
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 17 November 2022
Sec. Veterinary Epidemiology and Economics
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1025282
 Camila Hamond1,2
Camila Hamond1,2 Katherine L. Dirsmith3
Katherine L. Dirsmith3 Karen LeCount1,2
Karen LeCount1,2 Fred V. Soltero3
Fred V. Soltero3 Sarai Rivera-Garcia3
Sarai Rivera-Garcia3 Patrick Camp1
Patrick Camp1 Tammy Anderson1,2
Tammy Anderson1,2 Jessica A. Hicks1
Jessica A. Hicks1 Renee Galloway4
Renee Galloway4 Graham Sutherland4
Graham Sutherland4 Ilana J. Schafer4
Ilana J. Schafer4 Marga G. A. Goris5
Marga G. A. Goris5 Hans van der Linden5
Hans van der Linden5 Tod Stuber1
Tod Stuber1 Darrell O. Bayles6
Darrell O. Bayles6 Linda K. Schlater1,2
Linda K. Schlater1,2 Jarlath E. Nally2,6*
Jarlath E. Nally2,6*Leptospirosis is one of the most common zoonotic diseases in the world and endemic in the Caribbean Islands. Bovine leptospirosis is an important reproductive disease. Globally, cattle are recognized as a reservoir host for L. borgpetersenii serovar Hardjo, which is transmitted via urine, semen, and uterine discharges, and can result in abortion and poor reproductive performance. The dairy industry in Puerto Rico comprises up to 25% of agriculture-related income and is historically the most financially important agricultural commodity on the island. In this study, we report the isolation of two different pathogenic Leptospira species, from two different serogroups, from urine samples collected from dairy cows in Puerto Rico: L. borgpetersenii serogroup Sejroe serovar Hardjo and L. santarosai serogroup Pyrogenes. Recovered isolates were classified using whole-genome sequencing, serotyping with reference antisera and monoclonal antibodies, and immunoblotting. These results demonstrate that dairy herds in Puerto Rico can be concurrently infected with more than one species and serovar of Leptospira, and that bacterin vaccines and serologic diagnostics should account for this when applying intervention and diagnostic strategies.
Leptospirosis is a worldwide zoonotic disease with an estimated 1.03 million cases and 58,900 human deaths annually (1). Leptospirosis is endemic in the Caribbean islands including Jamaica, Martinique, Haiti, Trinidad and Tobago (2, 3), and the U.S. Virgin Islands (4–6). Leptospirosis is increasingly being diagnosed after hurricane events in Central America and Puerto Rico (7, 8). In Puerto Rico, the incidence and prevalence of human leptospirosis is largely underestimated since clinical signs are associated with other febrile diseases, including dengue, malaria, and Zika (8, 9). A total of 93 leptospirosis cases in humans were reported in Puerto Rico in 2019 (10). The most frequent reactive serogroups among human patients were Icterohaemorrhagiae, Autumnalis, Mini, Ballum, Australis, Bataviae, and Canicola (8, 11). Analysis of environmental samples in Puerto Rico has confirmed the presence of multiple and diverse species of leptospires, including a recently identified new pathogenic species that represents a novel serogroup (12, 13).
Bovine leptospirosis can cause abortion, infertility, stillbirths, weak offspring, and decreased milk production. Globally, cattle are recognized as a reservoir host for L. borgpetersenii serovar Hardjo (14). The dairy industry in Puerto Rico comprises up to 25% of agriculture-related income and is historically the most financially important agricultural commodity on the island (15). Livestock farming and associated abattoir workers have occupational risk factors for exposure to Leptospira species from cattle due to shared environments and high levels of animal contact (16, 17). Multiple species, including L. interrogans, L. kirschneri, L. borgpetersenii, L. santarosai, and L. noguchii, are associated with bovine leptospirosis in South America (18–23); L. kirschneri and L. santarosai in Mexico (24) and L. interrogans, L. kirschneri and L. borgpetersenii in the U.S. (25–27).
Though the culture of pathogenic Leptospira from animals is inherently difficult due to their fastidious growth requirements (28), a recovered isolate is essential for accurate epidemiology and the ability to perform comprehensive genome analysis and serotyping (25, 29, 30). This in turn informs better diagnostics and bacterin vaccine strategies. Here, we describe the isolation and characterization of both L. borgpetersenii and L. santarosai from dairy cows in Puerto Rico.
USDA Veterinary Services staff in Puerto Rico operated an approximately year-long tick treatment program in cattle in response to the discovery of a multi-acaricide resistant tick (Rhipicephalus microplus). Response activities included tick treatment of cattle at regular intervals at various dairy farms throughout Puerto Rico. During these treatment visits, opportunistic sampling for leptospirosis was performed in which free catch urine samples were collected on a selected number of animals (N = 49) as time and weather conditions allowed. These samples were then sent to the National Center for Animal Health (NCAH) Leptospira working group in Ames, Iowa, for PCR and fluorescent antibody testing (FAT) as previously described (4, 6).
