- 1National Veterinary Services Laboratories, Animal Plant and Health Inspection Service (APHIS), U.S. Department of Agriculture, Ames, IA, United States
- 2National Center for Animal Health (NCAH) Leptospira Working Group, U.S. Department of Agriculture, Ames, IA, United States
- 3Fort Wayne Children's Zoo, Fort Wayne, IN, United States
- 4Infectious Bacterial Diseases Research Unit, Agricultural Research Service (ARS), U.S. Department of Agriculture, Ames, IA, United States
A 1-year-old female red panda started showing symptoms of illness, including lethargy, anorexia, abdominal discomfort, and vomiting, shortly after transfer to a new zoo. Serum was tested for leptospirosis using the microscopic agglutination test, and a titer of 1:25,600 to serogroup Grippotyphosa was detected. Antimicrobial treatment with doxycycline was initiated. After completion of treatment and resolution of clinical symptoms, a urine sample was collected to ensure clearance of leptospires and cessation of urinary shedding prior to co-housing with other red pandas. A repeat serum sample taken 13 days later had a lower titer of 1:6,400 to serogroup Grippotyphosa. A sample of the animal's urine was cultured in HAN media and was culture positive for Leptospira. The recovered isolate was completely characterized by whole genome sequencing and serotyping with reference antisera, and the isolate was classified as Leptospira kirschneri serogroup Grippotyphosa serovar Grippotyphosa strain RedPanda1.
Introduction
Leptospirosis is a zoonotic disease of worldwide concern. It is caused by spirochetes of the genus Leptospira that are typically excreted via urine and transmitted through contact with infected animals or their contaminated habitats (1, 2). Zoo animals are at risk of leptospirosis from interactions with wildlife reservoir hosts, such as small mammals and rodents, and from other exotic animal species (3). Employees are also at risk of infection from handling infected animals or cleaning enclosures. Several cases of leptospirosis have been documented in zoo animals (4–7).
In 2001, two eastern black rhinoceros (Diceros bicornis michaeli) were diagnosed with leptospirosis using the fluorescent antibody test (FAT) and the microscopic agglutination test (MAT). The disease was hypothesized to be a result of infection with Leptospira kirschneri serovar Grippotyphosa through wallow contamination by wild raccoons (6). Multiple animal species, including non-human primates and a desert bighorn sheep (Ovis canadensis mexicana), were diagnosed with leptospirosis that was associated with relocation during exhibit construction and grounds renovation in an urban zoo (5). In both cases, serovar Grippotyphosa was inferred as the cause of infection, and as the cause of death in a young adult male titi monkey (Callicebus moloch). A juvenile south-central black rhinoceros (Diceros bicornis minor) showed evidence of infection from serogroup Autumnalis. Feral trapped rats were negative for leptospirosis, but feral trapped squirrels showed evidence of exposure to both serogroup Grippotyphosa and Autumnalis (5). In 1978, two zoo workers were diagnosed with leptospirosis based on high MAT titers against serogroup Icterohaemorrhagiae and isolation of Icterohaemorrhagiae from the urine of one patient (4). Both workers had contact with bear cubs that also had titers for Icterohaemorrhagiae.
In 2022, MAT and real-time polymerase chain reaction (RT-PCR) was used to diagnose a Sumatran tiger (Panthera tigris sumatrae) residing in a zoo with L. kirschneri serovar Grippotyphosa (7). Clinical signs of illness resolved after treatment, but a urine sample collected at 595 days after initial diagnosis was PCR positive. Leptospirosis is typically treated with doxycycline; however, antimicrobial treatment or clinical resolution does not guarantee cessation of shedding. Detection of Leptospira after clinical resolution has been documented in both human and animal cases. A 10-year old girl shed Leptospira in her urine for several weeks after clinical resolution (8). A dog that was treated with intravenous penicillin, and 4 days later treated with doxycycline, continued to be positive for leptospires in its urine by dark-field microscopy 1 week later (9). In another case, L. interrogans serovar Canicola was isolated from a dog after both vaccination and treatment with penicillin, and in the absence of a positive titer in the MAT. This dog was not showing any acute illness but showed signs of liver dysfunction in routine bloodwork that lead to the further testing for leptospirosis (10).
