
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 11 October 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1028866
Bovine Viral Diarrhea Virus (BVDV) is one of the main pathogens that affects ruminants worldwide, generating significant economic losses. Like other RNA viruses, BVDV is characterized by a high genetic variability, generating the emergence of new variants, and increasing the risk of new outbreaks. The last report on BVDV genotypes in France was in 2008, since which there have been no new information. The goal of this study is to determine the genetic diversity of BVDV strains currently circulating in France. To this aim, samples of cattle were taken from different departments that are part of the main areas of livestock production during the years 2018 to 2020. Using the partial sequence of the 5'UTR region of the viral genome, we identified and classified 145 samples corresponding to Pestivirus A and one sample corresponding to Pestivirus D. For the Pestivirus A samples, the 1e, 1b, 1d, and 1l genotypes, previously described in France, were identified. Next, the 1r and 1s genotypes, not previously described in the country, were detected. In addition, a new genotype was identified and was tentatively assigned as 1x genotype. These results indicate an increase in the genetic diversity of BVDV in France.
Bovine Viral Diarrhea Virus (BVDV) is a globally distributed cattle pathogen, which causes significant economic losses (1). This virus also affects other domestic animal species such as sheep, goat, camelids and swine (2), and wild animals including cervids and giraffes (3). BVDV is responsible for a wide spectrum of symptoms in cattle, which include immunosuppression, persistent infection, mucosal disease, respiratory syndromes and reproductive dysfunction, generating major economic losses for the cattle industry (4). BVDV is a member of the genus Pestivirus within the family Flaviviridae. This genus also includes other viruses affecting domestic livestock such as Pestivirus C also known as Classical Swine Fever Virus (CSFV) and Pestivirus D known as Border Disease Virus (BDV) (5). The BVDV genome consists of a single strand of positive-sense RNA (ssRNA+) ~12.3-kb-long, which is flanked by a 5′ untranslated region (UTR) and a 3′UTR. This genome encodes 11–12 structural and non-structural proteins (Npro, C, Erns, E1, E2, P7, NS2/3, NS4A, NS4B, NS5A, NS4B) (1). The 5′UTR being one of the most conserved genomic regions of BVDV, it is widely used for phylogenetic classification. Three main species of the genus Pestivirus have been identified in cattle: Pestivirus A (known as BVDV-1), which so far includes 23 genotypes (1a−1w), Pestivirus B (BVDV-2), including 4 genotypes (2a−2d) (6–8), and Pestivirus H (BVDV-3, also known as HoBi-like virus) for which 4 genotypes (3a−3d) have been described (8–10). A fourth species that affects cattle less frequently is Pestivirus D, also known as Border Disease Virus (BDV) (5). It mainly affects sheep, producing prenatal and postnatal infections in lambs. Interspecies transmission by Pestivirus D has been observed in cattle, goats, and pigs (11, 12).
Due to their ability or not to exhibit a cytopathogenic effect, BVDV isolates have been categorized into 2 biotypes, cytopathogenic (CP) and non-cytopathogenic (NCP) (1). Pestivirus A is the most widely distributed species globally, corresponding to 88% of the isolates worldwide (6). Within this species, several dominant genotypes have been described by region, such as 1a genotype in Africa, 1c in Australia, while in Asia, America and Europe 1b predominates. This high genetic variability is described throughout the worldwide distribution of the virus (6).
In the case of France, in the latest studies from 2001 and 2008, 73 viral strains were described, which were classified as 1e (n = 46) and 1b (n = 15) genotypes, the two dominant genotypes, followed by 1a (n = 3), 1d (n = 3) and 1l (n = 3). For the Pestivirus B species, only 3 sequences were identified (13, 14).
The Pestivirus A genotypes currently circulating in France are unknown. Pestiviruses are known for their rapid evolution that can lead to genetic drift. Here we provide an update on the phylogenetic classification of Pestivirus A strains recently collected from bovines throughout various regions of France.
In this study, 211 samples of whole blood from cattle were analyzed, collected from 38 departments in France during the years 2018 to 2020, and confirmed as positive for BVDV by RT-PCR by departmental diagnostic laboratories or sent by veterinary practitioners after having confirmed the positivity of the animal by antigen detection on blood (Idexx SNAP BVDV, Westbrook, USA). The samples were sent to the laboratory of Pathology, Faculty of Veterinary Medicine, University of Liege, for further processing. The map of the geographical distribution of the samples was designed using the QGIS 3.24 software (15). The samples used in this study were obtained from Departmental diagnostic laboratories in the context of the surveillance plan against BVDV in France. Getting access to these samples did not require the study to be reviewed or approved by an Ethics Committee.