Out of those animals that yielded PCR-positive urine samples, two to three animals from each farm were selected for urine sample collection to be used for culture of leptospires. In addition, a serum sample was collected for the microscopic agglutination test (MAT). The farm owners were contacted and informed of positive PCR results and were asked to separate the two to three pre-selected, PCR-positive animals from the rest of the herd to facilitate urine sample collection. On the day of sampling, the selected cows were moved into an alley or chute one by one. For each animal sampled, an assistant held the tail up and to the side. A 30 cc syringe was filled with sterile water and was used to thoroughly clean the vulva and surrounding area to remove all visible debris. Clippers were used to remove hair from the vulvar region. The vulvar region was again washed with sterile water. If any debris remained, a sterile alcohol wipe (soaked with 70% EtOH) was used to remove debris, and the wash with sterile water was repeated. The vulvar region was then sprayed with 70% EtOH. This process was often repeated multiple times to clean the area. The sample collector then stimulated the cow to urinate by rubbing the region distal to the vulva. In the other hand, which remained clean, a sterile specimen collection cup was held below the vulva and a first void urine sample was collected, which contained ~200 mL of urine. The sample collector then used a new, sterile specimen collection cup to collect a second void of urine. Then, the sample collector switched gloves and inoculated culture media with each void of urine. For this process, a sterile pipette was used to transfer 1 mL of urine from the first void sample into each of three conical tubes containing 9 mL of HAN transport media. The same process was followed for inoculation of the transport media with the second void urine sample. Any remaining urine from each void was transferred to a 50 mL conical tube, and the lid was sealed with parafilm and submitted for PCR analysis.
Samples were collected from two dairy cow farms in Puerto Rico. The first farm was in Yabucoa and although exact numbers of cattle present at the farm at any given time varied during the year-long tick treatment program, this farm comprised 629 dairy cattle at the end of the treatment program. Additionally, there were 145 beef cattle and 11 horses also housed at this dairy farm at the end of the program. The second farm was in Quebradillas and at the end of the treatment period, comprised 349 dairy cows. Animals at either farm had not been vaccinated against leptospirosis and did not show any clinical signs compatible with leptospirosis as a reproductive problem.
The microscopic agglutination test (MAT) was performed using a panel of 18 antigens representative of 15 serogroups (Supplementary Table 1). A titer was considered positive at ≥1:100 (31).
A 45 mL aliquot of urine was centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was removed, and the resultant pellet was resuspended in 1 mL PBS. The resuspended sample was centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was removed until approximately 150 μL remained and this was resuspended in 1 mL PBS. The pellet was again harvested by centrifugation at 12,000 × g for 10 min at 4°C. The supernatant was removed until ~150 μL remained.
A 10 μL aliquot of the 150 μL that remained was placed on a glass slide within a 7 mm well, in duplicate, and FAT was performed as previously described (27). DNA was extracted from the remaining sample using the Maxwell RSC Purefood Purification Pathogen kit (Promega Corporation, Madison, Wisconsin, USA), following manufacturer's instructions, but using 1 h of incubation with 200 μL lysis buffer A and a 100 μL elution volume. PCR for lipL32 was performed as previously described (25, 32, 33).
A 1 mL aliquot of freshly collected urine was immediately inoculated into 9 mL of transport HAN media which was transported by overnight delivery services at ambient temperature to the National Animal Disease Center, USDA, Ames, Iowa. On arrival, a 200 μL aliquot of inoculated transport HAN media was used to inoculate 5 mL HAN semi-solid and 5 mL T80/40/LH semi-solid media. Inoculated T80/40/LH media was incubated at 29°C and inoculated HAN media was incubated at both 29°C without CO2 as well as 37°C in 3% CO2 (28, 34). Semi-solid cultures were observed using a lighted black background to examine for the appearance of a Dinger's zone (DZ), and if noted, were confirmed as positive by dark-field microscopy (DFM), at days 3 and 5, weekly for 1 month, and monthly thereafter for 6 months.
Cultured bovine urine isolates of Leptospira were serotyped by the MAT method using a panel of polyclonal rabbit reference antisera representing thirteen serogroups; Australis, Autumnalis, Ballum, Bataviae, Canicola, Grippotyphosa, Hebdomadis, Icterohaemorrhagiae, Mini, Pomona, Pyrogenes, Sejröe, and Tarassovi (National Veterinary Services Laboratories, APHIS, USDA, Ames, Iowa) (Supplementary Table 2). The isolates were further typed to the serovar level by performing MAT with panels of monoclonal antibodies (mAbs) that characteristically agglutinate serovars from the serogroup Sejroe and Pyrogenes as previously described (35).
For Illumina Sequencing, DNA was extracted from independent 5 mL cultures of each isolated strain using the Maxwell RSC Purefood Purification Pathogen kit (Promega Corporation, Madison, WI), following manufacturer's instructions. For Nanopore Sequencing, DNA was extracted from independent 5 mL culture of the same three strains using the Nanobind CBB Big DNA Kit—Beta Handbook v1.8 (07/2019) (Circulomics, Baltimore, MD). The genomic DNA concentration for all preparations was determined by Qubit (Qubit dsDNA Broad Range Assay Kit, Qubit 3.0 fluorometer, Invitrogen, Carlsbad, CA, USA) to ensure that there was a minimum of 25 ng/μL for Nanopore and 1 ng/μL for Illumina sequencing. Genomic DNA purity was assessed using the NanoPhotometer Pearl® (IMPLEN).
Illumina whole-genome sequence was obtained (MiSeq Desktop Sequencer, 2x250 v2 paired-end chemistry and the Nextera XT DNA Library Preparation Kit, Ilumina, San Diego, CA, USA) per manufacturer's instructions. Prior to Nanopore sequencing, purified DNA was passed through the Circulomics Short Read Eliminator Kit XS following manufacturer's instructions. DNA was again quantified using the Qubit dsDNA Broad Range Assay Kit and 1 μg was used. The Native barcoding genomic DNA Kit was used following the manufacturer's instructions. Samples were pooled in equal amounts and loaded onto a Nanopore flowcell FLMIN106. The flowcell was run for 12 h.
