
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 23 September 2022
Sec. Livestock Genomics
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1008497
This article is part of the Research Topic Genomic and Epigenomic Applications in Animal and Veterinary Sciences View all 12 articles
The present study was designed to evaluate the association of polymorphisms in bovine trafficking protein particle complex subunit 9 (TRAPPC9) and cluster of differentiation 4 (CD4) genes with milk production and mastitis resistance phenotypic traits in a different cattle population. Three single nucleotide polymorphisms (SNPs) (SNP1 Position: Chr14:2484891, SNP2 (rs110017379), SNP3 Position: Chr14:2525852) in bovine TRAPPC9 and one SNP (Position: Chr5:104010752) in CD4 were screened through Chinese Cow's SNPs Chip-I (CCSC-I) and genotyped in a population of 312 Chinese Holsteins (156: Mastitis, 156: Healthy). The results were analyzed using the general linear model in SAS 9.4. Our analysis revealed that milk protein percentage, somatic cell count (SCC), somatic cell score (SCS), serum cytokines interleukin 6 (IL-6) and interferon-gamma (IFN-γ) were significantly (P < 0.05) associated with at least one or more identified SNPs of TRAPPC9 and CD4 genes. Furthermore, the expression status of SNPs in CD4 and TRAPPC9 genes were verified through RT-qPCR. The expression analysis showed that genotypes GG in SNP3 of TRAPPC9 and TT genotype in SNP4 of CD4 showed higher expression level compared to other genotypes. The GG genotype in SNP2 and TT genotype in SNP3 of TRAPPC9 were associated with higher bovine milk SCC and lower IL6. Altogether, our findings suggested that the SNPs of TRAPPC9 and CD4 genes could be useful genetic markers in selection for milk protein improvement and mastitis resistance phenotypic traits in dairy cattle. The CCSC-I used in current study is proposed to be validate in different and large population of dairy cattle not only in China but also in other countries. Moreover, our analyses recommended that besides SCC and SCS, the association of genetic markers could also be considered with the serum cytokines (IL-6, IFN-γ) while selecting genetically mastitis resistance dairy cattle.
Bovine mastitis is the inflammation of udder tissues with a marked decrease in milk quantity and quality in dairy cattle (1, 2). Bovine mastitis is one of the most costly diseases affecting dairy cattle's health and welfare globally (3, 4). This disease caused around $2 billion in losses to the US dairy industry annually (5). Because of mammary gland inflammation, the polymorphonuclear leukocytes from blood rush toward the site and results in a marked increase in milk's somatic cells content (6).
The somatic cell count (SCC) and somatic cell score (SCS) are the key indicators for susceptibility and resistance of a cow to mastitis (7–10). Due to the positive genetic correlation (0.4–0.8) between mastitis and SCC or SCS (11, 12), the strategy to minimize the risk of this disease by selecting dairy cattle against higher SCS is a worthy approach. However, the SCC and SCS are not constant and are influenced by many environmental factors (13, 14); therefore, in current research, we targeted serum cytokines (IL-6 and IFN-γ) in combination with SCC and SCS as mastitis resistance phenotypic traits.
Cytokines have dual nature, i.e., either activate or repress the inflammatory response and thus play a vital role in mastitis development (15). In addition, the increased levels of IFN-γ, IL-6 and IL-17 have been documented in acute mastitis (16). Consistently, a study showed that the detection of IL-6 in milk indicated subclinical mastitis earlier than SCC (17). Although the mentioned phenotypic traits are good indicators of mastitis, however, due to low heritability, mastitis resistance will yet be a challenge for animal breeders (18). Therefore, the association of mastitis resistance phenotypic traits with a polymorphism in genes is the research of interest in the modern dairy industry for control of mastitis.
The researchers rely on a marker-assisted selection strategy for mastitis resistance (19, 20). The candidate gene approach, which takes in account of SNPs in the genes that are associated with these traits, is a widely used method to control mastitis (19). Being quantitative traits, mastitis and milk production traits are controlled by many genes (21). The TRAPPC9 and CD4 are the key genes that play an important role in developing innate immunity and milk production traits.
