operational taxonomic unit (OTU) abundance. (B) Assessment of alpha diversity. (C) Rumen microbiota taxonomic profiling at the phylum level. (D) Relative abundances of Bacteroidetes, Firmicutes, Proteobacteria, Tenericutes, and Melainabacteria. Bars with different letters (a, b) indicate significant differences (p < 0.05) among different groups (the same below). (E) Rumen microbiota taxonomic profiling at the genus level. (F) Relative abundances of representative and significant difference genera. (G) LDA score of rumen microbiota composition according to LEfSe analysis by three treatments.
A total of 22 bacterial phyla were identified by taxonomic analysis in the rumen samples. The relative abundance of more than 1% were Bacteroidetes, Firmicutes, Proteobacteria, and Fibrobacteres (Figures 1C,D). Notably, the relative abundance of Bacteroidetes and Firmicutes was the richest in the three trial groups (Figures 1C,D). Additionally, the relative abundance of Tenericutes (p = 0.015) and Melainabacteria (p = 0.025) was significantly lower in group A than that in group B and group C, respectively, at the phyla level (Figure 1D).
A total of 122 bacterial genera were detected at the genus level. Twenty representative genera were elucidated in all the rumen samples (Figure 1E). Among these genera, the relatively high abundance (>1%) unidentified_Bacteroidales belonged to Bacteroidetes in the phylum, Fibrobacter is one of Fibrobacteres, unidentified_Ruminococcaceae, and unidentified_Lachnospiraceae belong to Clostridia in members of Firmicutes. Unidentified_Prevotellaceae belongs to Bacteroidetes, Succiniclasticum is also a member of Firmicutes. The relative abundances of Papillibacter (p = 0.008), Anaeroplasma (p = 0.017), and Anaerovorax (p = 0.043) had significant differences among the three groups, and the relative abundance of unidentified_Rikenellaceae (p = 0.088) also had a notable change in group B in the genus level (Figure 1F). Additionally, LEfSe analysis results showed the dominant bacteria species for each group by LDA score, the Prevotellaceae and Rikenellaceae showed statistical differences and were considered as biomarkers between groups A and B, the Prevotellaceae, Ruminococcaceae, and Rikenellaceae were the biomarkers between groups A and C, and the Rikenellaceae was the unique biomarker among the three groups (Figure 1G). Furthermore, the phylogenetic tree of the top 100 genera was obtained through multisequence alignment, as shown in Figure 2, in which the phylogenetic relationship of rumen bacteria species at the genus level could be presented more intuitively among the three groups.
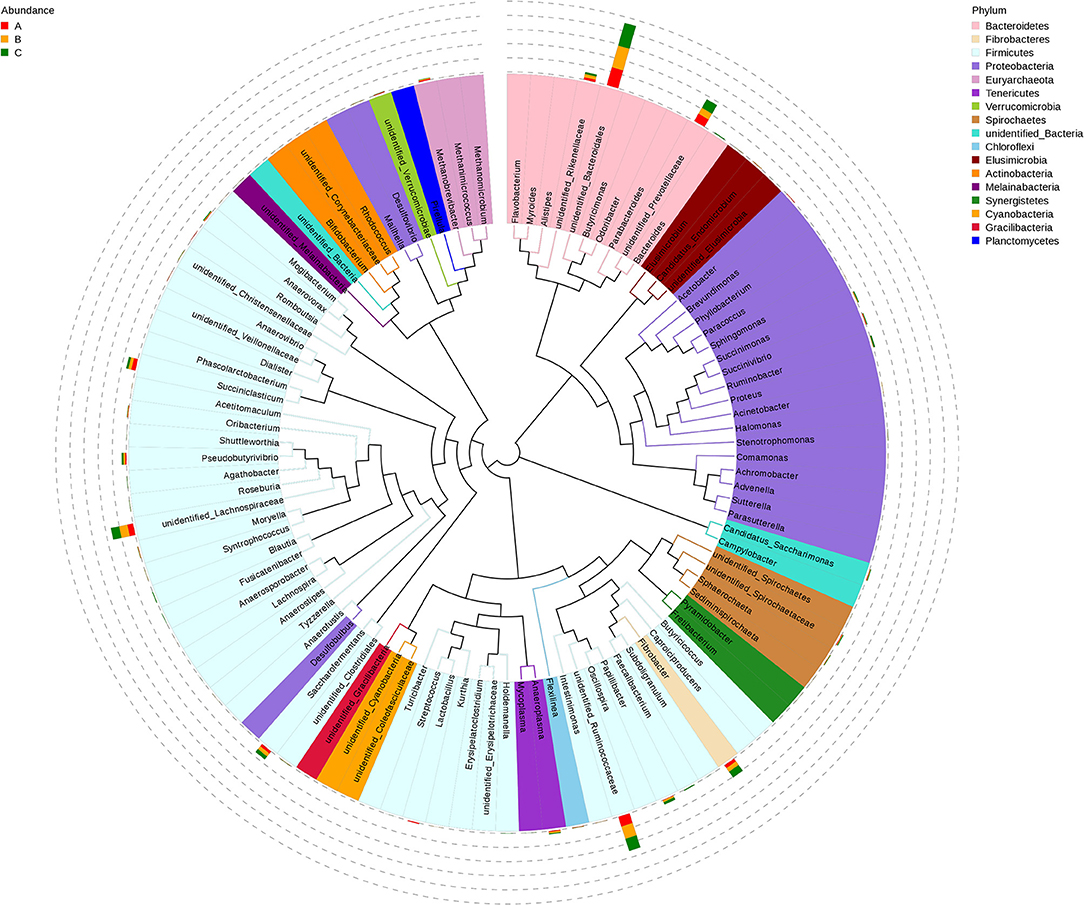