The case study presented here diagnosed leptospirosis in a female red panda (Ailurus fulgens) while in routine quarantine after movement from another zoo. Continued evaluation post-antimicrobial treatment confirmed urinary shedding of leptospires, highlighting continued risk of transmission. Urine samples were cultured, and recovery of an isolate allowed for complete characterization by serotyping and genome sequencing.
Case description
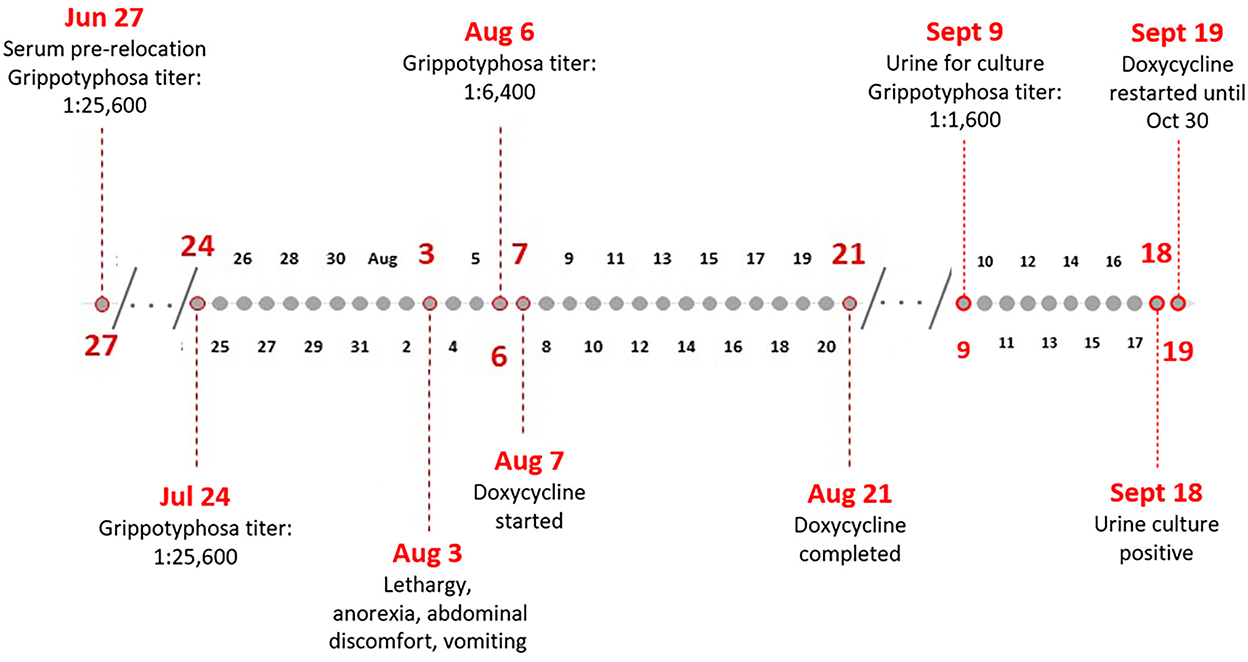
In mid-2019, a female Ailurus fulgens, commonly known as a red panda, aged 1 year, appeared clinically normal when transferred to a new zoo. On July 24, 2019, the red panda underwent a quarantine exam, routine diagnostics, and vaccinations including having blood drawn for Leptospira MAT and CBC/chemistries. Testing for leptospirosis, as well as other illnesses, is a part of the routine surveillance at the zoo. On exam, the panda was thin but otherwise appeared healthy. Several of the routine tests showed atypical results. Bloodwork indicated increased BUN:Creatine ratio, 43.3 ratio (US) (expected range based on best available match: 3.7–18.8). The MAT was performed at the National Veterinary Services Laboratories (NVSL) as previously described with a starting test dilution of 1:100 (11). The end point was identified as the last well with 50% or more agglutination. The sera were tested against 15 serovars (Supplementary Table 1). At that time, a titer of 1:25,600 to serogroup Grippotyphosa and lower titers to serogroups Icterohaemorrhagiae and Autumnalis (1:400 and 1:200, respectively), were detected. The high titer to Grippotyphosa was noted and additional samples were planned to be collected for follow-up testing at the next examination. Multiple fecal samples were collected since arrival at the zoo and several showed signs of cryptosporidium on ELISA testing but were negative on PCR. Proteus mirabilis was also present in a few of the fecal exams. Per the zoo policies, the red panda was scheduled to stay in quarantine for 30 days.
