- 1Department of Health Management, Atlantic Veterinary College, University of Prince Edward Island, Charlottetown, PE, Canada
- 2Aquatic Diagnostic Services, Atlantic Veterinary College, University of Prince Edward Island, Charlottetown, PE, Canada
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) allows rapid and reliable identification of microorganisms. The accuracy of bacterial identification using MALDI-TOF MS depends on main spectral profiles (MSPs) provided in a quality-assured commercial reference library, which requires ongoing improvement. This study aimed to develop and validate an in-house MALDI-TOF MS MSP to rapidly identify Yersinia ruckeri isolated from Atlantic salmon (Salmo salar). The novel MSP was prepared using an isolate of Y. ruckeri recovered from Atlantic salmon and confirmed by 16S rRNA gene sequencing. Subsequently, a validation set which comprises 29 isolates of Y. ruckeri were examined from three fishes: Atlantic salmon (Salmo salar) (n = 26), American eel (Anguilla rostrata) (n = 1), and Atlantic cod (Gadus morhua) (n = 2). These isolates were randomly selected from the Atlantic Veterinary College, Aquatic Diagnostic Services Bacteriology Laboratory's culture collection to validate the novel MSP. Analytical sensitivity of MALDI-TOF MS using the novel MSP to identify the validation set was 86.2%. Repeatability was assessed by acquiring spectra from 30 different spots of a randomly-selected isolate of Y. ruckeri, and analyzed spectra from each spot were compared against the novel MSP. The coefficient of variation was 3.3%. The novel MSP clustered with Bruker MSPs (n = 3) of Y. ruckeri in the reference library and did not falsely identify any closely related bacteria to Y. ruckeri. This study reports the development of a novel MSP of high analytical sensitivity and specificity for rapid identification of Y. ruckeri using MALDI-TOF MS.
Introduction
Yersinia ruckeri, the causative agent of enteric redmouth disease (ERM), is a bacterial pathogen that affects all life stages of salmonids as well as other fin-fish species, although the clinical disease is more acute in fry and fingerlings than older life stages (1, 2). Clinically, ERM is characterized by hemorrhagic septicemia, which may ultimately lead to mortality, and cumulative mortality can increase from 25 to 75% in the absence of therapeutic interventions (3, 4). Y. ruckeri was first isolated in rainbow trout (Oncorhynchus mykiss) but has since been detected in other salmonids such as Atlantic salmon (Salmo salar), lake trout (Salvelinus namaycush), brown trout (Salmo trutta), brook trout (Salvelinus fontinalis), Coho salmon (Oncorhynchus kisutch), cutthroat trout (Oncorhynchus clarkii), sockeye (Oncorhynchus nerka), Chinook (Oncorhynchus tshawytscha), and other species of fish like common carp (Cyprinus carpio), catfish (Silurus glanis), goldfish (Carassius auratus), European eel (Anguilla anguilla), coalfish (Pollachius virens), and perch (Perca fluviatilis) (5, 6). Over the past two decades across Atlantic Canada, Y. ruckeri has been one of the most commonly-detected bacterial pathogens from salmonid samples submitted to the Atlantic Veterinary College, Aquatic Diagnostic Services Bacteriology Laboratory (7).
Early detection of Y. ruckeri can hasten intervention and circumvent mortality events during ERM outbreaks in farmed salmonids (8, 9). The traditional method of identifying this pathogen in the laboratory is by culture and isolation using general-purpose media, usually followed by a series of biochemical tests to determine the genus and species of the bacterium (10). The time for identification of this bacterium in the laboratory can be 4 to 7 days if traditional methods are used (11). Biochemical tests are sometimes prone to species-level misidentification because Y. ruckeri is phylogenetically and biochemically similar to other species of the genus Yersinia. In addition, Y. ruckeri can also be misidentified or other members of the order Enterobacterales, such as Hafnia alvei, which is a common misidentification whenever Y. ruckeri is analyzed using the Analytical Profile Index® biochemical testing system (12, 13). Other Yersinia species that have been detected in Atlantic salmon and trout include Y. enterocolitica, Y. intermedia, Y. frederiksenii, while Y. kristensenii, Y. bercovieri, Y. mollaretii, Y. rohdei, and Y. aldovae have not been reported in salmonids (14–16). Molecular techniques such as polymerase chain reaction (PCR) and sequencing of genes such as 16S ribosomal RNA (rRNA) provide a higher specificity to differentiate Yersinia to species-level. However, these can be expensive or resource-consuming processes (and therefore not routinely available in most clinical laboratories). Molecular techniques can take an additional 5–24 h before species-level identification results are available (17, 18).
