
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 24 January 2023
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1026470
This article is part of the Research TopicRespiratory Diseases and Management in LivestockView all 7 articles
Introduction: Bacterial bronchopneumonia (BP) has been associated with purchasing cattle through auction markets. However, whether auction markets are a source of BP-associated bacterial pathogens is unknown. This study evaluated prevalence, antimicrobial susceptibility, and genetic relatedness (using pulsed-field gel electrophoresis, PFGE) of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from cattle either transported to an auction market prior to feedlot placement (AUC), or directly to a feedlot from a farm (RANC).
Methods: Two groups of cattle were enrolled (N = 30 per group) from two separate farms with 15 animals from an individual farm designated as AUC or RANC. Deep nasal swab (DNS) and trans-tracheal aspirates (TTA) were collected on day 0 at weaning (T0) and on day 2 at on-arrival processing at the feedlot (T1). The DNS were also collected on day 9 (T2) and day 30 (T3) after arrival at the feedlot.
Results and discussion: In both TTA and DNS, prevalence of bacteria did not differ between AUC and RANC groups (P > 0.05). None of the bacteria isolated at T0 were resistant to antimicrobials and diversity of all bacteria was greatest at T0 and T1. In Group 1 cattle, 100% of P. multocida isolated at T2 and T3 were multi-drug resistant. These isolates were highly related (>90%) according to PFGE, with most being clones. Though limited in size, results for animals evaluated in this study suggested that auction markets were not a major source of resistant BP pathogens, however, horizontal transmission of a multi-resistant strain of P. multocida occurred in a feedlot. Spread of resistant P. multocida was likely due to the selective pressures imposed by feedlot antimicrobial use and encoded resistance by the bacteria.
Bacterial bronchopneumonia (BP) remains a challenging health issue facing the North American feedlot industry, causing significant economic losses [over US $500 million annually (1)]. It affects 16.2% of cattle placed in North American feedlots, with ~4% of feedlot cattle dying from this condition (2). The main bacterial pathogens associated with BP are Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, and Mycoplasma bovis. Although BP is ultimately caused by bacterial pathogens, it is considered a multifactorial disease triggered by a combination of predisposing factors (3, 4). Bacterial pathogens causing BP typically reside in the upper respiratory tract of healthy cattle as commensals (5). However, under specific conditions stressors can suppress host immunity and facilitate proliferation of bacterial pathogens in the upper respiratory tract (URT), followed by colonization and potential infection of the lower respiratory tract (LRT) (6, 7). This explains why beef cattle mostly develop BP during the first 50 days after feedlot placement, as they are exposed to various respiratory viruses (bovine herpes virus 1, bovine respiratory syncytial virus, bovine para-influenza 3 virus, bovine viral diarrhea virus, etc.) at a time when stressors [such as weaning, transportation, co-mingling, adaptation to new diet, surgical procedures (e.g., castration and dehorning)] negatively affect their immune defenses (6, 8).
Trading cattle through auction markets is part of an efficient marketing system in the North American feedlot industry and represents 60% of cattle purchases in Western Canada (9). There is evidence to suggest that cattle purchased through auction markets are at higher risk of developing BP early in the feeding period than cattle coming directly from other beef operations (6). For example, Step et al. (10) reported 42% morbidity related to BP during the first 42 days after arrival in feedlot cattle purchased through auction markets vs. only 11% BP morbidity in ranch-direct cattle. The reasons for this are unclear but may partially be related to auction market cattle being exposed to a greater density of pathogens due to co-mingling and thus an increased risk of pathogen transmission and colonization (11). Furthermore, auction market cattle are exposed to additional stressors (e.g., handling, transportation, co-mingling, and dehydration) compared to cattle directly sourced from ranches.
In addition to stress and pathogen transmission, administration of antimicrobials may alter the bacterial communities of the respiratory tract by reducing prevalence of susceptible bacteria. In contrast, colonization by resistant bacteria could potentially alter the prevalence and diversity of pathogens in the respiratory tract, with resistant strains outcompeting those that are susceptible to the antimicrobial administered. More recently, we reported high levels of resistance against tulathromycin and oxytetracycline in M. haemolytica and P. multocida (64.1 and 66.7%, respectively) isolated from feedlot cattle (12). In that study, it was not possible to evaluate the source of resistant bacteria as cattle were sampled at a single time point. While a subsequent study showed lower levels of antimicrobial-resistant BRD pathogens when calves were sampled on farms, compared to feedlot (13), there is still a paucity of information on the prevalence and resistance in BP-associated bacterial pathogens in cattle prior to feedlot arrival. Thus, whether farms or auction markets could potentially be a source of resistant bacteria, is currently unknown. The objective of this study was therefore to compare the prevalence and resistance of BP bacterial pathogens in beef cattle that were sent either directly to a feedlot or co-mingled at an auction market before transportation to a feedlot.
This study was performed in strict accordance with the recommendations of the Canadian Council of Animal Care (14). All procedures were reviewed and approved by the University of Calgary Veterinary Sciences Animal Care Committee (Protocol AC14-0192).
Two groups of 30 angus-crossed beef heifers (n = 60) sourced from two cow-calf ranches were enrolled in the study and sampled from weaning to 28 days after entrance into a feedlot. The feedlots of each group of animals also differed, with Group 1 calves being placed in a high-capacity feedlot housing ~4,000 cattle (Feedlot-1) and Group 2 calves being placed in a low-capacity feedlot housing 600 cattle (Feedlot-2) (Figure 1).