On August 3, the red panda began to show abnormal behavior and was reported as lethargic, anorexic, with abdominal discomfort and vomiting (Figure 1). Three days later, she was examined again, and samples collected for additional testing. On this examination, she had signs of mild gastroenteritis, the likely cause of the abdominal pain and anorexia. Bloodwork showed increased basophil percentage, 7.0% (expected range: 0–4.0), neutrophil count 8.05 × 103 cells/μL (expected range: 0.970–7.913), basophil count, 0.85 × 103 cells/μL (expected range: 0–0.424), and total CO2 27 mmol/L (expected range: 8.8–24.3) (12). Eosinophils were within normal range. The urinalysis showed a slightly elevated specific gravity at 1.042 ratio (expected range: 1.004–1.040). Serum was collected for paired serum testing in the MAT, and urine was collected by cystocentesis for RT-PCR to confirm leptospirosis. The RT-PCR was performed at an external commercial diagnostic laboratory and results were reported as negative. A repeated MAT, on August 6, showed a decrease in titer, positive at 1:6,400 to Grippotyphosa and at 1:200 to Icterohaemorrhagiae. On August 7, treatment with doxycycline was initiated at 16.5 mg twice per day for 14 days with suspected leptospirosis infection, and continued lethargy, abdominal discomfort, diarrhea, and vomiting. The symptoms resolved after the full treatment of doxycycline. On August 29, 2019, serum that was collected on June 27, 2019, prior to the red panda being relocated, was also tested on the MAT for Leptospira titers. Results showed positive titers of 1:25,600 and 1:100 (serogroups Grippotyphosa and Icterohaemorrhagiae, respectively).
On September 9, the red panda was again examined and noted as still mildly thin but otherwise within normal limits. Bloodwork showed increased albumin 4.2 g/dL (expected range: 2.3–4.1) and MAT results showed a titer of 1:1,600 to Grippotyphosa. Since clinical resolution does not guarantee clearance of infection and cessation of urinary shedding of leptospires (7, 9), urine samples were also collected by cystocentesis to perform fluorescent antibody testing (FAT) and culture. Two tubes each of HAN and T80/40/LH semi-solid media provided to the zoo (13, 14) were inoculated with 2–4 drops of urine on site as soon as collected. All samples were shipped overnight to the National Centers for Animal Health (NCAH) Leptospira working group. HAN media cultures were incubated at 37°C in 3% CO2, and T80/40/LH media cultures were incubated at 29°C (13, 14). FAT was performed as previously described (15). The FAT was negative; however, on September 18, 2019, the urine was culture positive in HAN media. Doxycycline, at 16.5 mg twice per day, was initiated on September 19 for 42 days. A single injection of 66,000 IU of Penicillin G was also given. On two subsequent MATs, a titer of 1:200 remained to Grippotyphosa. On May 26, 2020, there were MAT titers of 1:100 to Ballum, Grippotyphosa, and Bratislava. The animal did not show any further symptoms of leptospirosis infection and fully recovered.
Diagnostic assessment
The isolate of Leptospira cultured from the red panda urine, designated strain RedPanda1, was genotyped and serotyped. The isolate was propagated in HAN liquid medium to a density of 2.5 × 108/ml. For short read sequencing, DNA was extracted from a 10 ml culture using the Maxwell RSC PureFood Pathogen Kit (Promega Corporation, Madison, WI) per the manufacturer's instructions. Library preparation and sequencing were performed using the Nextera XT DNA Library Preparation Kit and the MiSeq Desktop Sequencer, 2 × 250 paired-end chemistry (Illumina, San Diego, CA) as per manufacturer's instructions. For long read sequencing, DNA was extracted from a 10 ml culture using the Circulomics Nanobind CBB Big DNA Kit (Circulomics, Baltimore, MD) using the gram-negative bacteria-high molecular weight DNA extraction protocol. The extracted DNA was quantified using the Thermo Fisher Qubit™ dsDNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA) and evaluated for quality and purity using a NanoPhotometer Pearl (Implen, Inc. Westlake Village, CA). The sequencing library beads were prepared and loaded onto Flongle flow cells for sequencing using the Oxford Nanopore Rapid Sequencing Kit (SQK-RAD004) following protocol version: RSE_9046_v1_revM_14Aug2019 (Oxford Nanopore Technologies, OX4 4DQ, UK). Sequencing was performed for a period of 24 h.