Many clinical laboratories increasingly utilize matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) as a less expensive and faster analytical technique. MALDI-TOF MS has a high level of accuracy for species-level identification of bacteria that affect human and animal health (19–21). MALDI-TOF MS has replaced or supplemented phenotypic and molecular methods of identifying bacteria in modern-day clinical microbiology (22–24). Identification of bacteria to the species-level using MALDI-TOF MS is achieved by matching the spectral profile of bacterial peptides against existing main spectral profile (MSP) in the reference library (widely used in veterinary laboratories is the commercial MALDI Biotyper® reference library manufactured by Bruker), which can be performed in < 1 h (25, 26). The reliability of the MALDI-TOF MS to correctly identify bacteria largely depends on the presence and quality of MSPs in the available reference library (27). Notwithstanding, bacterial identification sometimes fails even with sufficient reference spectra due to the poor quality of the spectral composition (28). Several research investigations have further augmented or supplemented the commercial reference library with custom MSPs of clinically significant bacteria to extend the existing library and increase MALDI-TOF MS's performance in bacterial identification (29, 30). However, the Bruker reference library was developed using bacterial species isolated from humans (31), so there could be difficulty in applying this reference library to aquatic pathogens (32). Variability in bacterial identification using MALDI-TOF MS is linked to several other factors: sample preparation, quality or type of matrix used, acid concentration in the matrix, presence of surfactant in matrix, the concentration of bacterial cells, and instrument conditions (33). This study aimed to develop and validate an in-house main spectral profile for rapidly identifying Y. ruckeri isolated from Atlantic salmon using MALDI-TOF MS.
Materials and methods
Bacterial isolates used for the study
Isolates of Y. ruckeri from Atlantic salmon (Salmo salar) (n = 27), Atlantic cod (Gadus morhua) (n = 2), and American eel (Anguilla rostrata) (n = 1) were randomly selected from the culture collection of the Atlantic Veterinary College, Aquatic Diagnostic Services Bacteriology Laboratory (AVC ADSBL) at the University of Prince Edward Island. These isolates were primarily obtained during bacteriological analyses of swabs, culture plates, tissues, and fluids samples submitted to AVC ADSBL by aquatic facilities across Atlantic Canada and Ontario. Y. ruckeri isolates were identified by Gram stain morphology (gram-negative bacillus), oxidase activity (negative), growth at 35°C (positive), hydrogen sulfide production (negative), and motility (most strains were positive at 22°C). Suspect isolates were confirmed using the Analytical Profile Index® 20E (bioMérieux Inc., NC, USA) and agglutination techniques (34). The electronic laboratory information management system at AVC ADSBL did not document the life-stage data such as fry, parr, smolt grow-out, and brood-stock of submitted samples from which these isolates were recovered.
Bacterial culture and sub-culturing of lyophilized isolates
Lyophilized isolates of Y. ruckeri were reconstituted using Tryptic Soy Broth (Becton Dickinson and Company, MD, USA) and aseptically plated onto Columbia agar (Oxoid Company, ON, Canada) supplemented with 5% sheep blood. The blood agar (BA) plates were incubated at 22°C for 24–48 h. Morphologically, the colonies separated well and appeared off-white with smooth edges on the culture plates after incubation. Non-distinct colonies with a mixture of bacterial growth were sub-cultured to BA plates to obtain pure colonies.
Strain selection and identification for main spectral profile creation
One isolate of Y. ruckeri was randomly selected from the culture collection of the Atlantic salmon group (n = 27) for the creation of the MSP. The DNA of the selected isolate was extracted using the InstaGene™ Matrix (BioRad), following the manufacturer's guidelines with the following modifications: a loopful of bacteria instead of one colony was collected for the starting material, and the incubation time at 56°C was increased from 30 to 45 min. The 16S rRNA gene was amplified by PCR using primers 27F and 1492R (35), amplicons visualized on agarose gel, and sent to a commercial laboratory for purification and Sanger sequencing. Sequences were trimmed and aligned using CLC Sequence Viewer 7, and sequence identity was determined using the National Center Biotechnology Information nucleotide collection (BLASTn).
Creation of main spectral profile for Yersinia ruckeri
The creation of MSP for Y. ruckeri was done following Bruker's recommended procedure for custom MALDI Biotyper® (MBT) MSP and library creation.
Protein extraction procedure
Three hundred microliters of deionized water were measured into a 1.5 ml Eppendorf microcentrifuge tube using a pipette, followed by placing a single, fresh (24–48 h old), and pure colony of Y. ruckeri into the same tube. This was vortexed thoroughly and added a total of 900 μL of absolute ethanol to the mixture in the microcentrifuge tube. The mixture was centrifuged at a maximum speed of 13,000 revolutions per minute (rpm) for 2 min. The supernatant was decanted, while the residue in the tube was centrifuged again to separate any trace of absolute ethanol. The remaining ethanol was carefully decanted by using a pipette not to disturb the pellet formed at the bottom of the tube. The pellet was left to air dry at room temperature for at least 5 min. Once dried, 20 μL of freshly prepared 70% formic acid (Honeywell Fluka, NC, USA) was added to the pellet and vortexed. A volume of 20 μL of acetonitrile (ACN) (Sigma-Aldrich Canada Co., ON, Canada) was added to the mixture and placed in a centrifuge for 2 minutes at a maximum speed of 13,000 rpm. One microliter of supernatant was pipetted onto a clean MALDI stainless target plate and was repeated for ten replicates. The spots were allowed to air dry at room temperature. Within 30 min after drying, spots were overlaid with 1 μL of α-cyano-4-hydroxycinnamic acid (HCCA) matrix solubilized in 50% acetonitrile (Sigma-Aldrich Canada Co., ON, Canada), 47.5% water, and 2.5% trifluoroacetic acid solution. The matrix was left to air dry at room temperature.