Illumina sequencing reads for each isolate were mapped to the reference genome L. borgpetersenii serovar Hardjo and L. santarosai serovar Alexi using the Burrows Wheeler Aligner (BWA) and Genome Analysis Toolkit (GATK); according to GATK best practices. Illumina WGS reads were taxonomically identified using Kraken (36) and visually displayed with a Krona graph (37). Reads were assembled with SPAdes (38) and verified by comparing the expected genome size with the total assembly and verifying contigs as Leptospira by BLAST (39) against the nucleotide database. The Nanopore sequence was processed using Guppy to perform basecalling from the fast5 files generated by the Minion. QCAT was used to demultiplex the pooled samples by barcode. Porechop was run on the demultiplexed samples to remove the nanopore adaptors from each sample. Unicycler was then used in conservative mode to perform a hybrid genome assembly from the Illumina MiSeq and Nanopore Minion data (40). The genome annotation was completed by the NCBI Prokaryotic Genome Annotation Pipeline (41).
Using kSNP3.0 (42), a reference-free phylogenetic analysis tool, assembled genomes were compared using the output maximum likelihood tree. The sequences used were downloaded from NCBI or from the NVSL in-house sequence repository. Sequences from NCBI are indicated by the accession number, while the NVSL in-house sequences are identified only by the species, serogroup, serovar, and strain.
Leptospires (mid-late log phase, 1–3 × 108 leptospires/mL) were harvested by centrifugation (10,000 × g, 4°C, 30 min), washed twice with PBS, and processed for one-dimensional (1-D) SDS-PAGE on 12% acrylamide gels (BioRad) as per manufacturer's guidelines. Proteins were visualized by staining with Sypro Ruby (Invitrogen, CA, USA) and lipopolysaccharide was visualized by staining with Pro-Q Emerald 300 (Invitrogen, CA) as per manufacturer's guidelines. For immunoblotting, samples were transferred by semi-dry transfer (Amersham TE77 PWR) to Immobilon-P transfer membrane (Millipore, 220 Bedford, MA) and blocked overnight at 4°C with Starting Block (PBS) blocking buffer (Thermo Scientific, CO) (25).
Membranes were individually incubated with indicated antisera diluted in blocking buffer (anti-LipL32 at 1:4,000, or anti-Alexi, anti-Hardjo at 1:1,000) followed by incubation with horseradish-peroxidase anti-rabbit immunoglobulin G conjugate diluted 1:4,000 in blocking buffer (Sigma, MO). Bound conjugates were detected using Clarity Western ECL substrate (BioRad, CA) and images acquired using a Bio-Rad ChemiDoc MP imaging system.
In our prescreen, 7/35 (20%) bovine urine samples from farm 1, and 3/14 (21.4%) bovine urine samples from farm 2, were positive for Leptospira by PCR. By FAT, 2/35 (5.7%) and 1/14 (7.1%) bovine urine samples were positive on farm 1 and farm 2, respectively. All samples positive by FAT were also positive by PCR. Three PCR-positive animals on farm 1, and two PCR-positive animals on farm 2, were selected for further sampling to facilitate culture (Table 1). Of these repeat samples, three (100%; 3/3) urine samples on farm 1 (DCP-009, DCP-017 and DCP-026) were PCR positive in both voids but only one (50%; 1/2) urine sample from farm 2 (DCP-041) was positive in both voids (Table 1).
A serum sample collected at the same time as urine samples for culture showed that only 3/5 animals had a positive MAT titer: On farm 1, DCP-009 had a titer of 1:100 to serogroup Sejroe and DCP-017 had a titer of 1:200 to serogroup Australis while on farm 2, DCP-041 had a titer of 1:200 to serogroup Sejroe (Table 1).
On farm 1, two animals were culture positive (DCP-009 & DCP-017) and one animal was culture positive on farm 2 (DCP-041). A single positive culture from the second void of DCP-009 and DCP-017 in farm 1 samples was obtained in HAN media incubated at 37°C in 3% CO2, but both samples were negative in HAN incubated at 29°C. In farm 2 samples, both void 1 and void 2 from DCP-041 were culture positive in HAN incubated at 37°C in 3% CO2, and in T80/40/LH incubated at 29°C; void 2 also was culture positive in HAN incubated at 29°C (Table 1).
Serotyping of strains DCP-009 and DCP-041 by MAT with reference antisera against serogroups indicated that both belong to serogroup Sejroe, but strain DCP-017 belongs to serogroup Pyrogenes, Table 1. Additional serotyping with monoclonal antibodies to identify serovar confirmed that strains DCP-009 and DCP-041 belonged to serogroup Sejroe serovar Hardjo, due to their similar reactivity patterns with the Hardjo reference strain (Figure 1A). A serovar determination for strain DCP-017 was not possible as serotyping with monoclonal antibodies was unable to discriminate whether it belonged to serovar Alexi, Guaratuba or Princestown (Figure 1B).
