
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Vet. Sci. , 17 January 2023
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1045088
This article is part of the Research Topic Glanders and Melioidosis: One Health Model View all 8 articles
 Himali S. Jayasinghearachchi1*†
Himali S. Jayasinghearachchi1*† Thilini A. Muthugama2
Thilini A. Muthugama2 Jayanthi Masakorala3
Jayanthi Masakorala3 Upeksha S. Kulasekara2
Upeksha S. Kulasekara2 Kumari Jayaratne2
Kumari Jayaratne2 D. A. Dasun N. Jayatunga2
D. A. Dasun N. Jayatunga2 Aruna D. De Silva2
Aruna D. De Silva2 Enoka M. Corea3*†
Enoka M. Corea3*†Burkholderia pseudomallei is the causative agent of the potentially fatal infection, melioidosis. This study provides the first evidence for the presence of B. pseudomallei in soil and water in Sri Lanka. Targeted sampling of soil and natural water sources was done between November 2019 and October 2020 over eight field visits encompassing the neighborhood of 28 culture and/or antibody-positive melioidosis patients in northwestern, western and southern Sri Lanka. A total of eight environmental isolates of B. pseudomallei (BPs-env1 to BPs-env8) were cultured from 116 soil and 117 natural water samples collected from 72 locations. The presence of B. pseudomallei in soil and natural water in these areas poses a risk of melioidosis for populations cultivating crops in such soils and using untreated water from these sources for drinking, bathing, and other domestic purposes. Identifying sites positive for B. pseudomallei may help to mitigate risk by raising public awareness of contaminated environmental sources and allowing soil and water remediation.
Melioidosis, a potentially fatal infection of humans and animals, is a tropical disease with a non-specific presentation, ranging from overwhelming sepsis to community acquired pneumonia to chronic localized infection. Multifocal involvement of more than one system and abscess formation, especially in the liver, spleen, and prostate, are common (1). Burkholderia pseudomallei, the causative agent of melioidosis, is an aerobic, non-spore-forming, non-fermenting, Gram-negative environmental bacterium widely distributed in the rhizosphere, soil and water (1). Infection is acquired via exposure to soil or water by inoculation, inhalation, or ingestion (2). Agricultural workers and rural populations in endemic areas are at high risk (3).
A number of studies have highlighted the potential hazards associated with the presence of B. pseudomallei in natural water bodies and its ability to survive and proliferate in low nutrient environments such as unchlorinated drinking water sources (4, 5) and the bacterium has been detected in environments adjacent to culture-positive melioidosis patients (6–8). Inglis and Sousa have drawn attention to the public health implications of melioidosis and recommended targeted environmental threat assessment (9).
Many studies have shown that B. pseudomallei is phylogenetically diverse. Multilocus sequence typing (MLST) is a useful epidemiological tool to investigate the biogeographic and phylogenetic features of B. pseudomallei and has been used widely to identify the population genetics of isolates from a particular geographic area and to analyze their relationship to isolates from other regions (1). Sequence type (ST) data can be further analyzed using phylogenetic tools such as PHYLOVIZ to elucidate the genetic relatedness of B. pseudomallei from distinct locations (10). MLST has also been used to identify the source of outbreaks although the restricted number of genes analyzed may limit discrimination as demonstrated by whole genome sequencing (WGS) of identical STs revealing non-clonality in a case cluster (11). MLST and WGS have been employed to explore the genetic relationship between clinical and environmental isolates of B. pseudomallei in Australia (12, 13). A correlation between ST and virulence has not been found to date.
Variably present molecular markers such as the Burkholderia thailandensis-like flagellum and chemotaxis gene cluster (BTFC) and Yersinia-like fimbrial (YLF) gene cluster have been used to characterize B. pseudomallei populations in different endemic regions. For example, Australian B. pseudomallei isolates are more likely to encode the ancestral BTFC gene cluster whereas isolates from Asia almost exclusively carry the YLF gene cluster (14). In addition, variably present virulence factors such as the actin-based motility gene, bimA, lipopolysaccharide (LPS) O antigen, and filamentous hemagglutinin gene (fhaB) have been found to be useful in molecular characterization of B. pseudomallei in different geographical regions. The bimA gene is found in two different allele variants (typical B. pseudomallei-like bimABp and atypical B. mallei-like bimABm) and a differential distribution of these two variants in endemic regions has been reported. In the Australian population, 12% of B. pseudomallei isolates contain bimABm whereas in Thailand bimABm has not been reported. The fhaB allele has been found in three different forms. All Thai strains contain the fhaB3 gene while only 83% of Australia strains are positive (15). LPS O-antigen of B. pseudomallei is highly diverse and grouped by serotype into types A, B, and B2 (16).