Instrument calibration
One microliter of Bacterial Test Standard (BTS; Bruker) was applied to a clean spot on the same MALDI stainless steel target and allowed to air dry. After drying, the BTS spot was overlaid with 1 μL of HCCA matrix solution within 30 min and allowed to air dry at room temperature.
The target was loaded into the microflex® LT mass spectrometer (Bruker) for spectral acquisition. A cumulative spectrum is generated using six sets of 40 laser shots for each spot (for a cumulative total of 240 shots per spot) with a nitrogen laser frequency of 60-Hz, a voltage of 20 kv, and mass to charge ratio of ions range of 2–20 k Dalton. The instrument was calibrated with the BTS using the AutoXecute algorithm in flexControl® software version 3.4 (Bruker) to manufacturer specifications. For calibration, the automatic laser position was manually overridden by moving the laser to the left side of the spot. The instrument was considered to pass calibration with eight reference mass peaks successfully assigned within ± 300 parts per million.
Main spectral profile creation and quality control
Spectra were acquired using the same laser conditions as the BTS from each of 10 supernatant spots and repeated three times, resulting in a collection of 30 cumulative spectra. The acquired spectra were saved and analyzed using the flexAnalysis® software version 3.4 (Bruker). Spectra underwent automatic smoothing and baseline subtraction. Visual assessments of spectra were performed to identify flatline spectra, outlier mass peaks, dramatic mass shifts, or anomalies. Flatline spectra and outliers were removed prior to MSP creation. Five of the 30 spectra were excluded as outliers (having non-uniform peaks). In comparison, 25 spectra were saved as the MSP to the reference library as “Yersinia ruckeri 15945-2008 AVC 2021”. Hereafter, the new library will be referred to as the “novel MSP.”
Validation of novel main spectral profile
The remaining 29 isolates of Y. ruckeri are referred to as “validation set” hereafter, for ease of description, in this study. The direct smear method, in which a colony of Y. ruckeri is smeared onto the MALDI plate using a toothpick, then overlaid with 1 μL of HCCA matrix within 30 min, followed by instrument calibration, spectra collection, and analysis were carried out for the validation set. The acquired spectra from the validation set were carefully analyzed. The analyzed spectra were compared against the novel MSP and the MBT reference library (which contains Bruker MSPs) in AVC ADSBL. The reliability of using the novel MSP and Bruker MSPs was based on the log score values recommended by the manufacturer for the MBT Compass® software: a log score of 2.0 to 3.0 indicates high confidence identification, 1.7–1.9 indicates low confidence identification, and log score value < 1.7 indicates no reliable identification.
Analytical sensitivities of MALDI-TOF MS using the novel MSP and the Bruker MSPs were estimated based on high confidence identification of Y. ruckeri (log score value ≥ 2.0) with 95% confidence intervals (CI) calculated using the exact method in Stata version 15.
One isolate from the validation set was randomly selected for repeatability testing. Thirty spots on the selected isolate were directly smeared on a MALDI target, followed by instrument calibration, spectral acquisition, and analysis. These procedures were performed by a single technician on the same day. The acquired spectra from each of the 30 spots were compared against the novel MSP to identify the isolate and evaluate the degree of variability in their log scores. The organism best matched and the log score of identification were recorded, and the mean, standard deviation, range, and coefficient of variation in the log score of the isolates obtained from 30 different spots on the plate were calculated using Stata version 15.
MSP dendrogram analysis was performed to determine the relatedness in the mass peptide fingerprint of the MBT reference library and the novel MSP using distance correlation and single linkage function in the MBT Compass Explorer. The analyzed spectra of the validation set were cross-validated with the MSPs of other Yersinia species (including Y. intermedia, Y. massilensis, Y. bercovieri, Y. mollareti, Y. aleksiciae, Y. aldovae, Y. frederiksenii, Y. rohdei, Y. kristensenii, Y. enterocolitica subsp. palearctica, Y. enterocolitica subsp. enterocolitica, and Y. pseudotuberculosis) in the MBT reference library. The analyzed spectra of the validation set were further cross-validated with the novel MSP and the MSPs of bacterium like Aeromonas spp. (common gram-negative bacteria of salmonids) and other closely related bacteria which belong to the order Enterobacterales, such as Serratia spp., Hafnia spp., and Edwardsiella spp. (all have cultural and morphological similarity to Y. ruckeri).
Results
The identity of the isolate selected for creating the main spectral profile was verified using 16S rRNA gene sequencing. The isolate selected for MSP creation had a 99.13% identity to type strain Yersinia ruckeri DSM® 18506™ (=ATCC® 29473™) using a 1380 base-pair sequence and the GenBank accession number was KJ606914.1.