Figure 2. Phylogenetic tree of genus level species among PSR silage totally or partially substituted the corn silage groups in Angus beef cattle (N = 5). Group A, fed 60% PSR silage; Group B, fed 30% PSR silage + 30% corn silage; and Group C, fed 60% corn silage. Branches and fan colors represent its corresponding phyla, the accumulation histogram outside the fan ring shows the relative abundance distribution information of the genus in different groups.
Predicted Metabolic Pathways and Functions of Rumen Bacterial Communities
Metabolic functions of rumen bacteria were predicted by PICRUSt in the present study (Figure 2). The result showed that “metabolism” was in the highest relative abundance with more than 49% of all sequence reads among three groups at KEGG level 1 (Figure 3A). At KEGG level 2, the most relatively abundant gene families (relative abundance > 0.10%) from all rumen samples are present in Figure 3B. Genes belonging to amino acid metabolism, carbohydrate metabolism, replication and repair, membrane transport, translation, and energy metabolism were the most relative abundant among the three groups (Figure 3B). Among these gene families, the genes associated with metabolism of cofactors and vitamins (p = 0.028), cellular processes and signaling (p = 0.049), metabolism (p = 0.001), biosynthesis of other secondary metabolites (p = 0.024), infectious diseases (p = 0.006), signaling molecules and interaction (p = 0.013), nervous system (p = 0.041), and digestive system (p = 0.014) were significantly decreased from groups A to C, amino acid metabolism (p = 0.066) and nucleotide metabolism (p = 0.057) showed the descent tendency, while lipid metabolism (p = 0.039) was dramatically increased, and signal transduction had an increasing tendency (p = 0.092) from groups A to C (Figure 3C). At KEGG level 3, the majority of relatively abundant pathways were transporters, general function prediction only, DNA repair and recombination proteins, ribosome, purine metabolism, and ABC transporters (Figure 3D). Notably, the relative abundance of 11 pathways showed significant variation among the three groups (Figure 3E). The pathways involved in the pyrimidine metabolism (p = 0.023), DNA replication proteins (p = 0.038), glycine, serine, and threonine metabolism (p = 0.019), arginine and proline metabolism (p = 0.015), other ion coupled transporters (p = 0.005), alanine, aspartate, and glutamate metabolism (p = 0.019), cysteine and methionine metabolism (p = 0.003), transcription machinery (p = 0.029), energy metabolism (p = 0.010), and general function prediction only (p = 0.042) were significantly increased, and DNA repair and recombination proteins (p = 0.094) and peptidases (p = 0.074) had increased trend in group A compared with the other two groups. Inversely, secretion system (p = 0.030) was significantly decreased, and pyruvate metabolism (p = 0.079) had a decreased trend in group A than in the other two groups B and C.
