
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 03 October 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1015717
This article is part of the Research Topic Epidemic Status and Prevention of Swine Infectious Diseases View all 11 articles
 Yesheng Shen1†
Yesheng Shen1† Yudong Yang1†
Yudong Yang1† Jun Zhao1
Jun Zhao1 Ningwei Geng1
Ningwei Geng1 Kuihao Liu1
Kuihao Liu1 Yiran Zhao1
Yiran Zhao1 Fangkun Wang1
Fangkun Wang1 Sidang Liu1
Sidang Liu1 Ning Li1*
Ning Li1* Fanliang Meng1,2*
Fanliang Meng1,2* Mengda Liu3*
Mengda Liu3*Responsible for the acute infectious disease porcine epidemic diarrhea (PED), PED virus (PEDV) induces severe diarrhea and high mortality in infected piglets and thus severely harms the productivity and economic efficiency of pig farms. In our study, we aimed to investigate and analyze the recent status and incidence pattern of PEDV infection in some areas of Shandong Province, China. We collected 176 clinical samples of PED from pig farms in different regions of Shandong Province during 2019–2021. PEDV, TGEV, and PORV were detected using RT-PCR. The full-length sequences of positive PEDV S genes were amplified, the sequences were analyzed with MEGA X and DNAStar, and a histopathological examination of typical PEDV-positive cases was performed. RT-PCR revealed positivity rates of 37.5% (66/176) for PEDV, 6.82% (12/176) for transmissible gastroenteritis virus, and 3.98% (7/176) for pig rotavirus. The test results for the years 2019, 2020, and 2021 were counted separately, PEDV positivity rates for the years were 34.88% (15/43), 39.33% (35/89), and 36.36% (16/44), respectively. Histopathological examination revealed atrophied, broken, and detached duodenal and jejunal intestinal villi, as typical of PED, and severe congestion of the intestinal submucosa. Moreover, the results of our study clearly indicate that the G2 subtype is prevalent as the dominant strain of PEDV in Shandong Province, where its rates of morbidity and mortality continue to be high. Based on a systematic investigation and analysis of PEDV's molecular epidemiology across Shandong Province, our results enrich current epidemiological data regarding PEDV and provide some scientific basis for preventing and controlling the disease.
Porcine epidemic diarrhea (PED) is a common viral disease in raising pigs, one with high morbidity, high mortality, and complex causes that are difficult to control (1–4). Among piglets up to 3 months old, PED is especially problematic, with a prevalence of up to 70%. Nevertheless, epidemiological information on PED in large-scale pig farms in recent years in Shandong Province is currently lacking.
PED virus (PEDV), a member of the genus A coronavirus in the Coronaviridae family, is a linear single-stranded, positive-strand RNA virus ~28 kb in length (5). In the virus, ORF4, ORF5, and ORF6 encode four structural proteins—the spine (S) protein, the envelope (E) protein, the membrane (M) protein, and the nucleocapsid (N) protein (6, 7)—among which S proteins, due to their high genetic variability, are often used as markers for phylogenetic analysis and epidemiological investigations (8). Based on the genetic characteristics of the PEDV S gene, it is often divided into two subtypes: G1 and G2 (9–11). Between them, the G2 subtype is the more prevalent genotype of PED in China, whereas the more commonly used vaccine strain CV777 is of the G1 subtype (12, 13).
In our study, we collected 176 clinical samples of PED from pig farms in different regions of In Shandong Province, China during 2019–2021 in order to investigate the prevalence of PEDV there. We performed phylogenetic analysis, comparative nucleotide analysis, and comparative analysis of deduced amino acid sequences with the aim of clarifying the epidemiology of PEDV in Shandong Province.
During 2019–2021, we collected 176 intestinal and fecal samples from pigs with diarrheal symptoms on pig farms in localities throughout Shandong Province (Figure 1). The collected samples were ground into tissue homogenates, transferred into 1.5 ml centrifuge tubes, frozen and thawed 3 times, and stored at −80°C for later use.
The RNA isolater Total RNA Extraction Reagent R401-01 (Vazyme) was added to the tissue homogenate supernatant, and total RNA was extracted from the intestinal and fecal samples according to the instructions. The primers were synthesized by Sangon; Table 1 shows the sequences of the primers used. Three viruses—PEDV, transmissible gastroenteritis virus (TGEV), and porcine rotavirus (PoRV)—were amplified by RT-PCR using the EasyScript One-Step RT-PCR SuperMix (TransGen Biotech). After 1% agarose gel electrophoresis was performed for 45 min, the results were observed under a gel imaging system. PEDV-positive RNA was reverse-transcribed to cDNA using the ReverTra-Ace First-Strand cDNA Synthesis Kit (Toyobo), after which RT-PCR amplification was performed on PEDV-positive samples using 2 × Phanta Max Master Mix (Vazyme). All steps were performed strictly according to the instructions of the manufacturers.