The novel main spectral profile was validated with acquired spectra from the validation set. The organisms that best match the validation set and log score values are represented using a dot map in Figure 1. Of the validation set compared against the novel MSP, 86.2% (n = 25) were identified as Y. ruckeri with high confidence (having log score values ≥ 2.0), 10.3% (n = 3) were identified as Y. ruckeri with low confidence (having log score values ≥ 1.7 < 2.0), and 3.5% (n = 1) was no reliable identification (having log score value < 1.7). Analytical sensitivity of MALDI-TOF MS using the novel MSP to identify the validation set was 86.2% (95% CI = 68.3–96.1) based on high confidence identification.
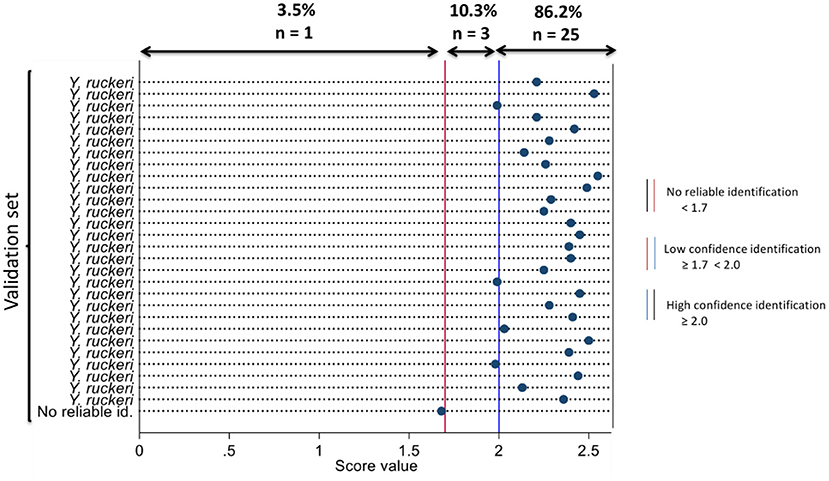
Figure 1. Dot diagram showing the organism that best matches the isolates in the validation set using the novel main spectral profile and their respective log scores at MALDI-TOF MS identification.
When the spectra of the validation set were compared against the MBT reference library and novel MSP, simultaneously; the novel MSP was the highest-ranked MSP which best identified Y. ruckeri isolates with high confidence for 16 individual isolates. The Bruker MSPs I and II were the highest-ranked MSPs that best identified Y. ruckeri isolates with high confidence for 10 and 2 individual isolates, respectively. The isolate classified as unreliable identification by the novel MSP was classified as Y. kristensenii with high confidence identification by the Bruker MSP III (Supplementary Figure 1). The log score values and organisms that best match the validation set using the Bruker MSPs and novel MSP at the MALDI-TOF MS identification are presented in Figure 2.
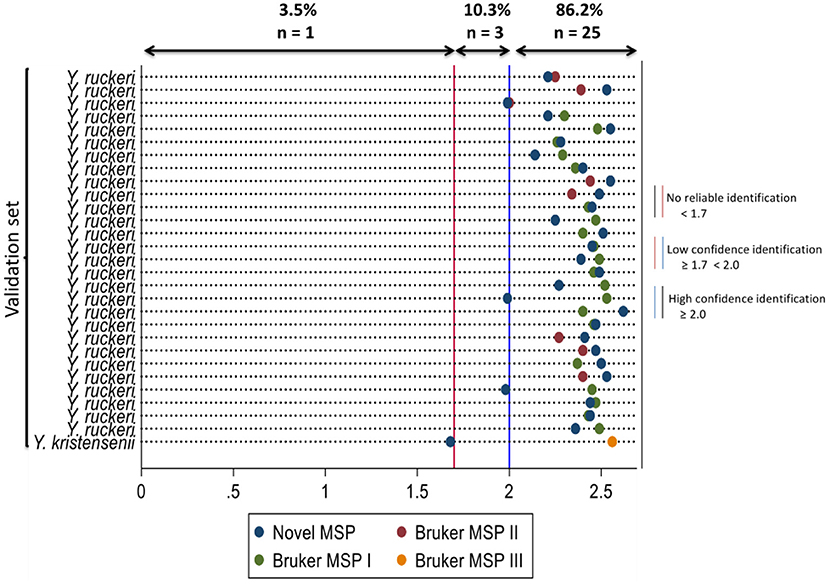
Figure 2. Dot diagram showing the organism that best matches the isolates in the validation set using the novel main spectral profile and the Bruker main spectral profiles with their respective log scores for MALDI-TOF MS identification. Novel MSP, Yersinia ruckeri 15945-2008 AVC 2021; Bruker MSP I, Yersinia ruckeri CCUG 21537 CCUG; Bruker MSP II, Yersinia ruckeri 3000 CVUA; Bruker MSP III, Yersinia kristensenii CCM 3562 CCM.
The species of fish from which the validation set was recovered, and the log score values obtained when the validation set were compared against the novel MSP are presented in (Supplementary Figure 2).
