
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 22 December 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1054316
Antibiotic resistance genes (ARGs) are emerging pollutants that have received extensive attention. Many different types of ARGs exist in livestock wastewater. If not effectively treated, they can threaten animal production, public health and the ecological safety of the surrounding environment. To address the high risk of livestock wastewater contamination by ARGs, the effects of different wastewater treatment processes on ARGs and their influencing factors and mechanisms are reviewed herein. Additionally, the current problems associated with removal of ARGs are discussed, and future research is proposed.
Antibiotics are widely used in animal husbandry and bacterial drug resistance appears to be substantial (1, 2). Research results have shown that the total amount of antibiotics used in China in 2013 was ~162,000 tons, which accounted for half of global consumption, of which 52%, or 84,000 tons, were veterinary antibiotics (3). According to a report of the U.S. Food and Drug Administration (FDA), 29.9 million pounds of antibiotics were used with farmed animals in 2011, which accounted for 80.5% of the total antibiotic consumption in the United States (4). Similarly, in Vietnam, more than 11 million pounds of antibiotics were used for growth promotion, 25 million pounds for disease prevention and 37 million pounds for therapeutic purposes in the pig industry (5). Use of antibiotics may select among microorganisms harbored in animal intestines for resistant strains, carrying antibiotic resistance genes (ARGs) (6, 7). In addition, veterinary antibiotics cannot be completely absorbed or degraded by livestock and poultry, and most of the residual antibiotics are excreted through urine and fecal material (manure) in the form of unmetabolized drugs and their metabolites (8). These residual antibiotics promote enrichment of ARGs and their host bacteria in feces and wastewater (9). The highly abundant ARGs can enter the surrounding environment through manure or application of reclaimed water. Moreover, the high abundant ARGs also enter other sensitive bacteria through horizontal gene transfer and threaten ecological security (10, 11) (Figure 1). In addition, pathogens are also important hosts of ARGs, and they cause widespread and severe problems duo to antibiotic-resistance contamination (12). In 2019, about ~ 4.95 million deaths worldwide were related to infections with antibiotic-resistant bacterial, of which 1.27 million people died directly from antibiotic resistance (13). The use of antibiotics as growth promoters have been banned in an increasing number of countries. However, the copy numbers of ARGs remain elevated in animal raising environment. Therefore, it is necessary to implement effective removal processes to prevent and control further spread of antibiotic resistance. Wastewater is one of the main livestock wastes and an important repository for ARGs (14). China produces ~ 3.8 billion tons of livestock waste every year (15). Among this, aquaculture wastewater exceeded 460 million tons (16). However, current livestock wastewater treatment processes are mainly aimed at treatment of conventional pollutants such as organic matter, ammonia nitrogen, phosphorus, antibiotics and heavy metals, but not ARGs or antibiotic resistant bacteria. Therefore, it is necessary to understand the ARGs removal effectiveness of different livestock wastewater treatment processes, clarify their influencing factors and mechanisms, and provide reference for prevention and control of ARGs and antibiotic-resistance pollution.
Livestock wastewater is an important repository for ARGs. Tetracyclines, aminoglycoside, sulfonamide, macrolide, streptomycin, bacitracin, β-lactam, chloramphenicol, quinolone, trimethoprim, fosmidomycin, polymyxin, and vancomycin resistance genes are frequently detected in wastewater, and the absolute abundance of ARGs is between 108 and 1010 copies/mL, with a relative abundance between 10−3 and 10−1 copies/16S rRNA genes (10, 17–19). Among them, the abundance of tetracycline, aminoglycoside, macrolide–lincosamide–streptogramin (MLS), sulfonamide and chloramphenicol resistance genes were the highest, and they account for 28.13, 23.64, 12.17, 11.53, 8.74 and 6.18% of total ARGs respectively, and their abundances can be up to 2.41 × 10−1, 2.03 × 10−1, 1.04 × 10−1, 9.90 × 10−2, 7.49 × 10−2 and 5.31 × 10−2 copies/16S rRNA genes, respectively (19). Based on different resistance mechanisms, ARGs can be divided into efflux pump, target protection (ribosome protection), enzyme modification (antibiotic inactivation) and other types of resistance genes (20–22). In addition, most ARGs exist within bacteria in livestock wastewater, while some exist in phages, and small amounts of ARGs are freely present in wastewater in the form of nucleic acids. One study reported that the absolute abundances of ARGs in bacteria and phages were 109 and 106 copies/mL, respectively (23).
Residual ARGs in livestock wastewater can enter farms and surrounding environments with efflux and utilization of the wastewater and threaten human health (9, 24). Yang et al. (10) found that the abundance of the blaTEM resistance gene in the downstream river water around a pig farm was 107 copies/mL, which was significantly higher than that in the upstream river water (105 copies/mL). In addition, the abundance of ARGs in downstream sediments around pig feedlots was 1.2-fold higher than that in the upstream sediments (25). This means that the ARGs that remained in livestock wastewater were enriched in the surrounding water and soil, and they can be carried by bacteria or phages, enter the bodies of aquatic animals and plants, and cause more severe antibiotic-resistance contamination throughout the food chain (26). Moreover, ARGs and resistant bacteria present in livestock wastewater can also enter the air in the form of aerosols (27). Sun et al. (28) found that the composition of ARGs in the fecal samples of students during 3-month internships at swine farms were consistent with that in the pig farm environment, indicating that the ARGs can enter the human gut through certain means and affect the composition of ARGs in the human gut. It should be noted that high-risk ARGs such as mcr-1, blaNDM and tetX were detected in livestock wastewater (29). ARGs such as blaCTX, blaCMYand qnrB, which have clinical resistance risks, were also detected in swine wastewater, and these can confer antibiotic resistance in pathogenic bacteria, making the treatment of infectious diseases more difficult (30, 31). Therefore, it is urgent to pay attention to the high abundance of residual ARGs in livestock waste.
Anaerobic treatment is a treatment technology that decomposes organic matter to generate biogas under anaerobic conditions with the help of anaerobic microorganisms. The four stages, hydrolysis, acidification, hydrogen and acetic acid production, and methane production, are completed by different anaerobic microorganisms (32). The up-flow anaerobic sludge blanket (UASB), anaerobic baffled reactor (ABR), buried biogas digester (BBD) and anaerobic filter (AF) are commonly used for treatment of livestock farm wastewater (33, 34). Another study showed that anaerobic treatment effectively reduced ARGs, among which tetX, ermB, mefA, ermF and sul2 showed average reductions of 1.34, 1.07, 1.03, 0.83, and 0.72 log, respectively (35). The removal rates of BBD and UASB anaerobic treatments based on the relative abundance of ARGs were 3.53 and 71.02% in another study, respectively (34) (Table 1). However, other studies have also shown that anaerobic treatments cannot effectively remove ARGs, and the relative abundances of bacterial ARGs were not decreased significantly after treatment (23, 36). Aerobic treatment refers to denitrification by facultative anaerobic denitrifying bacteria in an anaerobic or aerobic state (dissolved oxygen DO < 0.5 mg/L). Aerobic treatment is similar to anaerobic treatment, and has a certain removal effect on ARGs. However, some functional microorganisms, such as anaerobic denitrifying bacteria, are ARG hosts (37). They may increase the abundance of ARGs in anaerobic tank, which may increase the risk of environmental pollution.