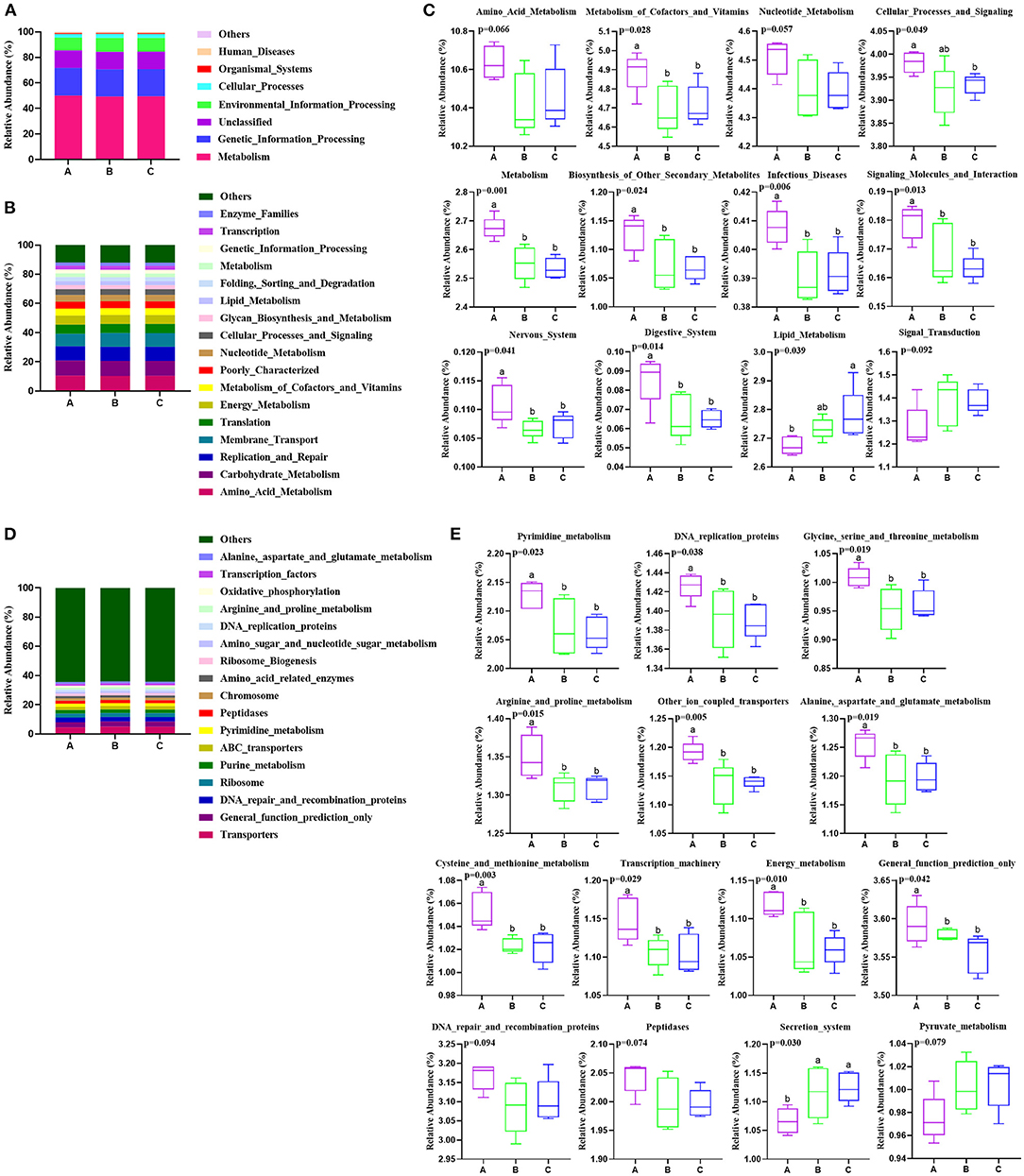
Figure 3. Effects of PSR silage totally or partially substituted the corn silage on the predicted functional composition of rumen bacterial community in Angus beef cattle (N = 5). Group A, fed 60% PSR silage; Group B, fed 30% PSR silage 30% corn silage; and Group C, fed 60% corn silage. (A) The majority of the gene sequences annotated to KEGG level 1. (B) The majority of the gene sequences annotated to KEGG level 2. (C) The gene families of significant differences at KEGG level 2. (D) The majority of the gene sequences annotated to KEGG level 3. (E) The relative abundant pathways with significant differences at KEGG level 3. Bars with different letters (a,b) indicate significant differences (p < 0.05) among different groups.