The viral RNA was extracted from blood samples using NucleoSpin® RNA kit (Macherey Nagel, Germany), following manufacturer's instructions. A 288-bp region of the region 5′UTR was amplified by end-point RT-PCR using the Pan-Pestivirus primers Forward 324: 5′-ATG CCC WTA GTA GGA CTA GCA-3′ and Reverse 326: 5′-TCA ACT CCA TGT GCC ATG TAC-3′ (14) and Luna Universal One-Step RT-qPCR Kit (New England BioLabs, USA). The results were visualized on a 1% agarose gel, amplicons of the expected size were purified using NucleoSpin Gel and PCR Clean-up kit® (Macherey Nagel, Germany) and then sequenced by the Sanger method (Eurofins Genomics®). The sequences obtained were aligned using ClustalW, implemented in the Geneious 10.2.3 software (Biomatters, New Zealand). The phylogenetic tree of the 5′UTR region was built using the Maximum Likelihood method with the Kimura 2-parameter model, Gamma distribution with Invariant Sites, as determined by a model prediction analysis, and 1,000 bootstrap replicates using the MEGA X software (16). Reference sequences of Pestivirus A, Band Hspecies and genotypes were retrieved from NCBI GenBank database and included in the phylogenetic analysis. Nucleotide similarity percentages were calculated using the distance matrix algorithm of the MEGA X software (16).
Using a Pan-Pestivirus end-point RT-PCR targeting the 5′UTR it was possible to obtain an amplicon of the expected size from 146 of the analyzed samples, from 31 departments (Figure 1). The amplicons were purified and subsequently sequenced.
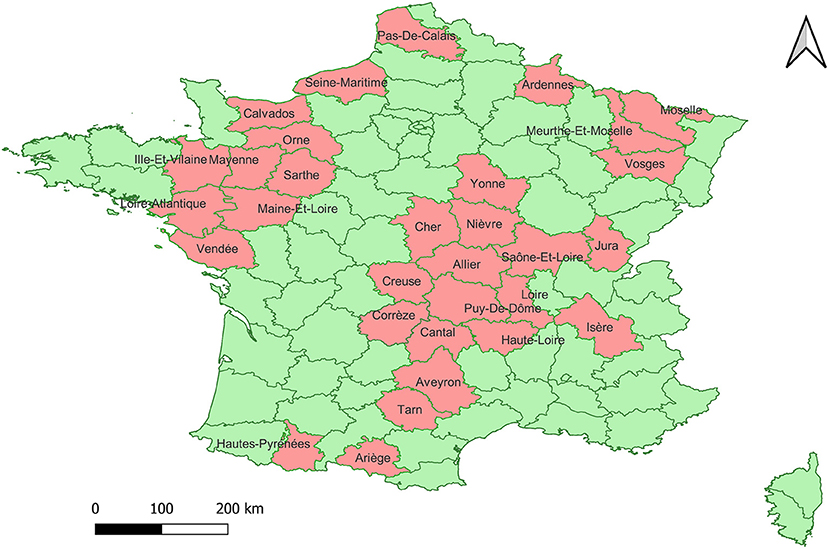
Figure 1. Map of the geographic distribution of the collection sites of the 146 samples included in the study. In pink the departments from which the samples have been collected.
These samples were compared with sequences from known genotypes of Pestivirus A, B and H species obtained from GenBank to build a phylogenetic tree. No sequence belonging to Pestivirus B and H were found. Except for sample ON693854, all the field sequences analyzed belonged toPestivirus A. Most of them corresponded to genotypes 1e (57.9%) (N = 84) and to 1b (32.4%) (N = 47), being identified in 26 and 20 of the sampled departments, respectively. To a lesser extent, samples of the genotypes 1d (3.4%) (N = 5) and 1l (2.1%) (N = 3) were detected, each in 2 different departments. In addition, samples corresponding to 1s (0.7%) (N = 1) and 1r (0.7%) (N = 1) were identified for the first time in the country. Finally, 4 (2.7%) sequences from 3 different departments clustered together and separately from currently known genotypes within Pestivirus A, forming a new genotype tentatively named 1x (Figure 2). The nucleotide similarity of the 5'UTR is greater than 95.4% between the 1x sequences. The closest genotypes are 1e and 1j, sharing a similarity of 90.7 and 90.4%, respectively (Figure 3). In the phylogenetic analysis, sample ON693854 clustered outside the sequences corresponding to BVDV. Therefore, it was analyzed with other reference sequences of the Pestivirus genus, where it clustered within Pestivirus Dspecies with sequences of the genotype 4 (data not shown).
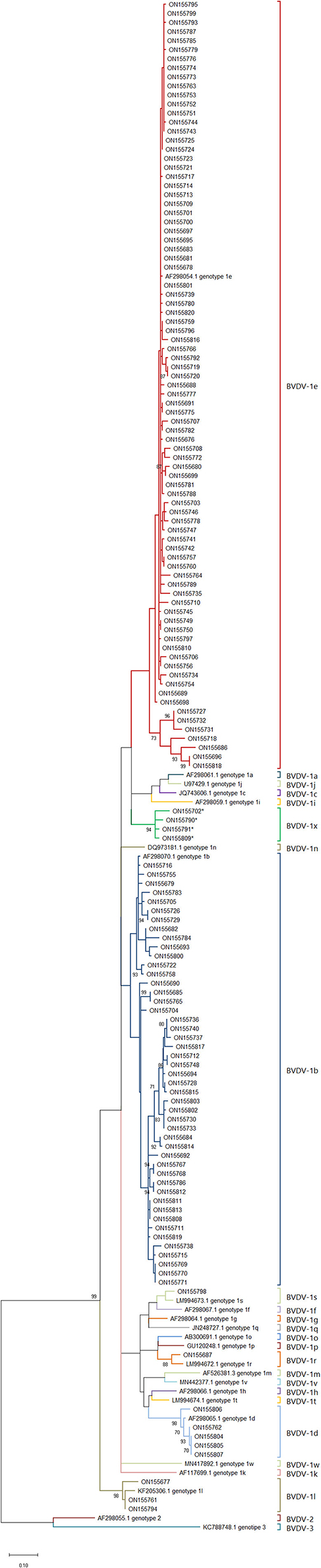
Figure 2. Phylogenetic tree of Pestivirus A genotypes circulating in France. Phylogenetic tree based on the BVDV 5′UTR fragment, using the Maximum Likelihood method and the two-parameter Kimura model (17). Reference strains were used for each known species/genotype. The GenBank accession number is indicated. Field strains described in this study are identified by the GenBank accession number (Table 1). The asterisk identifies the samples that correspond to the new 1x genotype.