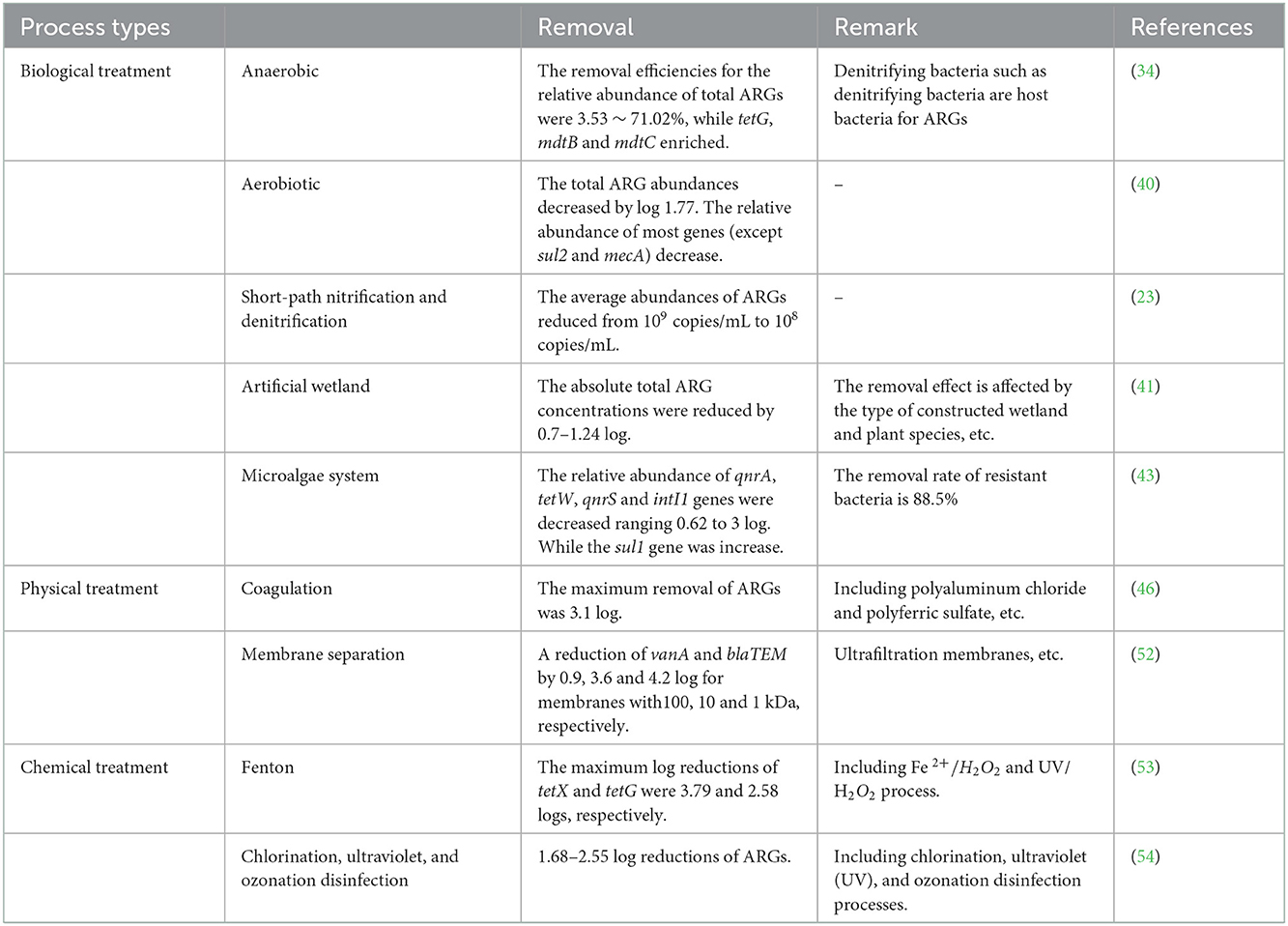
Table 1. Effects of common livestock and poultry wastewater treatment processes on antibiotic resistance genes removal.
Certain methods and equipment are used to force air into the wastewater so it is oxygenated by contact with the air, and the liquid is stirred to accelerate oxygen transfer from the air to the liquid. Aeration prevents the wastewater suspension from sinking and strengthens the contacts among organic matter, microorganisms and the dissolved oxygen in the wastewater. When sufficient dissolved oxygen is present, aerobic microorganisms oxidatively decompose the organic matter in wastewater. Studies have shown that aeration was beneficial for the reduction of ARGs (38, 39). Another study found that the abundance of total ARGs was decreased by 1.77 log copies/mL during aeration of the tank, but a change in sul1 abundance was not observed (40).
In addition to the above processes, there are also biological treatment processes such as constructed wetlands, short-cut nitrification and denitrification, and microalgae-bacteria symbiosis technology. Constructed wetland is primarily divided into surface flow constructed wetland (SFCW), horizontal subsurface flow wetland (HSFW) and vertical subsurface flow wetland (VSFW). It was found that the efficiencies for removal of tetM from wastewater by constructed wetlands was 70.9–97%, the removal rates for sul1 was 49.5–92.9%, and the removal rate for ermB was 58.2–96.7% (41). The short-path nitrification and denitrification process showed substantial levels of nitrogen and phosphorus removal. The average abundance of bacterial/phage ARGs in the effluent of a short-range nitrification/denitrification system was significantly lower than that in the influent (23). After treatment with microalgae-bacteria, the abundance of blaTEM and ermB in wastewater decreased by 0.56 and 1.75 log, respectively (42). In a microalgae treatment system, the relative abundance of qnrA, tetW, and qnrS decreased significantly, but the relative abundance of sul1 increased (43).
In a pig farm wastewater treatment system, the physical treatment processes mainly include coagulation and membrane separation technology. Under the action of a coagulation, the suspended solids present in wastewater are aggregated into larger particles and precipitate, thereby removing many suspended solids from the wastewater and achieving wastewater treatment. Chemical coagulants include polyaluminum chloride, polyaluminum sulfate, aluminum hydroxide iron, polyferric chloride and ferric chloride (44, 45). Studies have shown that treatment by coagulation and sedimentation provide successful removal of most ARGs in wastewater and that the abundance of ARGs can be reduced by 0.5–4.7 log (46, 47). However, different coagulants also have different effects on ARGs removal. The average removal of sul1 by polyferric chloride was 2.3 log, and the average removal of sul1 by ferric chloride was 2.6 log.
Membrane separation is a method used to separate, purify and concentrate different components through selective separation. In wastewater treatment, membrane separation is mainly achieved by using pressure as a driving force with the pore size, electrostatic effect, and diffusion effect of the membrane (48). These processes mainly include reverse osmosis (RO), nanofiltration (NF), microfiltration (MF), and ultrafiltration (UF), which have better removal capacities with ARGs (49–51). The removal rate of ARGs by membrane separation is inversely proportional to the membrane pore size (52).