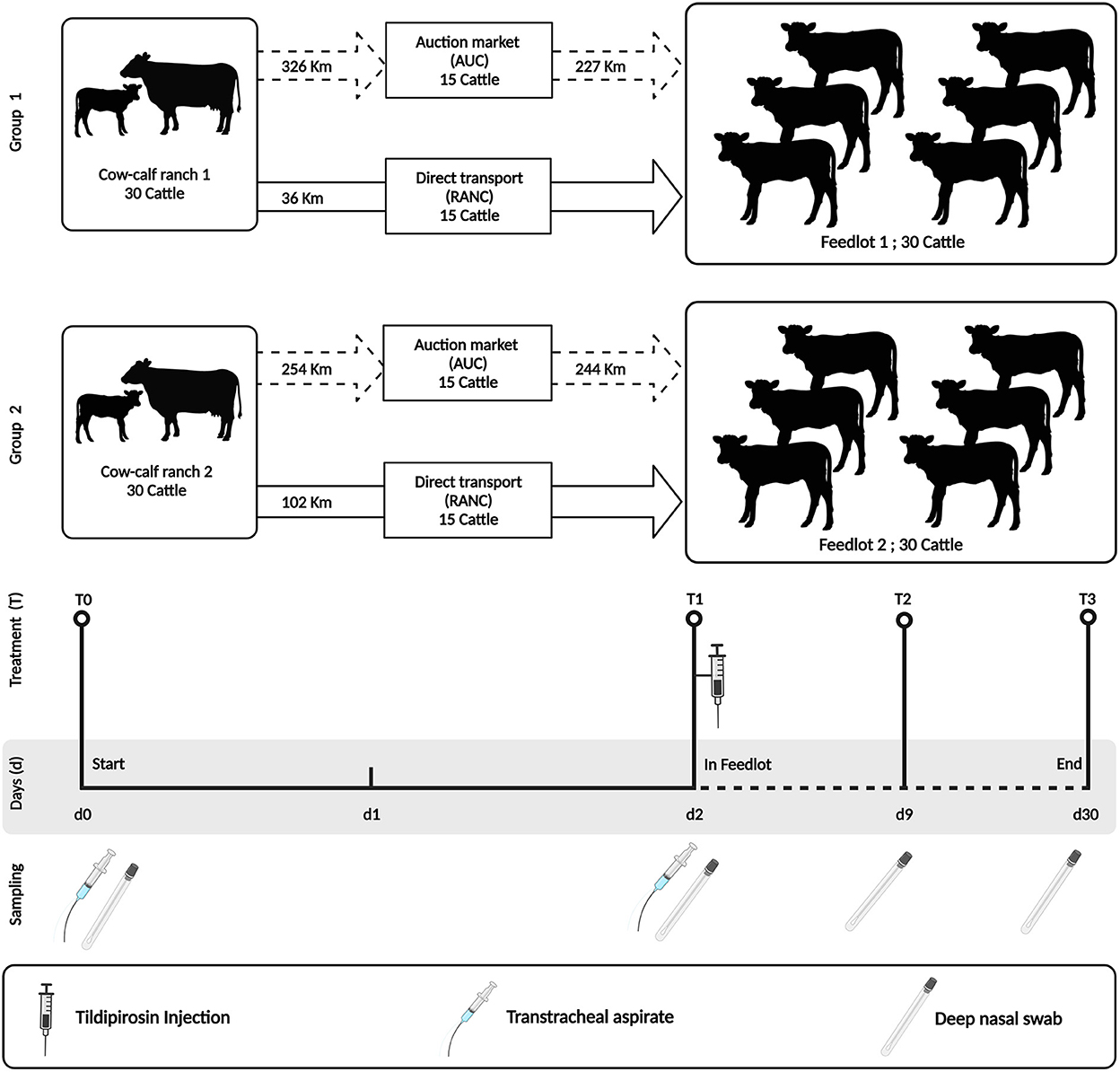
Figure 1. Study design indicating sampling time points and antimicrobial treatment. The study was performed using two groups of cattle (N = 30 per group). Group 1 cattle were placed in a high-capacity feedlot housing ~4,000 cattle (Feedlot-1). Group 2 cattle were placed in a low-capacity feedlot housing 600 cattle (Feedlot-2). Both groups were injected with tildipirosin at feedlot entry. All cattle were sampled similarly by deep nasal swab (DNS) and/or trans tracheal aspiration (TTA) at the indicated time points (T0–T3). This figure is created with BioRender.com.
At weaning on the cow-calf ranches (day 0 = T0), heifers were separated from their dams and their upper and lower respiratory tracts were sampled using a guarded deep nasopharyngeal swab (DNS) and trans-tracheal aspiration (TTA) kit, respectively. Thereafter, heifers were randomized into two groups of 15 animals and were transported either to a commercial auction market (AUCT) over a distance of 326 km (Group 1) and 254 km (Group 2), respectively. Heifers were kept at the auction market for 24 h in contact with other cattle, and then transported to respective feedlots over a distance of 227 km (Group 1) and 244 km (Group 2) on day 1. The other groups of 15 heifers were transported directly (RANC) from the cow-calf ranch to the feedlot over a distance of 36 km (Group 1) and 102 km (Group 2), respectively. During all steps of transportation, the heifers were not co-mingled with other cattle on the truck.
On day 2 (T1), all heifers (RANC and AUCT) were re-sampled by DNS and TTA. They also received subcutaneous injections of the macrolide tildipirosin (4 mg/kg, Zuprevo, Merck animal health, Kirkland, QC, Canada), vaccines against bovine rhinotracheitis virus, bovine viral diarrhea virus (types 1 and 2), bovine parainfluenza 3 virus, bovine respiratory syncytial virus, and Mannheimia haemolytica toxoid (Pyramid FP 5 + Presponse SQ, Boehringer Ingelheim, Burlington, ON, Canada), and clostridial disease and Histophilus somni (Vision 8 somnus, Merck animal health, Kirkland, QC, Canada) and were dewormed using topical moxidectin (Cydectin, Boehringer Ingelheim, Burlington, ON, Canada). Group 1 received two pulses of chlortetracycline at a dosage of 5 g per cattle per day from days 1–5 and days 8–12 (Aureomycin 220, Zoetis, Kirkland, QC, Canada), whereas Group 2 did not receive in-feed chlortetracycline.
Deep nasal swabs and TTA were also collected on day 9 (T2) and day 30 (T3). Heifers with visual BP signs (e.g., depression, nasal or ocular discharge, cough or dyspnea) and a rectal temperature ≥ 40.0°C were defined as a BP case and treated with enrofloxacin (12.5 mg/kg, Baytril 100, Bayer Inc., Mississauga, ON, Canada).
Deep nasal swab (DNS) and trans-tracheal aspirates (TTA) were collected at weaning (T0) and at on-arrival processing at the feedlot (T1) (Figure 1). Deep nasal swab sampling was then repeated on days 9 and 30 after arrival at the feedlot (T2 and T3, respectively).
Deep nasal swabs were collected as described (11) using long guarded swabs (27 cm) with a rayon bud (MW 124, Medical Wire & Equipment, Corsham, UK). Trans-tracheal aspirates were collected as described (7) using a 75-cm long catheter with an outside diameter of 2 mm (Centracath, Vygon, Ecouen, France). Immediately after sampling, deep nasal swabs were placed into sterile liquid Amies and refrigerated at 4°C. Fluid recovered from TTA (on average, 5–10 mL) was transferred into sterile plain tubes and also refrigerated at 4°C. Both DNS and TTA samples were shipped overnight to the Lethbridge Research and Development Center, Agriculture and Agri-Food Canada (AAFC) and processed within 12 h after arrival.
Upon receipt, the nasal swabs were removed from the transport medium and placed individually into 250 μl of brain heart infusion (BHI) broth and glycerol (60:40). For TTA, 250 μl of a sample was suspended in 250 μl of brain heart infusion (BHI) broth and glycerol (60:40). For bacterial isolation, the samples were vortexed and a 100-μl aliquot of sample suspension was used for culturing. The nasal swabs and the remaining amount of the TTA suspension were stored at −80°C.
For M. haemolytica and P. multocida, a 100-μL aliquot of the DNS and TTA suspensions were plated onto tryptic soy agar (TSA) containing 5% sheep blood and 15 μg/mL of bacitracin [to limit the growth of Gram-positive bacteria; (15)] and were incubated for 24 h at 37°C. For culturing of H. somni, 100 μL of the DNS and TTA suspension was plated onto TSA-blood plates without bacitracin and incubated for 48 h in a 10% CO2-enriched environment at 37°C. Up to three colonies from a plate displaying morphology indicative of M. haemolytica (white-gray, round, medium-sized, non-mucoid, exhibiting β-haemolysis), P. multocida (translucent, grayish in color, and mucoid in consistency), and H. somni (yellowish hue, haemolytic) were confirmed by PCR, as described previously (16). Positive isolates of M. haemolytica, P. multocida, and H. somni were then stored at −80°C for further characterization.