Repeatability testing of an isolate compared against the novel MSP yielded high confidence identification as Y. ruckeri for all the repeated runs (n = 30) with an average log score value of 2.38 (range, 2.2 to 2.6) from the 30 different spots of the isolate and has a low coefficient of variation (3.3%).
The novel MSP was clustered with the Bruker MSPs (n = 3) of Y. ruckeri and is clearly distinguished from other Bruker MSPs of other species of Yersinia (Supplementary Figure 3). During cross-validation of the validation set against MSPs of other Yersinia species (except Y. ruckeri) present in the MBT reference library, 96.6% (n = 28) were matched with other Yersinia species with low confidence identification. In contrast, an isolate from the validation set was classified as Y. kristensenii with high confidence identification (Supplementary Table 1). Cross-validation of the novel MSP, MSP of Aeromonas spp., MSPs of other close-related bacterial species such as Serratia spp., Hafnia spp., Edwardsiella tarda, and the validation set, yielded 89.7% (n = 26) high confidence identification of Y. ruckeri, 6.9% (n = 2) low confidence identification of Y. ruckeri, and 3.4% (n = 1) unreliable identification with the novel MSP only. In comparison, the validation set resulted in no reliable identification (100%, n = 29) when matched to MSPs of Aeromonas spp., Serratia spp., Hafnia spp., and Edwardsiella tarda (Supplementary Table 2).
Discussion
This study describes the development of a main spectral profile (MSP) to rapidly identify Yersinia ruckeri isolated from salmonid samples using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). This study has also provided the framework for MSP validation for other clinically important bacterial pathogens that affect salmonids aquaculture, especially in Canada, such as Renibacterium salmoninarum, Aeromonas salmonicida, and Streptococcus iniae (36).
The validation set used in this study was highly represented by Y. ruckeri isolates from Atlantic salmon than Atlantic cod and American eel. Furthermore, the novel MSP was developed using an isolate from the Atlantic salmon group because the novel MSP was primarily developed to identify Y. ruckeri in Atlantic salmon accurately. Y. ruckeri has been mostly detected from Atlantic salmon samples submitted for bacterial culture and isolation in the Atlantic Veterinary College, Aquatic Diagnostic Services Bacteriology Laboratory (AVC ADSBL) (7). However, the novel MSP successfully identified Y. ruckeri isolates from Atlantic salmon, Atlantic cod, and American eel in this study.
The direct smear method was used to validate the novel MSP as opposed to the protein extraction method because it is the same method used routinely by AVC ADSBL and most diagnostic labs for bacterial identification. The direct smear method is simpler to perform, faster, and it is not inferior to the protein extraction method for MALDI-TOF MS identification (37).
There are multiple MSPs available for identifying different bacterial species in the commercial reference library developed by Bruker to increase coverage of MALDI-TOF MS for identifying bacteria of various strains or serotypes (38, 39). In the present study, the analytical sensitivity of MALDI-TOF MS using the novel MSP provided high confidence identification for most of the Y. ruckeri isolates evaluated from Atlantic salmon. The novel MSP was the best to secure high confidence identification for 16 Y. ruckeri isolates evaluated compared to other individual MSPs developed by Bruker, which is in agreement with the report of Piamsomboon et al. (30). This finding gives credence to the extension or supplementation of the Bruker reference library with MSPs of aquatic origin to improve the identification of aquatic pathogens.
The Y. ruckeri isolate that could not be identified reliably with the novel MSP was identified as Y. kristensenii by Bruker MSP using the MALDI Biotyper® (MBT) reference library and during cross-validation with other species of Yersinia (except Y. ruckeri). This suggests that the traditional identification method (including Analytical Profile Index® 20E and or antisera agglutination method) used to confirm the validation set misclassified the isolate as Y. ruckeri and the Bruker MSP provided an accurate identification. Moreso, the isolate was detected and stored in 1990 in the AVC ADSL, and there is a possibility of misclassification, especially with the traditional identification methods. Gene sequencing of this isolate would have been recommended to provide a confirmatory identification; however, considering that the novel MSP classified the same isolate as no reliable identification twice, this was not pursued. Laboratories using traditional techniques to confirm bacteria not only stand the risk of bacterial misclassification at the species-level but also could overestimate or underestimate clinically relevant pathogens (40). On the other hand, misidentification of fish bacteria can have a severe economic impact. For instance, false-positive results may halt fish transfer from hatchery to production site and promote inappropriate use of antimicrobials for treatments which may lead to antimicrobial resistance (2). Misclassification error using the API 20E system for phenotypic identification of fish bacteria has been reported by several studies (41, 42). In addition, cross-reaction of bacterial isolates with antisera could potentially lead to bacterial misidentification (43). However, MALDI-TOF MS matches bacterial peptides from culture plates against spectra in the reference library, and the chance of misidentification is lower than the traditional method of bacterial identification (44).
