
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 13 October 2022
Sec. Animal Reproduction - Theriogenology
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1028867
This article is part of the Research TopicReproduction-related Non-coding RNAs in AnimalsView all 6 articles
 Kai Wang1
Kai Wang1 Ying Cheng1
Ying Cheng1 Tong Guo2
Tong Guo2 Xiangqian Guo1
Xiangqian Guo1 Hongzhi Zhang1
Hongzhi Zhang1 Xiaoyan Ma1
Xiaoyan Ma1 Yangyang Pan1
Yangyang Pan1 Ermias Kebreab3
Ermias Kebreab3 Dong Wang4*
Dong Wang4* Lihua Lyu1*
Lihua Lyu1*Cross-talk between competitive endogenous RNAs (ceRNAs) may play a critical role in revealing potential mechanism of bovine follicular cysts. Ovarian cyst has always been an intractable scientific problem and has led to considerable economic losses to bovine breeding industry. However, its pathogenesis and molecular mechanisms are still not well understood. Here, this study aimed to investigate the role of non-coding RNAs (ncRNAs) and the ceRNA networks in bovine follicular cyst. Whole transcriptome sequencing of bovine follicular granulosa cells (GCs) was conducted to obtain the expression profiles of mRNAs, lncRNAs and miRNAs. The results for the identified expressions of 8,003 mRNAs, 579 lncRNAs and 205 miRNAs were often altered between cystic and normal follicular GCs. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed on these differentially expressed mRNAs. Furthermore, the ceRNA network combining mRNAs, miRNAs, and lncRNAs using several bioinformatics methods based on co-expression analysis between the differentially expressed RNAs was conducted. Finally, the lncRNA NONBTAT027373.1-miR-664b-HSD17B7 pathway was verified by dual-luciferase reporting assay and RNA binding protein immunoprecipitation (RIP) assay. LncRNA NONBTAT027373.1 sponged miR-664b in GCs and prevented miR-664b from binding to the HSD17B7 3′-UTR. These results indicated that genes and lncRNAs related to steroid hormone synthesis and energy metabolism could play important roles in the formation of bovine cystic follicles through the ceRNA mechanism and represent candidate targets for further research. This can be used as a practical guideline for promoting healthy and highly efficient development in the bovine industry.
The source of oocytes restricts animal reproduction and the development of gamete and embryo bioengineering. Only a few oocytes ovulate to fertilize and produce offspring during livestock reproduction (1). Domestic animals form oocyte repository with many primitive follicles during embryogenesis and shortly after birth (2). After birth, with the animal growth and development, the majority of follicles undergo atresia and degeneration (3). Post-puberty only a few follicles develop to mature, but the absolute number of follicles decreases with age (4). If female animal has ovarian disease, the limited resources cannot be used effectively, which causes significant losses to the breeding industry. Ovarian cyst is a common disease in livestock species, particularly in bovine (5). The issue increases with age, which seriously limits the effective utilization of bovine oocytes. The incidence of follicular cysts in bovine has declined since the advent of accurate diagnosis by B-ultrasound. However, under intensive breeding conditions, follicular cyst caused by feeding management and genetic breeding is still increasing gradually (6). Therefore, solving the occurrence of bovine follicular cysts can shorten calving interval, reduce the frequency of mating, improve bovine reproductive performance, and increase the economic benefits of the cattle farm.
Studies have been focused on the effects of hormones such as luteinizing hormone (LH), follicle stimulating hormone (FSH), estrogen (E2) and granulosa cell receptor proteins on the occurrence of follicular cysts (7). For example, recent study has elucidated that changes in the adrenergic system and the contractile force of the follicular wall may contribute to the development of follicular cysts (8, 9). The reduced number of granulosa cell layers in cystic follicles is associated with reduced expression of cell adhesion factors such as cadherin (cell-cell adhesion) and integrin (cell-extracellular matrix adhesion) (10). IGFBP3 is a key regulator of IGF-1 in animals with cystic follicles (11). Moreover, the decreased expression of ovarian matrix metallo proteinase 1/2 (MMP1/2) is an important marker of the occurrence of follicular cysts in cows (12). The low expression of BMP6 and GATA6 in GCs and the high level of intra-follicular lipopolysaccharide (LPS) play an important role in the formation of follicular cysts (13). Reduced hepatic 17β-estradiol gluconaldehyde and liver disease are also associated with the occurrence of follicular cysts in dairy cows (14). Local changes in some metabolic sensors in follicles produce an unfavorable microenvironment for the recovery of ovarian activity, leading to the persistence of follicles and the occurrence of follicular cysts (15). The early development of cystic follicles in dairy cows coincides with changes in the expression of some vascular endothelial growth factors (EGFs) (16).
mRNA has been the traditional research focus as a template for protein synthesis, while ncRNA has been regarded as a large number of byproducts of transcription with little biological significance (17). Since the 1950s, ribosomal RNA (rRNA), transport RNA (tRNA) and a variety of RNAs have appeared successively providing a new understanding of ncRNA (18). High-throughput sequencing technology has maximized the impact of ncRNA in the biological area. More than 90% of mammalian genomes are transcribed, of which only about 2% are involved in protein-coding functions (19, 20), the rest of the non-coding genomes are roughly divided into microRNAs (miRNAs, 18–22 nt), long non-coding RNAs (lncRNAs, 200–100,000 nt) and some ncRNAs of variable length. All these include circular RNA (circRNA), enhancer RNA (eRNA), and small interfering RNA (siRNA) etc., (21, 22). Although studies on ncRNAs are scarce, new breakthroughs have been achieved such as miRNA, lncRNA and circRNA since 2006. Studies have shown ncRNAs are involved in a variety of biological processes in humans and animals, including muscle growth (23), adipogenesis (24), fat deposition (25), and hair follicle regulation (26). In recent years, studies on ncRNA in bovine are mainly focused on the proliferation, differentiation and related signaling pathways of muscle cells (27), while the studies on reproduction, especially on gametogenesis are still lagging behind.
With the wide application of total transcriptome sequencing technology, ncRNA has been rapidly developed in the molecular biological area. Total transcriptome sequencing is considered as using high-throughput sequencing technology to detect RNA sequences, and the expression levels and differences of RNA are reflected to form a map of ncRNA expression (28). Specifically, total transcriptome sequencing is performed by building two libraries: 1 miRNA library and 1 rRNA chain-de-specific library (mRNA, lncRNA and circRNA). Sequence information of four types of RNA (miRNA, lncRNA, mRNA and circRNA) are obtained. GO enrichment, KEGG pathway and cluster analysis are performed on the expressive map. Finally, the regulatory relationship between ncRNA and coding RNA is integrated and analyzed, mainly including the interactions between the two: miRNA-lncRNA, miRNA-circRNA, miRNA-mRNA, and among the three: LncRNA-miRNA-mRNA and circRNA-miRNA-mRNA, in which the regulatory relationship (ceRNA regulatory network) is an important means to comprehensively analyze scientific problems at the molecular level. The ceRNA principle has been widely proved to be a common mechanism for the expression of nodular genes launching new research fields and resulting in the enhancement of the ceRNA network databases predicted by bioinformatics (29–32).
Nowadays, investigation of the occurrence of bovine follicular cysts is mainly concentrated in granulosa cell receptor protein and the negative balance of nutrition and energy (33). At the molecular level, studies on the genetic regulation mechanism of bovine follicular cysts, especially the molecular regulation mechanism of ncRNAs is relatively delayed. The lack to fully revealing the rules of the occurrence of bovine follicular cysts and the molecular genetic regulation mechanism seriously influences the exertion of bovine reproductive potential, and further restricts the application of bovine artificial insemination, OPU-IVF (34), G6G-timing sperm insemination (35) and B-ultrasound pregnancy diagnosis (36), and ultimately affects the development of bovine industry and the development of gamete (ovum) and embryo bioengineering. This study aimed to take the bovine follicular (normal and cyst) as the research object, obtain bovine granulosa cells (GCs) with high purity through cell biology techniques such as microanatomy and in vitro cell culture, and introduce total transcriptome sequencing technology to complete transcriptome sequencing of GCs. Based on the differentially expressed (DE) mRNAs and ncRNAs, the related ceRNA networks were constructed to identify the key genomes and related ncRNAs led to follicular cysts, and the related ncRNAs and the key genes led to the formation of follicular cysts were overexpressed and silenced at the cellular level through transfection and other molecular methods. In order to prove its regulation and target genes, the results are expected to identify the key genes and related ncRNAs mediated by clear ceRNA, further providing a scientific basis for understanding the genetic regulatory mechanism of bovine follicular cysts. It has important guiding significance for promoting the healthy and efficient development of bovine industry and gamete- and embryo-bioengineering.
All animal procedures were conducted according to the guidelines of the China Council on Animal Care. All experimental protocols were approved by the Ethics Committee of Shanxi Agricultural University [Permit NO: 2020(058)].
Hereford cattle were raised under similar conditions, with free access to food and water and under natural lighting. Individual follicles were considered follicular cysts when the following symptom occurred: at least 25 mm in diameter with no luteal tissue found uni- or bilaterally in the ovaries detected by B-ultrasound successively for 7–10 d. The samples were obtained from Hereford cattle over 2 years of age, at least 45 days after calving (3.3 ± 1.5 lactations, range 2–7, mean 21 ± 5.5 kg of milk per day at diagnosis). Their ovaries were obtained after slaughter, cystic follicles and normal follicles were collected, which were transported to the laboratory in Dulbecco's phosphate-buffered saline solution containing antibiotics (100 IU/mL penicillin and 0.1 mg/mL streptomycin) at 4 °C. According to the selection criteria, 3 cystic (≥25 mm) follicles and 3 normal (10–15 mm) follicles were selected from different individuals. GCs from cystic and normal follicles were collected using microsurgery. After liquid nitrogen treatment, samples were placed in −80 °C freezer.
Total RNA was extracted from GCs using TRIzol RNA purification reagent, and genomic DNA was removed using RNase-free rDNase I (Takara, Japan), according to the manufacturer's instructions (Takara, Japan). The RNA quality was verified using the 2100 Bioanalyzer (Agilent, USA) and ND-2000 (NanoDrop Technologies). Only high-quality RNA samples (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RIN ≥8, 28S:18S ≥1.0, >10 μg) were used to construct a sequencing library.