For each bacterial species, one randomly selected isolate (when more than one isolate was banked) for nasal and tracheal samples from a single animal was analyzed to determine antimicrobial susceptibility. Antimicrobial susceptibility testing was performed by microdilution (Sensititre, Thermofisher Scientific, Nepean, ON, Canada) using a commercially available panel (BOPO6F custom bovine plates, TREK diagnostic systems, Cleveland, OH, USA) as described (17). Plates were automatically read using Vizion Digital MIC viewing (TREK diagnostics). Antimicrobials, range of concentrations tested, and minimum inhibitory concentrations (MIC) are listed in Supplementary Table S1 for M. haemolytica, Supplementary Table S2 for P. multocida, and Supplementary Table S3 for H. somni. Isolates were defined as resistant to antimicrobials according to MIC defined by the Clinical and Laboratory Standards Institute for ampicillin (≥0.25 μg/mL), ceftiofur (≥8 μg/mL), enrofloxacin (≥2 μg/mL), florfenicol (≥8 μg/mL), penicillin (≥1 μg/mL), oxytetracycline (≥8 μg/mL), tilmicosin (≥32 μg/mL, for M. haemolytica), and tulathromycin (≥64 μg/mL), [VET01S-Ed5, (18)]. CLSI breakpoints were not available for clindamycin, chlortetracycline, danofloxacin (for H. somni), gentamycin, tiamulin, tilmicosin (for H. somni), trimethoprim/sulfamethoxazole, tylosin, and neomycin. Therefore, susceptibility designations were not assigned for these compounds, with the exception of neomycin. For neomycin, resistance was defined as an MIC ≥ 32 μg/ml according to Klima et al. (17). Reference strains Staphylococcus aureus ATCC 29213, Escherichia coli ATCC 25922, and Enterococcus faecalis ATCC 29212 served as quality controls.
Isolates were typed by PFGE according to the standardized protocol for Molecular subtyping from Pulsenet (19) and previously described (20) using a CHEF DR II electrophoresis unit (Bio-Rad Laboratories, Mississauga, ON). Restriction digestions were completed using SalI for M. haemolytica, ApaI for P. multocida, and SacII for H. somni. Electrophoresis gels were run at 6 V/cm and electrophoresis conditions were a 4.0 s initial switch time and 40 s final switch time for 20 h at 12°C. Resulting patterns were analyzed using Bionumerics Version 7.1 software (Applied Maths Inc., Austin, TX). Dendrogram and minimum spanning tree (MST) analysis were based on similarity matrices generated from UPGMA clustering of Dice coefficient values with 1.0% tolerance and 0.5% optimization. Bin size for MST was set to 3.0% and complexes were generated with a maximum neighbor distance of 2. Isolates with ≥90% similarity according to BioNumerics analysis were grouped as a pulsotype.
Isolation rates of pathogens from DNS and TTA sample types between groups (AUC and RANC) at T0 were compared using chi-square tests in Microsoft Excel®. Effects of auction market on the isolation rates of pathogens from DNS sample types were assessed using a mixed effects logistic regression model, performed in R (R Core Team, Vienna, Austria). Variables included in this model were AUC and RANC cattle and different time points (T1, 2, 3). Deep nasal swabs and TTA were used as the experimental unit. Effects of auction market on the isolation rates of pathogens from TTA sample types were compared using a chi-square test in Microsoft Excel®. RANC and AUC samples were analyzed separately at each time point and in each group. For all analyses, a P-value of 0.05 was considered significant.
A schematic of the study design is shown in Figure 1. Across all time points and respiratory sample types, P. multocida was the most frequently isolated pathogen from RANC and AUC cattle in Group 1 (62.2%) and Group 2 (40.0%) compared to M. haemolytica [Group 1 (5.0%), Group 2 (12.5%)] or H. somni [Group 1 (16.0%), Group 2 (15.8%)]. No difference in prevalence of pathogens isolated from DNS (P > 0.05) or TTA (P > 0.05) was observed between AUC and RANC at T0 in both groups. Isolation rates of P. multocida (P > 0.05), M. haemolytica (P > 0.05), and H. somni (P > 0.05) from DNS samples did not differ between AUC and RANC at T1, T2, and T3 in both groups (Figure 2). Similarly, isolation rates of P. multocida (P > 0.05), M. haemolytica (P > 0.05), and H. somni (P > 0.05) from TTA samples did not differ between the AUC and RANC groups at T0 and T1 in both groups (Figure 3). Prevalence of P. multocida, M. haemolytica and H. somni isolated from TTA samples did not change from the cow-calf ranch to arrival at the feedlot (between T0 and T1) (P > 0.05) in both groups (Figure 3). In Group 2 cattle, the prevalence of P. multocida isolated from DNS decreased in RANC and AUC cattle from T0 (73.3 and 66.7%, respectively) and T1 (73.3 and 80.0%, respectively) to time point T2 (0%) and T3 (0%) (Figure 2).
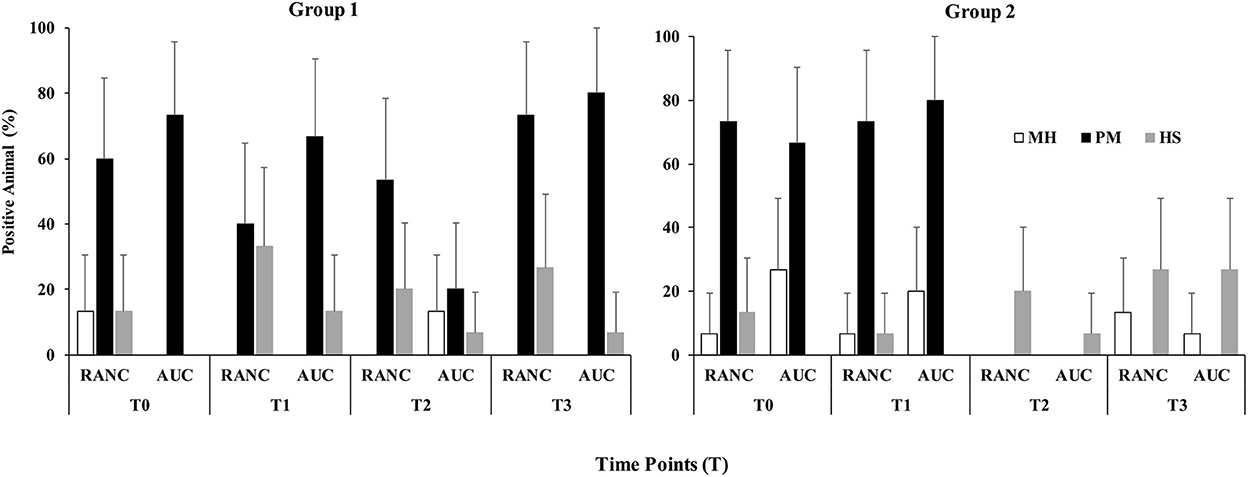
Figure 2. Prevalence of M. haemolytica (MH), P. multocida (PM), and H. somni (HS) in DNS samples from auction-derived (AUC) and ranch-direct (RANC) cattle for Group 1 and Group 2. Isolation rates of M. haemolytica (P > 0.05), P. multocida (P > 0.05), and H. somni (P > 0.05) did not differ between AUC and RANC at T1, T2, and T3 in both feedlots. The error bars represent 95% confidence interval.