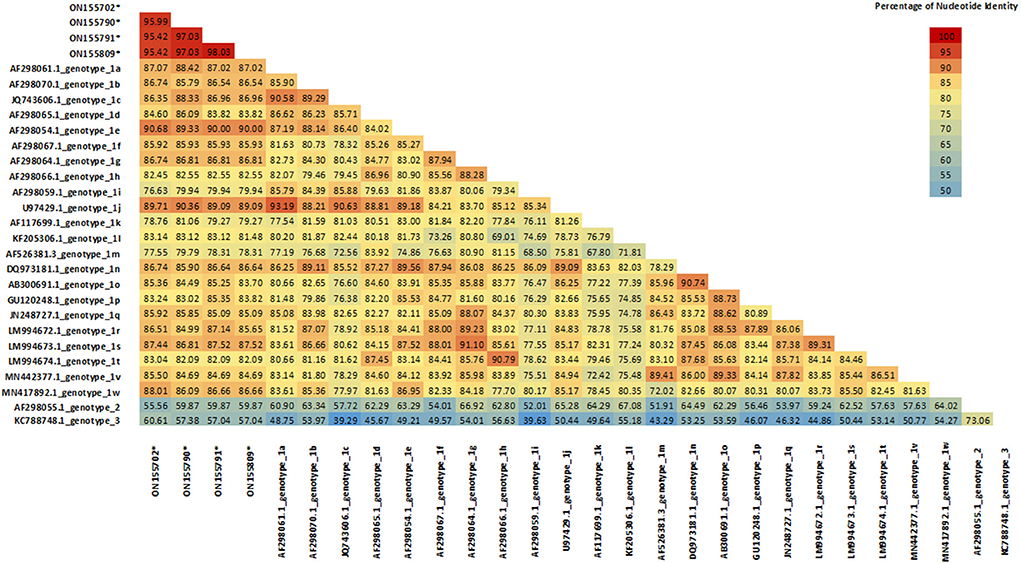
Figure 3. Sequence Identity Matrix of 1x strains compared with currently known Pestivirus A genotypes. Values generated from the 221-bp alignment of the 5′UTR segment. Sequences belonging to 1x genotype were compared with 24 reference strains of BVDV (one per known genotype).
The list of the samples analyzed is summarized in Table 1, where the distribution of the samples according to the department is detailed.
In this study we evaluated the genetic diversity of BVDV strains currently circulating in France. Viral sequences amplified by RT-PCR were genotyped based on the analysis of the 5' UTR. Out of 211 samples initially detected as positive for BVDV in Departmental laboratories or by veterinary practitioners, only 146 proved positive using the 5′UTR RT-PCR in our laboratory and yielded sequences of sufficient quality for further analyses. The delay between the initial detection and the reception of the samples in our laboratory and poor preservation conditions (samples stored at −20°C in the field laboratories, freeze-thaw cycles) likely contributed to explain such a difference.
The positive samples came from 31 departments that are part of the livestock production area with the highest numbers of animals per department in France (18). The samples corresponding to the Pestivirus A species were classified into 7 genotypes, in order of abundance: 1e, 1b, 1d, 1x, 1l, 1r and 1s. In previous studies carried out in 2001 and 2008, the circulation of the genotypes 1e (N = 46), 1b (N = 15), 1a (N = 3), 1d (N = 3), 1l (N = 3) and Pestivirus B genotype 2a (N = 3) genotypes was detected in France (13, 14). Our data confirm that, as previously reported, 1e and 1b are the most abundant genotypes in France (13, 14).
Between 2001 and 2008 1e was the dominant genotype in France, representing 65% of the strains identified (13, 14). In our study the distribution remained in the ranges previously observed, decreasing slightly to 57.9% among the strains identified. The 1e genotype has been described mostly in Europe. In America 1e isolates have been reported in cattle in Brazil (19) and recently in Argentina (20). In Europe 1e genotype is distributed throughout the entire continent, mainly in France, where it is the dominant strain (13) and in Italy (21) and Switzerland (22), where it is the second most abundant strain. The 1e genotype is estimated to have originated in northern Italy in the Lombardy region between 1957 and 1988, where a large number of isolates are concentrated (23). From this region it spread to the rest of the country and to neighboring countries between 1990 and 2000 following the movements of cattle (21). In Switzerland, the highest concentration of 1e is in the west of the country, bordering the Lombardy region, associated with Felckvieh cattle (22). In France, the first cases of 1e were detected in 1993 (14). In this work, a geographic cluster of 1e was not observed. On the contrary, this genotype was found throughout the French territory, similar to the current propagation of this genotype in Italy (21).