PEDV-positive RT-PCR amplification products were sent to Sangon for sequencing. The sequences were compared using MegAlign in DNAStar and analyzed for nucleotide and deduced amino acid homology with a selection of reference strains entered in the NCBI's database (Table 2). MEGA X was used to construct a genetic evolutionary tree based on the PEDV S gene, while Protean in DNAStar was used to analyze the antigenic epitopes of the isolates.
The digestive tracts of PEDV-positive pigs were fixed in 10% formalin and removed after 48 h to produce conventional paraffin sections for pathological histological examination using hematoxylin-eosin (HE) staining as a means to observe lesions.
In practice, severe diarrhea with vomiting (Figure 2A), severe dehydration, and wasting (Figure 2B) can be observed in piglets affected by PED. During autopsies, signs of congestion and hemorrhaging in the intestinal mucosa and mesentery were observed as well (Figure 2C). The walls of the small intestine were thin and transparent, the intestinal lumen was filled with milky or yellow slurry-like contents, and the stomach was clearly dilated and filled with curd-like contents (Figure 2D).
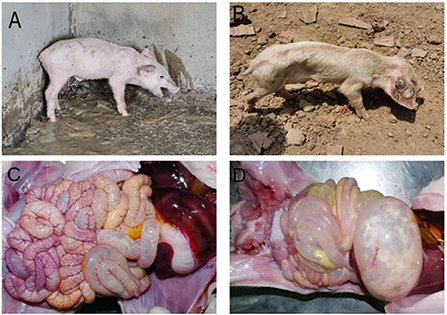
Figure 2. Clinical symptoms and pathological change. (A) Diarrhea accompanied by vomiting; (B) Dehydration and emaciation; (C) Intestinal mucosa and mesenteric hyperemia and bleeding; (D) The intestinal wall is clear and filled, and the stomach is dilated.
RT-PCR revealed positivity rates of 37.5% (66/176) for PEDV, 6.82% (12/176) for TGEV, and 3.98% (7/176) for PoRV. Among them, one case of mixed infection of PEDV and TGEV was found, but no case of mixed infection of PEDV and PoRV was found. When the test results for the years 2019, 2020, and 2021 were counted separately, PEDV positivity rates for the years were 34.88% (15/43), 39.33% (35/89), and 36.36% (16/44), respectively (Figure 3).
A total of 12 PEDV S gene sequences were obtained after sequencing (Table 3), all of which were subsequently uploaded to the NCBI's database.
A comparative nucleotide analysis of the 12 isolates with the classical vaccine strain CV777 using MegAlign in DNAStar revealed that all isolates had varying degrees of base mutations, deletions, and insertions. In particular, all had three consecutive nt (TTG) insertions at position 164, all had nine consecutive nt (GGGTGTTAA) insertions at position 177, and all had one G insertion at position 206. The SDHY-YT strain had three consecutive nt (ACC) insertions at position 419, whereas the other 11 strains all had AAC insertions. All isolates had an A deletion at position 218 and three consecutive nt (GAA) deletions at positions 480–482 (Figure 4).
A comparison of the deduced amino acid sequences of the 12 isolates with those of CV777 using MegAlign revealed multiple deduced amino acid mutations, deletions, and insertions as well. By contrast, all isolates had four consecutive deduced amino acid (QGVN) insertions at positions 59–62, the SDHY-YT strain had a T insertion at position 140, and all other isolates had an N insertion. All 12 isolates additionally had a G deletion at position 160 (Figure 5).
Analysis using MegAlign in DNAStar revealed that the nucleotide homology among the 12 isolates ranged from 97.2 to 99.9%, while the deduced amino acid homology ranged from 95.9 to 99.7%. Compared with the reference strains, the nucleotide homology was in the range of 93.0–99.1%, while the deduced amino acid homology was in the range of 90.9–99.4%. Compared with the classical vaccine strain CV777, the nucleotide sequence homology was in the range of 93.1–93.9%, while the deduced amino acid sequence homology was in the range of 91.2–93.0% (Table 4).