The longRNA-seq transcriptome library was prepared using the TruSeq total RNA kit (Illumina, San Diego, CA, USA) with 5 μg of total RNA. The ribosomal RNA (rRNA) was depleted instead of poly (A) purification using the Ribo-Zero Magnetic kit, which was then fragmented using fragmentation buffer. Next, random hexamer primers were used to synthesize first-strand cDNA. The RNA template was then removed, and a replacement strand was synthesized to replace dTTP with dUTP, thereby generating ds-cDNA. During polymerase amplification, the addition of dUTP quenched the second strand, preventing binding to the nucleotide. ds-cDNA was isolated from the second chain reaction mixture using AMPure XP beads. To prevent these blunt fragments from annealing together in the adapter ligation reaction, one “A” nucleotide was added to the 3′ end. Finally, the double-stranded cDNA was tagged with multiple indexing adapters. The library was 200–300 bp in size and amplified using 2% low range ultra-agarose, followed by amplification using 15 PCR cycles through Phusion DNA polymerase (NEB, Ipswich, MA, USA). After quantification using TBS380, the Illumina HiSeq XTEN //NovaSeq6000 (2 × 150 bp read length) was used to sequence the longRNA-seq library at the paired end. The sequencing adapter of the TruSeq Small RNA Sample Prep Kit (Illumina, USA) was also connected to 3 μg total RNA. The cDNA library was established by reverse transcription and PCR amplification using 12 PCR cycles. The deep sequencing was performed in Shanghai Majorbio Bio-Pharm Biotechnology Co. Ltd. (Shanghai, China).
The raw paired end reads were trimmed and quality controlled by SeqPrep (37) (https://github.com/jstjohn/SeqPrep) and Sickle (37) (https://github.com/najoshi/sickle) with default parameters. Then clean reads were separately aligned to reference genome (http://asia.ensembl.org/Bos_taurus/Info/Index) with orientation mode using HIASAT (38) (https://ccb.jhu.edu/software/hisat2/index.shtml) software. The mapped reads of each sample were assembled by StringTie (39) (https://ccb.jhu.edu/software/stringtie/index.shtml?t=example) in a reference-based approach.
To identify DE transcripts (mRNAs, lncRNAs and miRNAs) between the normal and cystic follicular GCs, transcripts per million reads (TPM) was used to calculate expression of each transcript. RSEM Abundance values (http://deweylab.biostat.wisc.edu/rsem/) for quantitative genes were analyzed. The mRNAs, lncRNAs and miRNAs with a p-value ≤ 0.05 were considered DE transcripts by DESeq2 (40). GO (41) and KEGG (42) analyses were also conducted to determine which metabolic pathways and GO terms of differentially expressed genes (DEGs) were significantly enriched at Bonferroni-corrected P ≤ 0.05, compared to the whole transcriptome background. Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) were used to perform GO functional enrichment and KEGG pathway analyses, respectively.
We constructed a regulatory network of ncRNA and mRNA to uncover the underlying interaction mechanisms. Miranda (43) (http://www.miranda.org/) and TargetScan databases (http://www.targetscan.org/) were used to predict pairs of miRNAs with lncRNAs and mRNAs. Using the Pearson's correlation coefficient (44), the correlations between DE miRNAs and DE lncRNAs/DE mRNAs were evaluated, and those with r < −0.8 were selected for further analysis. Cytoscape (45) was used to build and visualize the interaction network.
RNAiso Plus reagent (Takara, Japan) was used to isolate RNA from normal and cystic follicular GCs. Then, 500 ng cDNA was extracted from the total RNA using PrimeScript™ RT kit, gDNA Eraser (Takara, Japan), and the housekeeping gene ACTB was used as an internal control for RT-qPCR amplification using TB Green Premix Ex TaqTM II (Takara, Japan) on a CFX96 real-time PCR system (Bio-Rad, USA). Briefly, the reaction was performed at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s. The first cDNA strand corresponding to miRNA was prepared using the M5 miRNA cDNA Synthesis Kit (Mei5bio, China) and amplified by RT-qPCR using the M5 miRNA qPCR Assay Kit (Mei5bio, China). The reaction was performed at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min. U6 was considered as an internal control gene. Supplementary Table 1 shows the primer sequences synthesized by Sangon (Shanghai, China).
Approximately 107 GCs were required for the total number of cells. ParisTM Kit (Invitrogen, USA) was used to extract GCs nuclear and cytoplasmic RNA, and RT-qPCR was conducted to analyze the expression of lncRNA NONBTAT027373.1 in the nucleus and cytoplasm. GAPDH and U6 were considered as internal control genes.
Serum-free long-term bovine follicular GCs culture medium containing Minimum Essential Medium (MEM) Alpha (1×; Gibco, USA) was configured, and HEPES (20 mM, Sigma, USA), sodium bicarbonate (10 mM, Sigma, USA), bovine serum albumin (1 g/L, Sigma, USA), penicillin-streptomycin liquid (100 IU/mL penicillin and 0.1 mg/mL streptomycin, Solarbio, China), MEM non-essential amino acids solution (1.1 mg/L, Gibco, USA), bovine insulin solution (10 μg/L, Sigma, USA), sodium selenite (4 μg/L, Sigma, USA), LR3-IGF-1 (1 μg/L, Sigma, USA), holo-transferrin (5 mg/L, Sigma, USA), androstenedione (10−6 M, Rhawn, China) and FSH (5 ng/mL, Solarbio, China) were added. GCs from those follicles with 10–15 mm diameter were washed with culture medium three times and resuspended in the medium. GCs were inoculated in 12-well cell culture plates (1 × 106 viable cells/well) and cultured in a humidified atmosphere (5% CO2, 37 °C), with 75% medium replaced every 48 h with fresh medium.
Mimics, inhibitor and negative control (NC) of miR-664b, in addition to the siRNA of lncRNA NONBTAT027373.1, were designed and synthesized. The sequences are listed in Supplementary Table 2. The GCs were transfected using LipoFilter 3.0 (Hanbio, China), according to the manufacturer's instructions. A total of 1 × 106 cells were inoculated into a 12-well plate containing complete medium, ensuring 60% confluence during transfection. We diluted 2.5 μL of 20 μM siRNA stock solution was diluted with 100 μL Opti-MEM and mixed gently. Opti-DMEM (100 μL) was added to the LipoFilter 3.0 reagent (3 μL) and incubated at 25 °C for 5 min. The mixture was gently mixed and incubated at 25 °C for 20 min before being added to each well. The plates were placed in an incubator at 37 °C with 5% CO2 for 48 h for subsequent experiments.
In a 12-well plate, GCs (1 × 106 cells/well) were seeded and transfected. Using the ClonExpress II one-step cloning kit (Vazyme, China), the HSD17B7 3′-UTR/lncRNA NONBTAT027373.1 fragment complementary to the miR-664b seed region was subcloned into the pmirGLO plasmid digested with XhoI and SalI to construct the wild-type (WT) plasmid. The mutant (MUT) plasmid was also constructed using the Mut Express II Fast Mutagenesis Kit V2 (Vazyme, China). All primer sequences are listed in Supplementary Table 3. miR-664b mimics/NC with recombinant plasmids were transfected into plasmid using LipoFilter 3.0 (Hanbio, China) according to the manufacturer's instructions. The luciferase activity was measured 48 h after transfection using a dual-luciferase reporter assay system (Promega, USA).
The RIP assay was performed according to the following methods. Briefly, washed GCs (4 × 107 cells) once with pre-chilled PBS buffer and collected the cells with a cell scraper into a 15 mL RNase-free centrifuge tube. Resuspended the cell pellet with immunoprecipitation lysis buffer (Beyotime, China) containing phenylmethylsulfonyl fluoride (PMSF) (BOSTER, China) and RNase inhibitors (Solarbio, China) and dissolved on ice for 30 min. Collected the lysate supernatant at 12,000 g/min for 20 min at 4 °C. Incubated A/G protein beads (MCE, USA) with Ago2 (dilution multiple 1:100) (BOSTER, China)/IgG (dilution multiple 1:100) antibody (ABclonal, China) overnight at 4 °C. The lysate was incubated with beads-antibody complexes at 4 °C for 6 h. The reaction mixture was centrifuged at 4 °C, 3,000 r/min for 2 min. Discarded the supernatant and washed the beads 5 times with RIP buffer (20 mM Tris-HCl, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 0.5% NP40, 0.5% Triton-X100). Incubated the beads with proteinase-containing k-eluate (Solarbio, China) for 45 min at 65 °C in a metal water bath. Took the supernatant, centrifuged at 4 °C, 3,000 r/min for 5 min. RNA was purified using chloroform-isoamyl alcohol (24:1) and precipitated overnight at −20 °C with glycogen (Solarbio, China). RNA was washed using 75% ethanol for 3 times, 10,000 r/min for 10 min at 4 °C. Thereafter, RNA was diluted in RNase-free water and the abundance of lncRNA NONBTAT027373.1 was analyzed by RT-qPCR.
All experiments were repeated at least thrice. The data were analyzed by an independent samples t-test in SPSS 22.0, and were presented as mean ± standard error of mean (SEM). All data were normally distributed and the variance was similar between the groups. The relative expression of mRNA and ncRNA were calculated using the 2−ΔΔCt method. A P < 0.05 indicated significant differences. A P < 0.01 indicated highly significant differences.
In a slaughterhouse, three cystic and three normal follicles were collected. Total RNA was extracted from qualified biospecimens and whole-transcriptome sequencing was performed. LongRNA-seq (lncRNA+mRNA) and smallRNA-seq (miRNA) data were obtained using an Illumina Hiseq2500 platform. After the redundant and low-quality reads were removed, the clean data obtained by longRNA-seq in all samples exceeded 14.9 GB, and the percentage of Q30 bases was >93.3% (Supplementary Tables 4, 5). Meanwhile, the raw reads obtained from smallRNA-seq in all samples exceeded 12.4 MB, and the percentage of Q30 bases was >95.3% (Supplementary Tables 6, 7). The datasets were submitted to the National Center of Biotechnology Information (accession number: PRJNA756563). In total, 3,443 lncRNAs and 1,282 miRNAs were identified on all chromosomes, and 2,667 lncRNAs were annotated in the bovine genome (Supplementary Table 8). Additionally, there was one lncRNA in mitochondrial DNA (mtDNA) (Figure 1A), and our data included intergenic, intronic, antisense and sense-overlapping lncRNAs. A majority (67%) of the lncRNAs were intergenic, while the minority (3%) were intronic (Figure 1B). The length of lncRNAs ranged from 66 to 26,123 bp. The majority (67%) of the lncRNAs were <1,500 bp (Figure 1C).