Discussion
Depending on the results of animal feeding trial, PSR silage group (group A), mixed group (group B), and corn silage (group C) affected the growth performance of Angus beef cattle. The ADG was significantly affected by PSR silage, which in PSR silage group was the highest, followed by the mixed group, equally 34.62% higher than corn silage group. Inversely, the FCR in the PSR silage group and mixed group was obviously lower than in the corn silage, 31.52 and 23.23% lower, respectively, while the ADFI had no significant difference among the three groups. These results indicated that PSR silage substitution for corn silage totally or partially could improve the growth performance of beef cattle, mainly by improving feed utilization efficiency to increase the ADG in the growing–fattening of Angus beef cattle. Thus, it can be observed that PSR silage might completely replace corn silage for beef cattle breeding. Therefore, the effective use of P. australis and other unconventional feed materials may not only expand feed sources and reduce feeding costs but also further improve the weight gain rate and meat production performance of beef cattle, and finally increase the economic benefits of breeding.
Nutrients are digested and absorbed by the body and carried through the bloodstream to tissues, organs, and cells. Therefore, blood biochemical indicators can be a good response to the intake of nutrition levels of the body. GLU concentration can reflect the energy metabolism of animals (20). In the current study, the concentration of GLU in the mixed group was significantly lower than in the PSR silage and corn silage treatments, while the higher ADG was observed in the mixed group. These results were inconsistent with previous studies, and this might be related to species and feed composition (11–15). The concentration of BUN is perceived as an effective indicator to measure the metabolism of protein and amino acid, low BUN level indicates high nitrogen metabolism capacity (21). Diets supplemented with PSR silage increased the concentrations of BUN in the present study, which might be related to the high protein content of P. australis shoots remainder.
Rumen is the most powerful digestive organ, in which complex microbial communities are closely related to degrade and convert plant materials in ruminants (22). More than 70% of the energy was provided by rumen bacteria fermentation to ensure the host growth and reproduction performance (23), and the compositions of rumen microorganism are influenced by diets (24). In the present study, the core microbiome accounts for more than 78% of total OTUs among the three groups, and this result was similar with other researches (11–13). The core microbiome plays crucial roles in maintaining the “functional redundancy” of rumen ecosystem, and this redundancy further guarantees its major functional properties (25). The present study showed that the observed species (OTUs), Shannon, Chao1, and ACE indices were significantly decreased in the PSR silage diet, indicating that the rumen microbiome was altered by PSR silage.
The present study systematically revealed the composition and structure of rumen microbiome in Angus beef cattle fed PSR silage. Bacteroidetes and Firmicutes were the most two relatively abundant phyla in the current study, which was referred to the degradation of protein and carbohydrates (26), and these findings were consistent with previous studies (27). de Menezes et al. (28) have also found that the dairy cows fed pasture or TMR diets did not have obvious differences on the rumen microbiome at the phylum level, with the sequences of Bacteroidetes and Firmicutes representing approximately 80% of the total rumen microbiome. Consistently, the relative abundance of Bacteroidetes and Firmicutes were over 80% in all three diets in the current study. Proteobacteria was detected as the third most relatively abundant phylum in this study, which was similar to previous studies (29). In general, the composition and structure of rumen microbial community might be related to the feed efficiency and animal breed. Studies have found that several members belonged to the phylum Tenericutes related to being animal pathogens and parasites, and the greater abundance of Tenericutes was associated with the reduced intramuscular fat deposition of longissimus in Angus steers (30, 31). The relative abundance of Tenericutes was the lowest in the PSR silage diet in the present study, indicating that PSR silage might improve gastrointestinal health and further promotes intramuscular fat deposition. Actually, the growth performance was the greatest by feeding PSR silage diet. Melainabacteria is a newly identified gut bacteria, whose relative abundance in the rectal contents of diabetic model rats was significantly increased (32). The relative abundance of Melainabacteria was significantly decreased (especially in group A) after adding PSR silage to the diets in the present study, which indicated that PSR silage-substituted corn silage might improve host sugar, fat, and protein metabolism.
At the genus level, the effects of PSR silage on the rumen bacterial population were further identified in this study. The relative abundance of Papillibacter in the PSR silage group was significantly lower than that in the corn silage and mixed groups. Papillibacter is known as a butyrate producer (33). The decreased abundance of Papillibacter in PSR silage indicated that the butyrate production was relatively decreased by rumen microbiota. The relative abundance of Anaeroplasma in the mixed group was significantly higher than the groups of corn silage and PSR silage. Anaeroplasma is a genus characterized by its anaerobic fermentation, which produces fatty acids as propionate (34). All of these taxa were previously reported as members of the regular and efficient microbiota from rumen, and their increased abundance may indicate an improved ability of digestion or, at least, a need for more specialized fermentation in rumen due to, for example, more food intake. Besides, Anaeroplasma was highly correlated with high weight gain, and may be important for cattle nutrition either individually or in a consortium (35).