The 1b genotype has a worldwide distribution, being the dominant genotype in America, Asia and most of Europe. In the latter it is the dominant strain in Italy and Spain, while it is the second most abundant genotype in France as in Germany (6). Until 2008, 21.4% of the samples identified in France corresponded to 1b (13, 14). In the current study this percentage increased to 32.4% of the samples. This rising trend has been described elsewhere over recent years for this genotype (24, 25).
The 1d genotype was identified with a lower frequency. This strain has recently been described in China (7) and Brazil (26). In Europe it is distributed throughout the continent, mainly in Sweden, where it is dominant (6). The 1d genotype had previously been described in France with a low presence of 4.3% in 2008 (13), as in this work, a frequency of 3.4% was observed for 1d, without great variation over time.
The 1l genotype is mainly found in Turkey, where it is the most abundant genotype, but it has also been identified in France, Italy and Spain with a very low proportion (6). In this study, only three sequences were identified as 1l corresponding to 2.1%, maintaining the same relative abundance as in previous studies where a frequency of 4.3% was observed (13).
Z-TEST statistical one-tailed analysis of the proportions of genotypes 1e, 1b, 1d and 1l between the previous studies in France (13, 14) and the current study, indicate that only the increase in genotype 1b represents a significant change (alpha < 0.05).
For the first time in France, the 1r and 1s genotypes were detected. 1r genotype has been described in Italy, Turkey (6, 27) and Poland (28), while 1s was described in Italy (27) and Poland (28). These genotypes are rare and show a low prevalence in the countries where they have been described (6, 28).
In this work, a new genotype formed by the sequences ON155702, ON155790, ON155791 and ON155809 was identified. These sequences have a nucleotide similarity >95.5% between them. With the closest genotypes 1e and 1j, the similarity was 90.7 and 90.4%, respectively. These percentages of similarity are within the ranges observed between currently known genotypes (14), suggesting that this cluster forms a new genotype different from those previously described. We decided to tentatively assign the name 1x to this new genotype, which follows 1w, the last genotype described (7). Although 1x genotype was previously used in Switzerland to describe the strain CH-01-08 in 2008 (29), this strain was finally reclassified as 1l (30). The appearance of new genotypes is an event that is classically observed with the evolution of BVDV. In China for example, two new genotypes 1v and 1w were recently described (7, 31). This is mainly attributed to the lack of proofreading function of the RNA polymerase during BVDV genome replication. This variability leads to a genetic drift, generating new genotypes and species over time (32).
In contrast, the 1a genotype, previously present in France, was not identified in this work. This genotype is the second most abundant globally, has been isolated on all continents and is used in most vaccines. In Europe it is mainly found in Ireland and the United Kingdom (6, 33). In France it was only identified in 2001 (14), but neither in the 2008 study (13), nor in the current study.
In this work, no strains belonging to the Pestivirus B species were identified. The latest reports are from 1994 with 2 samples typed as Pestivirus B genotype 2a (14, 34) and 2008, with a sample from Nièvre typed as Pestivirus B (13). These samples represented 1.3% of the 218 BVDV samples previously identified in the country (13, 14). The absence of this species in this work and the low frequency in previous studies suggest that the presence of Pestivirus B in France is sporadic as seen in Italy (35) or that it has disappeared.
Sequences belonging to the Pestivirus H species were not detected either. This emerging group was detected for the first time in contaminated fetal bovine serum from Brazil, since then it has been detected in America, Asia and Europe (36). So far in Europe, it has only been detected in Italy associated with an outbreak of abortions in 2011 (37).
In addition, in this work, the sample ON693854 was classified as Pestivirus D genotype 4. This sample was obtained from a calf belonging to a BVDV-free farm where cattle and sheep herds were kept together. Interestingly, the infection coincides with the recent introduction of sheep to the farm from the South of France. Cross-species infection is common in Pestivirus D and has been documented in cows housed with sheep in the same barn or on pasture (38). In France, Pestivirus D has been previously described, with 3, 4, 5, 6 and Tunisian and Tunisian-like genotypes (39–41). The genotype 4 is the dominant in sheep flocks in Spain (12, 42), and has been associated with several outbreaks in chamois (Rupicapra pyrenaica pyrenaica) in the Pyrenees throughout Spain, France and Andorra (40). To our knowledge, Pestivirus D genotype 4 infection in cattle has only been evidenced by viral neutralization test (43), so this could be the first case of molecular characterization of genotype 4 in cattle documented in the literature.
Since 2020, a decree has made it mandatory to monitor and control BVDV throughout France in order to eradicate it. Screening is based on individual virus search on all calves before 20 days of age or serological monitoring of bulk tank milk or blood from a sample of randomly selected animals. When analysis results are not favorable, the herd is declared BVDV infected and sanitary measures are implemented. In particular, the testing and elimination of all PI confirmed animals (44).