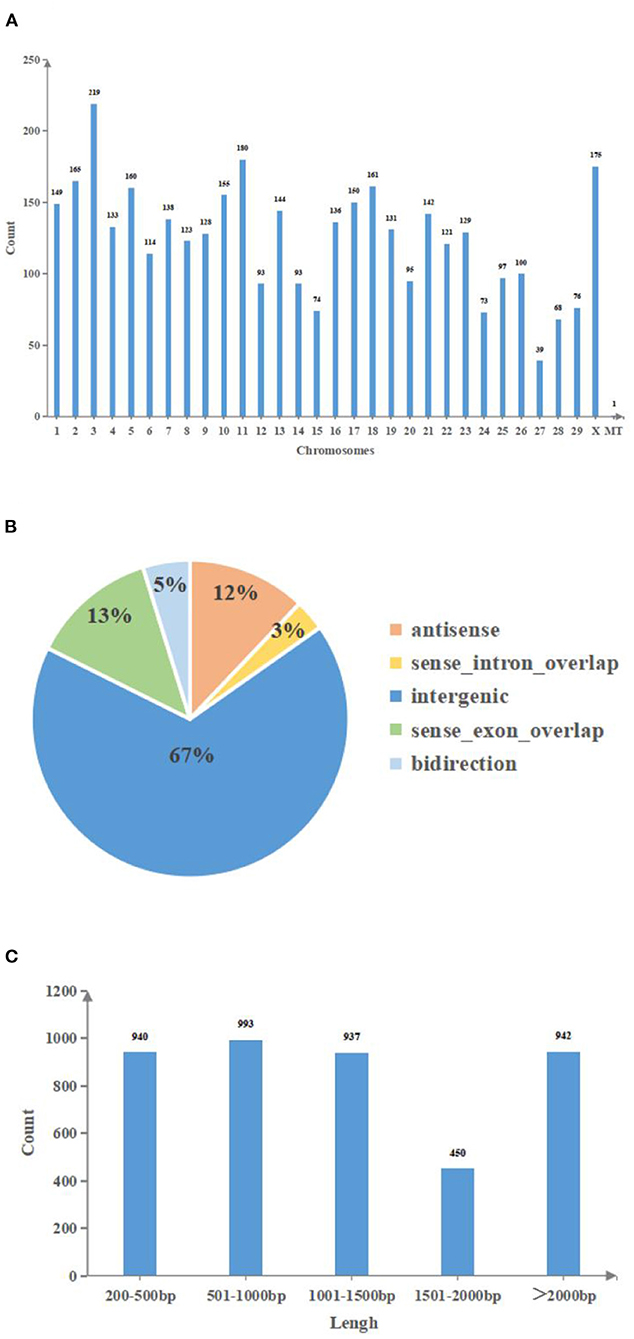
Figure 1. The features of lncRNAs in bovine follicular GCs. (A): Chromosomes distribution of lncRNAs. (B): Genomes distribution of lncRNAs. (C): Length distribution of lncRNAs. MT, mitochondria; Count, Number of lncRNA transcripts; Chromosomes, Chromosome number; Antisense, lncRNA that overlaps with the antisense chain of coding genes; Sense intron overlap, lncRNA locates in the intronic region of coding genes; Intergenic, lncRNA locates in the intergenic region of coding genes; Sense exon overlap, lncRNA that overlaps with the exon region of coding genes; Bidirection, lncRNA locates in the 1 kb upstream region of coding genes, the transcription direction is opposite to coding genes; Lengh, The base number of lncRNA.
The correlation between the selected samples were shown in Figure 2A and Supplementary Table 9. The DE ncRNAs (miRNAs and lncRNAs) and mRNAs in cystic and normal follicles were analyzed with cutoff fold changes ≤0.5 or ≥ 2, P < 0.05 and false discovery rate (FDR) <0.05 (Supplementary Tables 10–12). The results illustrated that 25,655 (83.6%) mRNAs, 2,596 (75.4%) lncRNAs and 822 (64.1%) miRNAs were shared by normal and cystic GCs (Figures 2B–D). The ncRNA and mRNA expressive abundances in bovine cystic follicular GCs were higher and lower than those in normal bovine follicular GCs, respectively (Figures 2E–G). DE mRNAs and ncRNAs in the samples are shown using a volcano plot (Figures 2H–J). The results elucidated there were 41 DE lncRNAs, 103 DE miRNAs and 4,205 DE mRNAs were downregulated in cystic follicular GCs compared with normal follicular GCs. In contrast, 538 DE lncRNAs, 102 DE miRNAs and 3,798 DE mRNAs were upregulated in cystic follicular GCs compared with normal follicular GCs. Hierarchical clustering of the DE ncRNAs and mRNAs revealed the expression patterns of the individuals for the same three comparisons (Figures 2K–M). We also discovered DE mRNAs and lncRNAs were distributed differently across the chromosomes (Supplementary Figure S1).
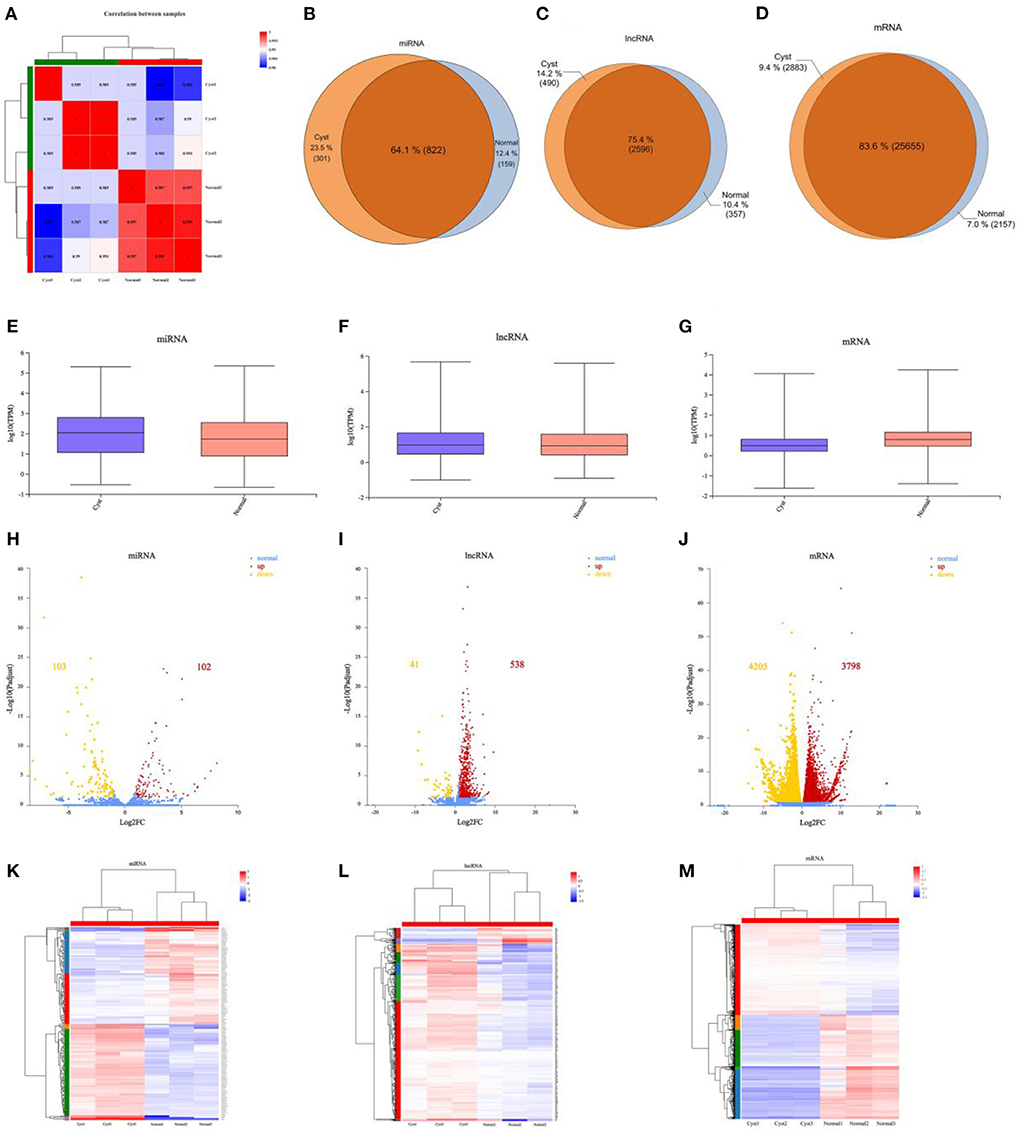
Figure 2. Comparative analysis of mRNAs, miRNAs and lncRNAs between normal and cystic follicular GCs. (A): Correlation analysis between samples. (B–D): The specific miRNAs, lncRNAs and mRNAs shared between normal and cystic follicular GCs. (E–G): Boxplots showing the expression distribution of miRNAs, lncRNAs and mRNAs. (H–J): Volcano plot of DE miRNAs, DE lncRNAs and DE mRNAs in normal and cystic follicular GCs. (K–M): Hierarchical clustering of DE miRNAs, DE lncRNAs and DE mRNAs in normal and cystic follicular GCs. The red points showed the up-regulated miRNAs, lncRNAs and mRNAs, the yellow points showed the downregulated miRNAs, lncRNAs and mRNAs, and the blue points showed the equally expressed miRNAs, lncRNAs and mRNAs in volcano plots. Normal: Bovine ovarian follicles between 10–15 mm in diameter. Cyst: Bovine ovarian follicles greater than 25 mm in diameter.