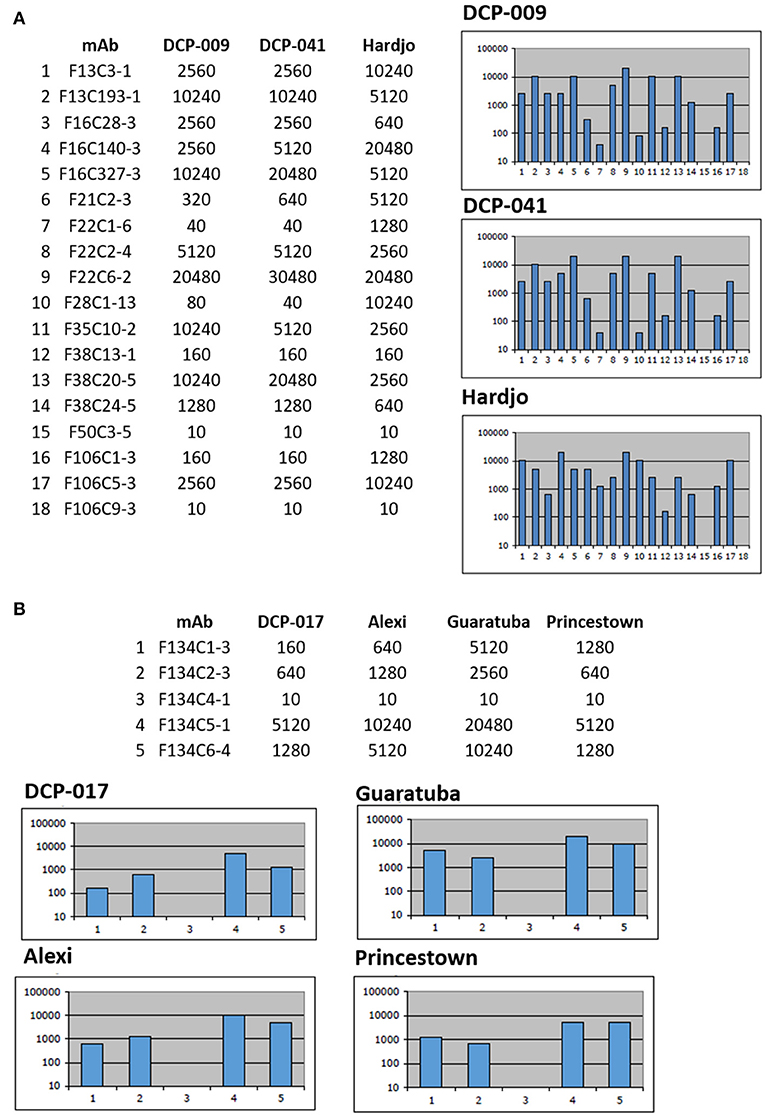
Figure 1. Serotyping with monoclonal antibodies (mAb) that characteristically agglutinate serovars from the serogroup Sejroe and serogroup Pyrogenes. Titers of reactivity for each mAb are provided for (A) L. borgpetersenii strains DCP-009, DCP-041 and reference strain L. interrogans serogroup Sejroe serovar Hardjo strain Hardjoprajitno and (B) L. santarosai strain DCP-017 and reference strains for serogroup Pyrogenes; L. santarosai serovar Alexi strain HS 616, L. interrogans serovar Guaratuba strain An 775 and L. santarosai serovar Princestown strain TRVL 112499. Reciprocal titers are shown on the y-axis; mAb number is shown on the x-axis.
Molecular typing indicated that strain DCP-009 and strain DCP-017 cultured from cows on farm 1 were L. borgpetersenii and L. santarosai, respectively. Strain DCP-041 cultured from farm 2 was L. borgpetersenii. Accession numbers for chromosome 1 and chromosome 2 for each of the three strains, as well as genome annotation features, are provided in Table 2.
Phylogenetic analysis based on complete whole genome sequence demonstrates clustering of DCP-009 and DCP-041 with L. borgpetersenii and that strain DCP-017 clusters with L. santarosai (Figure 2).
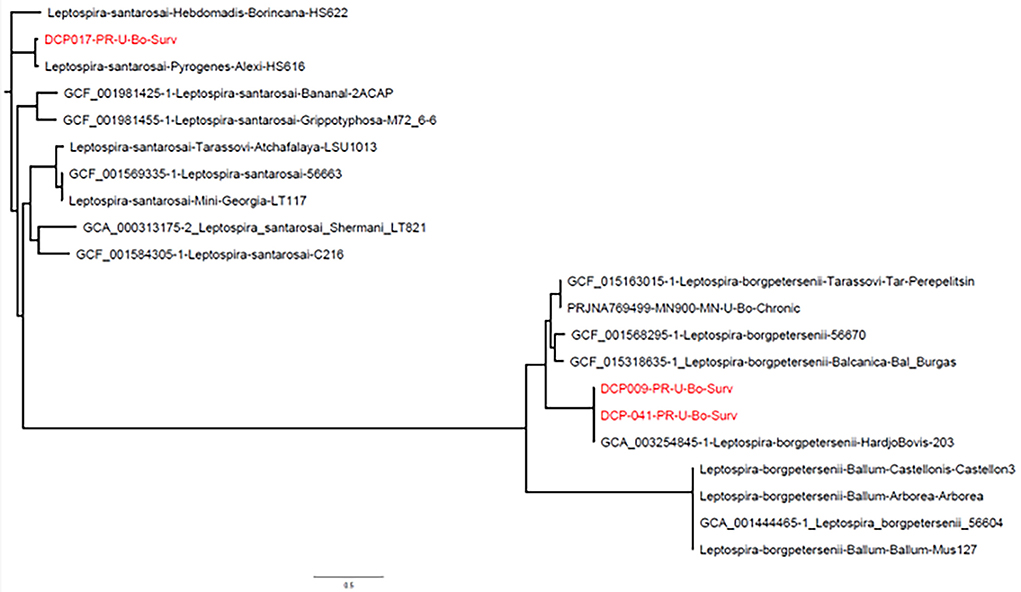
Figure 2. Phylogeny of L. borgpetersenii strain DCP-009 and strain DCP-041 and L. santarosai strain DCP-017 based on complete whole genome sequence analysis. Genome sequences from GenBank are preceded by an accession number, while the NVSL in-house sequences are identified only by the species, serogroup, serovar, and strain.