Melioidosis is endemic in Sri Lanka with a wide geographic and demographic distribution. A large number of B. pseudomallei clinical strains have been isolated from melioidosis patients throughout the country (3). Sequence typing of 193 B. pseudomallei clinical isolates from Sri Lanka revealed 71 distinct STs with ST1137, ST1136 and ST1132 being the most frequent. In addition, examination of variably present genetic markers i.e., bimA, fhaB3, YLF, and BTFC gene clusters and LPS type A among 310 isolates showed a diversity of genotypes, intermediate between Southeast Asia and Oceania (10).
Successful culture isolation of B. pseudomallei from soil and water is fraught with difficulties including the presence of the bacterium in the environment in a viable but non-culturable state and overgrowth by other soil bacteria, even with the use of selective media. Other factors influencing culture isolation include the depth of soil sampling, timing of sampling and volume of sample (1). A consensus guideline for soil sampling has been proposed (17) but requires a soil sampling depth of 30 cm that is both time and labor intensive. There is no guideline for isolation of B. pseudomallei from water but bacterial concentration by filtration has been used successfully (2).
In 2015, a cluster of melioidosis cases caused by genetically diverse strains was reported after heavy rainfall in the eastern part of Sri Lanka (11). A similar cluster followed strong monsoonal winds in 2017 (18). The occurrence of these case clusters indicates the presence of B. pseudomallei in the environment. However, the bacterium has never been isolated from soil or water in Sri Lanka. Knowledge on the distribution of B. pseudomallei in natural environments and its environmental persistence is of importance to understand the epidemiology of melioidosis and identification of B. pseudomallei contaminated environments is essential to determine potential risk areas, identify occupational groups that are frequently exposed to such environments and mitigate risk from such exposures.
This study reports the first isolation of B. pseudomallei from soil and natural water in the western, northwestern and southern parts of Sri Lanka employing targeted sampling of potential environment sources around the homes and workplaces of melioidosis patients and examines the genetic relatedness of the environmental isolates to the corresponding clinical isolates and to environmental isolates from other endemic regions.
Targeted sampling of soil and natural water suspected to be the source of infection was done in the course of eight field visits to 28 culture and/or antibody-positive melioidosis patients between November 2019 and October 2020. The patients (or relatives in the case of deceased individuals) were interviewed to identify the suspected source of exposure and appropriate soil and water specimens were obtained. If the household well or natural spring was identified as the likely source, ~250 mL of water was collected into pre-labeled sterilized polypropylene bottles. If the source was a natural water ditch, the water sample was collected using a hand spade. Soil sampling was done from the surface at depths varying from 0 to 30 cm, with ~100 g of soil from each sampling point collected into sterile, pre-labeled, polypropylene bags while one teaspoonful of soil was inoculated into 10 mL of modified Ashdown's broth in situ (19). Sample collection utensils were washed with bottled water, disinfected with 70% ethanol and dried between samples. Soil and water samples were transported to the laboratory on the same day under ambient conditions, protected from direct sunlight. Global positioning system (GPS) coordinates of each sampling location were recorded using GPS Tracker and mapped on Google Earth using ArcGIS 10.2.
Soil enrichment broth cultures were incubated at 37°C for 72 h and a loopful of broth, from the surface was inoculated on Ashdown's selective agar (20). Water samples were filtered through a 0.45 μm nitrocellulose membrane filter using a Millipore Filtration Unit and the filter membrane was aseptically transferred into a sterile 50 ml conical tube containing 10 ml of modified Ashdown's broth using a sterile forceps and incubated at 37°C in ambient air for 3–4 days. A loopful of broth, taken from the surface, was spread on Ashdown's selective agar and incubated at 37°C in ambient air for 3–4 days. B. pseudomallei colonies were identified based on their characteristic colony morphology and presumptive colonies were sub-cultured on fresh plates. Cultures were confirmed using a real-time polymerase chain reaction (RT-PCR) lpxO assay using DNA extracted by heat lysis (21) with DNA from a B. pseudomallei clinical isolate previously confirmed by whole genome sequencing as the positive control (22). Isolates were further confirmed by latex agglutination (23).