Chemical treatment processes include ozonation, chlorination and Fenton oxidation. Physicochemical mainly include UV irradiation and electrolysis. Many studies have shown that chemical oxidations remove ARGs from livestock wastewater. It was found that Fenton oxidation reduced selected ARG levels by 2.58 ~ 3.79 log (53). With an ozone mass concentration of 177.6 mg/L, ARGs decreased by 1.68 ~ 2.55 log; under a UV dose of 12.477 J/cm2, ARGs decreased by 2.48 ~ 2.74 log (54). With UV doses of 249.5 and 62.4 mJ/cm2, tetX decreased by 0.58 and 0.23 log, respectively (3). Studies have shown that microbial fuel cells constitute promising alternatives for enhanced removal of ARGs (55, 56). Recently, a study showed that the relative abundance of sul1 in the biofilm and effluent of microbial fuel cells ranged from 4.70 × 102 to 7.80 × 101, and the relative abundance of sul2 ranged from 4.21 × 105 to 7.61 × 103, which was lower than those in natural water bodies and in drinking water (57).
The factors influencing ARG removal are divided into direct driving factors and indirect influencing factors. The direct driving factors include factors that directly affect changes in ARGs, such as microbial communities, mobile genetic elements (MGEs), and wastewater physicochemical factors (58). Indirect influencing factors refer to those that indirectly remove of ARGs through factors affecting wastewater quality and microbial communities, such as breed management and operating parameters.
Microbial communities, including those of bacteria, viruses (bacteriophages), archaea and fungi, are the main direct drivers of ARG changes. Among them, bacteria are also important ARG hosts. Most of the ARGs present in livestock wastewater exist in bacterial cells. Studies have shown that Escherichia coli in livestock wastewater is a host for a variety of ARGs, including tetracycline, aminoglycoside, MLS, β-lactam and sulfonamide resistance genes (59–61). Enterococcus is the main host for vancomycin resistance genes, and Klebsiella and Enterobacter are hosts for fosfomycin resistance genes (62). In addition, another study showed that phages also carry a high abundance of ARGs, including multiple types of ARGs, reaching 2.01 ± 0.21 log (23). The microbial community acts as a carrier for ARGs. Therefore, the ARG composition changes with changes in the composition of the microbial community (11, 58). ARGs are spread by the movement of microorganisms. Therefore, during treatment of livestock wastewater, changes in the composition of the microbial community directly affect the process of ARG removal. The more complex the host composition of ARGs in livestock wastewater, the more difficult they may be to remove.
MGEs mainly include insertion sequences, transposons, integrons, plasmids, and bacteriophages (63, 64). The ARGs in livestock wastewater can be horizontally transferred by MGEs (65, 66), such that ARGs can be transferred among the same or even different microorganisms (67). Due to horizontal transfer of ARGs, microorganisms that do not originally have antibiotic resistance can acquire antibiotic resistance, which increases the complexity of antibiotic-resistant contamination in livestock wastewater (68, 69). Transposons can be copied or broken off from the original site, then cyclized and inserted into other sites. Integrators can capture and integrate foreign genes with their unique structure. Plasmids can carry genes and spread them among different cells.
Here, physicochemical factors mainly refer to the physicochemical properties of livestock and poultry breeding wastewater and include temperature, pH, electrical conductivity (EC), total nitrogen (TN), total phosphorus (TP), ammonia nitrogen, nitrate nitrogen, organic matter and heavy metals, which indirectly affect antibiotic resistance mainly by affecting the compositions of microbial communities. Studies have shown that total nitrogen, ammonia nitrogen and total phosphorus were significantly and positively correlated with the abundance of sul1, sul2, tetA, tetB, tetC and qnsR (23, 70, 71); total organic carbon was significantly and positively correlated with fexA, fexB, cfr, sul1, tetW, tetO, tetQ and tetS (72). In addition, physicochemical factors can indirectly affect the abundance of ARGs by affecting the morphology of free ARGs. One study showed that temperature affected the degradation efficiency of free nucleic acids, and free nucleic acids decayed as the temperature was increasesed (73).
Breed environment, manure removal methods, disinfection methods and other operations in breed management affect the removal of ARGs from livestock and poultry breeding wastewater. Antibiotics and heavy metals are widely used in animal husbandry (74–76). However, antibiotics and heavy metals that cannot be fully absorbed and metabolized can lead to antibiotic-resistance contamination (77, 78). A recent study found that when the ambient temperature decreased, the abundance of ARGs in feces and cecum contents decreased significantly (79). Common methods of manure removal in farms include artificial dry manure removal and mechanical manure removal. Among them, artificial dry manure removal has the best effect on the removal of ARGs (23). During disinfection processes in farms, disinfectants have a certain reduction effect on free ARGs and resistant bacteria. For example, chlorine disinfection can effectively kill resistant bacteria, but it cannot effectively remove ARGs (80). In addition, a recent study showed that after the use of disinfectants, the chloramines and free chlorine present in the wastewater increase the conversion rates of free ARGs (81).
Different treatment parameters for wastewater treatment process, such as water intake, residence time, temperature and aeration amount, affect the treatment effect of livestock and poultry breeding wastewater, and thereby affect the quality of the effluent (physicochemical factors such as total nitrogen, ammonia nitrogen, total phosphorus and organic matter). These physicochemical factors can indirectly affect the removal of ARGs by affecting the composition of the host flora (77, 82), and they can also directly affect the removal of free ARGs. In addition, treatment parameters such as temperature and the aeration rate can also directly or indirectly affect the removal of ARGs. Diehl (83) found that the abundance of tetA, tetO, tetW, and tetX decreased with increasing temperature in anaerobic reactors. However, it was found that although the ARG abundances in wastewater treatment systems were higher in winter than in summer, the removal rates ARGs such as tetG, tetM, and tetX were higher in winter than in summer (84).
Based on previous research, this review summarizes the removal mechanisms active in common wastewater treatment processes in livestock farms in China; these include physical, chemical, physical-chemical and biological removal mechanisms (Figure 2) (85).