The TRAPPC9 gene, residing on bovine chromosome 14 is the vital member of the nuclear factor kappa B (NF-κB) family which has an essential role in inflammation and innate immunity (22–24). The elevated level of TRAPPC9 gene enhances the activity of NF-κB signaling during mastitis development in dairy cattle (22). The associations of polymorphisms in CD4 and TRAPPC9 genes with milk production and mastitis resistance traits have been documented in previous reports (2, 25–27). Similarly, our in our previous study by using transcriptomic screening, we reported that TRAPPC9 gene was significantly associated with milk SCC and bovine mastitis susceptibility (28). Recently, a study have reported the association of TRAPPC9 gene with milk fat and immunity in Ayrshire and Jersey dairy cattle (29). In addition, the increase in milk CD4+ T cells was documented to be correlated with non-specific mastitis which suggested their link with low bacterial shedding (30).
Keeping in view the importance of these two immunity-associated genes (TRAPPC9, CD4); we selected three polymorphisms (SNP1 Position: Chr14:2484891, SNP2 (rs110017379), SNP3 Position: Chr14:2525852) in bovine TRAPPC9 and one SNP (Position: Chr5:104010752) in CD4 from our previous studies and validate them in a new and large Chinese Holstein population. For this purpose, these SNPs in TRAPPC9 and CD4 genes were detected by a new technique, i.e., Chinese Cow's SNPs Chip-I (CCSC-I) and genotyped in a different and a bit larger Chinese Holsteins population to explore their association with mastitis resistance and milk production phenotypic traits.
All animal procedures were performed according to the regulation approved by the ethical committee of the College of Animal Science and Technology, China Agriculture University, Beijing, PR China [Permission number: DK996]. All the data was collected from China Agriculture University dairy farm and no consent was needed from farmers.
We randomly selected a total of 312 Chinese Holstein cows (156: Mastitis, 156: Healthy) in parities ranging 1–3 from a single dairy farm in Beijing China. In addition, based on SCC level, the cows were confirmed as mastatic (cattle with SCC higher than 200,000/ml) or healthy (cattle with SCC lower than 200,000/ml). The blood samples were collected from the caudal vein of all the selected population of Chinese Holsteins in 9 mL of 3 tubes including one each for DNA extraction (EDTA coated tube), RNA extraction, and serum isolation (non-EDTA tube). For serum isolation, the blood samples were placed at room temperature for 30 min to enable blood coagulation and then centrifuged at 3,000 rpm for 10 min to separate serum. The serum samples were stored at 4°C and sent to the Beijing Huaying Biological Technology Research Institute within 24 h to detect the concentration of IL-6 and IFN-γ. The milk SCC data were obtained from the Beijing Jinyindao Dairy Farm data record section, while SCS was calculated using the formula: SCS = log2 (SCC / 100,000) + 3.
Genomic DNA was isolated from blood samples of 312 Chinese Holstein using Tiangen Blood DNA Kit (Tiangen Biotech Co., China) following the manufacturer's instructions. The quantity and quality of DNA were measured using NanoDrop ND-2000c Spectrophotometer (Thermo Scientific, Chelmsford, MA, USA) and gel electrophoresis. After confirmation of quality and quantity, all the DNA samples were sent to Capital Bio Technology Co., Ltd, Beijing, China, for identification of SNPs and genotyping with Chinese Cow's SNPs Chip-I. The selected SNPs in TRAPPC9 and CD4 genes were genotyped in the different and bit large population of 312 Chinese Holstein.
Total RNA extraction from the Holstein cattle's white blood cells was carried out through the standard TRIzol method (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocols. RNase-Free DNase Set (QIAGEN) was used to purify RNA and to ensure genomic DNA elimination. The quantity and quality of RNA were measured by using a NanoDropTM ND-2000c Spectrophotometer (Thermo Scientific, Inc.), and the integrity of RNA was monitored on 1% agarose gel.
According to the manufacturer's instructions, reverse transcription was performed using PrimeScript 1st Strand cDNA Synthesis kit (TaKaRa, Dalian, China). The PCR primers for the bovine CD4, TRAPPC9, and a housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were designed by Primer-Blast on NCBI and synthesized by Beijing Genomics Institute Tech, based on the golden rules for real-time reverse transcription PCR (RT-PCR). The amplification efficiency of these primer pairs was tested by RT-qPCR initially, and the mRNA expression of the two genes was normalized against the housekeeping gene GAPDH by the cDNA in the corresponding samples. Three pairs of primers designed for GAPDH, TRAPPC9, and CD4 are given in Table 1. For mRNA expression analysis, four samples for each SNP were run in triplicate.