L. borgpetersenii serogroup Sejroe serovar Hardjo strain DCP-009 and L. santarosai serogroup Pyrogenes strain DCP-017 have a similar protein profile (Figure 3A) and both express the pathogen-associated outer membrane protein LipL32 (Figure 3C). In contrast, and as expected with strains of pathogenic leptospires belonging to different serogroups and serovars, each strain presented with a unique lipopolysaccharide profile (Figure 3B) as confirmed by immunoblotting with antisera specific for serovar Hardjo and serovar Alexi (Figures 3D,E, respectively).
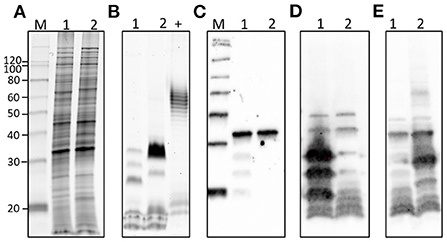
Figure 3. Representative images of (1) L. borgeptersenii serovar Hardjo strain DCP-009 and (2) L. santarosai serogroup Pyrogenes strain DCP-017 showing (A) total protein profiles, (B) total lipopolysaccharide profiles, (C) immunoblotting with anti-LipL32, (D) immunoblotting with anti-serovar Hardjo and (E) immunoblotting with anti-serovar Alexi. Five microgram of each strain was loaded per lane. +ve; postive control for LPS staining comprising 5 μg of E. coli serotype 055:B5. Molecular mass markers are indicated.
There is limited understanding of bovine leptospirosis in Puerto Rico. This study cultured and characterized isolates of Leptospira from dairy cows in Puerto Rico to determine what species and serovars are circulating in local dairy herds to aid in developing vaccines and diagnostics, and to understand the zoonotic risks of disease transmission.
L. borgpetersenii serovar Hardjo was isolated from two different dairy herds in Puerto Rico: though these represent the first serovar Hardjo isolates from dairy cows in Puerto Rico to our knowledge, it is consistent with the global distribution of leptospirosis in bovines which act as reservoir hosts for L. borgpetersenii serovar Hardjo, often with no apparent clinical signs of infection (14). In addition, L. santarosai serogroup Pyrogenes was also isolated in Puerto Rico, and from the same dairy herd shedding L. borgpetersenii serovar Hardjo. We were unable to serotype L. santarosai serogroup Pyogenes strain DCP-017 to the serovar level since monoclonal antibodies could not distinguish whether it belonged to serovar Alexi, Princestown, or Guaratuba. However, the reference strains for serovar Alexi and Princestown are also L. santarosai compared to serovar Guaratuba which belongs to the species L. interrogans. Serovar Alexi was originally isolated from a human patient in Puerto Rico in 1951 and, after comparison with serovars Pyrogenes and Zanoni (both of the serogroup Pyrogenes), the reference strain HS 616 was recognized as a new serovar (43). This new serovar first appears in the WHO list of 1965 and the strain was submitted to factor analysis by Kmety (44), who confirmed its status and thus inclusion in the subgroup Pyrogenes (45). Serovar Princestown was originally isolated from the blood of a 15-year-old boy from Princestown, Trinidad, West Indies, and described as a new serovar named Princestown, reference strain TRVL 112499, in the Pyrogenes group (46). In contrast, serovar Guaratuba reference strain An 7705 was originally isolated from an opossum in Brazil (47). Collectively, this would suggest the isolate from DCP-017 is more likely closer to serovar Alexi. Two strains are said to belong to different serovars if, after cross absorption with adequate amounts of heterologous antigen, more than 10% of the homologous titer regularly remains in at least one of the two antisera in repeated tests (48); the cross agglutination absorption test (CAAT) is required to determine if strain DCP-017 represents a new serovar (45).
L. santarosai is commonly associated with infection of humans, and domestic and wildlife animals in Latin America. This species has been isolated from cattle in Brazil (18, 20–22), Mexico (24), and Peru (23). Recently, L. santarosai was isolated from the uterus of a sub-fertile cow, highlighting a potential role in bovine genital leptospirosis and poor reproductive performance (21, 49). Clinical symptoms of human patients infected with L. santarosai range from mild to severe, including Weil's syndrome and liver failure (50). This coupled with the isolation of L. santarosai serovar Alexi strain HS 616 (serogroup Pyrogenes) and L. santarosai serovar Borincana strain HS622 (serogroup Hebdomadis) from human patients in Puerto Rico (43) highlights the zoonotic risk of infection.
Culture of leptospires provides a definitive diagnosis as well as an isolate that can be completely characterized at the genotypic and phenotypic level, included in an MAT diagnostic antigen rack, and potentially used for bacterin-based vaccination (51). Culture of leptospires is a technically difficult and laborious task but the detection of PCR-positive dairy cows prior to culture provides a screening tool to prioritize efforts and optimize successful outcomes. The recent development of new media formulations to support the growth of fastidious leptospires has enabled collection of samples which can now be transported long distances prior to processing (5, 6, 28). Both L. borgpetersenii and L. santarosai have similar protein profiles and express the pathogen-associated outer membrane protein LipL32. The characterization of these strains to serovar level highlights the significantly different expression of lipopolysaccharide (LPS). The O-antigen of LPS is a protective antigen and thus our results provide insight into which serovars should be considered for more efficacious bovine vaccine strategies, to limit animal disease as well as zoonotic transmission.