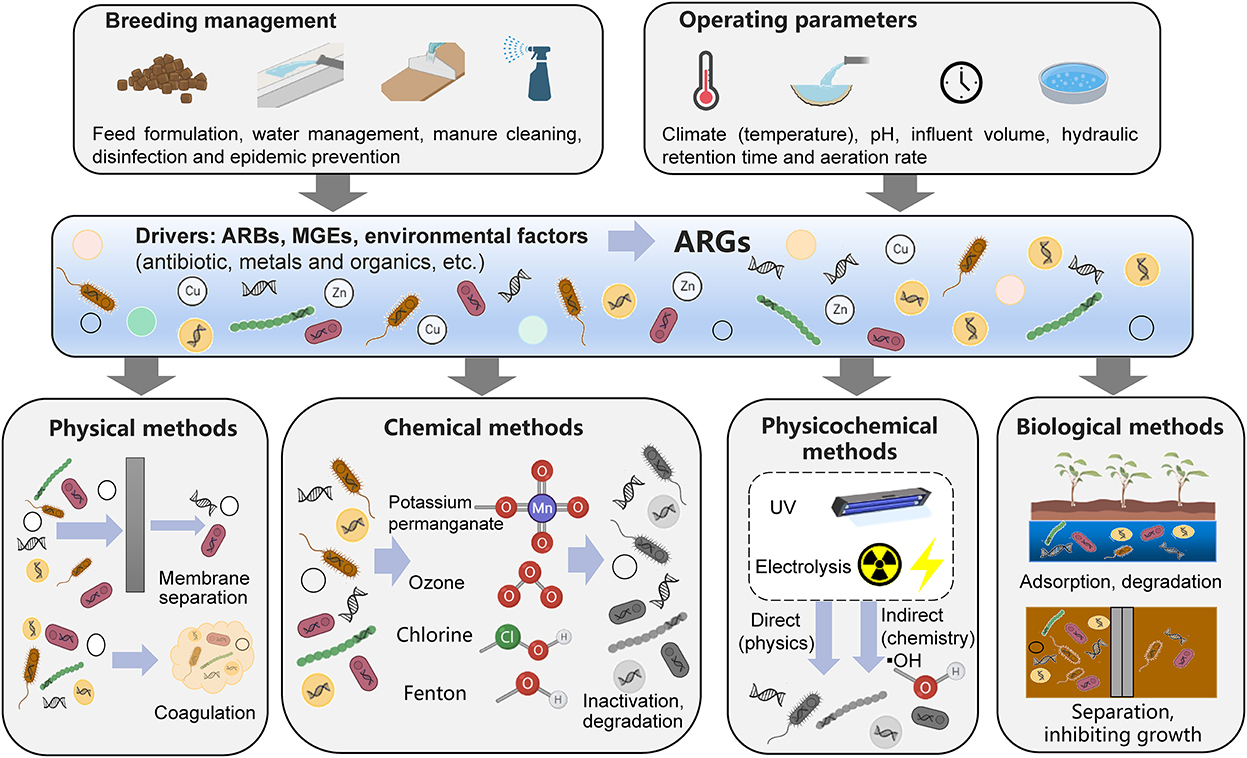
Figure 2. Removal mechanisms for antibiotic resistance genes from livestock wastewater treatment processes.
The ARGs in livestock wastewater are mainly derived from livestock manure. Physical methods mainly refer to the technical processes of coagulation sedimentation and membrane separation to separate the larger solid residues from livestock wastewater, thereby reducing the abundance of ARGs. Therefore, when a physical method is used to separate solid residues such as feces, most of the ARGs that remain in the feces will be separated to achieve ARG removal, but this method cannot remove free ARGs and their hosts in the wastewater. In addition, filters of small pore sizes (< 0.22 μm) can retain bacteria carrying antibiotic resistance gene in wastewater. The removal rate of ARGs is inversely proportional to the membrane pore size. In addition, the removal effects of different filter membrane materials on ARGs are also different (85, 86). The removal effect of polyethersulfone ultrafiltration membranes on ARGs is better than that of polyvinylidene fluoride ultrafiltration membranes.
Chemical oxidation is a method that reduces ARGs by decomposing antibiotic-resistant bacteria through strong oxidants. There are two mechanisms of chemical oxidation. In one, the oxidants react with antibiotic-resistant bacteria, dissolve cell walls and cell membranes, and then react with the cytoplasm and nucleic acid substances to kill bacteria. SecondS, the oxidants produce hydroxyl radicals, which increase pH and decompose resistant bacteria (87, 88). Ozonolysis produces large number of hydroxyl radicals, which react with components such as peptidoglycan in the cell membrane and cell wall, destroy the cell envelope structure, change cell permeability, and then degrade intracellular DNA and remove ARGs (87, 89). Chlorine alters the permeability of cells by disrupting cell surface structures, allowing their internal components to be broken down. At the same time, chlorine decompose into ClO− and HClO, which can enter antibiotic-resistant bacteria and further oxidize components such as nucleic acids (90, 91).
Physical-chemical methods mainly include ultraviolet methods and electrolysis methods. Ultraviolet light can be absorbed by intracellular or extracellular photosensitizers of antibiotic-resistant bacteria, producing large amounts of hydroxyl radicals and then oxidizing various cellular components to kill bacteria (54). In general, the UV intensity is proportional to the removal rate of ARGs (3). In addition, the use of UV in combination with other oxidants, such as Fenton and H2O2 can further improve the removal rate of ARGs (92, 93), which may be because the combination of different oxidation reactions can generate higher concentrations of oxidative radicals to improve oxidation efficiency. Iron-carbon microelectrolysis is commonly used, which uses the potential difference generated by iron and carbon components to form multiple tiny galvanic cells in water to achieve redox reactions and the coagulation, adsorption, and electrodeposition of pollutants and antibiotic-resistant bacteria (94, 95).
Biological methods include processes such as anoxic/aerobic processes, short-range nitrification and denitrification processes, constructed wetlands and microalgae-bacteria symbiosis. Biofilms or activated sludge in anoxic/aerobic processes and short-range nitrification and denitrification processes can adsorb suspended solid organic matter and other substances in livestock wastewater and simultaneously remove nitrogen, phosphorus and other substances necessary for microbial growth. Therefore, they can not only isolate ARG host bacteria that are attached to solid fecal residues in the wastewater by membrane separation and sedimentation but also inhibit the growth of ARG hosts by reducing nutrients to realize the removal of ARGs. The removal of pollutants in wastewater from constructed wetlands is achieved through the synergistic action of plant roots and rhizomes, microorganisms and solid medium components. These different fractions provide different microenvironments for various removal processes, such as mechanical filtration, adsorption, photolysis, volatilization, chemical degradation, plant uptake and microbial metabolism (96). Among them, adsorption, chemical degradation, and plant uptake play important roles in the removal of ARGs, but the mechanism for plant uptake of ARGs is still unclear and needs further research (97). The removal effect of constructed wetlands on ARGs is affected by the type of constructed wetland and plant species. Microalgae mainly utilize nitrogen and phosphorus in wastewater through photosynthesis to reduce the total bacterial abundance of the wastewater, thereby reducing the abundance of individual ARGs (43). However, it should be noted that microalgae may adsorb individual bacteria, which may lead to the enrichment of individual ARGs (42, 43).
Different processes have different removal effects on ARGs. Removal mainly proceeds via physical, chemical and biological mechanisms, which are influenced by major drivers such as microbial communities, MGEs and physicochemical factors. However, current treatment processes are mainly aimed at removal of conventional pollutants such as COD, ammonia nitrogen and total phosphorus and are not designed for removal of ARGs. Therefore, the current wastewater treatment processes cannot completely remove ARGs from wastewater, and some individual processes even have the potential to increase the abundance of individual ARGs. More importantly, overuse of antibiotics can produce arb and arg. Therefore, proper control of antibiotic usage in animal husbandry is an important way to reduce the spread of ARBs and ARGs through wastewater and feces. Future research on the removal of ARGs from livestock wastewater is anticipated to show the following trends:
(1) The focus of current research is to demonstration of ARGs and specific mechanisms for different treatment processes. At present, the rates and main mechanisms for removal of ARGs by typical wastewater treatment processes have been elucidated, but the removal effect and specific removal mechanisms of individual processes are still unclear.