Canu v. 2.0 (16) with the default parameters was used with 101 × of Oxford Nanopore reads to assemble the complete genome consisting of two circular chromosomes. The chromosomes were initially iteratively error corrected using Pilon v. 1.23 (17) (with options “—fix bases—mindepth 5—flank 1”) and 105 × Illumina MiSeq paired-end reads until no more corrections were made. The genome chromosomes were subsequently trimmed of overlapping sequence, rotated to begin at defined start genes, and again iteratively error corrected with Pilon (using the aforementioned options) and the Illumina reads until no more corrections were made. A phylogenetic tree was made using kSNP (18). The assembled genomes were compared using the maximum likelihood output.
The final completed assembly of its genome comprised two circular genomes of 4,025,345 and 351,295 bp, with a G+C content of 36%. The genome annotation was completed by the NCBI Prokaryotic Genome Annotation Pipeline (19) and accession numbers are CP085133 and CP085134 for chromosome 1 and chromosome 2, respectively, with the following features: genes (total) 3,597, CDSs (total) 3,553, genes (coding) 3,257, genes (RNA) 44, rRNAs 1, 2, 2 (5S, 16S, 23S), tRNAs 37, ncRNAs 2, and pseudo genes (total) 296.
The complete genome sequence was compared to other genomes of L. kirschneri including strains of L. kirschneri belonging to serogroups Grippotyphosa and Cynopteri, as well as genomes of L. interrogans with strains of L. interrogans belonging to serogroup Grippotyphosa and Icterohaemorrhagiae using kSNP analysis (18) (Figure 2). The L. interrogans strain Lai, L. kirschneri strain UT130, L. kirschneri strain RM52 and L. kirschneri strain Moskva genome sequences were obtained from the National Center for Biotechnology Information (NCBI) database, accession numbers: GCA_000092565, GCF_000246675, GCF_000243615 and GCF_000243855 respectively. The NVSL in-house genome sequences for L. interrogans serogroup Grippotyphosa strain Andaman, L. kirschneri serogroup Grippotyphosa strain Moskva V and L. kirschneri serogroup Cynopteri were also included. The genome of strain RedPanda1 aligned most closely with L. kirschneri isolates, specifically strains belonging to serogroup Grippotyphosa serovar Grippotyphosa (Figure 2).

Figure 2. Phylogenetic tree using kSNP analysis shows strain RedPanda1 grouping with Leptospira kirschneri serovar Grippotyphosa and separate from L. interrogans isolates. For those genome sequences downloaded from GenBank, accession numbers are provided followed by species_serogroup_serovar_strain. NVSL in-house genome sequences are indicated with an asterisk (*) followed by species_serogroup_serovar_strain.
To determine the serogroup, the recovered isolate was sub-cultured in P-80 Leptospira growth media (NVSL, Ames, IA) to a density of 60–70% transmittance at 400 nm wavelength, after centrifugation to remove any auto-agglutination (600 × g for 15 min). The isolate was initially serotyped by MAT using Leptospira polyvalent reference antisera representative of 14 serogroups (Supplementary Table 1). Agglutination observed as 50% or greater was marked as positive and titrated to determine which representative serogroup had the highest titer. Once the serogroup was determined, the isolate was further serotyped to identify serovar by MAT testing with a panel of polyvalent reference antisera from serovars within that serogroup (Table 1) (20). Reference antisera determined that strain RedPanda1 belonged to the serogroup Grippotyphosa, which had a positive MAT titer of 51,200 compared to all other serogroups tested which were MAT negative. Additional serotyping with reference antisera against serovars within the serogroup Grippotyphosa predicts the isolate belongs to serovar Grippotyphosa (Table 1). Thus, the recovered isolate is typed as L. kirschneri serogroup Grippotyphosa serovar Grippotyphosa strain RedPanda1.