Repeatability testing of a randomly-selected isolate against the novel MSP evaluated the variability in bacterial identification and the log score values. Each analyzed spectrum was correctly identified with high confidence as Y. ruckeri. The small variation in log scores from this analysis demonstrates a high confidence level in the result provided by the novel MSP. It should also be noted that sources of variation in log score could be from smear preparation and laser positioning, which might also explain why three of the validation set (Y. ruckeri from Atlantic salmon and American eel) evaluated were only identified with low confidence by the novel MSP.
During cross-validation, the novel MSP did not falsely identify any closely related bacterial species in the order Enterobacterales despite the relatedness, suggesting that the novel MSP was highly specific to identifying Y. ruckeri (45, 46). Furthermore, the MSP dendrogram clustered the novel MSP with Bruker MSPs of Y. ruckeri, and it clearly discriminates the collective MSPs of Y. ruckeri from others.
Individually, none of the Bruker MSPs were perfect to best identify all the isolates in the validation set evaluated in this study. It required three Bruker MSPs to identify the validation set with high confidence collectively. While it is prudent to supplement the Bruker reference library with new MSPs of aquatic pathogen, the novel MSP in this study has been uploaded and updated to the MBT reference library for rapid identification of Y. ruckeri from samples submitted for bacterial culture and isolation in AVC ADSBL and the novel MSP can be shared and used by other aquatic animal laboratories around the world.
Conclusion
This study has developed and validated a protein spectrum for the rapid detection of Yersinia ruckeri in Atlantic salmon using the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Study limitations
All the isolates in the validation set utilized for this study were not gene-sequenced because of cost implications. Molecular sequencing could have been used as a gold standard, and this would have identified the isolate in the validation set that was classified as Y. kristensenii by the Bruker MSP. In addition, strain diversity of the isolates used for creating the novel MSP and the validation set were unknown. This could have provided clearer insight into the serotype of Y. ruckeri identified by the novel MSP. Lastly, a single highly trained technician was used to conduct all experiments, and a repeatability assessment using two or more technicians in the same laboratory might have identified additional variability in results.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
RO, IG, DG, MS, SS, and KT conceived the study idea and designed the experiment. RO and MS conducted the experiment. RO wrote the first draft of the manuscript. IG and KT supervised the work. All authors have read, edited, and approved the final manuscript.
Acknowledgments
We thank Angela Abraham of Bruker for providing quality training on main spectral profile development and validation using the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Also, we thank Jan Giles and other technologists in the Atlantic Veterinary College, Aquatic Diagnostic Services Bacteriology Laboratory, University of Prince Edward Island, for their assistance in reviving the lyophilized isolates of Yersinia ruckeri utilized in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1031373/full#supplementary-material
References
1. Tobback E, Decostere A, Hermans K, Haesebrouck F, Chiers K. Yersinia ruckeri infections in salmonid fish. J Fish Dis. (2007) 30:257–68. doi: 10.1111/j.1365-2761.2007.00816.x
2. Mimeault C, Wade, J, Boily, F, Johnson, S, Jones, SRM, Aubry, P. Assessment of the risk to Fraser River Sockeye Salmon due to Yersinia ruckeri transfer from Atlantic Salmon farms in the Discovery Islands area, British Columbia. DFO Canadian Science Advisory Secretariat Research Document 2019/023 (2020). p. Viii + 44. Available online at: https://waves-vagues.dfo-mpo.gc.ca/Library/40947798.pdf (accessed October 10, 2022).
4. Roberts RJ. Fish Pathology Fourth Edition. Idaho: Blackwell Publishing Ltd. (2012). Vol. 4, p. 367–9.
5. Noga EJ. Fish Disease: Diagnosis and Treatment. Wilmington, NJ: John Wiley and Sons (2010). doi: 10.1002/9781118786758
6. Huang Y, Runge M, Michael GB, Schwarz S, Jung A, Steinhagen D. Biochemical and molecular heterogeneity among isolates of Yersinia ruckeri from rainbow trout (Oncorhynchus mykiss, Walbaum) in northwest Germany. BMC Vet Res. (2013) 9:215. doi: 10.1186/1746-6148-9-215
7. Ojasanya RA, Gardner IA, Groman DB, Saksida S, Saab ME, Thakur KK. Antimicrobial susceptibility profiles of bacteria commonly isolated from farmed salmonids in Atlantic Canada (2000–2021). Vet Sci. (2022) 9:159. doi: 10.3390/vetsci9040159
8. Chang WH, Yang SY, Wang CH, Tsai MA, Wang PC, Chen TY, et al. Rapid isolation and detection of aquaculture pathogens in an integrated microfluidic system using loop-mediated isothermal amplification. Sensors Actuat B Chem. (2013) 180:96–106. doi: 10.1016/j.snb.2011.12.054
9. Mishra SS, Das R, Sahoo SN, Swain P. Biotechnological tools in diagnosis and control of emerging fish and shellfish diseases. In: Genomics and Biotechnological Advances in Veterinary, Poultry, and Fisheries. Oxford: Academic Press (2020). p. 311–60. doi: 10.1016/B978-0-12-816352-8.00014-X
10. Ummey S, Khan S, Vijayakumar PPN, Ramya A. Enteric Red Mouth disease and its causative bacterium, Yersinia ruckeri, in Indian Major Carps from culture ponds in Andhra Pradesh, India. Aquacult Fisheries. (2021) 6:289–99. doi: 10.1016/j.aaf.2020.05.009
11. Kim JS, Lee GG, Park JS, Jung YH, Kwak HS, Kim, et al. A novel multiplex PCR assay for rapid and simultaneous detection of five pathogenic bacteria: Escherichia coli O157: H7, Salmonella, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus. J Food Prot. (2007) 70:1656–62. doi: 10.4315/0362-028X-70.7.1656
12. Austin DA, Robertson PAW, Austin B. Recovery of a new biogroup of Yersinia ruckeri from diseased rainbow trout (Oncorhynchus mykiss, Walbaum). Syst Appl Microbiol. (2003) 26:127–31. doi: 10.1078/072320203322337416
13. Stock I, Wiedemann B. Natural antimicrobial susceptibilities and biochemical profiles of Yersinia enterocolitica-like strains: Y. frederiksenii, Y intermedia, Y kristensenii and Y. rohdei. FEMS Immunol Med Microbiol. (2003) 38:139–52. doi: 10.1016/S0928-8244(03)00179-2