Amplification of seven housekeeping genes (ace-gltB-gmhD-lepA-lipA-narK-ndh) was performed using established oligonucleotide primer sequences as reported previously (24). PCR products were submitted for amplicon purification and SANGER sequencing to Macrogen Inc., Seoul, South Korea. Each DNA fragment was sequenced in forward and reverse directions using the same oligonucleotide primers that were used for the initial PCR amplification. The batch sequence (7 loci) from each isolate was queried in the MLST database (http://pubmlst.org/bpseudomallei.mlst.net/) to determine the allelic profile. The allelic profile of each isolate was queried for a match to the existing STs on the MLST database. PHYLOVIZ 2.0 available at PubMLST (http://pubmlst.org/bpseudomallei.mlst.net/) was used to determine the genetic relationship of STs of Sri Lankan environmental isolates of B. pseudomallei in relation to environmental STs (n = 454) from Australia (n = 220), Thailand (n = 163), Vietnam (n = 25), Laos (n = 19), China (n = 11), Cambodia (n = 8), Singapore (n = 4), India (n = 2), Taiwan (n = 1), and Bangladesh (n = 1).
Genotyping to detect the variably present gene markers YLF) and BTFC, fhaB3, bimA gene variants bimABP/bimABM and LPS O-antigen type A (LPSA) was done using RT-PCR assays with gene-specific oligonucleotide primers (14–16). BRYT green qRT-PCR master mixture (Promega, USA) was used in this study. Previously confirmed B. pseudomallei isolates were used as positive controls (22).
A total of eight environmental isolates of B. pseudomallei (BPs-env1to BPs-env8) were cultured from 116 soil and 117 natural water samples collected from 72 sites in northwestern, western, and southern Sri Lanka (Figure 1). Six isolates were from water, giving an isolation rate of 6/116 (5%), with three samples being from unchlorinated domestic wells and one each from still water in a paddy field, stagnant water from a garden and a perennial spring (Figure 2). Both soil isolates (isolation rate of 2/117, 1.7%) were from paddy fields. All the isolates were obtained from the environs of seven patients, six of whom survived (Table 1).
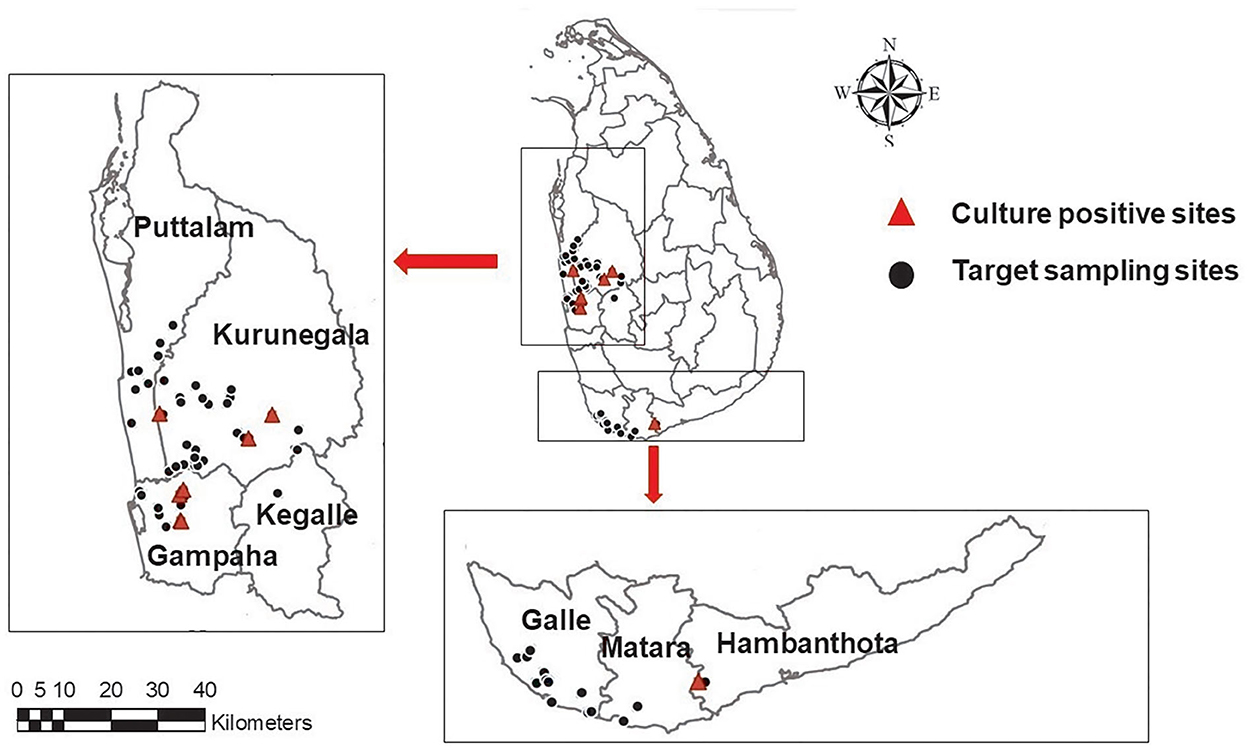
Figure 1. A map showing the geographical distribution of targeted sampling sites in western, northwestern, and southern parts of Sri Lanka; ( )-target sampling sites, (
)-target sampling sites, ( ) culture positive sites. Maps of the study region were generated using ArcGIS software version 10.3.1.