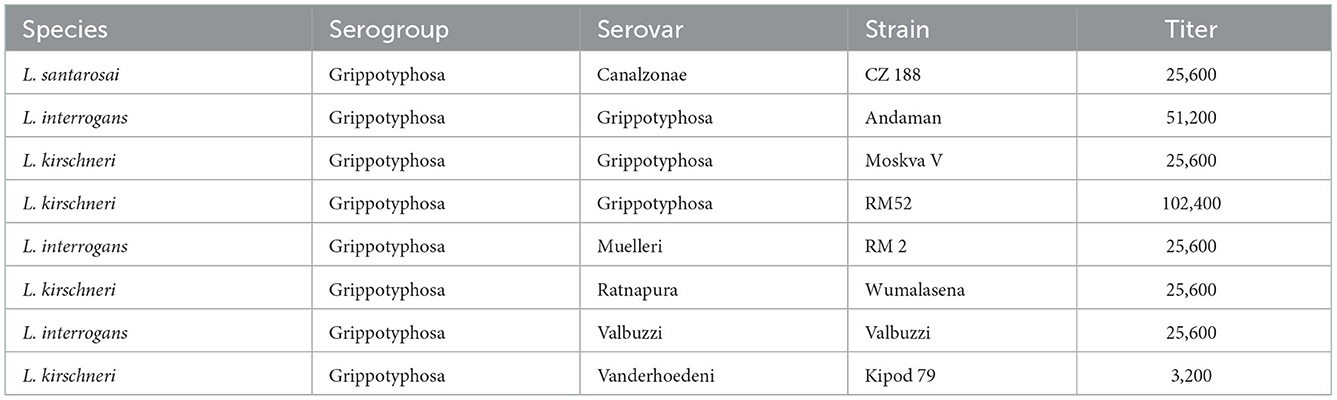
Table 1. MAT titers of the red panda isolate when tested with reference antisera against serovars within the serogroup Grippotyphosa.
Discussion
Pathogenic Leptospira comprise more than 38 pathogenic species and hundreds of serovars, and new species and serovars continue to be identified (21–24). All mammals in the zoo setting are at risk of disease since leptospires can infect such a vast number of different animal species. The zoo setting creates unique opportunities for transmission and exposure not only between exotic species, but from invasive and local wildlife. Leptospirosis is also a zoonotic disease and thus an occupational hazard in the zoo environment. Quarantine and routine testing are essential to help reduce risk of disease transmission (20). Leptospirosis in exotic animals presents a unique situation since there is little animal species-specific research on the infecting species and/or serovars of Leptospira, or the (sub) clinical manifestation of infection which can range from persistent urinary shedding of leptospires in asymptomatic reservoir hosts to acute fulminant infection. In this case report, a red panda was diagnosed with leptospirosis during quarantine thus enabling antimicrobial treatment to facilitate clinical resolution as well as cessation of shedding prior to cohabitating with other animals and allowing zoo staff to properly protect themselves.
Isolation and culture of pathogenic leptospires is inherently difficult due to their fastidious growth requirements and is not routinely practiced. Though some have inferred the infecting serovar based on the highest titer in an MAT reaction, this is not correct and serological results should not be confused with serovar identification. Several studies have demonstrated that inference of serovar based on patient serology is often incorrect due to paradoxical reactions, cross-reactivity, and the potential exclusion of the infecting serovar from the MAT antigen panel employed (25–27). The advent of newer media formulations has enhanced the ability to culture leptospires from animal tissues (14, 15, 28, 29). Recovery of an isolate of Leptospira from animal samples is definitive and essential for high resolution epidemiological typing, inclusion in antigen panels for diagnostic purposes (MAT), and to ensure bacterin-based vaccine strategies contain the relevant serovar to prevent infection and continued disease transmission.
Typing of pathogenic leptospires has evolved from serotyping to genotyping, which represent two different and unique methods for classifying leptospires that do not correlate well with each other (30). Multiple examples exist whereby the same serovar may belong to different species, e.g., serovar Hardjo may belong to either L. interrogans or L. borgpetersenii, serovar Grippotyphosa may belong to L. interrogans, L. kirschneri or L. santarosai (Table 1), and serovar Pomona may belong to L. interrogans, L. kirschneri, L. santarosai, or L. noguchii (31). In our case, strain RedPanda1 was genotyped as L. kirschneri, and serotyped as serovar Grippotyphosa (Table 1, Figure 2). L. kirschneri is often associated with animal and human disease, but few genomes have been sequenced. By performing complete genome sequencing, comparative genome analysis confirmed the species assignment for strain RedPanda1 which is now available for future high resolution molecular epidemiological studies and can be used to facilitate studies to further understand pathogenic mechanisms of leptospirosis.