(2) While paying attention to the removal of ARGs, it is necessary to pay attention to the impact of the process on the removal of total nitrogen and other substances and on the reduction of greenhouse gas emissions, and their relationship to ARGs. Total nitrogen removal and carbon emissions reduction will become new requirements for livestock wastewater treatment. However, the influence and relationships between total nitrogen removal, greenhouse gas emissions reduction and the removal of ARGs is yet to be clarified.
(3) Improving ARGs removal by existing wastewater treatment processes will be the focus of future research, and development of new processes will be an important trend. Currently employed wastewater treatment processes will be difficult to replace with new processes on a large scale and within a short period of time. Upgrade and transformations of the original processes are the most cost-effective options.
(4) The risk assessment system for antibiotic resistance must be improved to assess ARG removal more accurately. At present, the risk for contamination by antibiotic resistance genes is mainly assessed by analyzing the abundance of ARGs. The resistance characteristics of ARGs are also affected by genetic factors and environmental factors, and as-yet unknown influencing factors may exist that could affect the accuracie of ARGs contamination assessment.
FH, YH, XL, and YY conceptualized the review. All authors contributed to the article and approved the submitted version.
The study was financially supported by the Project funded by National Natural Science Foundation of China (32202734), China Postdoctoral Science Foundation (2021M700047, 2022T150226), Basic and Applied Basic Research Foundation of Guangdong Province (2021A1515110335), and Guangdong Provincial Science and Technology Plan Project (2020B1212060060).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. He Y, Yuan Q, Mathieu J, Stadler L, Senehi N, Sun R, et al. Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. NPJ Clean Water. (2020) 3:1–11. doi: 10.1038/s41545-020-0051-0
2. Bengtsson B, Greko C. Antibiotic resistance—consequences for animal health, welfare, and food production. Upsala J Med Sci. (2014) 119:96–102. doi: 10.3109/03009734.2014.901445
3. Zhang Q-Q, Ying G-G, Pan C-G, Liu Y-S, Zhao J-L. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ Sci Technol. (2015) 49:6772–82. doi: 10.1021/acs.est.5b00729
4. Leavey-Roback SL, Krasner SW, Suffet IMH. Veterinary antibiotics used in animal agriculture as NDMA precursors. Chemosphere. (2016) 164:330–8. doi: 10.1016/j.chemosphere.2016.08.070
5. Cheng D, Ngo H, Guo W, Liu Y, Zhou J, Chang S, et al. Bioprocessing for elimination antibiotics and hormones from swine wastewater. Sci Total Environ. (2018) 621:1664–82. doi: 10.1016/j.scitotenv.2017.10.059
6. Munk P, Knudsen BE, Lukjancenko O, Duarte ASR, Van Gompel L, Luiken RE, et al. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat Microbiol. (2018) 3:898–908. doi: 10.1038/s41564-018-0192-9
7. Zhu Y-G, Johnson TA, Su J-Q, Qiao M, Guo G-X, Stedtfeld RD, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci. (2013) 110:3435–40. doi: 10.1073/pnas.1222743110
8. Massé DI, Cata Saady NM, Gilbert Y. Potential of biological processes to eliminate antibiotics in livestock manure: an overview. Animals. (2014) 4:146–63. doi: 10.3390/ani4020146
9. He L-Y, Ying G-G, Liu Y-S, Su H-C, Chen J, Liu S-S, et al. Discharge of swine wastes risks water quality and food safety: antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ Int. (2016) 92:210–9. doi: 10.1016/j.envint.2016.03.023
10. Yang Y, Liu Z, Xing S, Liao X. The correlation between antibiotic resistance gene abundance and microbial community resistance in pig farm wastewater and surrounding rivers. Ecotoxicol Environ Saf. (2019) 182:109452. doi: 10.1016/j.ecoenv.2019.109452
11. Zhu N, Jin H, Ye X, Liu W, Li D, Shah GM, et al. Fate and driving factors of antibiotic resistance genes in an integrated swine wastewater treatment system: from wastewater to soil. Sci Total Environ. (2020) 721:137654. doi: 10.1016/j.scitotenv.2020.137654
12. Raza S, Shin H, Hur H-G, Unno T. Higher abundance of core antimicrobial resistant genes in effluent from wastewater treatment plants. Water Res. (2022) 208:117882. doi: 10.1016/j.watres.2021.117882
13. Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. (2022) 399:629–55. doi: 10.1016/S0140-6736(21)02724-0
14. Yuan Q-B, Zhai Y-F, Mao B-Y, Hu N. Antibiotic resistance genes and intI1 prevalence in a swine wastewater treatment plant and correlation with metal resistance, bacterial community and wastewater parameters. Ecotoxicol Environ Saf. (2018) 161:251–9. doi: 10.1016/j.ecoenv.2018.05.049
15. Zhang Z, Wang Y-B, editors. The application of excrement energy engineering in breeding industry. In: Proceedings of the 2nd Annual International Conference on Energy, Environmental and Sustainable Ecosystem Development (EESED 2016). Paris: Atlantis Press (2016). doi: 10.2991/eesed-16.2017.90
16. Liu R, Chen L, Song X, Wei D, Zheng W, Qiu S, et al. Treatment of digested piggery wastewater with a membrane bioreactor. Environ Eng Manag J. (2016) 15:2181–8. doi: 10.30638/eemj.2016.236
17. Cheng D, Ngo HH, Guo W, Chang SW, Nguyen DD, Liu Y, et al. Contribution of antibiotics to the fate of antibiotic resistance genes in anaerobic treatment processes of swine wastewater: a review. Bioresour Technol. (2020) 299:122654. doi: 10.1016/j.biortech.2019.122654
18. Jia S, Zhang X-X, Miao Y, Zhao Y, Ye L, Li B, et al. Fate of antibiotic resistance genes and their associations with bacterial community in livestock breeding wastewater and its receiving river water. Water Res. (2017) 124:259–68. doi: 10.1016/j.watres.2017.07.061
19. Zhang R-M, Liu X, Wang S-L, Fang L-X, Sun J, Liu Y-H, et al. Distribution patterns of antibiotic resistance genes and their bacterial hosts in pig farm wastewater treatment systems and soil fertilized with pig manure. Sci Total Environ. (2021) 758:143654. doi: 10.1016/j.scitotenv.2020.143654
20. Ouyang W-Y, Huang F-Y, Zhao Y, Li H, Su J-Q. Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China. Appl Microbiol Biotechnol. (2015) 99:5697–707. doi: 10.1007/s00253-015-6416-5