) culture positive sites. Maps of the study region were generated using ArcGIS software version 10.3.1.

Figure 2. Natural water sources of Burkholderia pseudomallei. (A, B) unchlorinated domestic well (C) Stagnant water in paddy field (D) perennial spring “Bubula”.
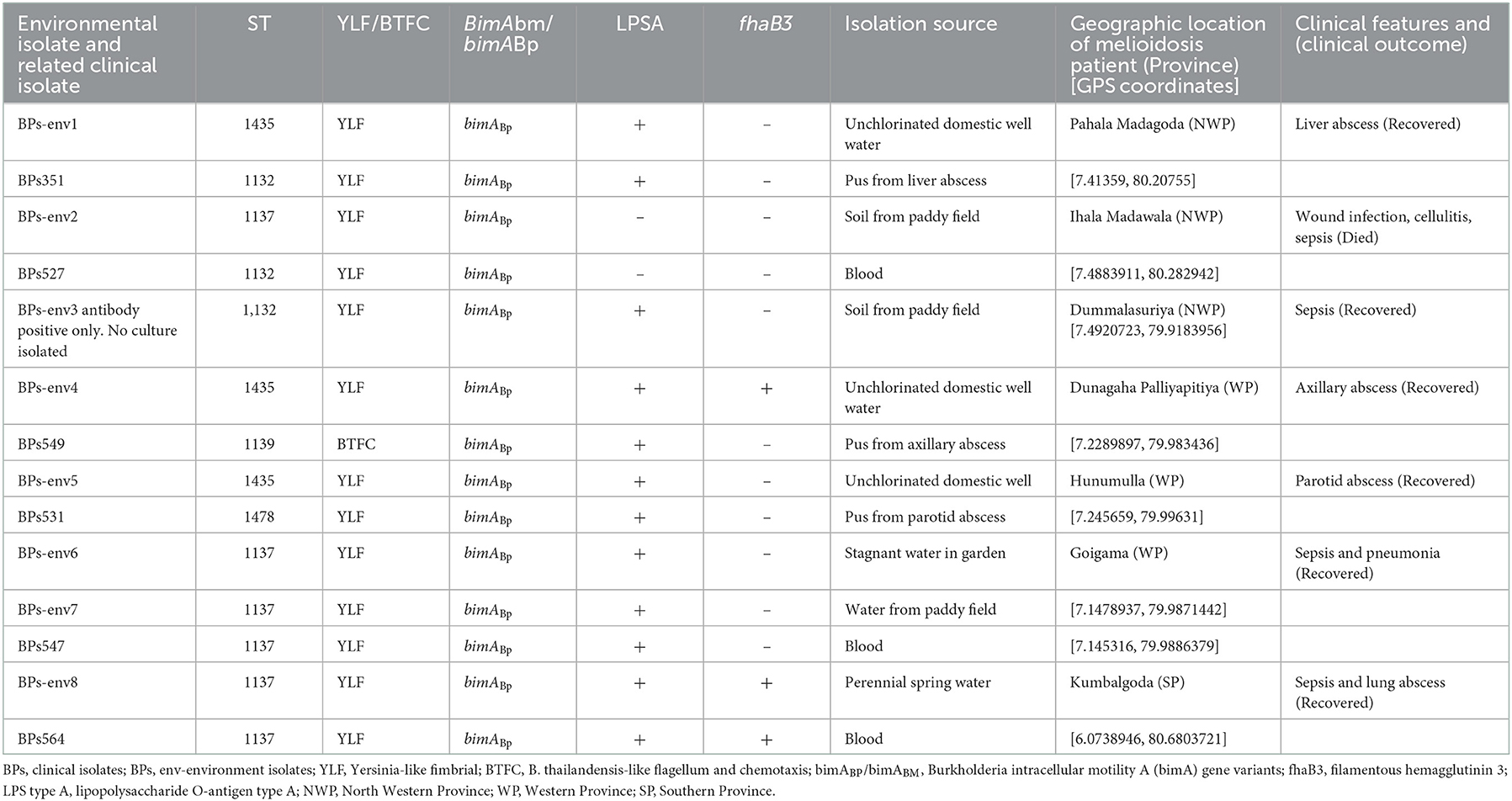
Table 1. Description of genotypes of environmental and corresponding clinical isolates, isolation source, geographic location, and clinical presentations.