As shown in Supplementary Table 13, the most downregulated lncRNA was lnc-RAP2A with a 10−3-fold change, and the most upregulated lncRNA was XR_003036450.1, with a 764-fold change. The most upregulated mRNA and miRNA were LRCH3 (4.3 × 106-fold change) and bta-miR-184 (287-fold change), respectively. The most downregulated mRNA and miRNA were ECE1 (6.39E-05-fold change) and bta-miR-33a (3 × 10−3-fold change), respectively. The top 10 downregulated and upregulated mRNAs and lncRNAs are described in Supplementary Tables 14, 15. The top five upregulated and downregulated miRNAs are listed in Supplementary Table 16. Six DE lncRNAs, six DE miRNAs and six DE mRNAs were randomly selected for this study. Reverse transcription quantitative PCR (RT-qPCR) was performed to detect the expression patterns of these transcripts in normal and cystic follicular GCs. The results were consistent with the RNA-seq data, indicating the reliability of the procedure (Supplementary Figure S2).
Gene Ontology (GO) analysis revealed these DE mRNAs in bovine cystic follicles GCs were mainly related to binding (molecular function), cell part (cellular component) and cellular (biological process), and more DE mRNAs related to metabolic process (biological process) were downregulated in cystic follicular GCs, indicating abnormal metabolism of cystic follicles, which would affect the synthesis of cholesterol, the raw material of ovarian steroid hormones, as explained in Figures 3A,B. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that multiple pathways were enriched in the DE mRNAs involved in bovine cystic follicular GCs. These genes were largely involved in steroid hormone biosynthesis pathways, oxidative phosphorylation, metabolic pathways, etc., all of which were important for follicular development (Supplementary Table 17; Figure 3C). Some DE mRNAs that could be associated with follicular cysts are shown in Figure 3D. Enrichment analyses of their target genes by GO and KEGG revealed the roles of DE lncRNAs. The 383 steroid hormone biosynthesis-associated DE lncRNAs were screened in the cystic vs. normal group (Supplementary Table 18). Furthermore, a co-expression network was constructed for the steroid hormone biosynthesis-associated DE lncRNAs (Supplementary Figure S3). This lays the foundation for investigating the potential function of lncRNAs in regulating target genes expression.

Figure 3. Functional analysis of DE mRNAs expressed in normal and cystic follicular GCs. (A): GO analysis of DE mRNAs upregulated in bovine cystic follicular GCs; (B): GO analysis of DE mRNAs downregulated in bovine cystic follicular GCs. (C) KEGG analysis. (D): Circos plot of DE mRNAs associated with steroid hormone biosynthesis and metabolic (the inner ring represents 1 to 29 bovine autosomal and X sex-chromosomes. The outer ring indicates the chromosomes gene density).
By the analysis strategy described in methods, the target mRNAs of these miRNAs were predicted based on miRanda and compared these target mRNAs with the DE mRNAs identified in the RNA-seq results. To fully explore the key genes related to follicular cysts and construct the networks more concise and effective, the key DE mRNAs closely related to bovine follicular cysts based on the literature and the results of KEGG pathway analysis in the above steps were further filtered. Then 22 mRNAs in the lncRNA-miRNA-mRNA networks were selected, which were involved in the nine pathways such as citrate cycle, estrogen signaling pathway, steroid biosynthesis, steroid hormone biosynthesis, ovarian steroidogenesis, DNA replication, pentose phosphate pathway, oxidative phosphorylation and glutathione metabolism. Then the lncRNA-miRNA-mRNA networks were constructed based on the selected mRNAs (Supplementary Table 19; Figures 4A,B). 115 lncRNA-miRNA-mRNA pathways were constructed including 40 lncRNAs, 31 miRNAs and 22 mRNAs. Among them, the expression levels of three mRNAs in cystic follicular GCs were significantly higher than that in normal follicular GCs, while the expression levels of 19 mRNAs in cystic follicular GCs were significantly lower than that in normal follicular GCs. These ceRNA regulatory relationships might be more important in the process of bovine follicular cysts formation considering the complexity among the ncRNAs and mRNAs.
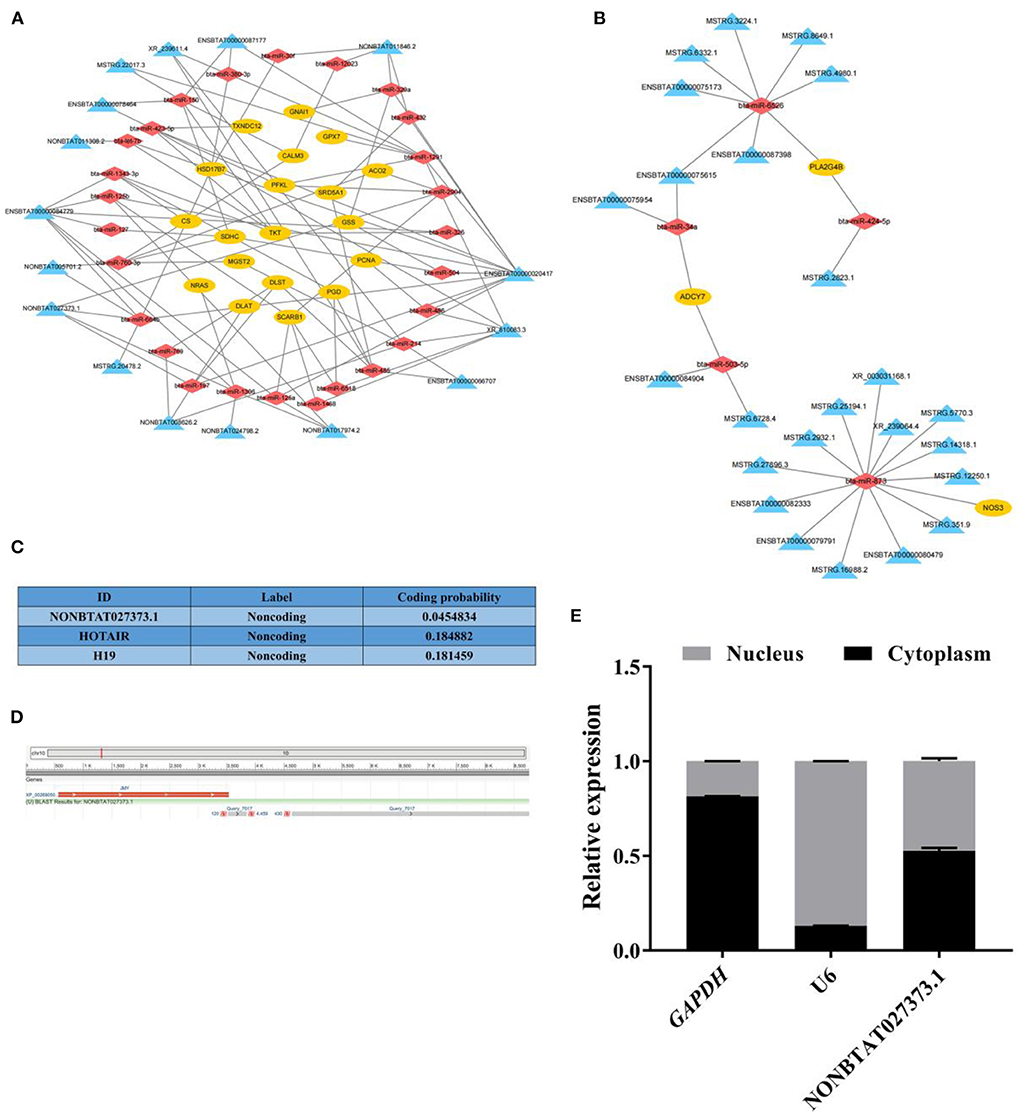
Figure 4. The network analysis of lncRNA-miRNA-mRNA and the basic information of lncRNA NONBTAT027373.1. (A): The lncRNA-miRNA-mRNA networks of DE mRNAs downregulated in cystic follicular GCs. (B): The lncRNA-miRNA-mRNA networks of DE mRNAs up-regulated in cystic follicular GCs. (C): The coding potential of lncRNA NONBTAT027373.1 and other RNAs predicted using computational approaches. (D): The sequence alignment of lncRNA NONBTAT027373.1. (E): The expression of lncRNA NONBTAT027373.1 in GCs nucleus and cytoplasm. Yellow nodes represent mRNAs; red nodes represent miRNAs and blue nodes represent lncRNAs in the networks of lncRNA-miRNA-mRNA.
In this study, the transcript NONBTAT027373.1 was obtained by longRNA-seq, and its sequence was identified as a known lncRNA with a total length of 4,889 nt by comparison with NONCODE database. The expression of this transcript in normal follicular GCs was significantly higher than that in cystic follicular GCs, which was a DE lncRNA. Coding potential analysis confirmed the non-coding nature of the transcript, similar to other well-characterized lncRNA, HOTAIR and H19 (Figure 4C). Furthermore, the transcript was located on the positive strand of DNA (location: 10381778-10387465) on bovine chromosome 10 and consisted of two exons, which overlap with the 3'-UTR of JMY (junction mediating and regulatory protein, p53 cofactor) (Figure 4D). This transcript was expressed in both the cytoplasm and nucleus of GCs (Figure 4E).