14. Carson J. Opportunistic infection by psychrotrophic bacteria of salmon. Bull Eur Assoc Fish Pathol. (1993) 3:49.
15. Sulakvelidze A. Yersiniae other than Y. enterocolitica, Y pseudotuberculosis, and Y pestis: the ignored species. Microbes Infect. (2000) 2:497–513. doi: 10.1016/S1286-4579(00)00311-7
16. Davies AR, Capell C, Jehanno D, Nychas GJ, Kirby RM. Incidence of foodborne pathogens on European fish. Food Control. (2001) 12:67–71. doi: 10.1016/S0956-7135(00)00022-0
17. Lazcka O, Del Campo FJ, Munoz FX. Pathogen detection: a perspective of traditional methods and biosensors. Biosen Bioelectron. (2007) 22:1205–17. doi: 10.1016/j.bios.2006.06.036
18. Calvez S, Gantelet H, Blanc G, Douet DG, Daniel P. Yersinia ruckeri biotypes 1 and 2 in France: presence and antibiotic susceptibility. Dis Aquat Organ. (2014) 109:117–26. doi: 10.3354/dao02725
19. Fernández-Álvarez C, Torres-Corral Y, Santos Y. Use of ribosomal proteins as biomarkers for identification of Flavobacterium psychrophilum by MALDI-TOF mass spectrometry. J Proteomics. (2018) 170:59–69. doi: 10.1016/j.jprot.2017.09.007
20. Müller A, Solem ST, Karlsen CR, Jørgensen TØ. Heterologous expression and purification of the infectious salmon anemia virus hemagglutinin esterase. Protein Expr Purif. (2008) 62:206–15. doi: 10.1016/j.pep.2008.08.006
21. Sy I, Margardt L, Ngbede EO, Adah MI, Yusuf ST, Keiser, et al. Identification of adult Fasciola spp. using matrix-assisted laser/desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. Microorganisms. (2021) 9:82. doi: 10.3390/microorganisms9010082
22. Alizadeh M, Yousefi L, Pakdel F, Ghotaslou R, Rezaee MA, Khodadadi E, et al. MALDI-TOF mass spectroscopy applications in clinical microbiology. Adv Pharmacol Pharm Sci. (2021) 2021:9928238. doi: 10.1155/2021/9928238
23. Jadhav SR, Shah RM, Palombo EA. Maldi-tof ms: a rapid methodology for identifying and subtyping Listeria monocytogenes. In: Listeria Monocytogenes. New York, NY: Humana (2021). p. 17–29) doi: 10.1007/978-1-0716-0982-8_2
24. Chai Z, Bi H. Capture and identification of bacteria from fish muscle based on immunomagnetic beads and MALDI-TOF MS. Food Chem X. (2022) 13:100225. doi: 10.1016/j.fochx.2022.100225
25. Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. (2015) 6:791. doi: 10.3389/fmicb.2015.00791
26. Hou TY, Chiang-Ni C, Teng SH. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J Food Drug Anal. (2019) 27:404–14. doi: 10.1016/j.jfda.2019.01.001
27. Paauw A, Jonker D, Roeselers G, Heng JM, Mars-Groenendijk RH, Trip H, et al. Rapid and reliable discrimination between Shigella species and Escherichia coli using MALDI-TOF mass spectrometry. Int J Med Microbiol. (2015) 305:446–52. doi: 10.1016/j.ijmm.2015.04.001
28. Veloo ACM, Knoester M, Degener JE, Kuijper EJ. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods for the identification of clinically relevant anaerobic bacteria. Clin Microbiol Infect. (2011) 17:1501–6. doi: 10.1111/j.1469-0691.2011.03467.x
29. Mougin J, Flahaut C, Roquigny R, Bonnin-Jusserand M, Grard T, Le Bris C. Rapid identification of Vibrio species of the harveyi clade using MALDI-TOF MS profiling with main spectral profile database implemented with an in-house database: luvibase. Front Microbiol. (2020) 11:586536. doi: 10.3389/fmicb.2020.586536
30. Piamsomboon P, Jaresitthikunchai J, Hung TQ, Roytrakul S, Wongtavatchai J. Identification of bacterial pathogens in cultured fish with a custom peptide database constructed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). BMC Vet Res. (2020) 16:52. doi: 10.1186/s12917-020-2274-1
31. Alispahic M, Christensen H, Hess C, Razzazi-Fazeli E, Bisgaard M, Hess M. MALDI-TOF mass spectrometry confirms clonal lineages of Gallibacterium anatis between chicken flocks. Vet Microbiol. (2012) 160:269–73. doi: 10.1016/j.vetmic.2012.05.032