Four isolates belonged to ST1137, three to ST1435 and one to ST1132 (Table 1). In the case of BPs-env1, BPs-env4, and BPs-env5 isolated from unchlorinated domestic drinking water wells and BPs-env2 from paddy soil, the sequence type (ST) of the environmental isolate differed from the corresponding clinical isolate. However, BPs-env6 and BPs-env7, isolated from stagnant garden water and paddy field water of a single patient, shared the same ST (ST1137) and possessed identical genotypes (YLF, bimABP, LPSA positive, and fhaB3 negative) with the related clinical isolate. BPs-env8 was isolated from a perennial spring, “Bubula.” Villagers use water from this spring for washing agricultural utensils and machinery. The corresponding melioidosis patient was residing approximately 150 m away and used water from the “Bubula” spring to wash his motorcycle. In this case, too, the clinical isolate was identical in terms of ST (1137) and genotypes (YLF, bimABP, LPSA positive, and fhaB3 positive) with the environmental isolate from the spring although soil and water samples obtained from his residence and domestic well were negative for B. pseudomallei.
PHYLOViZ analysis of the phylogenetic relationship of Sri Lankan environmental isolates with other selected environmental isolates revealed two distinct clusters i.e., South Asian strains and Australian strains. Sri Lankan environmental isolates clustered with Australian environmental isolates (Figure 3).
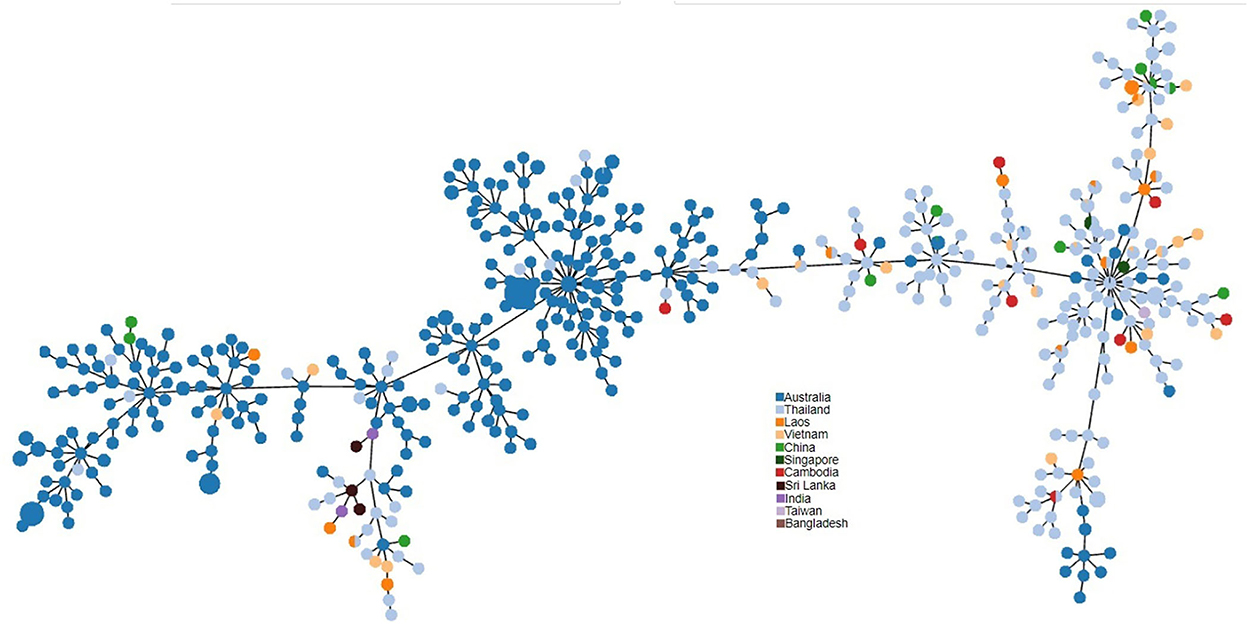
Figure 3. PHYLOViZ analysis of STs showing the phylogenetic relationship of Sri Lankan environment isolates (n = 3 STs) with environment isolates from Australia (n = 220), Thailand (n = 163), Vietnam (n = 25), Laos (n = 19), China (n = 11), Cambodia (n = 8), Singapore (n = 4), India (n = 2), Taiwan (n = 1), and Bangladesh (n = 1).
Melioidosis may be acquired by direct contact with contaminated soil, ingestion of untreated water or inhalation of dust or contaminated aerosols. Individuals with underlying diseases such as diabetes, renal, or liver disease and various forms of immunosuppression are most at risk (1). The bacterium is commonly found in the soil and stagnant water of paddy fields and wetlands in endemic regions and contaminated soil and water has been shown to be the most important source of infection (12). An association between melioidosis and paddy cultivation and the presence of B. pseudomallei in drinking water has been shown (2).