KEGG analysis showed that HSD17B7 (hydroxysteroid 17-beta dehydrogenase 7) was a key enzyme in the steroid hormones synthesis of bovine ovarian, and its mRNA expression in cystic follicular GCs was significantly lower than that in normal follicular GCs. As shown in Figure 5A, in HSD17B7-related ceRNA networks, there were 11 nodes and 10 connections between one mRNA, three miRNAs and seven lncRNAs. The expression of lncRNAs, miRNAs and HSD17B7 in the constructed ceRNA network were detected using RT-qPCR in normal and cystic follicular GCs (Figures 5B–D). The expression of miR-664b was increased in cystic follicular GCs. In contrast, the expression of its target genes including HSD17B7 and lncRNA NONBTAT027373.1 were decreased. Moreover, an RNA-binding protein immunoprecipitation (RIP) assay revealed lncRNA NONBTAT027373.1 was enriched in the RNA-induced silencing complex miR-664b-Ago2. The enrichment of miR-664b and lncRNA NONBTAT027373.1 in the Ago2 group increased simultaneously, which was significantly higher than that in the IgG group, confirming the interaction between lncRNA NONBTAT027373.1 and miR-664b (Figure 5E). Software analysis demonstrated the miR-664b seed sequence was complementary to the HSD17B7 3'-UTR and lncRNA NONBTAT027373.1 (Figure 5F). Luciferase reporter plasmids containing the HSD17B7-WT and lncRNA NONBTAT027373.1-WT miRNA response element (MRE) motif or the MUT versions were constructed (Figures 5G,H). miR-664b significantly decreased luciferase activity of the reporter containing the WT-MRE motif, while the reporter activity of the MUT-MRE motif did not change (Figures 5I,J). These results indicated HSD17B7 and lncRNA NONBTAT027373.1 could bind to the miR-664b seed region. In GCs, we also examined the transfection effects of miR-664b mimics and inhibitor (Figure 5K). Compared with the mimics NC group, the expression of HSD17B7 was significantly downregulated after transfection with miR-664b mimics. In contrast, the expression of HSD17B7 was significantly upregulated in GCs transfected with the miR-664b inhibitor (Figure 5L). The si-lncRNA NONBTAT027373.1-1 (termed si-lncRNA NONBTAT027373.1) with the highest inhibitory effect was selected for further investigation (Figure 5M). After transfecting si-lncRNA NONBTAT027373.1 into GCs, there was a significantly decrease in HSD17B7 mRNA expression. However, co-transfection of miR-664b inhibitor and si-lncRNA NONBTAT027373.1 in GCs rescued HSD17B7 mRNA expression (Figure 5N). We found that the miR-664b inhibitor could rescue the regulatory effect of si-lncRNA NONBTAT027373.1 on HSD17B7, suggesting that lncRNA NONBTAT027373.1 may regulate the function of GCs by sponging miR-664b-HSD17B7.
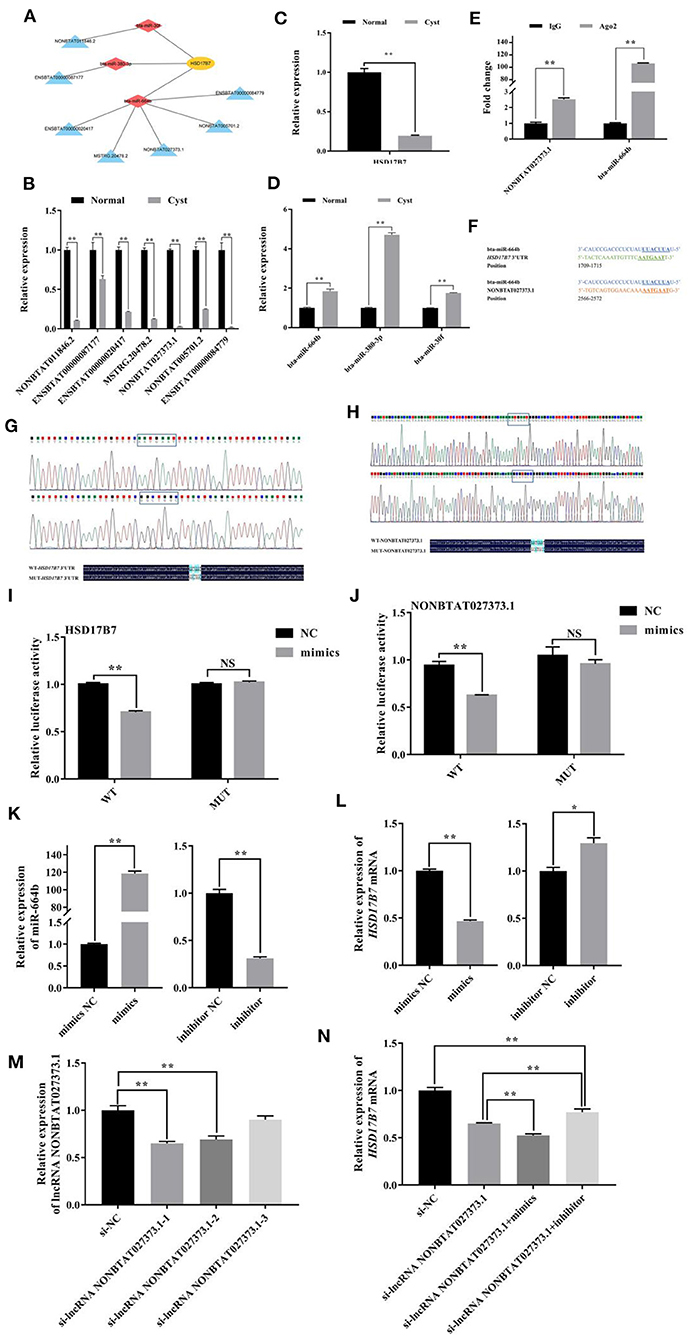
Figure 5. Analysis of miR-664b sponge HSD17B7 3'-UTR and lncRNA NONBTAT027373.1. (A): The networks of lncRNA-miRNA-HSD17B7 (B): The expression of DE lncRNAs in lncRNA-miRNA-HSD17B7 networks. (C): The expression of HSD17B7 mRNA. (D): The expression of DE miRNAs in lncRNA-miRNA-HSD17B7 networks. (E): Interaction between lncRNA NONBTAT027373.1 and miR-664b through Ago2 protein investigated using the RIP assay. lncRNA NONBTAT027373.1 and miR-664b expression quantified by RT-qPCR. (F): Schematic view of the miR-664b seed region, the motif of HSD17B7 3'-UTR and lncRNA NONBTAT027373.1. (G,H): Construction of WT and MUT plasmid vectors of HSD17B7 3'-UTR and lncRNA NONBTAT027373.1. (I): Dual-luciferase reporter gene analysis of the regulatory relationship between HSD17B7 3'-UTR and miR-664b. (J): Dual-luciferase reporter gene analysis of the regulatory relationship between lncRNA NONBTAT027373.1 and miR-664b. (K): Transfection efficiency of miR-664b after overexpression and knockdown of miR-664b. (L): miR-664b regulates the expression of HSD17B7 mRNA. (M): RT-qPCR analysis of the effect on silence of lncRNA NONBTAT027373.1 expression by siRNA. (N): Relative expression of HSD17B7 mRNA in groups. NS, Non-significant. Values represent means ± SEM for three individuals. *P < 0.05, **P < 0.01. Yellow nodes represent HSD17B7; red nodes represent miRNAs and blue nodes represent lncRNAs in lncRNA-miRNA-HSD17B7 networks. WT, Wild Type; MUT, Mutant; Mimics, Overexpression of miR-664b; Inhibitor, Knockdown of miR-664b; NC, Negative control.
Ovarian cyst has always been a difficult scientific problem for the bovine industry. Therefore, its molecular mechanism is worth studying. In this study, mRNA, miRNA, and lncRNA expression profiles were compared in normal and cystic bovine follicular GCs to identify the key factors involved in bovine follicular cysts formation. In addition, we constructed and verified a potential lncRNA-miRNA-mRNA regulatory network, which provides new insights into the molecular mechanism of bovine follicular cysts formation.
When bovine follicular cysts, estrus is abnormal, estrus cycle becomes short, and estrus period is prolonged, or there is sustained and strong estrus phenomenon defined as nymphomania (46). Follicular cysts are single or multiple, in one or both ovaries, with diameter at least 2.5 cm (47). The ovarian wall is thin and dense, filled with cystic fluid, and slightly fluctuating. The inner granular layer is atrophy, the inner wall is only a layer of flat cells, and sometimes even disappears (48). Bovine follicular cysts most commonly occur on the 15–45 days of early postpartum lactation, an important stage in the transition from non-estrus to estrus cycle (49). Generally, its formation is due to hypothalamus-pituitary-ovarian axis dysfunction caused by multiple factors, including genetic, phenotypic, and environmental factors (50). The pathogenesis of follicular cysts is divided into primary defects at the hypothalamic-pituitary hydropin level and primary defects at the ovary-follicle level. However, the formation of follicular cysts is co-determined by both ovary-follicular and hypothalamic-pituitary defects (51). Affected hypothalamic-pituitary hormone release or altered follicle growth and development may result in abnormal hypothalamic-pituitary gonadal axis function (51). High milk production and the associated negative energy balance in the early postpartum period in cattle also lead to follicular cysts. During this time the energy demand maintaining milk production is higher than the energy intake, resulting in NEB, which is accompanied by a variety of hormonal and metabolic changes that affect ovarian function (52). Some animals can compensate for higher milk production by consuming more dry matter, thus reducing the impact of milk production on energy balance (53).
This study is the first to propose how mRNAs, pseudogenes, lncRNAs, and circular RNAs (circRNA) interact via microRNA (miRNA)-response elements (54). Many lncRNAs and circRNAs act as ceRNAs, sponging miRNAs and regulating mRNA expression, and cell differentiation in animals. For example, the ceRNA regulation network related to intramuscular fat deposition in yak was revealed through the whole-transcriptome landscape of muscle and adipose tissues to elucidate the function of genes associated with intramuscular fat deposition (55). RNA sequencing was used to identify ceRNA which may regulate lipid metabolism in bovine liver, and some key genes were found to affect lipid metabolism through insulin, PI3K-Akt, MAPK, AMPK, mTOR and PPAR signaling pathways (56). Studies have analyzed the differential expression patterns of mRNA, lncRNA, circRNA and their ceRNA networks in longissimus dorsi muscle of two different pig species (Chinese Huainan pigs and Western commercial Duroc pigs). A potential lncRNAs/circRNAs-miRNAs-mRNAs regulatory network was found to share MYOD1, PPARD, miR-423-5p and miR-874, which are associated with muscle proliferation, differentiation/regeneration and adipogenesis in skeletal muscle (57). A complete transcripome analysis of the toxic effects of zearalenone exposure on porcine GCs has been performed and a ceRNA network has been constructed. In addition, it was observed that ZEA exposure to the G2/M phase of porcine GCs blocked the process of cell cycle, and non-coding RNA (ncRNAs) played an important role in this process by regulating the expression of genes related to cell cycle arrest (58). The construction of ceRNA networks associated with skeletal muscle development in cattle revealed the co-expressed lncRNAs were significantly enriched in biological processes and Wnt signaling pathways associated with muscle development (59).