Some studies indicate that Anaerovorax can generate more SCFAs to provide additional energy sources and maintain feed efficiency (36). Unexpectedly, the relative abundance of Anaerovorax in the PSR silage group is significantly lower than that in the corn silage and mixed groups in the present study. The relative abundance of unidentified_Rikenellaceae in the corn silage group was obviously lower than that in the PSR silage and mixed groups. To date, all cultured members of the family Rikenellaceae are described as anaerobic, mesophilic, and rod-shaped bacteria that usually ferment carbohydrates or proteins. Su et al. (37) isolated a carbohydrate-fermenting and hydrogen-producing Rikenellaceae from a reed swamp in China (37). It remains to identify and examine the functions of the unidentified Rikenellaceae to understand the roles in the present study. Additionally, the PSR silage diet increased the relative proportion of Rikenellaceae at the family level, and Rikenellaceae might be one of the biomarkers between PSR silage feed and corn silage feed using LEfSe.
The composition of rumen microorganisms affects host metabolic function and physiological health. The relative abundance of the dominant microbial phyla is stable in ruminants. The dominant three microbial phyla were Bacteroidetes, Firmicutes, and Proteobacteria in the rumen among the three groups, indicating that the rumen microbiota in cattle was also relatively stable at the phyla level. These results were in agreement with previous studies (38, 39), and the most dominant phyla Firmicutes and Bacteroidetes were closely related to carbohydrate and protein metabolism (40, 41). At the genus level, the dominant four genera were unidentified Bacteroidales, Fibrobacter, unidentified Ruminococcaceae, and unidentified Prevotellaceae, and their relative abundance all did not have significant difference among different diets in the present study, which was similar to previous studies (41, 42). Menni et al. (43) found that the abundance of Ruminococcaceae might play a significant role in energy and lipid metabolism, and which was negatively associated with vascular sclerosis. These results indicated that the PSR silage diet might not affect host health in cattle.
The PICRUSt prediction results showed that amino acid metabolism, carbohydrate metabolism, replication and repair, membrane transport, translation, and energy metabolism were the dominant gene families at KEGG level 2, all of which are essential for survival, growth, and reproduction of gastrointestinal microbes (44). These results were similar to our previous studies in sheep (11–15). Among these gene families, unexpectedly, the genes associated with metabolism of cofactors and vitamins, cellular processes and signaling, metabolism, biosynthesis of other secondary metabolites, infectious diseases, signaling molecules and interaction, nervous system, and digestive system were significantly higher in the PSR silage diet than in the mixed and corn silage diets, while lipid metabolism was dramatically lower in the PSR silage diet than in the corn silage diet. Furthermore, the majority of gene families were transporters, general function prediction only, DNA repair, and recombination proteins, ribosome, purine metabolism, and ABC transporters at KEGG level 3. Notably, the relative abundance of 11 pathways showed significant variation among the three groups. The pathways involved in the pyrimidine metabolism, DNA replication proteins, glycine, serine, and threonine metabolism, arginine and proline metabolism, other ion coupled transporters, alanine, aspartate, and glutamate metabolism, cysteine and methionine metabolism, transcription machinery, energy metabolism, and general function prediction only were significantly higher, and secretion system was significantly lower in the PSR silage diet than in the mixed and corn silage diets. These results indicated an enhanced fermentation activity performed by rumen microorganisms in the PSR silage diet. The current study also implied that feeding only a roughage of PSR silage diet altered the ruminal microbial functions.
In conclusion, the present study mainly investigated that the growth performance, blood biochemical parameters, and the composition and function of rumen microbiota of PSR silage feed totally or partially substituted the corn silage in Angus beef. The results suggest that the PSR silage diet and mixed diet increase ADG and decrease FCR, reduce serum glucose levels, and alter the rumen microbiota and inferred metabolic functions. These findings indicated that PSR silage could partially substitute corn silage for beef cattle breeding, replacing 30% of corn silage in the diet has good feeding effect in cattle.
Conclusions
Feeding different PSR silage level diets improved growth performance, changed the contents of serum glucose and urea nitrogen and, furthermore, might affect the energy and protein metabolic efficiency of Angus beef. Moreover, the rumen bacterial diversity indices decreased significantly by feeding PSR silage, the relative abundances of Tenericutes and Melainabacteria were significantly reduced by feeding PSR silage, and Papillibacter, Anaeroplasma, and Anaerovorax had significantly decreased by feeding PSR silage at the genus level, and furthermore, the metabolic pathways were significantly influenced by related bacteria for PSR silage. The results indicated that feeding PSR silage could improve the growth performance and alter the rumen bacteria diversity and the corresponding function, and PSR silage could partially substitute (30%) corn silage for beef cattle breeding.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR15662882-SRR15662896.