21. Kapoor G, Saigal S, Elongavan A. Action and resistance mechanisms of antibiotics: a guide for clinicians. J Anaesthesiol Clin Pharmacol. (2017) 33:300. doi: 10.4103/joacp.JOACP_349_15
22. Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. (2018) 4:482. doi: 10.3934/microbiol.2018.3.482
23. Yang Y, Xing S, Chen Y, Wu R, Wu Y, Wang Y, et al. Profiles of bacteria/phage-comediated ARGs in pig farm wastewater treatment plants in China: association with mobile genetic elements, bacterial communities and environmental factors. J Hazard Mater. (2021) 404:124149. doi: 10.1016/j.jhazmat.2020.124149
24. Wei R, Ge F, Huang S, Chen M, Wang R. Occurrence of veterinary antibiotics in animal wastewater and surface water around farms in Jiangsu Province, China. Chemosphere. (2011) 82:1408–14. doi: 10.1016/j.chemosphere.2010.11.067
25. Fang H, Han L, Zhang H, Long Z, Cai L, Yu Y. Dissemination of antibiotic resistance genes and human pathogenic bacteria from a pig feedlot to the surrounding stream and agricultural soils. J Hazard Mater. (2018) 357:53–62. doi: 10.1016/j.jhazmat.2018.05.066
26. Bengtsson-Palme J. Antibiotic resistance in the food supply chain: where can sequencing and metagenomics aid risk assessment? Curr Opin Food Sci. (2017) 14:66–71. doi: 10.1016/j.cofs.2017.01.010
27. Gaviria-Figueroa A, Preisner EC, Hoque S, Feigley CE, Norman RS. Emission and dispersal of antibiotic resistance genes through bioaerosols generated during the treatment of municipal sewage. Sci Total Environ. (2019) 686:402–12. doi: 10.1016/j.scitotenv.2019.05.454
28. Sun J, Liao X-P, D'Souza AW, Boolchandani M, Li S-H, Cheng K, et al. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat Commun. (2020) 11:1–11. doi: 10.1038/s41467-020-15222-y
29. Yang F, Gu Y, Zhou J, Zhang K. Swine waste: a reservoir of high-risk blaNDM and mcr-1. Sci Total Environ. (2019) 683:308–16. doi: 10.1016/j.scitotenv.2019.05.251
30. Meng L, Wang J, Li X. Insight into effect of high-level cephalexin on fate and driver mechanism of antibiotics resistance genes in antibiotic wastewater treatment system. Ecotoxicol Environ Saf. (2020) 201:110739. doi: 10.1016/j.ecoenv.2020.110739
31. Ibekwe A, Murinda SE, DebRoy C, Reddy GB. Potential pathogens, antimicrobial patterns and genotypic diversity of E. coli isolates in constructed wetlands treating swine wastewater. FEMS Microbiol Ecol. (2016) 92:fiw006. doi: 10.1093/femsec/fiw006
32. Wainaina SL, Kumar Awasthi M, Taherzadeh MJ. Bioengineering of anaerobic digestion for volatile fatty acids, hydrogen or methane production: a critical review. Bioengineered. (2019) 10:437–58. doi: 10.1080/21655979.2019.1673937
33. Liu R, Zhao Y, Doherty L, Hu Y, Hao X. A review of incorporation of constructed wetland with other treatment processes. Chem Eng J. (2015) 279:220–30. doi: 10.1016/j.cej.2015.05.023
34. Zhang M, Liu Y-S, Zhao J-L, Liu W-R, Chen J, Zhang Q-Q, et al. Variations of antibiotic resistome in swine wastewater during full-scale anaerobic digestion treatment. Environ Int. (2021) 155:106694. doi: 10.1016/j.envint.2021.106694
35. Sui Q, Zhang J, Chen M, Tong J, Wang R, Wei Y. Distribution of antibiotic resistance genes (ARGs) in anaerobic digestion and land application of swine wastewater. Environ Pollut. (2016) 213:751–9. doi: 10.1016/j.envpol.2016.03.038
36. Tao C-W, Hsu B-M, Ji W-T, Hsu T-K, Kao P-M, Hsu C-P, et al. Evaluation of five antibiotic resistance genes in wastewater treatment systems of swine farms by real-time. PCR Sci Total Environ. (2014) 496:116–21. doi: 10.1016/j.scitotenv.2014.07.024
37. Yang Y, Wu R, Hu J, Xing S, Huang C, Mi J, et al. Dominant denitrifying bacteria are important hosts of antibiotic resistance genes in pig farm anoxic-oxic wastewater treatment processes. Environ Int. (2020) 143:105897. doi: 10.1016/j.envint.2020.105897
38. Knecht CA. Fate of Antibiotic Resistant Bacteria and Antibiotic Resistance Genes in Constructed Wetlands. Leipzig: Helmholtz Centre for Environmental Research Press (2020). p. 134.
39. Guo B, Yu N, Weissbrodt DG, Liu Y. Effects of micro-aeration on microbial niches and antimicrobial resistances in blackwater anaerobic digesters. Water Res. (2021) 196:117035. doi: 10.1016/j.watres.2021.117035
40. McConnell MM, Hansen LT, Jamieson RC, Neudorf KD, Yost CK, Tong A. Removal of antibiotic resistance genes in two tertiary level municipal wastewater treatment plants. Sci Total Environ. (2018) 643:292–300. doi: 10.1016/j.scitotenv.2018.06.212
41. Abou-Kandil A, Shibli A, Azaizeh H, Wolff D, Wick A, Jadoun J. Fate and removal of bacteria and antibiotic resistance genes in horizontal subsurface constructed wetlands: effect of mixed vegetation and substrate type. Sci Total Environ. (2021) 759:144193. doi: 10.1016/j.scitotenv.2020.144193
42. da Silva Rodrigues DA, da Cunha CCRF, do Espirito Santo DR, de Barros ALC, Pereira AR, de Queiroz Silva S, et al. Removal of cephalexin and erythromycin antibiotics, and their resistance genes, by microalgae-bacteria consortium from wastewater treatment plant secondsary effluents. Environ Sci Pollut Res. (2021) 28:67822–32. doi: 10.1007/s11356-021-15351-x
43. Cheng X, Delanka-Pedige HM, Munasinghe-Arachchige SP, Abeysiriwardana-Arachchige IS, Smith GB, Nirmalakhandan N, et al. Removal of antibiotic resistance genes in an algal-based wastewater treatment system employing Galdieria sulphuraria: a comparative study. Sci Total Environ. (2020) 711:134435. doi: 10.1016/j.scitotenv.2019.134435
44. Ezeuko AS, Ojemaye MO, Okoh OO, Okoh AI. Technological advancement for eliminating antibiotic resistance genes from wastewater: a review of their mechanisms and progress. J Environ Chem Eng. (2021) 9:106183. doi: 10.1016/j.jece.2021.106183
45. Jian Y, He Y, Zhu J, Long D, Tan Q, Xu W, et al. Removal of microorganisms and antibiotic resistance genes from swine wastewater: a comparison between polyaluminum chloride (PAC), polyaluminum sulfate (LST), and aluminum hydroxide iron (LT). J Environ Sci Health B. (2022) 2022:1–8. doi: 10.1080/03601234.2022.2058844
46. Li N, Sheng G-P, Lu Y-Z, Zeng RJ, Yu H-Q. Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Res. (2017) 111:204–12. doi: 10.1016/j.watres.2017.01.010