32. Kurokawa S, Kabayama J, Fukuyasu T, Hwang SD, Park CI, Park SB, et al. Bacterial classification of fish-pathogenic Mycobacterium species by multigene phylogenetic analyses and MALDI Biotyper identification system. Mar Biotechnol. (2013) 15:340–8. doi: 10.1007/s10126-012-9492-x
33. Williams TL, Andrzejewski D, Lay JO, Musser SM. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J Am Soc Mass Spectrom. (2003) 14:342–51. doi: 10.1016/S1044-0305(03)00065-5
34. Romalde JL, Magarinos B, Fouz B, Bandin I, Nunez S, Toranzo AE. Evaluation of BIONOR Mono-kits for rapid detection of bacterial fish pathogens. Dis Aquat Organ. (1995) 21:25–34. doi: 10.3354/dao021025
36. Canadian Food Inspection Agency. Annually Notifiable Diseases. (2014). Available online at: https://inspection.canada.ca/animal-health/aquatic-animals/diseases/annually-notifiable/eng/1322950242720/1322950357496 (accessed July 25, 2022).
37. Fournier R, Wallet F, Grandbastien B, Dubreuil L, Courcol R, Neut C, et al. Chemical extraction versus direct smear for MALDI-TOF mass spectrometry identification of anaerobic bacteria. Anaerobe. (2012) 18:294–7. doi: 10.1016/j.anaerobe.2012.03.008
38. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. (2009) 49:543–51. doi: 10.1086/600885
39. Bizzini A, Durussel C, Bille J, Greub G, Prod'Hom G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. (2010) 48:1549–54. doi: 10.1128/JCM.01794-09
40. Petti CA, Polage CR, Schreckenberger P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J Clin Microbiol. (2005) 43:6123–5. doi: 10.1128/JCM.43.12.6123-6125.2005
41. Romalde JL, Toranzo AE. Evaluation of the API-20E system for the routine diagnosis of the enteric redmouth disease. Bull Eur Assoc Fish Pathol. (1991) 11:147.
42. Coz-Rakovac R, Strunjak-Perovic I. Commercial phenotypic tests (API 20E) in diagnosis of fish bacteria: a review. Vet Med. (2007) 52:49. doi: 10.17221/2058-VETMED
43. Buller NB. Bacteria From Fish and Other Aquatic Animals: A Practical Identification Manual. South Perth: CABI Publishing. (2004). doi: 10.1079/9780851997384.0000
44. Rodrigues NMB, Bronzato GF, Santiago GS, Botelho LAB, Moreira BM, Coelho IDS, et al. The Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) identification versus biochemical tests: a study with enterobacteria from a dairy cattle environment. Braz J Microbiol. (2017) 48:132–8. doi: 10.1016/j.bjm.2016.07.025
45. Ewing WH, Ross AJ, Brenner DJ, Fanning GR. Yersinia ruckeri sp. nov, the redmouth (RM) bacterium. Int J Syst Evol Microbiol. (1978) 28:37–44. doi: 10.1099/00207713-28-1-37
Keywords: MALDI-TOF MS, main spectral profile, Yersinia ruckeri, aquaculture, validation
Citation: Ojasanya RA, Gardner IA, Groman D, Saksida S, Saab ME and Thakur KK (2022) Development and validation of main spectral profile for rapid identification of Yersinia ruckeri isolated from Atlantic salmon using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Front. Vet. Sci. 9:1031373. doi: 10.3389/fvets.2022.1031373
Received: 29 August 2022; Accepted: 05 October 2022;
Published: 20 October 2022.
Edited by:
Om P. Dhungyel, The University of Sydney, AustraliaReviewed by:
Yasmine Hasanine Tartor, Zagazig University, EgyptNing Xu, Yangtze River Fisheries Research Institute (CAFS), China
Copyright © 2022 Ojasanya, Gardner, Groman, Saksida, Saab and Thakur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rasaq A. Ojasanya, cmFvamFzYW55YUB1cGVpLmNh
 Rasaq A. Ojasanya
Rasaq A. Ojasanya Ian A. Gardner
Ian A. Gardner David Groman
David Groman Sonja Saksida
Sonja Saksida Matthew E. Saab
Matthew E. Saab Krishna K. Thakur
Krishna K. Thakur