To understand the current genotypes of the dominant strains of PEDV in Shandong Province, we constructed a genetic evolutionary tree (Figure 6) using the neighbor-joining method in MEGA X (14) between the 12 isolates and the reference strains. The results showed that the 12 isolates, all belonging to the G2 genotype, were located in the same branch and in close genetic proximity.

Figure 6. A phylogenetic tree based on the nucleotide construction of the PEDV S gene. “Purple” represents G1-1 subtype, “green” represents G1-2 subtype and “blue” represents G2 subtype. “▴” represents the isolates obtained in this study.
Next, our antigenic index prediction analysis of the vaccine strain CV777 S protein and the 12 isolated S proteins using the Jameson–Wolf method with Protean in DNAStar revealed that the S protein's overall antigenic index was high, with a significant difference in antigenicity within ~20–280 aa and a less significant difference elsewhere (Figure 7).
Histopathological analysis revealed that the duodenal villi had atrophied and detached, and that the submucosa was congested (Figure 8A). The colonic mucosa showed an increased secretion of cupped cells, vacuolation, and the necrosis of epithelial cells (Figure 8B). The jejunal epithelium was necrotic, the submucosa was congested (Figure 8C), the jejunal villi were broken and severely detached (Figure 8D), and the jejunal intestinal canal, filled with necrotic detached epithelial cells, was severely congested and bleeding (Figure 8E). The lamina propria of the gastric mucosa was congested and edematous and showed inflammatory cell infiltration (Figure 8F).
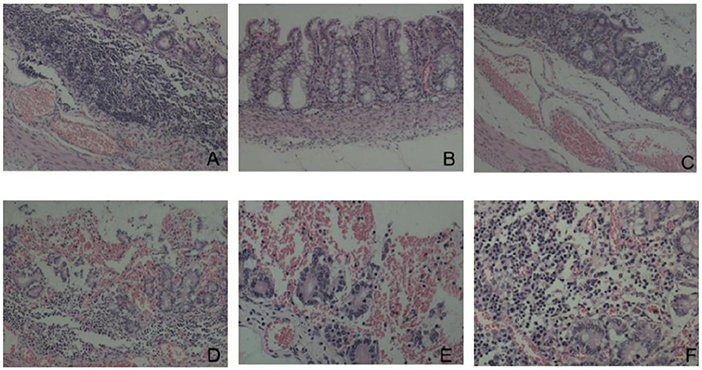
Figure 8. Histopathological changes (HE dyeing). (A) Duodenal villi had atrophied and detached, submucosa was congested, ×100; (B) increased goblet cells and necrosis of epithelial cells, ×200; (C) Necrosis of jejunal mucosal epithelial cells and submucosal hyperemia, ×100; (D) rupture and shedding of jejunal villi, ×100; (E) jejunum full of necrotic and exfoliated epithelial cells, ×100; (F) congestion and inflammatory cell infiltration in the lamina propria of gastric mucosa, ×200.
Coping with PED continues to be a daunting challenge for pig farming in China (15). Studies have revealed PEDV positivity rates ranging from 30–45%, with slight differences between northern and southern China (16–19). In our study, PEDV positivity was 37.5% (66/176), TGEV positivity was 6.82% (12/176), and PoRV positivity was 3.98% (7/176); however, PEDV was the primary pathogen responsible for diarrhea in the piglets. By year, PEDV positivity in 2019, 2020, and 2021 was respectively 34.88% (15/43), 39.33% (35/89), and 36.36% (16/44). Overall, PED was shown to be moderately to highly prevalent in some areas of Shandong Province, with relatively stable trends in prevalence. By furnishing those findings, our study has filled gaps in epidemiological data concerning PEDV in Shandong Province in recent years.
In past research, the S gene of PEDV, the primary structural protein gene, has shown significant variation. The S gene sequences of the prevalent strains were systematically analyzed to accurately capture current trends in the strains and identify markers of PEDV variation (20). The results showed that all 12 isolates had varying degrees of base mutations, deletions, and insertions in the S gene, with changes occurring primarily in the N-terminal structural domain (NTD) region of S1 (21). Homology analysis of nucleotides and of deduced amino acid sequences showed that although the 12 isolates were closely related to the Chinese isolate N12-GD2017 from 2017, the American isolates MN and IA2 from 2013, and the Chinese strain HLJ-2012 from 2012, they were more closely related to the classical Chinese vaccine strain CV777, the Japanese strain KH, and the weak Japanese vaccine strain 83P-PEDV. Meanwhile, S1-IDMP were dominated by S1-IDMP S58_S58insQGVN-N135dup-D158_I159del. By contrast, in our study, S1-IDMP were dominated by S1-IDMP S58_S58insQGVN-N135dup-D158_I159del. Taken together, the deduced amino acid sequence mutations in the isolates primarily concentrated in the NTD region of S1. We observed a highly conserved SS2 antigenic epitope sequence (i.e., Y748SNIGVCK) within PEDV; however, two AA substitutions (i.e., 764 at L → S and 766 at D → S) occurred at the SS6 antigenic epitope S764QSGQVKI. Antigenic index predictions showed a more pronounced difference in antigenicity at 20 aa to 280 aa, with little difference at the remaining positions, which suggests that PEDV S protein changes primarily concentrated in the S1 gene region. In general, the current epidemic strains of PEDV in Shandong Province have high affinity and more obvious mutations in the S1 gene.