In France between 1982 and 2015, according to online data from the national food safety agency (ANSES) (45), 11 vaccines, including three with the fetal protection claim, have been approved for use in cattle against BVDV. These are composed of mono- and polyvalent vaccines with inactivated or modified live viruses (MLV) of genotypes 1a, 1b and 2a (45). The humoral protection of vaccines against homologous genotypes is well documented and is superior to that against heterologous genotypes (25, 46–48). In the case of the most abundant genotype in this study, i.e., the 1e genotype, cross-reactivity tests with vaccines composed of 1a and 1b genotypes and a strain belonging to this genotype have shown a complete absence of neutralizing antibodies (48), possibly due to the significant antigenic differences against this strain (29, 49). Nevertheless, data are not sufficient to generalize this finding to all the BVD-1e strains. In the case of the 1b genotype, cross-protection studies with vaccines containing 1a showed contradictory results. While some studies showed high levels of cross-reactivity (46, 48, 50), other studies suggested moderate to insufficient protection against 1b genotype (47, 48, 51, 52). This could be due to the high diversity observed in the 1b genotype and the use of 5′UTRs for phylogenetic classification rather than key viral antigenic protein sequences (25).
On the other hand, a protective cellular immune response triggered by vaccines can occur even before or without the presence of antibodies (53–55). This response is mostly observed with MLV. Unlike inactivated vaccines, MLV have the ability to replicate in the cytoplasm and synthesize viral proteins, such as NS2/3 (56) and NS3, that are highly conserved among Pestiviruses A, B, H (57). These proteins generate reactivity of CD4+, CD8+ T cells, which is speculated to be responsible for the good “inter-genotype” cross-protection conferred by Pestivirus A vaccines (57).
T-cell and B-cell response levels approaching sterilizing immunity are required for the prevention of fetal infections (32), which are mostly observed in animals vaccinated with MLV. In studies of animals challenged with BVDV, those that were vaccinated with MLV presented greater fetal protection (50, 58, 59) even against other species such as Pestivirus H (60), compared to inactivated vaccines that present variable levels of protection (51) or lack of protection from fetal infection, as it has been reported in recent studies (61, 62).
Due to the great genetic diversity of BVDV and the constant appearance of new genotypes, it is important to consider, in the application of eradication plans, the use of vaccines that generate proven cross-protection against heterologous strains present in the country. Moreover, these new genotypes can also affect the diagnosis of Pestiviruses, as it was the case for Pestivirus H, for which it was necessary to develop a new RT-qPCR assay to increase the detection efficiency of this species (63). For these reasons, it is important to maintain regular follow-up of the genetic evolution of BVDV, allowing the implementation of adequate eradication plans, as well as their monitoring to effectively control the disease.
In summary, in this work the genetic diversity of BVDV in France was updated, showing an increase in the variability of Pestivirus A strains. Only genotypes belonging to species Pestivirus A were found in circulation, the most abundant being 1e and 1b. Furthermore, genotypes 1r, 1s and 1x are described for the first time in France, the latter being a new genotype.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The samples used in this study were obtained from Departmental Diagnostic Laboratories in the context of the surveillance plan against BVDV in France. Getting access to these samples did not require the study to be reviewed or approved by an Ethics Committee.
JR participated to the laboratory analyses and wrote the original draft. CD helped in the construction of figures. AH was the technical support for the RNA extractions and RT-PCR. M-MG and PG designed the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was conducted between Ceva Santé Animale and the Department of Pathology of the FMV of the University of Liège without specific funding.
We would like to express our warm gratitude to the French Departmental Diagnostic Laboratories and field veterinary practitioners for the sample collection.
Author PG was employed by Ceva Santé Animale.
The authors declare that this study received funding from Ceva Santé Animale. The funder was not involved in the laboratory work, analysis and interpretation of the data.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
BDV, Border Disease Virus; BVDV, Bovine Viral Diarrhea Virus; CP, Cytopathogenic; CSFV, Classical Swine Fever Virus; MLV, modified live viruses; NCP, non-cytopathogenic; ssRNA+, single strand of positive-sense RNA; UTR, untranslated region.