In this study, both cows (DCP-009 and DCP-041) shedding serovar Hardjo were seropositive by MAT, but the cow (DCP-017) shedding serogroup Pyrogenes was seronegative for serogroup Pyrogenes. Though seronegative animals can shed leptospires (27, 52), the efficacy of the MAT is based in part on inclusion of appropriate serovars representative of each serogroup within a geographical locale. In addition, as recommended by the WOAH-Manual, the sensitivity of the MAT can be improved by using local isolates. The use of local isolates from Puerto Rico can be used on larger seroprevalence studies to determine levels of exposure by dairy cows to pathogenic leptospires.
The dairy industry is the most financially important agricultural commodity in Puerto Rico (15). Since bovine leptospirosis can result in significant economic costs (53), it is important to accurately define the epidemiological aspects of this disease to ensure efficacious intervention strategies, and to determine whether to target transmission of disease within the herd, from other domestic animals (54), or invasive small mammals (16). Our results demonstrate that both L. borgpetersenii serovar Hardjo and L. santarosai serogroup Pyrogenes infect dairy cows in Puerto Rico and highlight the need to consider multiple species and serovars to mitigate domestic animal infection and limit zoonotic transmission of leptospirosis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP097243, CP097244, CP097245, CP097246, CP096186, CP096185.
All sample collection was conducted in accordance with protocols as reviewed and approved by the Animal Care & Use Committee at the CDC, protocol 2879SALMULX-A3. Informed consent was obtained from the owners for the participation of their animals in this study.
CH, IS, and JN: conceptualization. CH, KD, KL, FS, SR-G, PC, TA, JH, RG, GS, IS, MG, HL, TS, DB, and JN: methodology. CH, KD, KL, TA, JH, MG, HL, TS, DB, and JN: formal analysis and writing—review and editing. FS, IS, MG, and LS: resources. CH, KL, TS, MG, HL, and JN: figures. CH and JN: writing—original draft preparation. All authors have read and agreed to the published version of the manuscript.
This research was supported by USDA and in part by an appointment to the Animal and Plant Health Inspection Service (APHIS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE-SC0014664.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
All opinions expressed in this paper are the author's and do not necessarily reflect the policies and views of USDA, DOE, ORAU/ORISE or CDC. USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1025282/full#supplementary-material
1. Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis. (2015) 9:e0003898. doi: 10.1371/journal.pntd.0003898
2. Desvars A, Cardinale E, Michault A. Animal leptospirosis in small tropical areas. Epidemiol Infect. (2011) 139:167–88. doi: 10.1017/S0950268810002074
3. Petrakovsky J, Bianchi A, Fisun H, Nájera-Aguilar P, Pereira MM. Animal leptospirosis in Latin America and the Caribbean countries: reported outbreaks and literature review (2002–2014). Int J Environ Res Public Health. (2014) 11:10770–89. doi: 10.3390/ijerph111010770
4. Cranford HM, Browne AS, LeCount K, Anderson T, Hamond C, Schlater L, et al. Mongooses (Urva auropunctata) as reservoir hosts of Leptospira species in the United States Virgin Islands, 2019–2020. PLoS Negl Trop Dis. (2021) 15:e0009859. doi: 10.1371/journal.pntd.0009859
5. Cranford HM, Taylor M, Browne AS, Alt DP, Anderson T, Hamond C, et al. Exposure and Carriage of Pathogenic Leptospira in Livestock in St. Croix, US Virgin Islands. Trop Med Infect Dis. (2021) 6:85. doi: 10.3390/tropicalmed6020085
6. Hamond C, Browne AS, de Wilde LH, Hornsby RL, LeCount K, Anderson T, et al. Assessing rodents as carriers of pathogenic Leptospira species in the U. S Virgin Islands and their risk to animal and public health. Sci Rep. (2022) 12:1132. doi: 10.1038/s41598-022-04846-3
7. Sanders EJ, Rigau-PÃ J, Smits HL, Deseda CC, Vorndam VA, Aye T, et al. Increase of leptospirosis in dengue-negative patients after a hurricane in Puerto Rico in 1996 [correction of 1966]. Am J Trop Med Hyg. (1999) 61:399–404. doi: 10.4269/ajtmh.1999.61.399
8. Briskin EA, Casanovas-Massana A, Ryff KR, Morales-Estrada S, Hamond C, Perez-Rodriguez NM, et al. Seroprevalence, risk factors, and rodent reservoirs of leptospirosis in an urban community of Puerto Rico, 2015. J Infect Dis. (2019) 220:1489–97. doi: 10.1093/infdis/jiz339