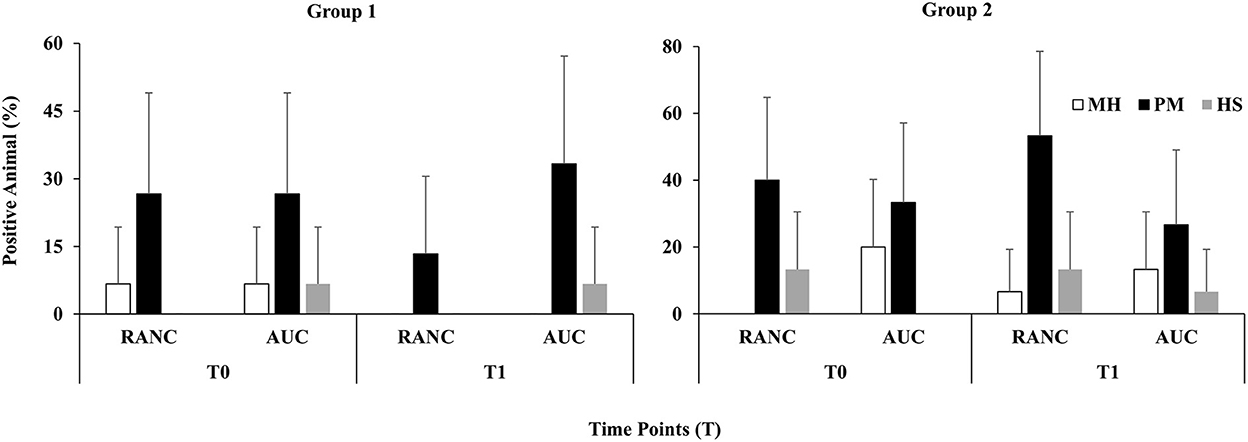
Figure 3. Prevalence of M. haemolytica (MH), P. multocida (PM), and H. somni (HS) in TTA samples from auction-derived (AUC) and ranch-direct (RANC) cattle for Group 1 and Group 2. Isolation rates of M. haemolytica (P > 0.05), P. multocida (P > 0.05), and H. somni (P > 0.05) did not differ between AUC and RANC at T0 and T1 in both feedlots. The error bars represent 95% confidence interval.
Genetic diversity of M. haemolytica, P. multocida, and H. somni did not appear to relate to whether cattle were direct or auction-derived, as most pulsogroups were identified in isolates from AUC and RANC cattle (Table 1). In addition, there were no strains that appeared specific to only the trachea, as most LRT isolates were from a pulsogroup that also included bacteria isolated from the URT. Three pulsogroups were observed for M. haemolytica and two for H. somni in Group 2 calves whereas one pulsogroup was observed for M. haemolytica and two for H. somni in Group 1 calves. The same pulsotype of H. somni was isolated from one animal in the AUC group and three cattle in the RANC group at T3 in Group 1. A total of 13 M. haemolytica (Group 1, N = 5; Group 2, N = 8) and 9 H. somni (Group 1, N = 5; Group 2, N = 4) were unique and did not share 90% genetic relatedness to any other isolates.
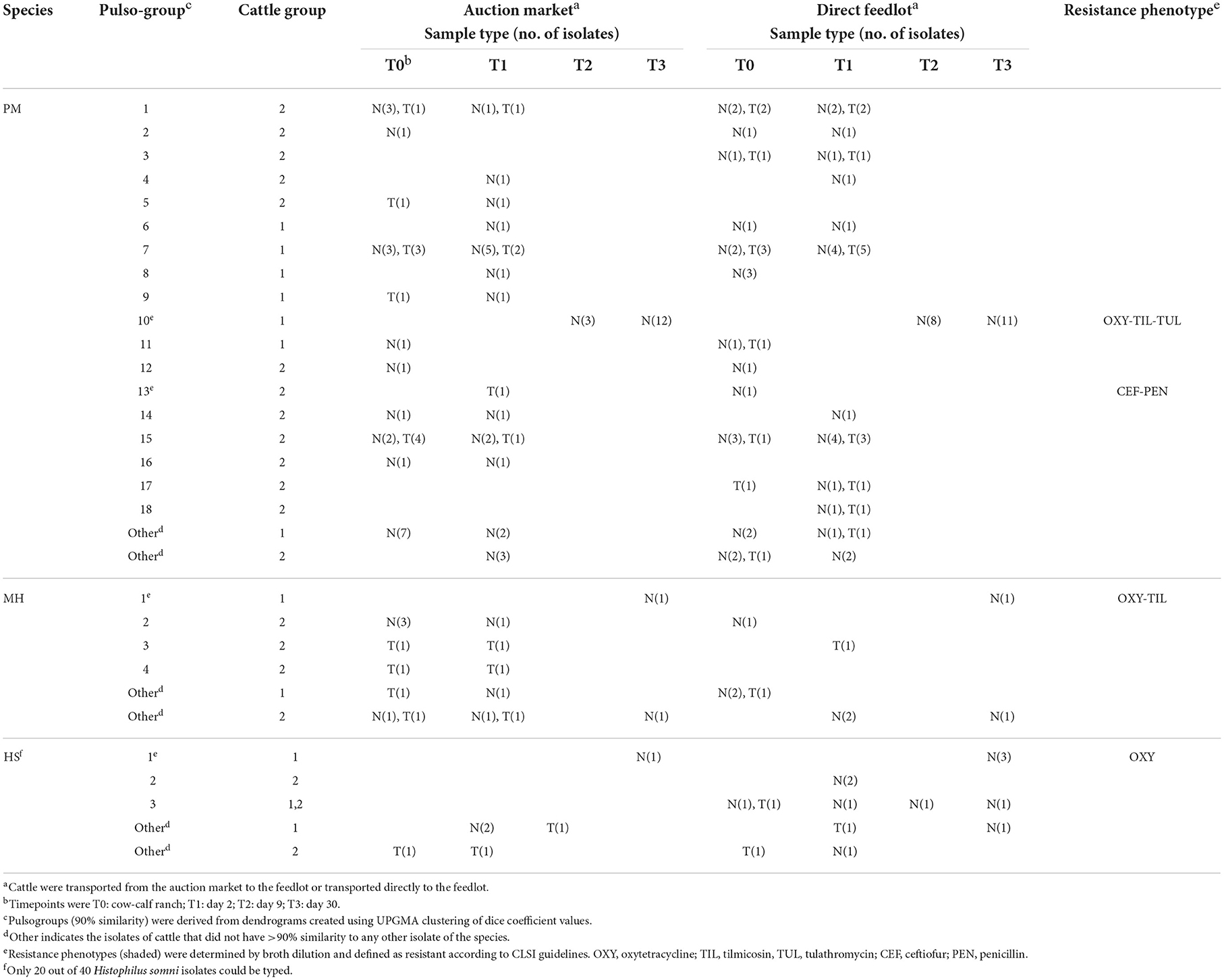
Table 1. Characterization of Pasteurella multocida (PM), Mannheimia haemolytica (MH), and Histophilus somni (HS) isolates from cattle sampled by deep nasal swab (N) or trans-tracheal aspiration (T).