1. Ridpath JF. Bovine Viral Diarrhea Virus: Global Status. Vol. 26, Veterinary Clinics of North America—Food Animal Practice. Amsterdam: Elsevier (2010). p. 105–21. doi: 10.1016/j.cvfa.2009.10.007
2. Krametter-Froetscher R, Duenser M, Preyler B, Theiner A, Benetka V, Moestl K, et al. Pestivirus infection in sheep and goats in West Austria. Vet J. (2010) 186:342–6. doi: 10.1016/j.tvjl.2009.09.006
3. Becher P, Orlich M, Shannon AD, Horner G, König M, Thiel HJ. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J General Virol. (1997) 78:1357–66. doi: 10.1099/0022-1317-78-6-1357
4. Blome S, Beer M, Wernike K. New leaves in the growing tree of pestiviruses. Adv Virus Res. (2017) 99:139–60. doi: 10.1016/bs.aivir.2017.07.003
5. Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Muerhoff AS, et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J General Virol. (2017) 98:2106–12. doi: 10.1099/jgv.0.000873
6. Yeşilbag K, Alpay G, Becher P. Variability and global distribution of subgenotypes of bovine viral diarrhea virus. Viruses. (2017) 9:128. doi: 10.3390/v9060128
7. Deng M, Chen N, Guidarini C, Xu Z, Zhang J, Cai L, et al. Prevalence and genetic diversity of bovine viral diarrhea virus in dairy herds of China. Vet Microbiol. (2020) 242:108565. doi: 10.1016/j.vetmic.2019.108565
8. Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Dempsey DM, et al. Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020). Arch Virol. (2020) 165:2737–48. doi: 10.1007/s00705-020-04752-x
9. Decaro N. HoBi-like pestivirus and reproductive disorders. Front Vet Sci. (2020). 7:622447. doi: 10.3389/fvets.2020.622447
10. Giammarioli M, Ridpath JF, Rossi E, Bazzucchi M, Casciari C, De Mia GM. Genetic detection and characterization of emerging HoBi-like viruses in archival foetal bovine serum batches. Biologicals. (2015) 43:220–4. doi: 10.1016/j.biologicals.2015.05.009
11. Vilček S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol. (1994) 136:309–23. doi: 10.1007/BF01321060
12. Righi C, Petrini S, Pierini I, Giammarioli M, De Mia GM. Global distribution and genetic heterogeneity of border disease virus. Viruses. (2021) 13:950. doi: 10.3390/v13060950
13. Jackova A, Novackova M, Pelletier C, Audeval C, Gueneau E, Haffar A, et al. The extended genetic diversity of BVDV-1: typing of BVDV isolates from France. Vet Res Commun. (2008) 32:7–11. doi: 10.1007/s11259-007-9012-z
14. Vilček Š, Paton DJ, Durkovic B, Strojny L, Ibata G, Moussa A, et al. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol. (2001) 146:99–115. doi: 10.1007/s007050170194
15. QGIS.org (2022). Available online at: https://www.qgis.org/en/site/ (accessed March 23, 2022).
16. Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
17. Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol. Evol. 16:111–20. doi: 10.1007/BF01731581
18. FranceAgriMer. Les filières de l'élevage français. Les cahiers de franceagrimer. Montreuil-sous-Bois: Éric Allain (2013). p. 87.
19. Silveira S, Weber MN, Mósena ACS, da Silva MS, Streck AF, Pescador CA, et al. Genetic diversity of brazilian bovine pestiviruses detected between 1995 and 2014. Transbound Emerg Dis. (2017) 64:613–23. doi: 10.1111/tbed.12427
20. Spetter MJ, Louge Uriarte EL, Verna AE, Leunda MR, Pereyra SB, Odeón AC, et al. Genomic diversity and phylodynamic of bovine viral diarrhea virus in Argentina. Infect Genet Evol. (2021) 96:105089. doi: 10.1016/j.meegid.2021.105089
21. Luzzago C, Decaro N. Epidemiology of bovine pestiviruses circulating in Italy. Front Vet Sci. (2021) 8:669942. doi: 10.3389/fvets.2021.669942
22. Stalder H, Bachofen C, Schweizer M, Zanoni R, Sauerländer D, Peterhans E. Traces of history conserved over 600 years in the geographic distribution of genetic variants of an RNA virus: Bovine viral diarrhea virus in Switzerland. PLoS ONE. (2018) 13:e0207604. doi: 10.1371/journal.pone.0207604
23. Ebranati E, Lauzi S, Cerutti F, Caruso C, Masoero L, Moreno A, et al. Highlighting priority areas for bovine viral diarrhea control in Italy: a phylogeographic approach. Infect Genet Evol. (2018) 58:258–68. doi: 10.1016/j.meegid.2018.01.006
24. Fulton RW, Briggs RE, Ridpath JF, Saliki JT, Confer AW, Payton ME, et al. Transmission of Bovine viral diarrhea virus 1b to susceptible and vaccinated calves by exposure to persistently infected calves. Can J Vet Res. (2005) 69:161.
25. Pecora A, Malacari DA, Ridpath JF, Perez Aguirreburualde MS, Combessies G, Odeón AC, et al. First finding of genetic and antigenic diversity in 1b-BVDV isolates from Argentina. Res Vet Sci. (2014) 96:204–12. doi: 10.1016/j.rvsc.2013.11.004