Ethics Statement
The experimental procedures of this study were approved by the Animal Care Committee of Hunan Normal University in reference to the Administration of Affairs Concerning Experimental Animals.
Author Contributions
HY and QW conceptualized the study. XZ and XL handled the methodology. PH and ZF were in charge of the software. MZ was responsible for the validation. YZ and YaW performed the formal analysis. QW conducted the investigation, prepared and wrote the original draft, and handled the project administration. HY handled the resources and acquired the funding. XW and JH were responsible for the data curation. MC and YiW assisted in the writing, review, and editing of the draft. MZ and JY handled the visualization. JL was in charge of the supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Hunan Provincial Key Laboratory of Animal Nutritional Physiology and Metabolic Process open fund projects (ISA2020113), Scientific Research Project of Hunan Education Department (No. 20B369), Young Elite Scientists Sponsorship Program by CAST (YESS20160086), and Hunan Province's Strategic and Emerging Industrial Projects (2018GK4035).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to the Yuanjiang Xinda Forestry and Animal Husbandry Technology Development Co., Ltd. of Yiyang (China) for the experimental animals and the research infrastructure.
References
1. Tanaka TS, Irbis C, Kumagai H, Inamura T. Timing of harvest of Phragmites australis (CAV.) Trin. ex Steudel affects subsequent canopy structure and nutritive value of roughage in subtropical highland. J Environ Manage. (2016) 166:420–8. doi: 10.1016/j.jenvman.2015.10.055
2. Lambertini C, Guo WY, Ye S, Eller F, Guo X, Li XZ, et al. Phylogenetic diversity shapes salt tolerance in Phragmites australis estuarine populations in East China. Sci Rep. (2020) 10:17645. doi: 10.1038/s41598-020-74727-0
3. Baran M, Varadyova Z, Krcacmar S, Hedvabný J. The common reed (Phragmites australis) as a source of roughage in ruminant nutrition. Acta Vet Brno. (2002) 71:e445–9.
4. Foroughi AR. Investigation of utilization of dry reed forage (Phragmites australis) in diet of fattening buffalo male calves. Iran Anim Sci Res J. (2008) 3:57–63.
5. Kadi SA, Quendi M, Slimani M, Selmani K, Bannelier C, Berchiche M, et al. Nutritive value of common reed (Phragmites australis) leaves for rabbits. In: Proceedings of 10th World Rabbit Congress (WRC). Sharm El-Sheikh (2012). p. 513–7.
6. Prajapati VS, Purohit HJ, Raje DV, Parmar N, Patel AB, Jones OAH, et al. The effect of a high-roughage diet on the metabolism of aromatic compounds by rumen microbes: a metagenomic study using Mehsani buffalo (Bubalus bubalis). Appl Microbiol Biot. (2016) 100:1319–31. doi: 10.1007/s00253-015-7239-0
7. Faubladier C, Julliand V, Danel J, Philippeau C. Bacterial carbohydrate-degrading capacity in foal faeces:changes from birth to pre-weaning and the impact of maternal supplementation with fermented feed products. Br J Nutr. (2013) 110:1040–52. doi: 10.1017/S0007114512006162
8. Ding S, Fang J, Liu G, Veeramuthu D, Abdullah AN, Yin Y. The impact of different levels of cysteine on the plasma metabolomics and intestinal microflora of sows from late pregnancy to lactation. Food Funct. (2019) 10:691–702. doi: 10.1039/c8fo01838c