A total of 18 pulsogroups (Group 1, N = 6; Group 2, N = 12) were observed for P. multocida, whereas 21 isolates were unique and did not share 90% genetic relatedness with other strains (Group 1, N = 13; Group 2, N = 8). Pulsogroup 10 was most frequently isolated (N = 34) and all bacteria within this group were from time points T2 and T3 in Group 1, whereas no other pulsogroups were observed after T1. Pasteurella multocida was not isolated from cattle in Group 2 after T1. Thus, high genetic diversity with up to 12 pulsogroups in Group 2 and six in Group 1 was observed prior to feedlot placement and at arrival at the feedlot in both auction-derived and ranch-direct cattle. The level of genetic diversity decreased with days on feed at the feedlot, as illustrated by a reduction in pulsogroups after T1. In support of this, when pulsotypes were analyzed using a similarity matrix and presented as a minimum spanning tree, it was clear that only one dominant clade was present at time points T2 and T3, whereas multiple clades were observed in Group 1 and 2 animals at T0 and T1 (Figure 4). The genetic relatedness of P. multocida pulsogroup 10 was high, with most isolates in this group having a common PFGE restriction pattern and indicating a clonal origin (100% related; Figure 5).
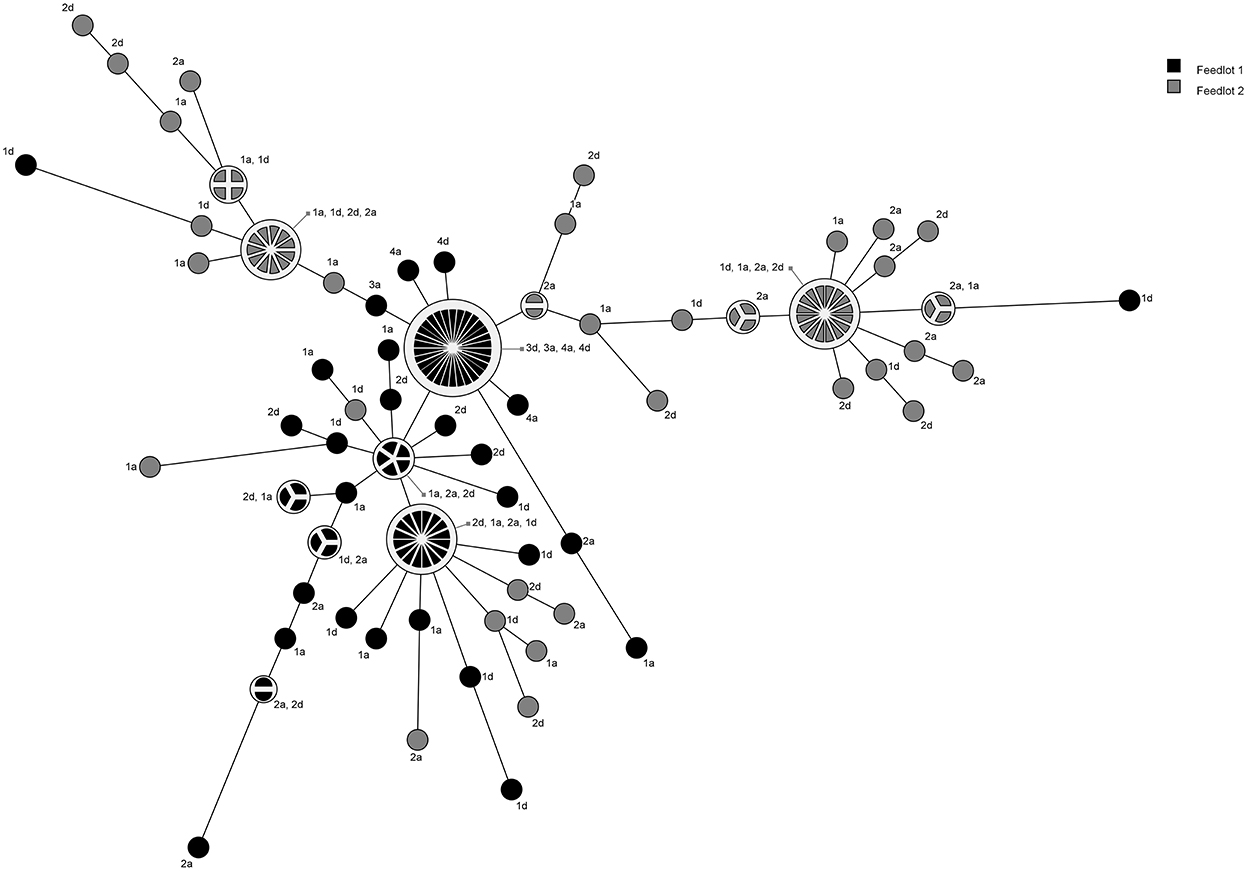
Figure 4. Minimum spanning tree based on a similarity matrix generated from pulsed-field gel elctrophoresis profiles of P. multocida isolates collected from cattle in Groups 1 and 2. Distance between isolates indicates the similarity. Shading indicates group of animals the bacteria were isolated from (black = Group 1, gray = Group 2). Numbers refer to sampling times (1 = day 0, 2 = day 2, 3 = day 9, 4 = day 30). Letters represent the treatment (D, direct transport; A, auction).