47. Yu K-F, Li P, Li H, Zhang B, Yang J, Huang F-Y, et al. Potential of coagulation to remove particle-associated and free-living antibiotic resistome from wastewater. J Hazard Mater. (2021) 406:124295. doi: 10.1016/j.jhazmat.2020.124295
48. Zhang T, Lv K, Lu Q, Wang L, Liu X. Removal of antibiotic-resistant genes during drinking water treatment: a review. J Environ Sci. (2021) 104:415–29. doi: 10.1016/j.jes.2020.12.023
49. Samanta P, Horn H, Saravia F. Removal of diverse and abundant ARGs by MF-NF process from pig manure and digestate. Membranes. (2022) 12:661. doi: 10.3390/membranes12070661
50. Guo Y editor editor Removal Ability of Antibiotic Resistant Bacteria (Arb) and Antibiotic Resistance Genes (Args) by Membrane Filtration Process. IOP Conference Series: Earth and Environmental Science. Bristol: IOP Publishing (2021). doi: 10.1088/1755-1315/801/1/012004
51. Leddy MB, Hasan NA, Subramanian P, Heberling C, Cotruvo J, Colwell RR. Characterization of microbial signatures from advanced treated wastewater biofilms. J Am Water Works Assoc. (2017) 109:E503–E12. doi: 10.5942/jawwa.2017.109.0116
52. Krzeminski P, Feys E, d'Auriac MA, Wennberg AC, Umar M, Schwermer CU, et al. Combined membrane filtration and 265 nm UV irradiation for effective removal of cell free antibiotic resistance genes from feed water and concentrate. J Membr Sci. (2020) 598:117676. doi: 10.1016/j.memsci.2019.117676
53. Zhang Y, Zhuang Y, Geng J, Ren H, Xu K, Ding L. Reduction of antibiotic resistance genes in municipal wastewater effluent by advanced oxidation processes. Sci Total Environ. (2016) 550:184–91. doi: 10.1016/j.scitotenv.2016.01.078
54. Zhuang Y, Ren H, Geng J, Zhang Y, Zhang Y, Ding L, et al. Inactivation of antibiotic resistance genes in municipal wastewater by chlorination, ultraviolet, and ozonation disinfection. Environ Sci Pollut Res. (2015) 22:7037–44. doi: 10.1007/s11356-014-3919-z
55. Yan W, Guo Y, Xiao Y, Wang S, Ding R, Jiang J, et al. The changes of bacterial communities and antibiotic resistance genes in microbial fuel cells during long-term oxytetracycline processing. Water Res. (2018) 142:105–14. doi: 10.1016/j.watres.2018.05.047
56. Zhang S, Song H-L, Yang X-L, Huang S, Dai Z-Q, Li H, et al. Dynamics of antibiotic resistance genes in microbial fuel cell-coupled constructed wetlands treating antibiotic-polluted water. Chemosphere. (2017) 178:548–55. doi: 10.1016/j.chemosphere.2017.03.088
57. Xue W, Li F, Zhou Q. Degradation mechanisms of sulfamethoxazole and its induction of bacterial community changes and antibiotic resistance genes in a microbial fuel cell. Bioresour Technol. (2019) 289:121632. doi: 10.1016/j.biortech.2019.121632
58. Yang Y, Li L, Huang F, Hu X, Cao X, Mi J, et al. The fate of antibiotic resistance genes and their association with bacterial and archaeal communities during advanced treatment of pig farm wastewater. Sci Total Environ. (2022) 2022:158364. doi: 10.1016/j.scitotenv.2022.158364
59. Sabaté M, Prats G, Moreno E, Ballesté E, Blanch AR, Andreu A. Virulence and antimicrobial resistance profiles among E. coli strains isolated from human and animal wastewater. Res Microbiol. (2008) 159:288–93. doi: 10.1016/j.resmic.2008.02.001
60. Dawangpa A, Lertwatcharasarakul P, Ramasoota P, Boonsoongnern A, Ratanavanichrojn N, Sanguankiat A, et al. Genotypic and phenotypic situation of antimicrobial drug resistance of E. coli in water and manure between biogas and non-biogas swine farms in central Thailand. J Environ Manag. (2021) 279:111659. doi: 10.1016/j.jenvman.2020.111659
61. Wandee S, Chan R, Chiemchaisri W, Chiemchaisri C. Alteration of antibiotic-resistant phenotypes and minimal inhibitory concentration of E. coli in pig farming: comparison between closed and open farming systems. Sci Total Environ. (2021) 781:146743. doi: 10.1016/j.scitotenv.2021.146743
62. Xiong W, Wang Y, Sun Y, Ma L, Zeng Q, Jiang X, et al. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome. (2018) 6:1–11. doi: 10.1186/s40168-018-0419-2
63. Sobecky PA, Hazen TH. Horizontal gene transfer and mobile genetic elements in marine systems. Horizontal Gene Transf. (2009) 2009:435–53. doi: 10.1007/978-1-60327-853-9_25
64. Domingues S, da Silva GJ, Nielsen KM. Integrons: vehicles and pathways for horizontal dissemination in bacteria. Mobile Genet Elements. (2012) 2:211–23. doi: 10.4161/mge.22967
65. Guo J, Li J, Chen H, Bond PL, Yuan Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. (2017) 123:468–78. doi: 10.1016/j.watres.2017.07.002
66. Wang F-H, Qiao M, Su J-Q, Chen Z, Zhou X, Zhu Y-G. High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ Sci Technol. (2014) 48:9079–85. doi: 10.1021/es502615e
67. Le Roux F, Blokesch M. Eco-evolutionary dynamics linked to horizontal gene transfer in Vibrios. Annu Rev Microbiol. (2018) 72:89–110. doi: 10.1146/annurev-micro-090817-062148
68. Zarei-Baygi A, Smith AL. Intracellular vs. extracellular antibiotic resistance genes in the environment: prevalence, horizontal transfer, and mitigation strategies. Bioresour Technol. (2021) 319:124181. doi: 10.1016/j.biortech.2020.124181
69. Sun D, Jeannot K, Xiao Y, Knapp CW. Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Lausanne: Frontiers Media SA (2019). p. 1933. doi: 10.3389/978-2-88963-157-5
70. Liang C, Wei D, Yan W, Zhang S, Shi J, Liu L. Fates of intracellular and extracellular antibiotic resistance genes during the cattle farm wastewater treatment process. Bioresour Technol. (2022) 344:126272. doi: 10.1016/j.biortech.2021.126272
71. Wang R, Ji M, Zhai H, Guo Y, Liu Y. Occurrence of antibiotics and antibiotic resistance genes in WWTP effluent-receiving water bodies and reclaimed wastewater treatment plants. Sci Total Environ. (2021) 796:148919. doi: 10.1016/j.scitotenv.2021.148919
72. He L-Y, Liu Y-S, Su H-C, Zhao J-L, Liu S-S, Chen J, et al. Dissemination of antibiotic resistance genes in representative broiler feedlots environments: identification of indicator ARGs and correlations with environmental variables. Environ Sci Technol. (2014) 48:13120–9. doi: 10.1021/es5041267
73. Eichmiller JJ, Best SE, Sorensen PW. Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ Sci Technol. (2016) 50:1859–67. doi: 10.1021/acs.est.5b05672