9. Bruce MG, Sanders EJ, Leake JA, Zaidel O, Bragg SL, Aye T, et al. Leptospirosis among patients presenting with dengue-like illness in Puerto Rico. Acta Trop. (2005) 96:36–46. doi: 10.1016/j.actatropica.2005.07.001
10. Nationally Notifiable Infectious Diseases and Conditions United States: Annual Tables. Atlanta, GA: CDC (2019). Available online at: https://wonder.cdc.gov/nndss/static/2019/annual/2019-table2i-H.pdf
11. Gorbea H, Garcia-Rivera EJ, Torres H, Lorenzi OD, Rivera A, Galloway RL, et al. Leptospirosis cases infected with uncommon serogroups, Puerto Rico, 2013–2015. Am J Trop Med Hyg. (2018) 98:258. doi: 10.4269/ajtmh.17-0538
12. Stone NE, Hall CM, Ortiz M, Hutton SM, Santana-Propper E, Celona KR, et al. Diverse lineages of pathogenic Leptospira species are widespread in the environment in Puerto Rico, USA. PLoS Neglect Trop Dis. (2022) 16:e0009959. doi: 10.1371/journal.pntd.0009959
13. Fernandes LGV, Stone NE, Roe CC, Goris MGA, van der Linden H, Sahl JW, et al. Leptospira sanjuanensis sp. nov., a pathogenic species of the genus Leptospira isolated from soil in Puerto Rico. Int J Syst Evol Microbiol. (2022) 72. doi: 10.1099/ijsem.0.005560
14. Ellis WA. Animal leptospirosis. Curr Top Microbiol Immunol. (2015) 387:99–137. doi: 10.1007/978-3-662-45059-8_6
15. Census of Agriculture Puerto Rico Data Release (2020). Available online at: https://www.nass.usda.gov/Newsroom/Executive_Briefings/2020/06-09-2020.pdf
16. Benavidez KM, Guerra T, Torres M, Rodriguez D, Veech JA, Hahn D, et al. The prevalence of Leptospira among invasive small mammals on Puerto Rican cattle farms. PLoS Negl Trop Dis. (2019) 13:e0007236. doi: 10.1371/journal.pntd.0007236
17. Yupiana Y, Vallee E, Wilson P, Collins-Emerson J, Weston J, Benschop J, et al. Emerging Leptospira strain poses public health risk for dairy farmers in New Zealand. Prev Vet Med. (2019) 170:104727. doi: 10.1016/j.prevetmed.2019.104727
18. Hamond C, Pinna M, Medeiros MA, Bourhy P, Lilenbaum W, Picardeau M. A multilocus variable number tandem repeat analysis assay provides high discrimination for genotyping Leptospira santarosai strains. J Med Microbiol. (2015) 64 (Pt 5):507. doi: 10.1099/jmm.0.000045
19. Martins G, Loureiro AP, Hamond C, Pinna MH, Bremont S, Bourhy P, et al. First isolation of Leptospira noguchii serogroups Panama and Autumnalis from cattle. Epidemiol Infect. (2015) 143:1538–41. doi: 10.1017/S0950268814002416
20. Guedes IB, de Souza GO, de Souza Rocha K, Cavalini MB, Neto MSD, de Paula Castro JF, et al. Leptospira strains isolated from cattle in the Amazon region, Brazil, evidence of a variety of species and serogroups with a high frequency of the Sejroe serogroup. Comp Immunol Microbiol Infect Dis. (2021) 74:101579. doi: 10.1016/j.cimid.2020.101579
21. Aymée L, Nogueira Di Azevedo MI, de Souza Pedrosa J, Loria de Melo JDS, Carvalho-Costa FA, Lilenbaum W. The role of Leptospira santarosai serovar Guaricura as agent of Bovine Genital Leptospirosis. Vet Microbiol. (2022) 268:109413. doi: 10.1016/j.vetmic.2022.109413
22. Loureiro A, Hamond C, Pinto P, Bremont S, Bourhy P, Lilenbaum W. Molecular analysis of leptospires from serogroup Sejroe obtained from asymptomatic cattle in Rio de Janeiro—Brazil reveals genetic proximity to serovar Guaricura. Res Vet Sci. (2016) 105:249–53. doi: 10.1016/j.rvsc.2016.02.012
23. Rivera P, Ticlla M, Balda L, Gonzalez D, Céspedes M. Genetic diversity of Peruvian isolates of Leptospira spp. through pulsed field gel electrophoresis. Rev Peruana Med Exp Salud Publica. (2012) 29:469–76. doi: 10.1590/S1726-46342012000400008
24. Carmona-Gasca CA, Lara LL, Castillo-Sánchez LO, Ramírez-Ortega JM, Palomera CL, de la Peña-Moctezuma A. Detection of Leptospira santarosai and L kirschneri in cattle: new isolates with potential impact in bovine production and public health. Vet Mexico. (2011) 42:277–88. Available online at: https://www.medigraphic.com/cgi-bin/new/resumenI.cgi?IDREVISTA=&IDARTICULO=31603&IDPUBLICACION=
25. Hamond C, LeCount K, Putz EJ, Bayles DO, Camp P, Goris MGA, et al. Bovine leptospirosis due to persistent renal carriage of leptospira borgpetersenii serovar tarassovi. Front Vet Sci. (2022) 9:848664. doi: 10.3389/fvets.2022.848664
26. Miller D, Wilson M, Beran G. Survey to estimate prevalence of Leptospira interrogans infection in mature cattle in the United States. Am J Vet Res. (1991) 52:1761–5.