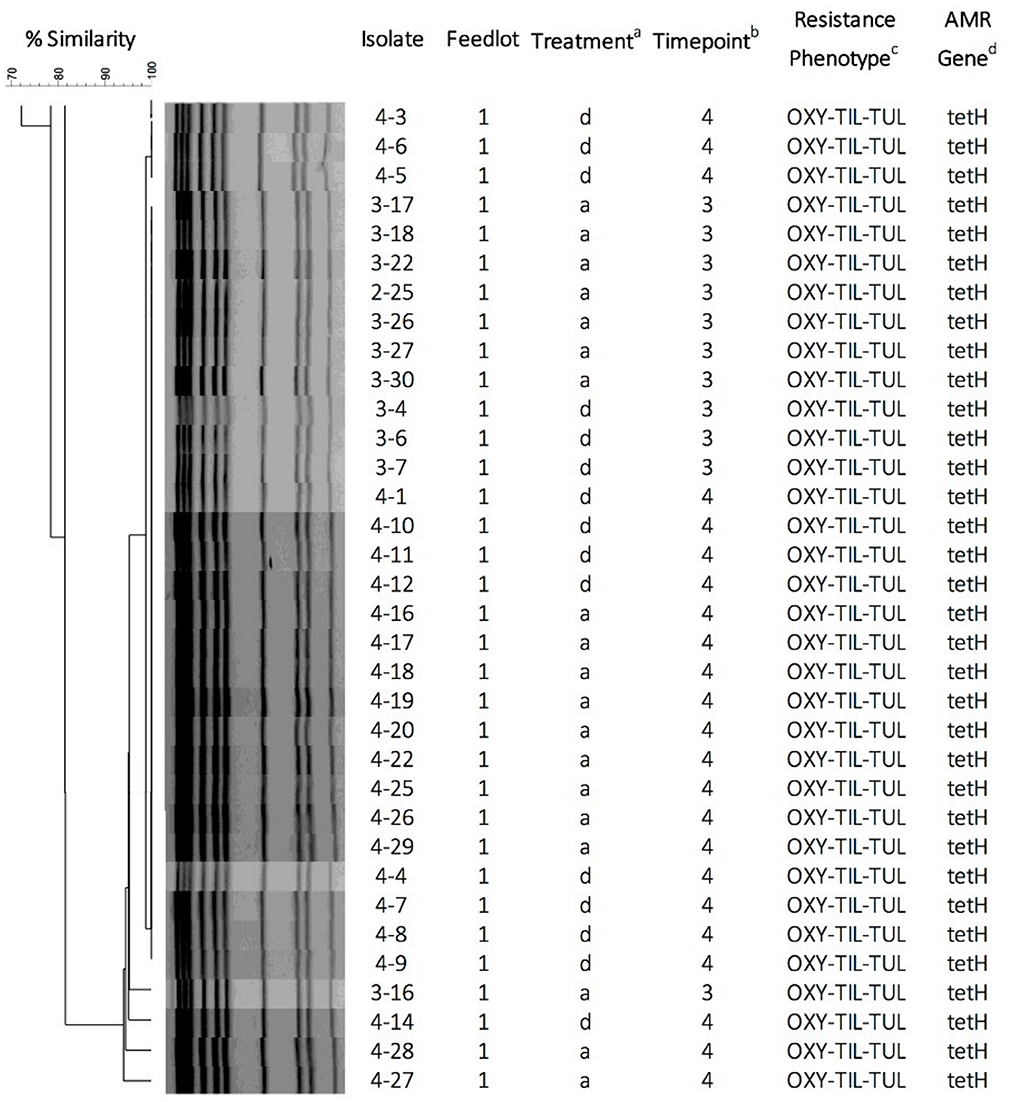
Figure 5. Pulsed-field gel electrophoresis dendrogram, cluster analysis, and antimicrobial susceptibility for 34 P. multocida isolates representing pulsogroup 10 (90% similarity threshold cutoff value). P. multocida were digested with ApaI. The dendrogram was created by UPGMA cluster analysis using Dice coefficients of similarity with optimization and tolerance settings of 0.5 and 1.0%, respectively. aTreatment: d, calves directly transported from cow-calf ranch to feedlot; a, calves transported to and co-mingled at auction markets before placement in a feedlot; bSampling timepoints: 3 = day 9; 4 = day 30; cPhenotypic resistance against antimicrobials OXY (oxytetracycline), TIL (tilmicosin), and TUL (tulathromycin). dAntimicrobial resistance genes detected by PCR.
None of the bacteria isolated at time points T0 and T1 were resistant to any of the antimicrobials tested (Table 1). Only one pulsogroup of M. haemolytica, containing two isolates, were resistant to oxytetracycline and tilmicosin. These isolates were from Group 1 AUC and RANC cattle and were recovered at T3. Similarly, only one pulsogroup of H. somni, containing four isolates, was resistant to oxytetracycline. These H. somni were also recovered from both AUC and RANC cattle in Group 1, at T3. Two isolates of P. multocida (pulsogroup 13) isolated from Group 2 cattle were resistant to ceftiofur and penicillin and were recovered at T0 and T1 from RANC and AUC cattle, respectively. All other isolates of P. multocida from T2 and T3 (Group 1 cattle) were within pulsogroup 10 and shared an oxytetracycline-tilmicosin-tulathromycin multidrug resistant phenotype. Pulsogroup 10 P. multocida isolates (N = 34) were cultured from 24 cattle in Group 1. An attempt was made to screen for genes conferring resistance to oxytetracycline [tet(H)] and macrolides [msr(E), mph(E), erm(42), erm(A), erm(B), erm(C), erm(F), erm(T) and erm(X)] in resistant isolates. All bacteria displaying oxytetracycline resistance harbored tet(H), whereas none of the macrolide resistance genes were detected in any of the macrolide-resistant isolates.
Nine of the 30 heifers (30%) in Group 1 were diagnosed and treated for BP at d 9; five heifers within RANC group and four heifers within AUC cattle. In Group 2, three heifers (10%) were diagnosed with BP and treated; one within RANC at d 2 and two within AUC cattle at d 2 and 8 of the study. No other diseases were observed.
No differences were observed between heifers that remained healthy and those diagnosed with BP in pathogen isolation rates of nasopharyngeal samples on any sampling day.
The hypothesis that auction market cattle are at high risk for BP at entrance into feedlots because they acquire pathogens during the process of co-mingling at auction markets was not supported in the present study. While some new strains for each of the pathogens were identified at T1 (feedlot entry), this was the case for both AUC and RANC cattle, and none of these strains persisted to T2 and T3 (Table 1). Thus, specific instances of a strain presenting in AUC cattle after co-mingling was not observed. While caution should be used in interpreting these data, due to the small sample size, this study suggests that previous reports indicating greater susceptibility to BP in auction market cattle may have been due to a multitude of stressors from the marketing process, such as sorting and loading, transportation, and co-mingling of cattle. These procedures result in increased stress periods which compromise the local and systemic immune defenses predisposing cattle to respiratory disease (21). It should be noted that viral agents were not investigated in the present study, and whether transfer occurs during co-mingling at the auction markets could not be determined.
Results of this study demonstrated that high heterogeneity of BP pathogens was present at the first two time points, prior to long-term feedlot placement. The level of genetic diversity decreased by days 9 and 30, and for P. multocida, and no isolates were recovered at the last two time points in Group 2 animals. The decrease was likely due to antimicrobial administration. Although limited information on the active concentrations of antibiotics in the upper respiratory tract of treated cattle exist, oxytetracycline has been shown to achieve therapeutic concentrations in oral fluid (22) and nasal secretions (23) of pigs after intramuscular administration. It is likely that some secretion of tildipirosin and chlortetracycline occurred, reducing the pathogens in the nasopharynx. However, this decline in genetic variation does not correlate with the findings reported by Klima et al. (24) where there was a greater extent of genetic diversity observed among isolates of M. haemolytica at exit of the feedlot. In that study however, cattle were administered metaphylactic doses of oxytetracycline or tulathromycin, and the sampling time was twice as long as our study, perhaps accounting for these differences observed.