The bacterium has been isolated from a wide range of environment niches including natural water environments and unchlorinated drinking water wells in endemic regions. The unusual ability of B. pseudomallei to thrive in harsh environments has been shown to contribute to its long-term persistence in such environments (25).
This study reports the first evidence for the presence of B. pseudomallei in soil and natural water in Sri Lanka. Notably, it demonstrates that taking a careful history of exposure from the patient can help to pinpoint sites where the bacterium is present.
The presence of B. pseudomallei in natural water sources in Sri Lanka poses a major public health issue as the proportion of households with access to pipe-borne treated water is currently below 50% (www.waterboard.lk). Water from household wells is commonly used for drinking, cooking and bathing, and the high proportion of melioidosis cases presenting with parotid and abdominal abscesses in Sri Lanka may be a reflection of transmission by ingestion of contaminated water (3).
In addition to dug wells, natural perennial bubbling springs, such as the “Bubula” sampled during this study, are often enclosed and used for community needs. Water from such springs continuously overflows into the surrounding environment, posing a high risk of contamination and spread of the pathogen to distant areas during flooding and via strong winds (26, 27).
MLST of the eight environmental isolates revealed only three STs, showing a low diversity among the environmental strains as has been noted in Singapore (1). There were no novel STs, when compared with the international B. pseudomallei database (https://pubmlst.org/organisms/burkholderia-pseudomallei) and all three sequence types had been recorded previously in clinical infections in Sri Lankan patients (10). PHYLOViZ analysis of selected environmental STs from the B. pseudomallei database divided them into two distinct populations, Australian and Southeast Asian. The Sri Lankan environmental isolates clustered with the Australian isolates. It is interesting to note that the Australian population is recognized as the most ancestral population (28). The reason for the close relationship between B. pseudomallei isolates from Sri Lanka and Australia is unknown but could be related to their original positions in Gondwanaland (10).
BPs-env6 and BPs-env7, isolated from the patient's immediate environs (stagnant water in the garden and paddy water, respectively), and BPs-env8 recovered from a community spring shared ST 1137 and genotypes with the clinical isolates from the corresponding patients which may indicate direct transmission of the pathogen from the environment (29–31). However, since 1137 is a widespread ST in Sri Lanka, whole genome sequencing will be required to confirm that the clinical and environmental isolates are, indeed, clonal in nature. The presence of non-identical genetic patterns among the clinical and environmental isolates in the other four cases, suggests significant environmental contamination with multiple strains, as reported previously (32).
The almost ubiquitous presence of B. pseudomallei in the environment of endemic countries and universal exposure of resident agricultural populations pose a challenge to the prevention and control of melioidosis. On a local scale, if contamination of water sources is confirmed by culture, preventive measures, such as chlorination of wells or filtering or boiling water before drinking can be recommended to minimize the risk of acquiring melioidosis (2).
This study confirms the presence of B. pseudomallei in soil and natural water in Sri Lanka, placing the populations using untreated water from such sources at risk of melioidosis. Although no vaccine is currently available, advising the public of the dangers associated with consumption of untreated water and raising awareness among clinicians that melioidosis should be considered in the differential diagnosis of febrile illness in areas without a pipe-borne, treated water supply may help to reduce transmission and enhance early diagnosis. Identifying specific environmental sites positive for B. pseudomallei may assist to mitigate risk via soil and water remediation to reduce the bacterial burden.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics approval for this study was obtained from the Ethics Review Committee, Faculty of Medicine, University of Colombo, Sri Lanka (EC-17-020).
HJ and EC contributed to the conceptualization and design of the study. TM, UK, KJ, JM, and DJ collected and organized the data. TM and HJ analyzed the data. HJ wrote the original draft. EC, HJ, and DJ contributed to the interpretation of the results and manuscript revision. All authors contributed to the article and approved the submitted version.
This study was supported by the National Research Council of Sri Lanka (Research Grant No. NRC18-079) and Colombo University Research Grant No. AP/3/2/2020/SG/13.