27. Nally JE, Hornsby RL, Alt DP, Bayles D, Wilson-Welder JH, Palmquist DE, et al. Isolation and characterization of pathogenic leptospires associated with cattle. Vet Microbiol. (2018) 218:25–30. doi: 10.1016/j.vetmic.2018.03.023
28. Hornsby RL, Alt DP, Nally JE. Isolation and propagation of leptospires at 37 degrees C directly from the mammalian host. Sci Rep. (2020) 10:9620. doi: 10.1038/s41598-020-66526-4
29. Nally JE, Arent Z, Bayles DO, Hornsby RL, Gilmore C, Regan S, et al. Emerging infectious disease implications of invasive mammalian species: the greater white-toothed shrew (Crocidura russula) is associated with a novel serovar of pathogenic Leptospira in Ireland. PLoS Negl Trop Dis. (2016) 10:e0005174. doi: 10.1371/journal.pntd.0005174
30. Nally JE, Bayles DO, Hurley D, Fanning S, McMahon BJ, Arent Z. Complete genome sequence of Leptospira alstonii serovar Room22 strain GWTS# 1. Genome Announc. (2016) 4:e01230–16. doi: 10.1128/genomeA.01230-16
31. Cole JR, Sulzer CR, Pursell AR. Improved microtechnique for the leptospiral microscopic agglutination test. Appl Microbiol. (1973) 25:976–80. doi: 10.1128/am.25.6.976-980.1973
32. Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. (2009) 64:247–55. doi: 10.1016/j.diagmicrobio.2009.03.014
33. Galloway RL, Hoffmaster AR. Optimization of LipL32 PCR assay for increased sensitivity in diagnosing leptospirosis. Diagn Microbiol Infect Dis. (2015) 82:199–200. doi: 10.1016/j.diagmicrobio.2015.03.024
34. Ellis W, Montgomery J, Cassells J. Dihydrostreptomycin treatment of bovine carriers of Leptospira interrogans serovar Hardjo. Res Vet Sci. (1985) 39:292–5. doi: 10.1016/S0034-5288(18)31716-8
35. Hartskeerl R, Smits H, Korver H, Goris M, Terpstra W. Manual International Course on Laboratory Methods for the Diagnosis of Leptospirosis. Amsterdam: KIT (2006).
36. Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. (2019) 20:1–13. doi: 10.1186/s13059-019-1891-0
37. Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. (2011) 12:1–10. doi: 10.1186/1471-2105-12-385
38. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinformat. (2020) 70:e102. doi: 10.1002/cpbi.102
39. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
40. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. (2017) 13:e1005595. doi: 10.1371/journal.pcbi.1005595
41. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. (2016) 44:6614–24. doi: 10.1093/nar/gkw569
42. Gardner SN, Slezak T, Hall BG. kSNP3 0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. (2015) 31:2877–8. doi: 10.1093/bioinformatics/btv271
44. Kmety E. Faktorenanalyse von Leptospiren der Icterohaemorrhagiae und einiger verwandter Serogruppen. Bratislava: Slovak Academy of Sciences (1967).
45. Dikken H, Kmety E. Serological typing methods of leptospires. Methods Microbiol. (1978) 11:259–307. doi: 10.1016/S0580-9517(08)70493-8
46. Green AE, Sulzer CR, Evarard CO, Jones WL. Four new leptospira serotypes from Trinidad. West Indian Med J. (1978) 27:117–26.
47. Rosa CAS, Sulzer CR, Giorgi W, da Silva AS, Yanaguita RM, Lobao AO. Leptospirosis in wildlife in Brazil: isolation of a new serotype in the pyrogenes group. Am J Vet Res. (1975) 36:1363–5.
48. Kmety E, Dikken H. Classification of the Species Leptospira interrogans and History of Its Serovars. Groningen: University Press Groningen (1993).
49. Loureiro AP, Lilenbaum W. Genital bovine leptospirosis: A new look for an old disease. Theriogenology. (2020) 141:41–7. doi: 10.1016/j.theriogenology.2019.09.011
50. Peláez Sanchez RG, Lopez JÁ, Pereira MM, Arboleda Naranjo M, Agudelo-Flórez P. Genetic diversity of Leptospira in northwestern Colombia: first report of Leptospira santarosai as a recognised leptospirosis agent. Memór Instituto Oswaldo Cruz. (2016) 111:737–44. doi: 10.1590/0074-02760160245
51. Putz EJ, Nally JE. Investigating the immunological and biological equilibrium of reservoir hosts and pathogenic Leptospira: balancing the solution to an acute problem? Front Microbiol. (2020) 11:2005. doi: 10.3389/fmicb.2020.02005
52. Nally JE, Mullen W, Callanan JJ, Mischak H, Albalat A. Detection of urinary biomarkers in reservoir hosts of leptospirosis by capillary electrophoresis-mass spectrometry. Proteom Clin Appl. (2015) 9:543–51. doi: 10.1002/prca.201400205
53. Bennett R, IJpelaar J. Updated estimates of the costs associated with thirty four endemic livestock diseases in Great Britain: a note. J Agric Econ. (2005) 56:135–44. doi: 10.1111/j.1477-9552.2005.tb00126.x
Keywords: Leptospira, borgpetersenii, santarosai, leptospirosis, cow, dairy, Puerto Rico
Citation: Hamond C, Dirsmith KL, LeCount K, Soltero FV, Rivera-Garcia S, Camp P, Anderson T, Hicks JA, Galloway R, Sutherland G, Schafer IJ, Goris MGA, van der Linden H, Stuber T, Bayles DO, Schlater LK and Nally JE (2022) Leptospira borgpetersenii serovar Hardjo and Leptospira santarosai serogroup Pyrogenes isolated from bovine dairy herds in Puerto Rico. Front. Vet. Sci. 9:1025282. doi: 10.3389/fvets.2022.1025282
Received: 22 August 2022; Accepted: 31 October 2022;
Published: 17 November 2022.
Edited by:
Max Francois Millien, Université Quisqueya, HaitiReviewed by:
Rigoberto Hernandez-Castro, Hospital General Dr. Manuel Gea Gonzalez, MexicoCopyright © 2022 Hamond, Dirsmith, LeCount, Soltero, Rivera-Garcia, Camp, Anderson, Hicks, Galloway, Sutherland, Schafer, Goris, van der Linden, Stuber, Bayles, Schlater and Nally. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jarlath E. Nally, amFybGF0aC5uYWxseUB1c2RhLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.