Interestingly, none of the bacteria from time points T0 and T1 were resistant to any antibiotics tested. This would suggest that antimicrobial use in feedlots selects for resistant pathogens, though it should be noted that the cow-calf-ranch use of antimicrobials for the two groups of cattle was unknown, and it cannot be ruled out that antimicrobial administration of calves prior to feedlot placement does not also select for resistant bacteria. Except for one isolate of P. multocida from a Group 2 calf (ceftiofur/penicillin-resistant phenotype), all resistant phenotypes observed corresponded to the classes of antimicrobials administered (i.e., tetracyclines and macrolides). It is interesting to note that no other resistant bacteria were isolated from Group 2 cattle. The feedlot that Group 1 calves were placed in was larger but also administered therapeutic doses of chlortetracycline. Thus, it cannot be ascertained as to whether feedlot size or in-feed chlortetracycline resulted in the differences observed between Group 1 and 2 cattle. The fact that a common pulsogroup of P. multocida colonized the majority of AUC and RANC Group 1 cattle after feedlot placement supports that antimicrobial treatment selected for a resistant strain in Group 1. As this strain was not observed prior to time point T2, it is possible that it was endemic to the feedlot. Transmission of P. multocida Type B:2 between buffalo calves was reported before (25). Furthermore, evidence of clonal spread and transmission of M. haemolytica within the feedlot has been previously observed (7, 17, 26). This highlights the importance of monitoring resistance in feedlots and making strong efforts to enlist management practices that do not promote the spread of resistant pathogens to new arrivals or between sick and healthy calves.
The antimicrobial resistant phenotype of the dominant pulsogroup (pulsogroup 10), which was found in Group 1 cattle, conferred macrolide and tetracycline resistance. Although tildipirosin was not available to test on the antimicrobial panel used in our study, it is likely that this antimicrobial can select for resistance to tulathromycin and tilmicosin, which were tested. In a previous longitudinal study (2007–2012) monitoring bacterial resistance, a lack of tulathromycin resistance was observed in cattle sampled at entry and at 60 DOF (27). However, several studies have now shown widespread macrolide resistance in P. multocida and M. haemolytica (28–30), indicating that rates of resistance are increasing. The dissemination of a single clone from 34 AUC and RANC cattle within 3 weeks of sampling indicates a high horizontal transmission rate of resistant isolates within the feedlot. That all of these isolates displayed oxytetracycline and tilmicosin/tulathromycin resistance, indicated that genes conferring their resistance may be harbored on a common element (16). It is interesting that previously identified macrolide resistance genes could not be detected in any of the isolates. This would suggest that an unknown mechanism is conferring macrolide resistance in these isolates.
Pasteurella multocida was identified as the predominant pathogen in the current study, which is consistent with results reported previously (12, 31). However, this finding suggests that this pathogen is emerging at higher frequencies and gaining dominance over M. haemolytica, which was considered the most prevalent pathogen causing BP over the last decades (32, 33). Reasons could be that currently used vaccines are more effective against M. haemolytica or current antimicrobial treatments are more effective for reduction of M. haemolytica (34). It seems that P. multocida is more likely to develop antimicrobial resistance and it is also possible that acquisition of antibiotic resistance genes by genetic drift could promote more resilient populations (35), as was suggested by our study. The antimicrobial usage history (metaphylactic, therapeutic, subtherapeutic) in cattle is likely correlated to newly acquired antimicrobial resistance strains present in feedlots and also likely contributes to reducing bacterial diversity.
Although the sample size was limited, we concluded that for the calves enrolled in this study, transportation to and co-mingling at an auction market for 24 h did not result in acquisition of major bacterial respiratory pathogens (e.g., M. haemolytica, P. multocida, and H. somni). Furthermore, horizontal transmission of a multi-resistant strain of P. multocida among calves can occur in the feedlot. Clonal spread of resistant bacteria was likely due to the selective pressures imposed by feedlot antimicrobial use and encoded resistance by the bacteria.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by University of Calgary Veterinary Sciences Animal Care Committee (Protocol AC14-0192).
CH was involved in study design, sample collection, laboratory work, and wrote the initial manuscript. ET and TA were involved in concept development, study design, and manuscript preparation. MU was involved in figure preparation and assisted with laboratory work. LG contributed in figure and manuscript preparation. All authors read and approved the final manuscript.
This research was supported by Results Driven Agricultural Research (RDAR; Project #2016R032R).
The authors thank Dr. Mike Jelinski from Veterinary Agri-Health Services Ltd., Airdrie, AB for his help with cattle access.
CH and ET were employed by Ceva Santé Animale.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1026470/full#supplementary-material
1. Miles DG. Overview of the North American beef cattle industry and the incidence of bovine respiratory disease (BRD). Anim Health Res Rev. (2009) 10:101–3. doi: 10.1017/S1466252309990090
2. USDA. Feedlot 2011 Part IV: Health and Health Management on U.S. Feedlots With a Capacity of 1000 or More Head. (2013). Available online at: https://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot2011/Feed2011_dr_PartIV.pdf (accessed January 20, 2022).
3. Duff GC, Galyean ML. Recent advances in management of highly stressed, newly received feedlot cattle. J Anim Sci. (2007) 85:823–40. doi: 10.2527/jas.2006-501
4. Griffin D, Chengappa MM, Kuszak J, McVey DS. Bacterial pathogens of the bovine respiratory disease complex. Vet Clin N Am Food Anim Pract. (2010) 26:381–94. doi: 10.1016/j.cvfa.2010.04.004
5. Confer AW. Update on bacterial pathogenesis in BRD. Anim Health Res Rev. (2009) 10:145–8. doi: 10.1017/S1466252309990193
6. Taylor JD, Fulton RW, Lehenbauer TW, Step DL, Confer AW. The epidemiology of bovine respiratory disease: what is the evidence for predisposing factors? Can Vet J. (2010) 51:1095–102.
7. Timsit E, Christensen H, Bareille N, Seegers H, Bisgaard M, Assié S. Transmission dynamics of Mannheimia haemolytica in newly-received beef bulls at fattening operations. Vet Microbiol. (2013) 161:295–304. doi: 10.1016/j.vetmic.2012.07.044
8. Hodgson PD, Aich P, Stookey J, Popowych Y, Potter A, Babiuk L, et al. Stress significantly increases mortality following a secondary bacterial respiratory infection. J Vet Res. (2012) 43:21. doi: 10.1186/1297-9716-43-21
9. Canfax. Estimate Direct Feeder Sales in Western Canada. Canfax Research Services (2015). Available online at: http://www.canfax.ca/research.aspx (accessed January 20, 2022).
10. Step DL, Krehbiel CR, DePra HA, Cranston JJ, Fulton RW, Kirkpatrick JG, et al. Effects of commingling beef calves from different sources and weaning protocols during a forty-two-day receiving period on performance and bovine respiratory disease. J Anim Sci. (2008) 86:3146–58. doi: 10.2527/jas.2008-0883
11. Timsit E, Holman DB, Hallewell J, Alexander TW. The nasopharyngeal microbiota in feedlot cattle and its role in respiratory health. Anim Front. (2016) 6:44–50. doi: 10.2527/af.2016-0022
12. Timsit E, Hallewell J, Booker C, Tison N, Amat S, Alexander TW. Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet Microbiol. (2017) 208:118–25. doi: 10.1016/j.vetmic.2017.07.013
13. Guo Y, McMullen C, Timsit E, Hallewell J, Orsel K, van der Meer F, et al. Genetic relatedness and antimicrobial resistance in respiratory bacteria from beef calves sampled from spring processing to 40 days after feedlot entry. Vet Microbiol. (2020) 240:108478. doi: 10.1016/j.vetmic.2019.108478
14. Olfert ED, Cross BM, McWilliam AA, Canadian Council on Animal Care. Guide to the Care and Use of Experimental Animals. Vol. 1. 2nd ed. Ottawa, ON: Canadian Council on Animal Care (1993). Available online at: http://www.ccac.ca/en/CCAC_Programs/Guidelines_Policies/GUIDES/ENGLISH/toc_v1.htm (accessed January 20, 2022).