74. Cheng D, Ngo HH, Guo W, Chang SW, Nguyen DD, Liu Y, et al. A critical review on antibiotics and hormones in swine wastewater: Water pollution problems and control approaches. J Hazard Mater. (2020) 387:121682. doi: 10.1016/j.jhazmat.2019.121682
75. Guo J, Selby K, Boxall AB. Assessment of the risks of mixtures of major use veterinary antibiotics in European surface waters. Environ Sci Technol. (2016) 50:8282–9. doi: 10.1021/acs.est.6b01649
76. Zhang M, Liu Y-S, Zhao J-L, Liu W-R, He L-Y, Zhang J-N, et al. Occurrence, fate and mass loadings of antibiotics in two swine wastewater treatment systems. Sci Total Environ. (2018) 639:1421–31. doi: 10.1016/j.scitotenv.2018.05.230
77. Wang P, Wu D, You X, Su Y, Xie B. Antibiotic and metal resistance genes are closely linked with nitrogen-processing functions in municipal solid waste landfills. J Hazard Mater. (2021) 403:123689. doi: 10.1016/j.jhazmat.2020.123689
78. Ji X, Shen Q, Liu F, Ma J, Xu G, Wang Y, et al. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J Hazard Mater. (2012) 235:178–85. doi: 10.1016/j.jhazmat.2012.07.040
79. Yang Y, Chen N, Sun L, Zhang Y, Wu Y, Wang Y, et al. Short-term cold stress can reduce the abundance of antibiotic resistance genes in the cecum and feces in a pig model. J Hazard Mater. (2021) 416:125868. doi: 10.1016/j.jhazmat.2021.125868
80. Sullivan BA, Vance CC, Gentry TJ, Karthikeyan R. Effects of chlorination and ultraviolet light on environmental tetracycline-resistant bacteria and tet (W) in water. J Environ Chem Eng. (2017) 5:777–84. doi: 10.1016/j.jece.2016.12.052
81. Zhang S, Wang Y, Lu J, Yu Z, Song H, Bond PL, et al. Chlorine disinfection facilitates natural transformation through ROS-mediated oxidative stress. ISME J. (2021) 15:2969–85. doi: 10.1038/s41396-021-00980-4
82. Yi X, Tran NH, Yin T, He Y, Gin KY-H. Removal of selected PPCPs, EDCs, and antibiotic resistance genes in landfill leachate by a full-scale constructed wetlands system. Water Res. (2017) 121:46–60. doi: 10.1016/j.watres.2017.05.008
83. Diehl DL, LaPara TM. Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids. Environ Sci Technol. (2010) 44:9128–33. doi: 10.1021/es102765a
84. Sui Q, Zhang J, Tong J, Chen M, Wei Y. Seasonal variation and removal efficiency of antibiotic resistance genes during wastewater treatment of swine farms. Environ Sci Pollut Res. (2017) 24:9048–57. doi: 10.1007/s11356-015-5891-7
85. Wang J, Chen X. Removal of antibiotic resistance genes (ARGs) in various wastewater treatment processes: an overview. Crit Rev Environ Sci Technol. (2022) 52:571–630. doi: 10.1080/10643389.2020.1835124
86. Suna L, Shib P, Zhangb Q, Lvc J, Zhanga Y. Effects of using different ultrafltration membranes on the removal efficiency of antibiotic resistance genes from secondsary effluent. Desalin Water Treat. (2019) 156:52–8. doi: 10.5004/dwt.2019.24255
87. Michael-Kordatou I, Karaolia P, Fatta-Kassinos D. The role of operating parameters and oxidative damage mechanisms of advanced chemical oxidation processes in the combat against antibiotic-resistant bacteria and resistance genes present in urban wastewater. Water Res. (2018) 129:208–30. doi: 10.1016/j.watres.2017.10.007
88. Sui Q, Chen Y, Yu D, Wang T, Hai Y, Zhang J, et al. Fates of intracellular and extracellular antibiotic resistance genes and microbial community structures in typical swine wastewater treatment processes. Environ Int. (2019) 133:105183. doi: 10.1016/j.envint.2019.105183
89. Chand R, Bremner DH, Namkung KC, Collier PJ, Gogate PR. Water disinfection using the novel approach of ozone and a liquid whistle reactor. Biochem Eng J. (2007) 35:357–64. doi: 10.1016/j.bej.2007.01.032
90. Bommer A, Böhler O, Johannsen E, Dobrindt U, Kuczius T. Effect of chlorine on cultivability of Shiga toxin producing E. coli (STEC) and β-lactamase genes carrying E. coli and Pseudomonas aeruginosa. Int J Med Microbiol. (2018) 308:1105–12. doi: 10.1016/j.ijmm.2018.09.004
91. Dodd MC. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment. J Environ Monit. (2012) 14:1754–71. doi: 10.1039/c2em00006g
92. Hou J, Chen Z, Gao J, Xie Y, Li L, Qin S, et al. Simultaneous removal of antibiotics and antibiotic resistance genes from pharmaceutical wastewater using the combinations of up-flow anaerobic sludge bed, anoxic-oxic tank, and advanced oxidation technologies. Water Res. (2019) 159:511–20. doi: 10.1016/j.watres.2019.05.034
93. Yoon Y, Chung HJ Di DYW, Dodd MC, Hur H-G, Lee Y. Inactivation efficiency of plasmid-encoded antibiotic resistance genes during water treatment with chlorine, UV, and UV/H2O2. Water Res. (2017) 123:783–93. doi: 10.1016/j.watres.2017.06.056
94. Ruan X-C, Liu M-Y, Zeng Q-F, Ding Y-H. Degradation and decolorization of reactive red X-3B aqueous solution by ozone integrated with internal micro-electrolysis. Separ Purif Technol. (2010) 74:195–201. doi: 10.1016/j.seppur.2010.06.005
95. Wu S, Qi Y, Gao Y, Xu Y, Gao F, Yu H, et al. Preparation of ceramic-corrosion-cell fillers and application for cyclohexanone industry wastewater treatment in electrobath reactor. J Hazard Mater. (2011) 196:139–44. doi: 10.1016/j.jhazmat.2011.09.003
96. Carvalho PN, Araújo JL, Mucha AP, Basto MCP, Almeida CMR. Potential of constructed wetlands microcosms for the removal of veterinary pharmaceuticals from livestock wastewater. Bioresour Technol. (2013) 134:412–6. doi: 10.1016/j.biortech.2013.02.027
Keywords: antibiotic resistance gene, wastewater treatment process, livestock wastewater, remove, mechanism
Citation: Huang F, Hong Y, Mo C, Huang P, Liao X and Yang Y (2022) Removal of antibiotic resistance genes during livestock wastewater treatment processes: Review and prospects. Front. Vet. Sci. 9:1054316. doi: 10.3389/fvets.2022.1054316
Received: 26 September 2022; Accepted: 08 December 2022;
Published: 22 December 2022.
Edited by:
Lixing Huang, Jimei University, ChinaReviewed by:
Marja-Liisa Hänninen, University of Helsinki, FinlandCopyright © 2022 Huang, Hong, Mo, Huang, Liao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiwen Yang,  eWl3ZW55QGZveG1haWwuY29t
eWl3ZW55QGZveG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.