We thank the technical staff at the Biomedical Laboratory, Kotelawala Defense University, and the Department of Medical Microbiology and Immunology, Faculty of Medicine, the University of Colombo for their valuable support. We also thank the patients and their families for assistance in identifying the locations for sample collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gassiep I, Armstrong M, Norton R. Human melioidosis. Clin Microbiol Rev. (2020) 33:e00006–19. doi: 10.1128/CMR.00006-19
2. Limmathurotsakul D, Kanoksil M, Wuthiekanun V, Kitphati R, deStavola B, Day NP, et al. Activities of daily living associated with acquisition of melioidosis in northeast Thailand: a matched case-control study. PLoS Negl Trop Dis. (2013) 7:e2072. doi: 10.1371/journal.pntd.0002072
3. Corea EM, de Silva AD, Thevanesam V. Melioidosis in Sri Lanka. Trop Med Infect Dis. (2018) 3:22. doi: 10.3390/tropicalmed3010022
4. Zimmermann RE, Ribolzi O, Pierret A, Rattanavong S, Robinson MT, Newton PN, et al. Rivers as carriers and potential sentinels for Burkholderia pseudomallei in Laos. Sci Rep. (2018) 8:8674. doi: 10.1038/s41598-018-26684-y
5. Mayo M, Kaesti M, Harrington G, Cheng AC, Ward L, Karp D, et al. Burkholderia pseudomallei in unchlorinated domestic bore water, Tropical Northern Australia. Emerg Infect Dis. (2011) 17:1283–5. doi: 10.3201/eid1707.100614
6. Parry CM, Wuthiekanun V, Hoa NT, Diep TS, Thao LT, Loc PV, et al. Melioidosis in Southern Vietnam: clinical surveillance and environmental sampling. Clin Infect Dis. (1999) 29:1323–6. doi: 10.1086/313479
7. Inglis TJ, Foster NF, Gal D, Powell K, Mayo M, Norton R, et al. Preliminary report on the northern Australian melioidosis environmental surveillance project. Epidemiol Infect. (2004) 132:813–20. doi: 10.1017/S0950268804002663
8. Rolim DB, Rocha MF, Brilhante RS, Cordeiro RA, Leitão NP Jr, Inglis TJ, et al. Environmental isolates of Burkholderia pseudomallei in Ceará State, northeastern Brazil. Appl Environ Microbiol. (2009) 75:1215–8. doi: 10.1128/AEM.01953-08
9. Inglis TJ, Sousa AQ. The public health implications of melioidosis. Braz J Infect Dis. (2009) 13:59–66. doi: 10.1590/S1413-86702009000100013
10. Jayasinghearachchi HS, Corea EM, Jayaratne KI, Fonseka RA, Muthugama TA, Masakorala J, et al. Biogeography and genetic diversity of clinical isolates of Burkholderia pseudomallei in Sri Lanka. PLoS Negl Trop Dis. (2021) 15:e0009917. doi: 10.1371/journal.pntd.0009917
11. Jayasinghearachchi HS, Francis VR, Sathkumara HD, Krishnananthasivam S, Masakorala J, Muthugama T, et al. Nonclonal Burkholderia pseudomallei population in melioidosis case cluster, Sri Lanka. Emerg Infect Dis. (2021) 27:2955–7. doi: 10.3201/eid2711.210219
12. McRobb E, Sarovich DS, Price EP, Kaestli M, Mayo M, Keim P, et al. Tracing melioidosis back to the source: using whole-genome sequencing to investigate an outbreak originating from a contaminated domestic water supply. J Clin Microbiol. (2015) 53:1144–8. doi: 10.1128/JCM.03453-14
13. Currie BJ, Price EP, Mayo M, Kaestli M, Theobald V, Harrington I, et al. Use of whole-genome sequencing to link Burkholderia pseudomallei from air sampling to mediastinal melioidosis, Australia. Emerg Infect Dis. (2015) 21:2052. doi: 10.3201/eid2111.141802
14. Tuanyok A, Auerbach RK, Brettin TS, Bruce DC, Munk AC, Detter JC, et al. A horizontal gene transfer event defines two distinct groups within Burkholderia pseudomallei that have dissimilar geographic distributions. J Bacteriol. (2007) 189:9044–9. doi: 10.1128/JB.01264-07
15. Sarovich DS, Price EP, Webb JR, Ward LM, Voutsinos MY, Tuanyok A, et al. Variable virulence factors in Burkholderia pseudomallei (melioidosis) associated with human disease. PLoS ONE. (2014) 9:e91682. doi: 10.1371/journal.pone.0091682
16. Tuanyok A, Stone JK, Mayo M, Kaestli M, Gruendike J, Georgia S, et al. The genetic and molecular basis of O-antigenic diversity in Burkholderia pseudomallei lipopolysaccharide. PLoS Negl Trop Dis. (2012) 6:e1453. doi: 10.1371/journal.pntd.0001453