Recently, there have been some studies on mammalian ovarian ceRNA such as human, mouse and sheep, most of which involve lncRNAs. In addition, there are almost fewer studies related to ncRNA and ceRNA in bovine ovary. Recent studies suggest the mouse Scd2 gene is activated by two lncRNAs and interdependent enhancers in ovarian follicular GCs, which provides a new insight into transcriptional activation in GCs (60). Studies have demonstrated FDNCR sponges miR-543-3p in GCs and prevents miR-543-3p from binding to the DCN 3′-UTR, resulting in DCN transactivation and TGF-β pathway inhibition and promotion of GC apoptosis in Hu sheep (61). PWNR2 plays an important role in oocyte nuclear maturation in PCOS by functioning as a ceRNA to reduce the availability of miR-92b-3p for TMEM120B target binding during oocyte maturation in PCOS (62). lncRNA MALAT1 regulated apoptosis and estradiol synthesis of mouse GCs through the miR-205/CREB1 axis (63). The long non-coding RNA Xist regulates oocyte loss via suppressing miR-23b-3p/miR-29a-3p maturation and upregulating STX17 in perinatal mouse ovaries (64). Our findings simultaneously identified the lncRNA, miRNA and mRNA expression profile in the normal and the cystic follicular GCs by the whole transcriptome RNA-seq, there were 579 lncRNAs, 205 miRNAs and 8,003 mRNAs differentially expressed with 2-fold change.
In our study, some genes participated in steroid hormone biosynthesis, such as HSD17B7 (hydroxysteroid 17-beta dehydrogenase 7), NSDHL (NAD(P) dependent steroid dehydrogenase-like) and CYP51A1 (cytochrome P450, family 51, subfamily A, polypeptide 1), and the genes associated with pentose phosphate pathway such as PGD (phosphogluconate dehydrogenase), PFKM (phosphofructokinase, muscle) and PGM1 (phosphoglucomutase 1). Meanwhile, in the GO analyses, the transition between the normal and cystic follicles for DE mRNAs was enriched. It has been well established energy metabolism is crucial to mammalian reproductive capacity, and the imbalance between nutrient intake and body storage during lactation is not sufficient to provide cholesterol as the raw material for steroid synthesis (65). NADPH+H produced by the pentose phosphate pathway provides a large amount of hydrogen for the synthesis of cholesterol and steroid hormones (66). Abnormal synthesis of steroid hormones, especially estrogen, may affect the feedback to the hypothalamus, leading to blocking synthesis of LH and failure to produce pre-ovulatory LH surge. Ultimately, ovulation fails, and the persistent existence of follicles causes follicular cysts (67). In addition, DEGs is also enriched in DNA replication, glutathione metabolism pathway and citrate cycle pathway. Therefore, the occurrence of follicular cysts could also be related to oxidative stress. The expression level of ITGB1 (integrin subunit beta 1) in cystic follicular GCs was significantly lower than that of normal ones, and the low expression of ITGB1 in GCs might be decreased cell-extracellular matrix adhesion (10) leading to the reduction of follicular granulosa cell layer and the decrease of estrogen level, which further affects ovulation and follicle persistence. PGD plays an important role in fat synthesis and metabolism in bovine adipocytes (68). Researchers have found that PGD, a cytoplasmic enzyme involved in carbohydrate metabolism, is necessary for the structural integrity of the endoplasmic reticulum (ER) and for protein secretion, and that PGD is the link between cytoplasmic carbohydrate metabolism and protein secretion. Moreover, chemical or genetic inhibition of PGD activity leads to cellular stress (69). The effects of different dietary protein and energy density levels on gene expression in adipose tissue of growing replacement lactating cows were studied using next-generation RNA sequencing technology (RNASeq), and the transcriptional regulation of energy metabolism of PGD was found and emphasized (70). In addition, phosphorylated PGD was found to be an important factor in the conversion of NADP+ to NAD+ (71). PGD is a key gene in the pentose phosphate pathway, which provides sufficient reduction equivalent for the synthesis of cholesterol, which is the raw material for the synthesis of steroid hormones, and the secretion level of steroid hormones affects the occurrence of bovine follicular cysts.
In the present study, we provided a systematic perspective on the potential function of ncRNAs in the bovine follicular cysts. The lncRNA-miRNA-mRNA regulatory networks in the bovine follicular cysts were comprehensively integrated and predicted on the basis of the screening and bioinformatics analysis. The mRNAs selected in the network conformed to the ceRNA rules and were also contained in known steroid hormone synthesis related genes or metabolic pathways. Then we constructed the lncRNA-miRNA-mRNA regulatory networks including 40 lncRNAs, 31 miRNAs and 22 mRNAs. Of them, there are 115 lncRNA-miRNA-mRNA pathways. These intertwined ceRNA networks suggests more important role in the follicular cysts regulation. In addition, to further demonstrate the credibility of our results, one predicted pathway, lncRNA NONBTAT027373.1/miR-664b/HSD17B7 was selected and verified by the RT-qPCR, RIP and dual luciferase reporter gene system.
In fact, lncRNA NONBTAT027373.1 has not been studied yet. MicroRNA transcriptome analysis was conducted on the mammary glands of cows infected with Staphylococcus aureus and normal mammary glands, and miR-664b was differently expressed (72). This provided an experimental basis for revealing the etiology and regulatory mechanism of mastitis, and also suggested miR-664b can be used as a biomarker for the diagnosis of mastitis in cows (73). Studies have revealed miR-664b acts as a central regulator of gene expression in response to Gram-positive bacterial infection, and significantly regulates the sentinel ability of bovine mammary epithelial cells to mobilize the innate immune system (74). However, there has been fewer studies of miR-664b in bovine ovaries. The rodent (75) and human (76) HSD17B7 enzyme catalyzes the conversion of E1 and E2 in vitro, and the enzyme produced by E2 is important based on the high expression of HSD7B7 in mouse ovaries. HSD17B7 has a similar expression pattern with a known cholesterol stimulating enzyme during mouse embryonic development, suggesting a role for mouse HSD17B7 in cholesterol biosynthesis (77). Analysis of cholesterol concentrations and various cholesterol biosynthesis intermediates in wild, heterozygous and HSD17B7 KO mouse embryos clearly suggests HSD17B7 is also essential for cholesterol biosynthesis in mice (78). In this study, we found lncRNA NONBTAT027373.1 and HSD17B7 expression were simultaneously decreased, whereas miR-664b expression was increased in the cystic follicular GCs. Therefore, we speculated decreasing HSD17B7 expression in bovine cyst follicles hindered the synthesis of steroid hormones in follicles and could not convert E1 to E2. The changes in the endocrine levels of sex hormones may cause the occurrence of follicular cysts. The validation of lncRNA NONBTAT027373.1-miR-664b-HSD17B7 pathway lays a foundation for the study of ncRNA function in bovine follicular GCs.
ncRNAs are involved in steroid hormone synthesis and energy metabolism in bovine follicular GCs, acting as ceRNAs for vital genes via functional miRNAs. Our findings provides a theoretical basis for further understanding the mechanism of ncRNA and ceRNA regulation in follicular development.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA756563.
The animal study was reviewed and approved by the National Institutes of Health.
KW and LL conceived and designed the experiments. YC, TG, and XM collected samples. KW and YC drafted the manuscript. XG, YP, and HZ discussed and contributed to the data analysis. LL, DW, and EK revised the paper. All authors read and approved the final manuscript.
This study was supported by Shanxi Youth Sanjin Talent Project, Shanxi Basic Research Plan Grant (Free Exploration, 20210302123393), Datong Key Research and Development (Agriculture) Grant (2021027) and the earmarked fund for Modern Agro-Industry Technology Research System in Shanxi Province.
Authors are grateful of Prof. George W. Smith in Michigan State University in the USA for his direction of this study. Authors are grateful of the senior experimentalist Mrs. Liying Qiao and Prof. Pengfei Li for their technical direction and help of the study. And authors would like to thank Editage (www.editage.cn) for English Language Editing and data analysis on the free online platform of Majorbio Cloud Platform (www.majorbio.com).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1028867/full#supplementary-material
BMP6, Bone morphogenetic protein 6; ceRNA, Competitive endogenous RNA; circRNA, Circular RNA; Cystic follicles, Bovine ovarian follicles >25 mm in diameter; CYP51A1, Cytochrome P450, family 51, subfamily A, polypeptide 1; CREB1, Cyclic AMP response element binding protein 1; DEGs, Differentially expressed genes; DCN, Decorin; E2, 17β-estradiol; E1, Estrone; ECE1, Endothelin converting enzyme 1; eRNA, enhancer RNA; FSH, Follicle-stimulating hormone; FC, Fold Change; FDR, False discovery rate; FDNCR, Follicular development-associated lncRNA; GO, Gene Ontology; GATA6, GATA binding protein 6; GCs, Granulosa cells; HSD17B7, Hydroxysteroid 17-beta dehydrogenase 7; IGFBP3, Insulin-like growth factor binding protein 3; IGF-1, Insulin-like growth factor 1; IVF, In-vitro fertilization; ITGB1, Integrin subunit beta 1; KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, Long non coding RNA; LH, Luteinizing hormone; LPS, Lipopolysaccharide; LRCH3, Leucine rich repeats and calponin homology domain containing 3; miRNA, Micro RNA; MMP1/2, Matrix metallo proteinase 1/2; mtRNA, Mitochondria RNA; MRE, miRNA response element; Normal follicles, Bovine ovarian follicles between 10–15 mm in diameter; ncRNA, Non-coding RNA; NEB, Negative energy balance; NSDHL, NAD(P) dependent steroid dehydrogenase-like; OPU, Ovum pick-up; PCC, Pearson correlation coefficient; PGD, Phosphogluconate dehydrogenase; PFKM, Phosphofructokinase muscle; PGM1, Phosphoglucomutase 1; RAP2A, RAP2A, member of RAS oncogene family; RIP, RNA-binding protein immunoprecipitation; siRNA, Small interfering RNA.