Reservoir hosts of leptospirosis can excrete leptospires via urine in the absence of a detectable MAT response (32, 33). Similarly, as clinical symptoms resolve in acutely ill patients and MAT titers decline, viable leptospires may continue to be excreted in urine (9). No single assay to detect urinary shedding of leptospires is considered optimal, and the use of more than one assay to detect leptospires in urine is recommended (34). As reported here, a urine sample from the red panda post treatment was FAT negative but culture positive, which may reflect greater sensitivity of newer media formulations. In any case, culture is definitive of a live viable leptospire excreted in urine which can be both genotyped and serotyped. Live leptospires detected by culture also indicate the risk for transmission of disease to other animals and humans (35). In this case, the red panda was still shedding live leptospires, so it was important to keep her quarantined until shedding ceased.
Bacterins for leptospirosis in animals are available, but their efficacy is dependent on inclusion of relevant serovars associated with animal host infection. Since bacterins do not cross protect against multiple serovars, isolation and culture of leptospires from animal samples is essential to determine appropriate serovars for inclusion in bacterins. Recently, serovar Tarassovi was isolated from a U.S. dairy cow, but no U.S. animal vaccine includes serovar Tarassovi (28). Given the limited epidemiological data on leptospirosis in exotic animals, serovar identification can provide guidance on selection of appropriate bacterins that may be useful to manage disease outbreaks in zoos. Serovar identification can also provide guidance on the source of exposure, for example, serovar Grippotyphosa is often associated with wildlife and environmental contamination (36). In this case it was important to identify the infecting serovar so an appropriate bacterin could be used to prevent future infections in this red panda and protect the other red pandas she would be housed with.
Animals maintained in zoos and wildlife parks are at risk from infection with pathogenic leptospires from a range of sources. Though the MAT can diagnose acute leptospirosis concurrent with clinical symptoms, it cannot identify animals shedding leptospires in urine. Detection of leptospires in urine from reservoir hosts, or from patients after clinical resolution of infection, requires alternative assays such as PCR, FAT, and/or culture to accurately assess excretion and continued risk of disease to other animal and human contacts.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, CP085133, CP085134.
Ethics statement
Ethical review and approval was not required for the animal study because samples collected were part of normal diagnostic procedures.
Author contributions
KL, KF, and JN: conceptualization. KL, KF, TA, DB, TS, JH, and JN: methodology. KL, KF, TA, DB, TS, JH, LS, and JN: formal analysis and writing—review and editing. KF, LS, and JN: resources. KL, TS, and JN: figures. KL and JN: writing—original draft preparation. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the USDA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1064147/full#supplementary-material
References
1. Ellis WA. Animal leptospirosis. Curr Top Microbiol Immunol. (2015) 387:99–137. doi: 10.1007/978-3-662-45059-8_6
2. Putz EJ, Nally JE. Investigating the immunological and biological equilibrium of reservoir hosts and pathogenic Leptospira: balancing the solution to an acute problem? Front Microbiol. (2020) 11:2005. doi: 10.3389/fmicb.2020.02005