9. NRC. Nutrient Requirements of Beef Cattle. 8th ed. Washington, DC: National Academy Press (2016).
10. Yin L, Li J, Wang H, Yi Z, Wang L, Zhang S, et al. Effects of vitamin B6 on the growth performance, intestinal morphology, and gene expression in weaned piglets that are fed a low-protein diet1. J Anim Sci. (2020) 98:skaa022. doi: 10.1093/jas/skaa022
11. Wang Y, Shen Q, Zhong S, Chen Y, Yang Y. Comparison of rumen microbiota and serum biochemical indices in white cashmere goats fed ensiled or sun-dried mulberry leaves. Microorganisms. (2020) 8:981. doi: 10.3390/microorganisms8070981
12. Wang Q, Wang Y, Hussain T, Dai C, Li J, Huang P, et al. Effects of dietary energy level on growth performance, blood parameters and meat quality in fattening male Hu lambs. J Anim Physiol Anim Nutr. (2020) 104:418–30. doi: 10.1111/jpn.13278
13. Wang Y, Wang Q, Dai C, Li J, Huang P, Li Y, et al. Effects of dietary energy on growth performance, carcass characteristics, serum biochemical index, and meat quality of female Hu lambs. Anim Nutr. (2020) 6:499–506. doi: 10.1016/j.aninu.2020.05.008
14. Wang Q, Zeng Y, Zeng X, Wang X, Wang Y, Dai C, et al. Effects of dietary energy levels on rumen fermentation, gastrointestinal tract histology and bacterial community diversity in fattening male Hu lambs. Front Microbiol. (2021) 12:695445. doi: 10.3389/fmicb.2021.695445
15. Wang X, Wang Y, Wang Q, Dai C, Li J, Huang P, et al. Effect of dietary protein on growth performance, and serum biochemical index in late pregnant Hu ewes and their offspring. Anim Biotechnol. (2021) 30:1–9. doi: 10.1080/10495398.2021.1939042
16. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. (2011) 17:10−2. doi: 10.14806/ej.17.1.200
17. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
18. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. doi: 10.1038/nmeth.2604
19. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41:D590–6. doi: 10.1093/nar/gks1219
20. Graugnard DE, Bionaz M, Trevisi E, Moyes KM, Salak-Johnson JL, Wallace RL, et al. Blood immunometabolic indices and polymorphonuclear neutrophil function in peripartum dairy cows are altered by level of dietary energy prepartum. J Dairy Sci. (2012) 95:1749–58. doi: 10.3168/jds.2011-4579
21. Song S, Wu J, Zhao S, Casper DP, Zhang L, He B, et al. The effect of periodic energy restriction on growth performance, serum biochemical indices, and meat quality in sheep. J Anim Sci. (2018) 96:4251–63. doi: 10.1093/jas/sky299
22. Li D, Zhang Y, Cui Z, He L, Chen W, Meng Q, et al. Effects of phytoecdysteroids (PEDS) extracted from Cyanotis arachnoidea on rumen fermentation, enzyme activity and microbial efficiency in a continuous-culture system. PLoS ONE. (2016) 11:e153584. doi: 10.1371/journal.pone.0153584
23. Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. (2007) 9:1101–11. doi: 10.1111/j.1462-2920.2007.01281.x
24. Henderson G, Cox F, Ganesh S, Jonker A, Young W, Janssen PH. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. (2015) 5:14567. doi: 10.1038/srep14567
25. Shade A, Handelsman J. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol. (2012) 14:4–12. doi: 10.1111/j.1462-2920.2011.02585.x
26. Ye H, Liu J, Feng P, Zhu W, Mao S. Grain-rich diets altered the colonic fermentation and mucosa-associated bacterial communities and induced mucosal injuries in goats. Sci Rep. (2016) 6:20329. doi: 10.1038/srep20329
27. Derakhshani H, Tun HM, Cardoso FC, Plaizier JC, Khafipour E, Loor JJ. Linking peripartal dynamics of ruminal microbiota to dietary changes and production parameters. Front Microbiol. (2017) 7:2143. doi: 10.3389/fmicb.2016.02143
28. de Menezes AB, Lewis E, O'Donovan M, O'Neill BF, Clipson N, Doyle EM. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol Ecol. (2011) 78:256–65. doi: 10.1111/j.1574-6941.2011.01151.x
29. Golder HM, Denman SE, Mcsweeney C, Wales WJ, Auldist MJ, Wright MM, et al. Effects of partial mixed rations and supplement amounts on milk production and composition, ruminal fermentation, bacterial communities, ruminal acidosis. J Dairy Sci. (2014) 97:5763–85. doi: 10.3168/jds.2014-8049