26. Porto GS, Dall Agnol AM, Leme RA, de Souza TCGD, Alfieri AA, Alfieri AF. Detection of pestivirus A (bovine viral diarrhea virus 1) in free-living wild boars in Brazil. Braz J Microbiol. (2021) 52:1037–42. doi: 10.1007/s42770-021-00449-8
27. Giammarioli M, Ceglie L, Rossi E, Bazzucchi M, Casciari C, Petrini S, et al. Increased genetic diversity of BVDV-1: recent findings and implications thereof. Virus Genes. (2015) 50:147–51. doi: 10.1007/s11262-014-1132-2
28. Mirosław P, Polak M. Increased genetic variation of bovine viral diarrhea virus in dairy cattle in Poland. BMC Vet Res. (2019) 15:1–12. doi: 10.1186/s12917-019-2029-z
29. Bachofen C, Stalder H, Braun U, Hilbe M, Ehrensperger F, Peterhans E. Co-existence of genetically and antigenically diverse bovine viral diarrhoea viruses in an endemic situation. Vet Microbiol. (2008) 131:93–102. doi: 10.1016/j.vetmic.2008.02.023
30. Casaubon J, Vogt HR, Stalder H, Hug C, Ryser-Degiorgis MP. Bovine viral diarrhea virus in free-ranging wild ruminants in Switzerland: low prevalence of infection despite regular interactions with domestic livestock. BMC Vet Res. (2012) 8:204. doi: 10.1186/1746-6148-8-204
31. Tian B, Cai D, Li W, Bu Q, Wang M, Ye G, et al. Identification and genotyping of a new subtype of bovine viral diarrhea virus 1 isolated from cattle with diarrhea. Arch Virol. (2021) 166:1259. doi: 10.1007/s00705-021-04990-7
32. Ridpath JF. Immunology of BVDV vaccines. Biologicals. (2013) 41:14–9. doi: 10.1016/j.biologicals.2012.07.003
33. Booth RE, Thomas CJ, El-Attar LM, Gunn G, Brownlie J. A phylogenetic analysis of Bovine Viral Diarrhoea Virus (BVDV) isolates from six different regions of the UK and links to animal movement data. Vet Res. (2013) 44:43. doi: 10.1186/1297-9716-44-43
34. Giangaspero M, Harasawa R, Weber L, Belloli A. Genoepidemiological evaluation of Bovine viral diarrhea virus 2 species based on secondary structures in the 5′ untranslated region. J Vet Med Sci. (2008) 70:571–80. doi: 10.1292/jvms.70.571
35. Luzzago C, Lauzi S, Ebranati E, Giammarioli M, Moreno A, Cannella V, et al. Extended genetic diversity of bovine viral diarrhea virus and frequency of genotypes and subtypes in cattle in Italy between 1995 and 2013. Biomed Res Int. (2014) 2014:147145. doi: 10.1155/2014/147145
36. Silveira S, Cibulski SP, Junqueira DM, Mósena ACS, Weber MN, Mayer FQ, et al. Phylogenetic and evolutionary analysis of HoBi-like pestivirus: insights into origin and dispersal. Transbound Emerg Dis. (2020) 67:tbed.13520. doi: 10.1111/tbed.13520
37. Decaro N, Lucente MS, Mari V, Sciarretta R, Pinto P, Buonavoglia D, et al. Hobi-like pestivirus in aborted bovine fetuses. J Clin Microbiol. (2012) 50:509–12. doi: 10.1128/JCM.05887-11
38. Braun U, Hilbe M, Peterhans E, Schweizer M. Border disease in cattle. Vet J. (2019) 246:12–20. doi: 10.1016/j.tvjl.2019.01.006
39. Dubois E, Russo P, Prigent M, Thiéry R. Genetic characterization of ovine pestiviruses isolated in France, between 1985 and 2006. Vet Microbiol. (2008) 130:69–79. doi: 10.1016/j.vetmic.2008.01.002
40. Luzzago C, Ebranati E, Cabezón O, Fernández-Sirera L, Lavín S, Rosell R, et al. Spatial and temporal phylogeny of border disease virus in pyrenean chamois (Rupicapra p. Pyrenaica). PLoS ONE. (2016) 11:e0168232. doi: 10.1371/journal.pone.0168232
41. Martin C, Duquesne V, Adam G, Belleau E, Gauthier D, Champion JL, et al. Pestiviruses infections at the wild and domestic ruminants interface in the French Southern Alps. Vet Microbiol. (2015) 175:341–8. doi: 10.1016/j.vetmic.2014.11.025
42. Valdazo-González B, Álvarez-Martínez M, Sandvik T. Genetic and antigenic typing of border disease virus isolates in sheep from the Iberian Peninsula. Vet J. (2007) 174:316–24. doi: 10.1016/j.tvjl.2006.10.002
43. Paniagua J, García-Bocanegra I, Arenas-Montes A, Berriatua E, Espunyes J, Carbonero A, et al. Absence of circulation of pestivirus between wild and domestic ruminants in southern Spain. Vet Record. (2016) 178:215. doi: 10.1136/vr.103490
44. Ministère de L'agriculture et de l'alimentation. Publications officielles - Journal officiel - Arrêté du 31 juillet 2019 fixant des mesures de surveillance et de lutte contre la maladie des muqueuses/diarrhée virale bovine (BVD) NOR : AGRG1920523A. J Officiel Républ Franç. (2019). p. 1–3. Available online at: https://www.legifrance.gouv.fr/download/pdf?id=UxVuUK_SInm5dKwr6bbw08eXG16_Zzx4qTH8vEQiEuo= (accessed Jun 24, 2022).
45. Agence nationale de sécurité sanitaire de l'alimentation, de l'environnement et du travail, Anses - Index des Médicaments vétérinaires autorisés en France. Available online at: http://www.ircp.anmv.anses.fr/results.aspx (accessed November 22, 2021).