17. Limmathurotsakul D, Dance DA, Wuthiekanun V, Kaestli M, Mayo M, Warner J, et al. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl Trop Dis. (2013) 7:e2105. doi: 10.1371/journal.pntd.0002105
18. Jayaweera LJ, Wickramasinghe D, Randeep K, Wijerathna P, Javahir IM, Sharjoon M, et al. Cluster of cases of pulmonary melioidosis presenting as acute fulminant pneumonia in the East of Sri Lanka. J Ceylon Coll Phys. (2021) 52:34–9. doi: 10.4038/jccp.v52i1.7929
19. Baker AL, Ezzahir J, Gardiner C, Shipton W, Warner JM. Environmental attributes influencing the distribution of Burkholderia pseudomallei in Northern Australia. PLoS ONE. (2015) 10:e0138953. doi: 10.1371/journal.pone.0138953
20. Ashdown LR. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology. (1979) 11:293–7. doi: 10.3109/00313027909061954
21. Merritt A, Inglis TJ, Chidlow G, Harnett G. PCR-based identification of Burkholderia pseudomallei. Rev Inst Med Trop São Paulo. (2006) 48:239–44. doi: 10.1590/S0036-46652006000500001
22. Jayasinghearachchi HS, Corea EM, Krishnananthasivam S, Sathkumara HD, Francis VR, Abeysekere TR, et al. Whole-genome sequences of eight clinical isolates of Burkholderia pseudomallei from melioidosis patients in eastern Sri Lanka. Microbiol Resour Announc. (2019) 8:e00645–19. doi: 10.1128/MRA.00645-19
23. Wuthiekanun V, Anuntagool N, White NJ, Sirisinha S. Short report: a rapid method for the differentiation of Burkholderia pseudomallei and Burkholderia thailandensis. Am J Trop Med Hyg. (2002) 66:759–61. doi: 10.4269/ajtmh.2002.66.759
24. Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, et al. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. (2003) 41:2068–79. doi: 10.1128/JCM.41.5.2068-2079.2003
25. Inglis TJ, Sagripanti JL. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl Environ Microbiol. (2006) 72:6865–75. doi: 10.1128/AEM.01036-06
26. Shaharudin R, Ahmad N, Kamaluddin MA, Veloo Y. Detection of Burkholderia pseudomallei from post-flood soil samples in Kelantan, Malaysia. Southeast Asian J Trop Med Public Health. (2016) 47:951–6.
27. Merritt AJ, Inglis TJJ. The role of climate in the epidemiology of melioidosis. Curr Trop Med Rep. (2017) 4:185–91. doi: 10.1007/s40475-017-0124-4
28. Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, et al. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. (2009) 7:78. doi: 10.1186/1741-7007-7-78
29. Chen YS, Lin HH, Mu JJ, Chiang CS, Chen CH, Buu LM, et al. Distribution of melioidosis cases and viable Burkholderia pseudomallei in soil: evidence for emerging melioidosis in Taiwan. J Clin Microbiol. (2010) 48:1432–4. doi: 10.1128/JCM.01720-09
30. Prakash A, Thavaselvam D, Kumar A, Kumar A, Arora S, Tiwari S, et al. Isolation, identification and characterization of Burkholderia pseudomallei from soil of coastal region of India. Springerplus. (2014) 16:438. doi: 10.1186/2193-1801-3-438
31. Currie BJ, Mayo M, Anstey NM, Donohoe P, Haase A, Kemp DJ. A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am J Trop Med Hyg. (2001) 65:177–179. doi: 10.4269/ajtmh.2001.65.177
Keywords: melioidosis, Burkholderia pseudomallei, Sri Lanka, natural water, environment
Citation: Jayasinghearachchi HS, Muthugama TA, Masakorala J, Kulasekara US, Jayaratne K, Jayatunga DADN, De Silva AD and Corea EM (2023) Burkholderia pseudomallei in soil and natural water bodies in rural Sri Lanka: A hidden threat to public health. Front. Vet. Sci. 9:1045088. doi: 10.3389/fvets.2022.1045088
Received: 15 September 2022; Accepted: 12 December 2022;
Published: 17 January 2023.
Edited by:
Chiranjay Mukhopadhyay, Manipal Academy of Higher Education, IndiaReviewed by:
Christopher Cote, United States Army Medical Research Institute of Infectious Diseases (USAMRIID), United StatesCopyright © 2023 Jayasinghearachchi, Muthugama, Masakorala, Kulasekara, Jayaratne, Jayatunga, De Silva and Corea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Himali S. Jayasinghearachchi,  amF5YXNpbmdoZWFyYWNoY2hpaHNAa2R1LmFjLmxr; Enoka M. Corea,
amF5YXNpbmdoZWFyYWNoY2hpaHNAa2R1LmFjLmxr; Enoka M. Corea,  ZW5va2FjQG1pY3JvLmNtYi5hYy5saw==
ZW5va2FjQG1pY3JvLmNtYi5hYy5saw==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.