3. Woolf D, Sanchez C, Gonzalez-Astudillo V, Navarro M, Tapia CC, Franco M, et al. Leptospira species status of captive nonhuman primates and free-ranging rodents at the barranquilla zoo, colombia, 2013. J Zoo Wildl Med. (2021) 51:780–8. doi: 10.1638/2019-0192
4. Anderson DC, Geistfeld JG, Maetz HM, Patton CM, Kaufmann AF. Leptospirosis in zoo workers associated with bears. Am J Trop Med Hyg. (1978) 27 (1 Pt. 1):210–1. doi: 10.4269/ajtmh.1978.27.210
5. Gamble K, Alvarado T, editors. Multi-species leptospirosis in an urban zoo: exhibit modification impact on a wildlife point source. In: Annual Conference-American Association of Zoo Veterinarians 2000. American Association of Zoo Veterinarians. International Association for Aquatic Animal Medicine (1998). Available online at: https://www.vin.com/apputil/content/defaultadv1.aspx?pId=11257&catId=32414&id=3864512
6. Neiffer DL, Klein EC, Wallace-Switalski C. Leptospira infection in two black rhinoceroses (Diceros bicornis michaeli). J Zoo Wildl Med. (2001) 32:476–86. doi: 10.1638/1042-7260(2001)032[0476:LIITBR]2.0.CO;2
7. Webb JK, Keller KA, Sander SJ, Allender MC, Sheldon JD. Clinical disease and treatment of Leptospira kirschneri sv Grippotyphosa in a sumatran tiger (Panthera tigris sumatrae). J Am Vet Med Assoc. (2022) 1:1–6. doi: 10.2460/javma.21.04.0185
8. Chow E, Deville J, Nally J, Lovett M, Nielsen-Saines K. Prolonged leptospira urinary shedding in a 10-year-old girl. Case Rep Pediatr. (2012) 2012:169013. doi: 10.1155/2012/169013
9. Juvet F, Schuller S, O'Neill E, O'Neill P, Nally J. Urinary shedding of spirochaetes in a dog with acute leptospirosis despite treatment. Vet Rec. (2011) 168:564. doi: 10.1136/vr.d740
10. Piredda I, Sechi S, Cocco R, Bertoldi L, Palmas B, Chisu V. Isolation of Leptospira interrogans serovar canicola in a vaccinated dog without clinical symptoms. Pathogens. (2022) 11:406. doi: 10.3390/pathogens11040406
11. Cole JR, Sulzer CR, Pursell AR. Improved microtechnique for the leptospiral microscopic agglutination test. Appl Microbiol. (1973) 25:976–80. doi: 10.1128/am.25.6.976-980.1973
12. Burrell C, Luo L, Jones MK, Lee A, Freeman E, Aitken-Palmer C. Hematology and serum biochemistry values of the red panda subspecies (Ailurus fulgens styani). J Zoo Wildl Med. (2018) 49:384–95. doi: 10.1638/2017-0104.1
13. Ellis W, Montgomery J, Cassells J. Dihydrostreptomycin treatment of bovine carriers of Leptospira interrogans serovar Hardjo. Res Vet Sci. (1985) 39:292–5. doi: 10.1016/S0034-5288(18)31716-8
14. Hornsby RL, Alt DP, Nally JE. Isolation and propagation of leptospires at 37 degrees C directly from the mammalian host. Sci Rep. (2020) 10:9620. doi: 10.1038/s41598-020-66526-4
15. Hamond C, Browne AS, de Wilde LH, Hornsby RL, LeCount K, Anderson T, et al. Assessing rodents as carriers of pathogenic Leptospira species in the U.S. Virgin Islands and their risk to animal and public health. Sci Rep. (2022) 12:1132. doi: 10.1038/s41598-022-04846-3
16. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. (2017) 27:722–36. doi: 10.1101/gr.215087.116
17. Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. (2014) 9:e112963. doi: 10.1371/journal.pone.0112963
18. Gardner SN, Slezak T, Hall BG. kSNP3. 0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. (2015). 31:2877–8. doi: 10.1093/bioinformatics/btv271
19. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. (2016) 44:6614–24. doi: 10.1093/nar/gkw569
20. Sykes JE, Reagan KL, Nally JE, Galloway RL, Haake DA. Role of diagnostics in epidemiology, management, surveillance, and control of leptospirosis. Pathogens. (2022) 11:395. doi: 10.3390/pathogens11040395