Despite the continued high-intensity vaccination of pigs against PED in Shandong Province, the disease can nevertheless occur after multiple vaccinations. In recent years, the incidence of PED in China has shown that the prevalent strains are the main cause of PED, while the incidence and mortality rates have increased year after year (22). In our study, genetic evolutionary analysis showed that all 12 isolates belong to the G2 subtype. However, most of the PEDV vaccines in Shandong Province currently use the CV777 strain of the G1 subtype as their source strain, which may partly explain the occurrence of PED on pig farms in the province. Therefore, we recommend using the more prevalent strain as the source strain for such vaccines, which may yield a more satisfactory immunization effect.
The results of this study clearly indicate that PEDV is the main pathogen causing diarrheal symptoms in pigs, the G2 subtype is the dominant strain of PEDV in Shandong Province, where its rates of morbidity and mortality continue to be high. At present, the most prevalent strain of PEDV in some areas of the province, however, is only distantly related to the vaccine strain CV777. Using the most prevalent strain for vaccines against PED is therefore recommended. In addition, all 12 PEDV strains S1-IDMP isolated in this study had S58_S58insQGVN-N135dup-D158_I159del-like mutations, which require ongoing attention.
The sequences of the isolated strains in this study have been uploaded to GenBanK (accession ON890168 to ON988085-ON988096).
All animal protocols and procedures were performed according to the Chinese Regulations of Laboratory Animals and were approved by the Animal Ethics Committee of Shandong Agricultural University.
YS and YY: writing—original draft preparation. NL, ML, and FM: writing—review and editing. KL and NG: software. YZ, KL, and JZ: investigation. SL: funding acquisition. All authors have read and agreed to the published version of the manuscript.
This study was funded by the Shandong Provincial Agricultural Major Application Technology Innovation Project (Establishment and Demonstration of Healthy Pig Breeding Technology Integration and Product Supply Chain Traceability System).
Author FM was employed by company Huayun (Shandong) Inspection and Quarantine Service Co.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Choudhury B, Dastjerdi A, Doyle N, Frossard JP, Steinbach F. From the field to the lab - An European view on the global spread of PEDV. Virus Res. (2016) 226:40–9. doi: 10.1016/j.virusres.2016.09.003
2. Li W, Li H, Liu Y, Pan Y, Deng F, Song Y, et al. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis. (2012) 18:1350–3. doi: 10.3201/eid1803.120002
3. Shibata I, Tsuda T, Mori M, Ono M, Sueyoshi M, Uruno K. Isolation of porcine epidemic diarrhea virus in porcine cell cultures and experimental infection of pigs of different ages. Vet Microbiol. (2000) 72:173–82. doi: 10.1016/S0378-1135(99)00199-6
4. Wang Q, Vlasova AN, Kenney SP, Saif LJ. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. (2019) 34:39–49. doi: 10.1016/j.coviro.2018.12.001
5. Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. (1978) 58:243–7. doi: 10.1007/BF01317606
6. Chen P, Wang K, Hou Y, Li H, Li X, Yu L, et al. Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011-2017. Infect Genet Evol. (2019) 69:153–65. doi: 10.1016/j.meegid.2019.01.022
7. Jung K, Saif LJ, Wang Q. Porcine epidemic diarrhea virus (PEDV): an update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. (2020) 286:198045. doi: 10.1016/j.virusres.2020.198045
8. Li N, Liu J, Qi J, Hao F, Xu L, Guo K. Genetic diversity and prevalence of porcine circovirus type 2 in China during 2000-2019. Front Vet Sci. (2021) 8:788172. doi: 10.3389/fvets.2021.788172