30. Zhan J, Liu M, Wu C, Su X, Zhan K, Zhao GQ. Effects of alfalfa flavonoids extract on the microbial flora of dairy cow rumen. Asian Australas J Anim Sci. (2017) 30:1261–9. doi: 10.5713/ajas.16.0839
31. Krause TR, Lourenco JM, Welch CB, Rothrock MJ, Callaway TR, Pringle TD. The relationship between the rumen microbiome and carcass merit in Angus steers. J Anim Sci. (2020) 98:skaa287. doi: 10.1093/jas/skaa287
32. Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife. (2013) 2:e01102. doi: 10.7554/eLife.01102
33. Xue Y, Lin L, Hu F, Zhu W, Mao S. Disruption of ruminal homeostasis by malnutrition involved in systemic ruminal microbiota-host interactions in a pregnant sheep model. Microbiome. (2020) 8:138. doi: 10.1186/s40168-020-00916-8
34. Wang B, Wang Y, Zuo S, Peng S, Wang Z, Zhang Y, et al. Untargeted and targeted metabolomics profiling of muscle reveals enhanced meat quality in artificial pasture grazing tan lambs via rescheduling the rumen bacterial community. J Agric Food Chem. (2021) 69:846–58. doi: 10.1021/acs.jafc.0c06427
35. de Freitas AS, de David DB, Takagaki BM, Roesch L. Microbial patterns in rumen are associated with gain of weight in beef cattle. Antonie van Leeuwenhoek. (2020) 113:1299–312. doi: 10.1007/s10482-020-01440-3
36. Lau S, Teng J, Chiu TH, Chan E, Tsang A, Panagiotou G, et al. Differential microbial communities of omnivorous and herbivorous cattle in Southern China. Comput Struct Biotechnol J. (2018)16:54–60. doi: 10.1016/j.csbj.2018.02.004
37. Su XL, Tian Q, Zhang J, Yuan XZ, Shi XS, Guo RB, et al. Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol. (2014) 64:2986–91. doi: 10.1099/ijs.0.063917-0
38. Wetzels SU, Mann E, Pourazad P, Qumar M, Pinior B, Metzler-Zebeli BU, et al. Epimural bacterial community structure in the rumen of Holstein cows with different responses to a long-term subacute ruminal acidosis diet challenge. J Dairy Sci. (2017) 100:1829–44. doi: 10.3168/jds.2016-11620
39. Plaizier JC, Li S, Tun HM, Khafipour E. Nutritional models of experimentally-induced subacute ruminal acidosis (SARA) differ in their impact on rumen and hindgut bacterial communities in dairy cows. Front Microbiol. (2017) 7:2128. doi: 10.3389/fmicb.2016.02128
40. Hook SE, Steele MA, Northwood KS, Dijkstra J, France J, Wright AD, et al. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol. (2011) 78:275–84. doi: 10.1111/j.1574-6941.2011.01154.x
41. Wang Y, Cao P, Wang L, Zhao Z, Chen Y, Yang Y. Bacterial community diversity associated with different levels of dietary nutrition in the rumen of sheep. Appl Microbiol Biotechnol. (2017) 101:3717–28. doi: 10.1007/s00253-017-8144-5
42. Wang W, Li C, Li F, Wang X, Zhang X, Liu T, et al. Effects of early feeding on the host rumen transcriptome and bacterial diversity in lambs. Sci Rep. (2016) 6:32479. doi: 10.1038/srep32479
43. Menni C, Lin C, Cecelja M, Mangino M, Matey-Hernandez ML, Keehn L, et al. Gut microbial diversity is associated with lower arterial stiffness in women. Eur Heart J. (2018) 39:2390–7. doi: 10.1093/eurheartj/ehy226
Keywords: Phragmites australis feed, growth performance, rumen microbiota, rumen bacterial function, beef cattle
Citation: Wang Q, Zeng X, Zeng Y, Liu X, Wang Y, Wang X, Li J, Wang Y, Feng Z, Huang P, Yin J, Huang J, Zhu M and Yang H (2022) Effects of Phragmites australis Shoot Remainder Silage on Growth Performance, Blood Biochemical Parameters, and Rumen Microbiota of Beef Cattle. Front. Vet. Sci. 9:778654. doi: 10.3389/fvets.2022.778654
Received: 17 September 2021; Accepted: 03 January 2022;
Published: 22 February 2022.
Edited by:
Tarique Hussain, Nuclear Institute for Agriculture and Biology, PakistanReviewed by:
Duanqin Wu, Institute of Bast Fiber Crops (CAAS), ChinaF. Capela e Silva, University of Evora, Portugal
Muhammad Saleem Kalhoro, Sindh Agriculture University, Pakistan
Copyright © 2022 Wang, Zeng, Zeng, Liu, Wang, Wang, Li, Wang, Feng, Huang, Yin, Huang, Zhu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingzhi Zhu, bXp6aHVjbkBob3RtYWlsLmNvbQ==; Huansheng Yang, eWhzQGh1bm51LmVkdS5jbg==
 Qiye Wang
Qiye Wang Xianglin Zeng1
Xianglin Zeng1 Yancan Wang
Yancan Wang Jia Yin
Jia Yin Mingzhi Zhu
Mingzhi Zhu Huansheng Yang
Huansheng Yang