21. Faine S, Adler B, Bolin, C, Perolat P. Leptospira and Leptospirosis. 2nd edition. Melbourne, VIC: MediSci (1999).
22. Nally JE, Arent Z, Bayles DO, Hornsby RL, Gilmore C, Regan S, et al. Emerging infectious disease implications of invasive mammalian species: the greater white-toothed shrew (Crocidura russula) is associated with a novel serovar of pathogenic Leptospira in Ireland. PLoS Negl Trop Dis. (2016) 10:e0005174. doi: 10.1371/journal.pntd.0005174
23. Vincent AT, Schiettekatte O, Goarant C, Neela VK, Bernet E, Thibeaux R, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl Trop Dis. (2019) 13:e0007270. doi: 10.1371/journal.pntd.0007270
24. Fernandes LGV, Stone NE, Roe CC, Goris MG, van Der Linden H, Sahl JS, et al. Leptospira sanjuanensis sp. nov., a pathogenic species of the genus Leptospira isolated from soil in Puerto Rico. Int J Syst Evol Microbiol. (2022) 72. doi: 10.1099/ijsem.0.005560
25. Smythe LD, Wuthiekanun V, Chierakul W, Suputtamongkol Y, Tiengrim S, Dohnt MF, et al. The microscopic agglutination test (MAT) is an unreliable predictor of infecting Leptospira serovar in Thailand. Am J Trop Med Hyg. (2009) 81:695–7. doi: 10.4269/ajtmh.2009.09-0252
26. Murray CK, Gray MR, Mende K, Parker TM, Samir A, Rahman BA, et al. Use of patient-specific Leptospira isolates in the diagnosis of leptospirosis employing microscopic agglutination testing (MAT). Trans R Soc Trop Med Hyg. (2011) 105:209–13. doi: 10.1016/j.trstmh.2010.12.004
27. Levett PN. Usefulness of serologic analysis as a predictor of the infecting serovar in patients with severe leptospirosis. Clin Infect Dis. (2003) 36:447–52. doi: 10.1086/346208
28. Hamond C, Lecount K, Putz EJ, Bayles DO, Camp P, Goris MG, et al. Bovine leptospirosis due to persistent renal carriage of Leptospira borgpetersenii serovar tarassovi. Front Vet Sci. (2022) 9:848664. doi: 10.3389/fvets.2022.848664
29. Hamond C, Dirsmith KL, LeCount K, Soltero FV, Rivera-Garcia S, Camp P, et al. Leptospira borgpetersenii serovar Hardjo and Leptospira santarosai serogroup pyrogenes isolated from bovine dairy herds in Puerto Rico. Front Vet Sci. (2022) 9:1025282. doi: 10.3389/fvets.2022.1025282
30. Sykes JE, Gamage CD, Haake DA, Nally JE. Understanding leptospirosis: application of state-of-the-art molecular typing tools with a One Health lens. Am J Vet Res. (2022) 83. doi: 10.2460/ajvr.22.06.0104
31. Picardeau M,. Leptospira. Bergey's Manual of Systematics of Archaea Bacteria. John Wiley & Sons, Inc. (2015). Available online at: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118960608.gbm01244.pub2
32. Nally JE, Hornsby RL, Alt DP, Bayles D, Wilson-Welder JH, Palmquist DE, et al. Isolation and characterization of pathogenic leptospires associated with cattle. Vet Microbiol. (2018) 218:25–30. doi: 10.1016/j.vetmic.2018.03.023
33. Nally JE, Mullen W, Callanan JJ, Mischak H, Albalat A. Detection of urinary biomarkers in reservoir hosts of leptospirosis by capillary electrophoresis-mass spectrometry. Proteomics Clin Applic. (2015) 9:543–51. doi: 10.1002/prca.201400205
34. Nally JE, Ahmed AA, Putz EJ, Palmquist DE, Goris MG. Comparison of real-time PCR, bacteriologic culture and fluorescent antibody test for the detection of Leptospira borgpetersenii in urine of naturally infected cattle. Vet Sci. (2020) 7:66. doi: 10.3390/vetsci7020066
35. Sykes JE, Haake DA, Gamage CD, Mills WZ, Nally JE. A global one health perspective on leptospirosis in humans and animals. J Am Vet Med Assoc. (2022) 260:1–8. doi: 10.2460/javma.22.06.0258
Keywords: Leptospira, leptospirosis, red panda, Ailurus fulgens, L. kirschneri, serovar Grippotyphosa, case report
Citation: LeCount K, Fox K, Anderson T, Bayles DO, Stuber T, Hicks J, Schlater LK and Nally JE (2023) Isolation of Leptospira kirschneri serovar Grippotyphosa from a red panda (Ailurus fulgens) after antimicrobial therapy: Case report. Front. Vet. Sci. 9:1064147. doi: 10.3389/fvets.2022.1064147
Received: 07 October 2022; Accepted: 28 December 2022;
Published: 02 February 2023.
Edited by:
Irene Iglesias, National Institute for Agricultural and Food Research and Technology, SpainReviewed by:
Fabrizio Bertelloni, University of Pisa, ItalyFernando Monroy, Northern Arizona University, United States
Copyright © 2023 LeCount, Fox, Anderson, Bayles, Stuber, Hicks, Schlater and Nally. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jarlath E. Nally,  amFybGF0aC5uYWxseUB1c2RhLmdvdg==
amFybGF0aC5uYWxseUB1c2RhLmdvdg==
†These authors have contributed equally to this work and share first authorship
 Karen LeCount
Karen LeCount Kami Fox3†
Kami Fox3† Darrell O. Bayles
Darrell O. Bayles Linda K. Schlater
Linda K. Schlater Jarlath E. Nally
Jarlath E. Nally