9. Antas M, Olech M, Szczotka-Bochniarz A. Molecular characterization of porcine epidemic diarrhoea virus (PEDV) in Poland reveals the presence of swine enteric coronavirus (SeCoV) sequence in S gene. PLoS ONE. (2021) 16:e0258318. doi: 10.1371/journal.pone.0258318
10. Chen J, Liu X, Shi D, Shi H, Zhang X, Li C, et al. Detection and molecular diversity of spike gene of porcine epidemic diarrhea virus in China. Viruses. (2013) 5:2601–13. doi: 10.3390/v5102601
11. Dortmans J, Li W, van der Wolf PJ, Buter GJ, Franssen PJM, van Schaik G, et al. Porcine epidemic diarrhea virus (PEDV) introduction into a naive Dutch pig population in 2014. Vet Microbiol. (2018) 221:13–8. doi: 10.1016/j.vetmic.2018.05.014
12. Bi J, Zeng S, Xiao S, Chen H, Fang L. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J Virol. (2012) 86:10910–1. doi: 10.1128/JVI.01919-12
13. Shi W, Hao H, Li M, Niu J, Hu Y, Zhao X, et al. Expression and purification of a PEDV-neutralizing antibody and its functional verification. Viruses. (2021) 13:472. doi: 10.3390/v13030472
14. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30:2725–9. doi: 10.1093/molbev/mst197
15. Turlewicz-Podbielska H, Pomorska-Mól M. Porcine coronaviruses: overview of the state of the art. Virol Sin. (2021) 36:833–51. doi: 10.1007/s12250-021-00364-0
16. Chen X, Zhang XX, Li C, Wang H, Wang H, Meng XZ, et al. Epidemiology of porcine epidemic diarrhea virus among Chinese pig populations: a meta-analysis. Microb Pathog. (2019) 129:43–9. doi: 10.1016/j.micpath.2019.01.017
17. Lee DU, Kwon T, Je SH, Yoo SJ, Seo SW, Sunwoo SY, et al. Wild boars harboring porcine epidemic diarrhea virus (PEDV) may play an important role as a PEDV reservoir. Vet Microbiol. (2016) 192:90–4. doi: 10.1016/j.vetmic.2016.07.003
18. Mai TN, Yamazaki W, Bui TP, Nguyen VG, Le Huynh TM, Mitoma S, et al. A descriptive survey of porcine epidemic diarrhea in pig populations in northern Vietnam. Trop Anim Health Prod. (2020) 52:3781–8. doi: 10.1007/s11250-020-02416-1
19. Reveles-Félix S, Carreón-Nápoles R, Mendoza-Elvira S, Quintero-Ramírez V, García-Sánchez J, Martínez-Bautista R, et al. Emerging strains of porcine epidemic diarrhoea virus (PEDv) in Mexico. Transbound Emerg Dis. (2020) 67:1035–41. doi: 10.1111/tbed.13426
20. Su M, Li C, Qi S, Yang D, Jiang N, Yin B, et al. A molecular epidemiological investigation of PEDV in China: characterization of co-infection and genetic diversity of S1-based genes. Transbound Emerg Dis. (2020) 67:1129–40. doi: 10.1111/tbed.13439
21. Wang E. Molecular epidemiological investigation and virus isolation and identification of S1 gene of porcine epidemic diarrhea virus in China from 2015 to 2016 (Master), Heilongjiang Bayi Agricultural University (2018).
Keywords: porcine epidemic diarrhea, epidemiological survey, S gene, genetic evolutionary analysis, prevention and control
Citation: Shen Y, Yang Y, Zhao J, Geng N, Liu K, Zhao Y, Wang F, Liu S, Li N, Meng F and Liu M (2022) Molecular epidemiological survey of porcine epidemic diarrhea in some areas of Shandong and genetic evolutionary analysis of S gene. Front. Vet. Sci. 9:1015717. doi: 10.3389/fvets.2022.1015717
Received: 10 August 2022; Accepted: 29 August 2022;
Published: 03 October 2022.
Edited by:
Lian-Feng Li, Harbin Veterinary Research Institute (CAAS), ChinaReviewed by:
Aibing Wang, Hunan Agricultural University, ChinaCopyright © 2022 Shen, Yang, Zhao, Geng, Liu, Zhao, Wang, Liu, Li, Meng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning Li, c2R0YWlhbmxpbmluZ0AxMjYuY29t; Fanliang Meng, MTg3NTQ4NzU5MjFAMTYzLmNvbQ==; Mengda Liu, bGl1bWVuZ2RhQGNhaGVjLmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.