1. Barros RG, Lodde V, Franciosi F, Luciano AM. In vitro culture strategy for oocytes from early antral follicle in cattle. J Vis Exp. (2020) 8:161. doi: 10.3791/61625
2. Krisher RL. The effect of oocyte quality on development. J Anim Sci. (2004) 82:E14–23. doi: 10.2527/2004.8213_supplE14x
3. McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. (2000) 21:200–14. doi: 10.1210/edrv.21.2.0394
4. Bhardwaj JK, Paliwal A, Saraf P, Sachdeva SN. Role of autophagy in follicular development and maintenance of primordial follicular pool in the ovary. J Cell Physiol. (2022) 237:1157–70. doi: 10.1002/jcp.30613
5. Díaz PU, Stangaferro ML, Gareis NC, Silvia WJ, Matiller V, Salvetti NR, et al. Characterization of persistent follicles induced by prolonged treatment with progesterone in dairy cows: an experimental model for the study of ovarian follicular cysts. Theriogenology. (2015) 84:1149–60. doi: 10.1016/j.theriogenology.2015.06.015
6. Vanholder T, Opsomer G, de Kruif A. Aetiology and pathogenesis of cystic ovarian follicles in dairy cattle: a review. Reprod Nutr Dev. (2006) 46:105–19. doi: 10.1051/rnd:2006003
7. Kengaku K, Tanaka T, Kamomae H. Changes in the peripheral concentrations of inhibin, follicle-stimulating hormone, luteinizing hormone, progesterone and estradiol-17beta during turnover of cystic follicles in dairy cows with spontaneous follicular cysts. J Reprod Dev. (2007) 53:987–93. doi: 10.1262/jrd.18107
8. Rizzo A, Spedicato M, Mutinati M, Minoia G, Pantaleo M, Sciorsci RL. In vivo and in vitro studies of the role of the adrenergic system and follicular wall contractility in the pathogenesis and resolution of bovine follicular cysts. Theriogenology. (2011) 76:1526–31. doi: 10.1016/j.theriogenology.2011.06.024
9. Paredes AH, Salvetti NR, Diaz AE, Dallard BE, Ortega HH, Lara HE. Sympathetic nerve activity in normal and cystic follicles from isolated bovine ovary: local effect of beta-adrenergic stimulation on steroid secretion. Reprod Biol Endocrinol. (2011) 9:66. doi: 10.1186/1477-7827-9-66
10. Ohira T, Murayama C, Shimizu T, Yoshimura Y, Isobe N. Comparison of cadherin and integrin localization in bovine cystic and healthy follicles. Anim Sci J. (2013) 84:303–9. doi: 10.1111/asj.12008
11. Rodríguez FM, Salvetti NR, Colombero M, Stangaferro ML, Barbeito CG, Ortega HH, et al. Interaction between IGF1 and IGFBPs in bovine cystic ovarian disease. Anim Reprod Sci. (2013) 140:14–25. doi: 10.1016/j.anireprosci.2013.04.012
12. Mutlag AM, Wang X, Yang Z, Meng J, Wang X, Zhang J, et al. Study on matrix metalloproteinase 1 and 2 gene expression and NO in dairy cows with ovarian cysts. Anim Reprod Sci. (2015) 152:1–7. doi: 10.1016/j.anireprosci.2014.11.006
13. Polat IM, Alçigir E, Pekcan M, Vural SA, Özenç E, Canatan HE, et al. Characterization of transforming growth factor beta superfamily, growth factors, transcriptional factors, and lipopolysaccharide in bovine cystic ovarian follicles. Theriogenology. (2015) 84:1043–52. doi: 10.1016/j.theriogenology.2015.06.003
14. Tanemura K, Ohtaki T, Kuwahara Y, Tsumagari S. Association between liver failure and hepatic UDP-glucuronosyltransferase activity in dairy cows with follicular cysts. J Vet Med Sci. (2017) 79:86–91. doi: 10.1292/jvms.15-0674
15. Gareis NC, Angeli E, Huber E, Salvetti NR, Rodríguez FM, Ortega HH, et al. Alterations in key metabolic sensors involved in bovine cystic ovarian disease. Theriogenology. (2018) 120:138–46. doi: 10.1016/j.theriogenology.2018.07.045
16. Stassi AF, Gasser F, Velázquez MML, Bolotti EM, Gareis NC, Rey F, et al. Contribution of the VEGF system to the follicular persistence associated with bovine cystic ovaries. Theriogenology. (2019) 138:52–65. doi: 10.1016/j.theriogenology.2019.07.002
17. Wajahat M, Bracken CP, Orang A. Emerging functions for snoRNAs and snoRNA-Derived fragments. Int J Mol Sci. (2021) 22:10193. doi: 10.3390/ijms221910193
18. Zhang P, Wu W, Chen Q, Chen M. Non-Coding RNAs and their integrated networks. J Integr Bioinform. (2019) 16:20190027. doi: 10.1515/jib-2019-0027
19. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. (2005) 309:1559–63. doi: 10.1126/science.1112014
20. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. (2012) 489:101–8. doi: 10.1038/nature11233
21. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. (2009) 458:223–7. doi: 10.1038/nature07672
22. Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. (2016) 17:207–23. doi: 10.1038/nrg.2016.4
23. Li H, Yang J, Jiang R, Wei X, Song C, Huang Y, et al. Long non-coding RNA profiling reveals an abundant MDNCR that promotes differentiation of myoblasts by sponging miR-133a. Mol Ther Nucleic Acids. (2018) 12:610–25. doi: 10.1016/j.omtn.2018.07.003
24. Raza SHA, Khan R, Cheng G, Long F, Bing S, Easa AA, et al. RNA-Seq reveals the potential molecular mechanisms of bovine KLF6 gene in the regulation of adipogenesis. Int J Biol Macromol. (2022) 195:198–206. doi: 10.1016/j.ijbiomac.2021.11.202
25. Yu L, Tai L, Zhang L, Chu Y, Li Y, Zhou L. Comparative analyses of long non-coding RNA in lean and obese pig. Oncotarget. (2017) 8:41440–50. doi: 10.18632/oncotarget.18269
26. Yang HY, Yang H, Shi GQ, Shen M, Yang JQ, Yang YL, et al. Expression profile analysis of microRNAs during hair follicle development in the sheep foetus. Biosci Biotechnol Biochem. (2019) 83:1045–61. doi: 10.1080/09168451.2019.1591261
27. Shen X, Tang J, Jiang R, Wang XG, Yang YZ, Lan XY, et al. CircRILPL1 promotes muscle proliferation and differentiation via binding miR-145 to activate IGF1R/PI3K/AKT pathway. Cell Death Dis. (2021) 12:142. doi: 10.1038/s41419-021-03419-y
28. Wu J, Yuan XH, Jiang W, Lu YC, Huang QL, Yang Y, et al. Genome-wide map of N6-methyladenosine circular RNAs identified in mice model of severe acute pancreatitis. World J Gastroenterol. (2021) 27:7530–45. doi: 10.3748/wjg.v27.i43.7530
29. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. (2014) 42:D92–7. doi: 10.1093/nar/gkt1248
30. Sarver AL, Subramanian S. Competing endogenous RNA database. Bioinformation. (2012) 8:731–3. doi: 10.6026/97320630008731
31. Yip DK, Pang IK, Yip KY. Systematic exploration of autonomous modules in noisy microRNA-target networks for testing the generality of the ceRNA hypothesis. BMC Genomics. (2014) 15:1178. doi: 10.1186/1471-2164-15-1178
32. Wang P, Zhi H, Zhang Y, Liu Y, Zhang J, Gao Y, et al. miRSponge: a manually curated database for experimentally supported miRNA sponges and ceRNAs. Database. (2015) 2015:bav098. doi: 10.1093/database/bav098
33. Wathes DC, Taylor VJ, Cheng Z, Mann GE. Follicle growth, corpus luteum function and their effects on embryo development in postpartum dairy cows. Reprod Suppl. (2003) 61:219–37. doi: 10.1530/biosciprocs.5.017
34. Sakaguchi K, Maylem ERS, Tilwani RC, Yanagawa Y, Katagiri S, Atabay EC, et al. Effects of follicle-stimulating hormone followed by gonadotropin-releasing hormone on embryo production by ovum pick-up and in vitro fertilization in the river buffalo (Bubalus bubalis). Anim Sci J. (2019) 90:690–5. doi: 10.1111/asj.13196
35. Waqas M, Mehmood MU, Shahzad Q, Kausar R, Sattar A, Naseer Z. Comparative efficacy of G6G and Ovsynch protocols on synchronization and pregnancy rate in Nili-Ravi buffalo. Anim Reprod Sci. (2016) 166:9–14. doi: 10.1016/j.anireprosci.2015.12.006
36. Zhao C, Bai Y, Fu S, Wu L, Xia C, Xu C. Comparison of metabolic alterations in serum and milk whey between inactive ovaries and Estrus dairy cows. Front Vet Sci. (2021) 7:609391. doi: 10.3389/fvets.2020.609391
37. Dozmorov MG. GitHub statistics as a measure of the impact of open-source bioinformatics software. Front Bioeng Biotechnol. (2018) 6:198. doi: 10.3389/fbioe.2018.00198
38. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. (2019) 37:907–15. doi: 10.1038/s41587-019-0201-4
39. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. (2015) 33:290–5. doi: 10.1038/nbt.3122
40. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
41. Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. (2010) 11:R14. doi: 10.1186/gb-2010-11-2-r14
42. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. (2011) 39:W316–22. doi: 10.1093/nar/gkr483
43. Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. (2003) 5:R1. doi: 10.1186/gb-2003-5-1-r1
44. Schober P, Boer C, Schwarte LA. Correlation coefficients: appropriate use and interpretation. Anesth Analg. (2018) 126:1763–8. doi: 10.1213/ANE.0000000000002864
45. Otasek D, Morris JH, Bouças J, Pico AR, Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. (2019) 20:185. doi: 10.1186/s13059-019-1758-4
46. Gümen A, Wiltbank MC. Follicular cysts occur after a normal estradiol-induced GnRH/LH surge if the corpus hemorrhagicum is removed. Reproduction. (2005) 129:737–45. doi: 10.1530/rep.1.00653
47. Çolakoglu HE, Küplülü S, Polat IM, Pekcan M, Özenç E, Baklaci C, et al. Association among lipopolysaccharide, the transforming growth factor-beta superfamily, follicular growth, and transcription factors in spontaneous bovine ovarian cysts. Domest Anim Endocrinol. (2020) 70:106398. doi: 10.1016/j.domaniend.2019.106398
48. Jeffcoate IA, Ayliffe TR. An ultrasonographic study of bovine cystic ovarian disease and its treatment. Vet Rec. (1995) 136:406–10. doi: 10.1136/vr.136.16.406
49. Marelli BE, Diaz PU, Salvetti NR, Rey F, Ortega HH. mRNA expression pattern of gonadotropin receptors in bovine follicular cysts. Reprod Biol. (2014) 14:276–81. doi: 10.1016/j.repbio.2014.08.002
50. Peter AT. An update on cystic ovarian degeneration in cattle. Reprod Domest Anim. (2004) 39:1–7. doi: 10.1046/j.0936-6768.2003.00466.x
51. Rey F, Rodríguez FM, Salvetti NR, Palomar MM, Barbeito CG, Alfaro NS, et al. Insulin-like growth factor-II and insulin-like growth factor-binding proteins in bovine cystic ovarian disease. J Comp Pathol. (2010) 142:193–204. doi: 10.1016/j.jcpa.2009.11.002
52. Beam SW, Butler WR. Effects of energy balance on follicular development and first ovulation in postpartum dairy cows. J Reprod Fertil Suppl. (1999) 54:411–24.