46. Alpay G, Yeşilbag K. Serological relationships among subgroups in bovine viral diarrhea virus genotype 1 (BVDV-1). Vet Microbiol. (2015) 175:1–6. doi: 10.1016/j.vetmic.2014.10.034
47. Ridpath JF, Fulton RW, Kirkland PD, Neill JD. Prevalence and antigenic differences observed between bovine viral diarrhea virus subgenotypes isolated from cattle in Australia and feedlots in the Southwestern United States. J Vet Diagn Investig. (2010) 22:184–91. doi: 10.1177/104063871002200203
48. Sozzi E, Righi C, Boldini M, Bazzucchi M, Pezzoni G, Gradassi M, et al. Cross-reactivity antibody response after vaccination with modified live and killed bovine viral diarrhoea virus (BVD) vaccines. Vaccines (Basel). (2020) 8:1–10. doi: 10.3390/vaccines8030374
49. Mosena ACS, Falkenberg SM, Ma H, Casas E, Dassanayake RP, Booth R, et al. Use of multivariate analysis to evaluate antigenic relationships between US BVDV vaccine strains and non-US genetically divergent isolates. J Virol Methods. (2022) 299:114328. doi: 10.1016/j.jviromet.2021.114328
50. Leyh RD, Fulton RW, Stegner JE, Goodyear MD, Witte SB, Lucas P, et al. Fetal protection in heifers vaccinated with a modified-live virus vaccine containing bovine viral diarrhea virus subtypes 1a and 2a and exposed during gestation to cattle persistently infected with bovine viral diarrhea virus subtype 1b. Am J Vet Res. (2011) 72:367–75. doi: 10.2460/ajvr.72.3.367
51. Walz PH, Riddell KP, Newcomer BW, Neill JD, Falkenberg SM, Cortese VS, et al. Comparison of reproductive protection against bovine viral diarrhea virus provided by multivalent viral vaccines containing inactivated fractions of bovine viral diarrhea virus 1 and 2. Vaccine. (2018) 36:3853–60. doi: 10.1016/j.vaccine.2018.04.005
52. Mosena ACS, Falkenberg SM, Ma H, Casas E, Dassanayake RP, Walz PH, et al. Multivariate analysis as a method to evaluate antigenic relationships between BVDV vaccine and field strains. Vaccine. (2020) 38:5764–72. doi: 10.1016/j.vaccine.2020.07.010
53. Young NJ, Thomas CJ, Thompson I, Collins ME, Brownlie J. Immune responses to non-structural protein 3 (NS3) of bovine viral diarrhoea virus (BVDV) in NS3 DNA vaccinated and naturally infected cattle. Prev Vet Med. (2005) 72:115–20. doi: 10.1016/j.prevetmed.2005.08.013
54. Endsley JJ, Ridpath JF, Neill JD, Sandbulte MR, Roth JA. Induction of T lymphocytes specific for bovine viral diarrhea virus in calves with maternal antibody. Viral Immunol. (2004) 17:13–23. doi: 10.1089/088282404322875421
55. Endsley JJ, Roth JA, Ridpath J, Neill J. Maternal antibody blocks humoral but not T cell responses to BVDV. Biologicals. (2003) 31:123–5. doi: 10.1016/S1045-1056(03)00027-7
56. Sangewar N, Waghela SD, Yao J, Sang H, Bray J, Mwangi W. Novel potent IFN-γ-inducing CD8 + T cell epitopes conserved among diverse bovine viral diarrhea virus strains. J Immunol. (2021) 206:1709–18. doi: 10.4049/jimmunol.2001424
57. Riitho V, Strong R, Larska M, Graham SP, Steinbach F. Bovine pestivirus heterogeneity and its potential impact on vaccination and diagnosis. Viruses. (2020) 12:1134. doi: 10.3390/v12101134
58. Meyer G, Deplanche M, Roux D, Moulignie M, Picard-Hagen N, Lyazrhi F, et al. Fetal protection against bovine viral diarrhoea type 1 virus infection after one administration of a live-attenuated vaccine. Vet J. (2012) 192:242–5. doi: 10.1016/j.tvjl.2011.05.011
59. Newcomer BW, Chamorro MF, Walz PH. Vaccination of cattle against bovine viral diarrhea virus. Vet Microbiol. (2017) 206:78–83. doi: 10.1016/j.vetmic.2017.04.003
60. Nardelli S, Decaro N, Belfanti I, Lucente MS, Giammarioli M, Mion M, et al. Do modified live virus vaccines against bovine viral diarrhea induce fetal cross-protection against HoBi-like Pestivirus? Vet Microbiol. (2021) 260:109178. doi: 10.1016/j.vetmic.2021.109178
61. Antos A, Miroslaw P, Rola J, Polak MP. Vaccination failure in eradication and control programs for bovine viral diarrhea infection. Front Vet Sci. (2021) 8:688911. doi: 10.3389/fvets.2021.688911
62. Klimowicz-Bodys MD, Polak MP, Płoneczka-Janeczko K, Bagnicka E, Zbroja D, Rypuła K. Lack of fetal protection against bovine viral diarrhea virus in a vaccinated heifer. Viruses. (2022) 14:311. doi: 10.3390/v14020311
Keywords: Bovine Viral Diarrhea Virus, cattle, genetic diversity, genotype, France
Citation: Rivas J, Hasanaj A, Deblon C, Gisbert P and Garigliany M-M (2022) Genetic diversity of Bovine Viral Diarrhea Virus in cattle in France between 2018 and 2020. Front. Vet. Sci. 9:1028866. doi: 10.3389/fvets.2022.1028866
Received: 26 August 2022; Accepted: 23 September 2022;
Published: 11 October 2022.
Edited by:
Guillermo Roberto Risatti, University of Connecticut, United StatesReviewed by:
Christina Topliff, University of Nebraska-Lincoln, United StatesCopyright © 2022 Rivas, Hasanaj, Deblon, Gisbert and Garigliany. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mutien-Marie Garigliany, bW1nYXJpZ2xpYW55QHVsaWVnZS5iZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.