15. Catry B, Decostere A, Schwarz S, Kehrenberg C, de Kruif A, Haesebrouck F. Detection of tetracycline-resistant and susceptible Pasteurellaceae in the nasopharynx of loose group-housed calves. Vet Res Commun. (2006) 30:707–15. doi: 10.1007/s11259-006-3347-8
16. Klima CL, Zaheer R, Cook S, Booker CW, Hendrick S, Alexander TW, et al. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J Clin Microbiol. (2014) 52:438–48. doi: 10.1128/JCM.02485-13
17. Klima CL, Alexander TW, Hendrick S, McAllister TA. Characterization of Mannheimia haemolytica isolated from feedlot cattle that were healthy or treated for bovine respiratory disease. Can J Vet Res. (2014) 78:38–45.
18. CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; Approved Standard. 5th ed. CLSI document VET01S. Wayne, PA: Clinical and Laboratory Standards Institute (2020).
19. Centers for Disease Control and Prevention. Standardized Molecular Subtyping of Food-Borne Bacterial Pathogens by Pulsed-Field Gel Electrophoresis: A Manual. National Center for Infectious Diseases, Atlanta, GA (1996).
20. Klima CL, Alexander TW, Selinger LB, et al. Comparison of repetitive PCR and pulsed-field gel electrophoresis for the genotyping of Mannheimia haemolytica. J Microbiol Methods. (2010) 81:39–47. doi: 10.1016/j.mimet.2010.01.020
21. Ives SE, Richeson JT. Use of antimicrobial metaphylaxis for the control of bovine respiratory disease in high-risk cattle. Vet Clin North Am Food Anim Pract. (2015) 31:341–50. doi: 10.1016/j.cvfa.2015.05.008
22. Gajda A, Jablonski A, Sikorska MG, Posyniak A. Correlation between oral fluid and plasma oxytetracycline concentrations after intramuscular administration in pigs. J Vet Pharmacol Ther. (2017) 40:e39–44. doi: 10.1111/jvp.12413
23. Bimazubute M, Cambier C, Baert K, Vanbelle S, Chiap P, Gustin P. Penetration of oxytetracycline into the nasal secretions and relationship between nasal secretions and plasma oxytetracycline concentrations after oral and intramuscular administration in healthy pigs. J Vet Pharmacol Ther. (2011) 34:176–83. doi: 10.1111/j.1365-2885.2010.01210.x
24. Klima CL, Alexander TW, Read RR, Gow SP, Booker CW, Hannon S, et al. Genetic characterization and antimicrobial susceptibility of Mannheimia haemolytica isolated from the nasopharynx of feedlot cattle. Vet Microbiol. (2011) 149:390–8. doi: 10.1016/j.vetmic.2010.11.018
25. Annas S, Zamri-Saad M, Jesse FFA, Zunita Z. New sites of localisation of Pasteurella multocida B:2 in buffalo surviving experimental haemorrhagic septicaemia. BMC Vet Res. (2014) 10:1–7. doi: 10.1186/1746-6148-10-88
26. Briggs RE, Frank GH, Purdy CW, Zehr ES, Loan RW. Rapid spread of a unique strain of Pasteurella haemolytica serotype 1 among transported calves. Am J Vet Res. (1998) 59:401–5.
27. Alexander TW, Cook S, Klima CL, Topp E, McAllister TA. Susceptibility to tulathromycin in Mannheimia haemolytica isolated from feedlot cattle over a 3-year period. Front Microbiol. (2013) 4:297. doi: 10.3389/fmicb.2013.00297
28. Anholt RM, Klima C, Allan N, Matheson-Bird H, Schatz C, Ajitkumar P, et al. Antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex in Alberta, Canada. Front Vet Sci. (2017) 4:207. doi: 10.3389/fvets.2017.00207
29. Stanford K, Zaheer R, Klima C, McAllister T, Peters D, Niu YD, et al. Antimicrobial resistance in members of the bacterial bovine respiratory disease complex isolated from lung tissue of cattle mortalities managed with or without the use of antimicrobials. Microorganisms. (2020) 8:288. doi: 10.3390/microorganisms8020288
30. Nobrega D, Andres-Lasheras S, Zaheer R, McAllister T, Homerosky E, Anholt RM, et al. Prevalence, risk factors, and antimicrobial resistance profile of respiratory pathogens isolated from suckling beef calves to reprocessing at the feedlot: a longitudinal study. Front Vet Sci. (2021) 8:764701. doi: 10.3389/fvets.2021.764701
31. Capik SF, White BJ, Lubbers BV, Apley MD, DeDonder KD, Larson RL, et al. Comparison of the diagnostic performance of bacterial culture of nasopharyngeal swab and bronchoalveolar lavage fluid samples obtained from calves with bovine respiratory disease. Am J Vet Res. (2017) 78:350–8. doi: 10.2460/ajvr.78.3.350
32. Booker CW, Abutarbush SM, Morley PS. Microbiological and histopathological findings in cases of fatal bovine respiratory disease of feedlot cattle in Western Canada. Can Vet J. (2008) 49:473–81.
33. Fulton RW, Blood KS, Panciera RJ. Lung pathology and infectious agents in fatal feedlot pneumonias and relationship with mortality, disease onset, and treatments. J Vet Diagn Invest. (2009) 21:464–77. doi: 10.1177/104063870902100407
34. Noyes NR, Benedict KM, Gow SP, Booker CW, Hannon SJ, McAllister TA, et al. Mannheimia haemolytica in feedlot cattle: prevalence of recovery and associations with antimicrobial use, resistance, and health outcomes. J Vet Intern Med. (2015) 29:705–13. doi: 10.1111/jvim.12547
Keywords: antimicrobial resistance, auction market, bacterial bronchopneumonia, bovine respiratory disease, cattle, feedlot
Citation: Hirsch C, Timsit E, Uddin MS, Guan LL and Alexander TW (2023) Comparison of pathogenic bacteria in the upper and lower respiratory tracts of cattle either directly transported to a feedlot or co-mingled at auction markets prior to feedlot placement. Front. Vet. Sci. 9:1026470. doi: 10.3389/fvets.2022.1026470
Received: 23 August 2022; Accepted: 29 November 2022;
Published: 24 January 2023.
Edited by:
Barbara Padalino, University of Bologna, ItalyReviewed by:
Chiara Francesca Magistrali, Experimental Institute of Zooprophylaxis of Umbria and Marche (IZSUM), ItalyCopyright © 2023 Christina Hirsch, Edouard Timsit, Le Luo Guan, and His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Trevor W. Alexander and Muhammed Salah Uddin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Trevor W. Alexander, dHJldm9yLmFsZXhhbmRlckBhZ3IuZ2MuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.