53. Lucy MC. Reproductive loss in high-producing dairy cattle: where will it end? J Dairy Sci. (2001) 84:1277–93. doi: 10.3168/jds.S0022-0302(01)70158-0
54. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP, A. ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
55. Wang H, Zhong J, Zhang C, Chai Z, Cao H, Wang J, et al. The whole-transcriptome landscape of muscle and adipose tissues reveals the ceRNA regulation network related to intramuscular fat deposition in yak. BMC Genomics. (2020) 21:347. doi: 10.1186/s12864-020-6757-z
56. Liang R, Han B, Li Q, Yuan Y, Li J, Sun D. Using RNA sequencing to identify putative competing endogenous RNAs (ceRNAs) potentially regulating fat metabolism in bovine liver. Sci Rep. (2017) 7:6396. doi: 10.1038/s41598-017-06634-w
57. Wang J, Ren Q, Hua L, Chen J, Zhang J, Bai H, et al. Comprehensive analysis of differentially expressed mRNA, lncRNA and circRNA and their ceRNA networks in the longissimus Dorsi Muscle of two different pig breeds. Int J Mol Sci. (2019) 20:1107. doi: 10.3390/ijms20051107
58. Li N, Liu XL, Zhang FL, Tian Y, Zhu M, Meng LY, et al. Whole-transcriptome analysis of the toxic effects of zearalenone exposure on ceRNA networks in porcine granulosa cells. Environ Pollut. (2020) 261:114007. doi: 10.1016/j.envpol.2020.114007
59. Yue B, Li H, Liu M, Wu J, Li M, Lei C, et al. Characterization of lncRNA-miRNA-mRNA network to reveal potential functional ceRNAs in bovine skeletal muscle. Front Genet. (2019) 10:91. doi: 10.3389/fgene.2019.00091
60. Mayama S, Hamazaki N, Maruyama Y, Matsubara S, Kimura AP. Transcriptional activation of the mouse Scd2 gene by interdependent enhancers and long noncoding RNAs in ovarian granulosa cells. J Reprod Dev. (2020) 66:435–44. doi: 10.1262/jrd.2019-161
61. Yao X, Gao X, Bao Y, El-Samahy MA, Yang J, Wang Z, et al. lncRNA FDNCR promotes apoptosis of granulosa cells by targeting the miR-543-3p/DCN/TGF-β signaling pathway in Hu sheep. Mol Ther Nucleic Acids. (2021) 24:223–40. doi: 10.1016/j.omtn.2021.02.030
62. Huang X, Pan J, Wu B, Teng X. Construction and analysis of a lncRNA (PWRN2)-mediated ceRNA network reveal its potential roles in oocyte nuclear maturation of patients with PCOS. Reprod Biol Endocrinol. (2018) 16:73. doi: 10.1186/s12958-018-0392-4
63. Sun L, Zhang P, Lu W. lncRNA MALAT1 Regulates mouse granulosa cell apoptosis and 17β-estradiol synthesis via regulating miR-205/CREB1 axis. Biomed Res Int. (2021) 2021:6671814. doi: 10.1155/2021/6671814
64. Zhou M, Liu X, Qiukai E, Shang Y, Zhang X, Liu S, et al. Long non-coding RNA Xist regulates oocyte loss via suppressing miR-23b-3p/miR-29a-3p maturation and upregulating STX17 in perinatal mouse ovaries. Cell Death Dis. (2021) 12:540. doi: 10.1038/s41419-021-03831-4
65. Cortes VA, Busso D, Maiz A, Arteaga A, Nervi F, Rigotti A. Physiological and pathological implications of cholesterol. Front Biosci. (2014) 19:416–28. doi: 10.2741/4216
66. Jamieson PM, Chapman KE, Edwards CR, Seckl JR. 11 beta-hydroxysteroid dehydrogenase is an exclusive 11 beta- reductase in primary cultures of rat hepatocytes: effect of physicochemical and hormonal manipulations. Endocrinology. (1995) 136:4754–61. doi: 10.1210/endo.136.11.7588203
67. Gümen A, Wiltbank MC. An alteration in the hypothalamic action of estradiol due to lack of progesterone exposure can cause follicular cysts in cattle. Biol Reprod. (2002) 66:1689–95. doi: 10.1095/biolreprod66.6.1689
68. Hudson NJ, Reverter A, Griffiths WJ, Yutuc E, Wang Y, Jeanes A, et al. Gene expression identifies metabolic and functional differences between intramuscular and subcutaneous adipocytes in cattle. BMC Genomics. (2020) 21:77. doi: 10.1186/s12864-020-6505-4
69. Li H, Ericsson M, Rabasha B, Budnik B, Chan SH, Freinkman E, et al. 6-Phosphogluconate dehydrogenase links cytosolic carbohydrate metabolism to protein secretion via modulation of glutathione levels. Cell Chem Biol. (2019) 26:1306–14.e5. doi: 10.1016/j.chembiol.2019.05.006
70. Wærp HKL, Waters SM, McCabe MS, Cormican P, Salte R. RNA-seq analysis of bovine adipose tissue in heifers fed diets differing in energy and protein content. PLoS ONE. (2018) 13:e0201284. doi: 10.1371/journal.pone.0201284
71. Huang R, Chen H, Zhong C, Kim JE, Zhang YH. High-throughput screening of coenzyme preference change of thermophilic 6-phosphogluconate dehydrogenase from NADP(+) to NAD(). Sci Rep. (2016) 6:32644. doi: 10.1038/srep32644
72. Wang X, Fan Y, He Y, Han Z, Gong Z, Peng Y, et al. Integrative analysis of miRNA and mRNA expression profiles in mammary glands of Holstein cows artificially infected with Staphylococcus aureus. Pathogens. (2021) 10:506. doi: 10.3390/pathogens10050506
73. Li R, Zhang CL, Liao XX, Chen D, Wang WQ, Zhu YH, et al. Transcriptome microRNA profiling of bovine mammary glands infected with Staphylococcus aureus. Int J Mol Sci. (2015) 16:4997–5013. doi: 10.3390/ijms16034997
74. Lawless N, Foroushani AB, McCabe MS, O'Farrelly C, Lynn DJ. Next generation sequencing reveals the expression of a unique miRNA profile in response to a gram-positive bacterial infection. PLoS ONE. (2013) 8:e57543. doi: 10.1371/journal.pone.0057543
75. Nokelainen P, Peltoketo H, Vihko R, Vihko P. Expression cloning of a novel estrogenic mouse 17 beta-hydroxysteroid dehydrogenase/17-ketosteroid reductase (m17HSD7), previously described as a prolactin receptor-associated protein (PRAP) in rat. Mol Endocrinol. (1998) 12:1048–59. doi: 10.1210/mend.12.7.0134
76. Törn S, Nokelainen P, Kurkela R, Pulkka A, Menjivar M, Ghosh S, et al. Production, purification, and functional analysis of recombinant human and mouse 17beta-hydroxysteroid dehydrogenase type 7. Biochem Biophys Res Commun. (2003) 305:37–45. doi: 10.1016/S0006-291X(03)00694-6
77. Laubner D, Breitling R, Adamski J. Embryonic expression of cholesterogenic genes is restricted to distinct domains and colocalizes with apoptotic regions in mice. Brain Res Mol Brain Res. (2003) 115:87–92. doi: 10.1016/S0169-328X(03)00094-9
78. Jokela H, Rantakari P, Lamminen T, Strauss L, Ola R, Mutka AL, et al. Hydroxysteroid (17beta) dehydrogenase 7 activity is essential for fetal de novo cholesterol synthesis and for neuroectodermal survival and cardiovascular differentiation in early mouse embryos. Endocrinology. (2010) 151:1884–92. doi: 10.1210/en.2009-0928
Keywords: bovine, lncRNA, miRNA, ceRNA, follicular cysts
Citation: Wang K, Cheng Y, Guo T, Guo X, Zhang H, Ma X, Pan Y, Kebreab E, Wang D and Lyu L (2022) Analyzing the interactions of mRNAs, miRNAs and lncRNAs to predict ceRNA networks in bovine cystic follicular granulosa cells. Front. Vet. Sci. 9:1028867. doi: 10.3389/fvets.2022.1028867
Received: 26 August 2022; Accepted: 26 September 2022;
Published: 13 October 2022.
Edited by:
Sayed Haidar Abbas Raza, Northwest A&F University, ChinaReviewed by:
Ayman Hassan Abd El-Aziz, Damanhour University, EgyptCopyright © 2022 Wang, Cheng, Guo, Guo, Zhang, Ma, Pan, Kebreab, Wang and Lyu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihua Lyu, bGlodWFsdnN4YXVAMTI2LmNvbQ==; Dong Wang, ZHdhbmdjbjIwMDJAdmlwLnNpbmEuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.