Real-time quantitative polymerase chain reaction (RT-qPCR) was performed to determine TRAPPC9 and CD4 genes expression levels. The reactions were performed in a total volume of 20 μL containing 2 μL cDNA, 1 μL each primer, 10 μL SYBR Green Master Mix (Roche, Penzberg, Germany), 6 μL nuclease-free water using the following amplification condition: 94°C for 10 min, followed by 44 cycles of 94°C for 15 s, 60°C for 10 s, 72°C for 10 s, and 72°C for 30 s. Fluorescence signals were collected at 60°C step. Mean was consequential from the two repeats for each sample. Light Cycler 480 RT-PCR system was used to perform amplification, detection and data analyses.
The allele and genotype frequencies were tested for deviations from proportions of Hardy–Weinberg equilibrium (HWE) by using Chi-square test (χ2). The association analysis of SNPs in TRAPPC9 and CD4 with milk production and mastitis-related traits were carried out by the least-squares method as applied in the GLM procedure of SAS (SAS Institute Inc., Cary, NC, USA) according to the following linear model.
where Pijkn indicates mastitis traits (SCC, SCS or serum concentration of cytokine IL-6 and IFN-γ) or milk production traits (fat percentage or protein percentage), μ is overall mean, fi is the fixed effect of the farm, pj is the fixed effect of parity, snpk is the fixed effect of genotype, eijkn is the random residual error.
The estimated genotype effects were further divided into additive effect (A) and dominant effect (D). The additive effect was the mean deviation of two homozygous genotypes (Formula 1), and the dominant influence was calculated by the deviation of the heterozygous genotype from the mean of two homozygous genotypes (Formula 2) (31).
Where, AA, AB and BB were least square means of genotype AA, AB and BB, respectively.
Student t-test was performed for RT-qPCR analyses for the comparison of mRNA expression levels of different genotypes of SNPs in the two genes (TRAPPC9 and CD4).
Three SNPs in TRAPPC9 and one SNP from CD4 gene were screened through CCSC-I (10) and genotyped in a total of 312 (156: Mastitis, 156: Healthy) Chinese Holstein population. It was found that except the SNP1, all SNPs' allele and genotypic frequencies were in Hardy–Weinberg equilibrium (P > 0.05). The observations, genotypic and allelic frequencies and values of Chi-square test (χ2) of the selected SNPs in the present study are summarized in Table 2.
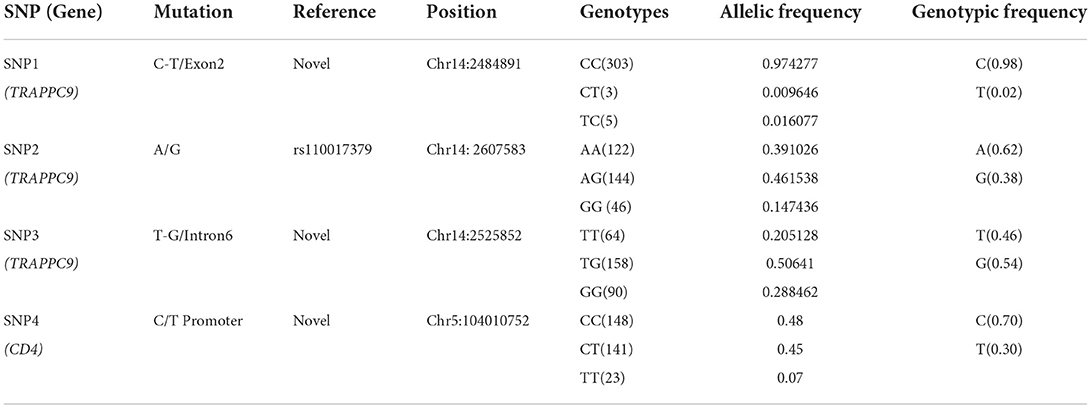
Table 2. The information of single nucleotide polymorphisms and their Allelic and genotypic frequencies in TRAPPC9 and CD4 genes.
The association of three SNPs in TRAPPC9 was evaluated with milk production and mastitis resistance traits. Our findings illustrated that SNP1 was significantly associated with milk protein, the SNP2 at position A/G 2607583 linked notably with SCS (P < 0.05), whereas the SNP3 (T/G 2525852) with SCC, SCS, and serum cytokine IL-6 (P < 0.05). Additionally, the association analysis revealed that the SNP in the CD4 gene (104010752C/T) did not show any link with milk production traits, however, revealed a significant association with IFN-γ and IL-6 (P < 0.05). Finally, the association analysis showed that the genotypes GG in SNP3 and homozygous TT (SNP4) were significantly associated with low SCC, SCS and a higher level of IL-6 (Table 3). Similarly, the analyses revealed that genotypes TT (SNP3) and CC (SNP4) were associated with low IL-6 and high SCC level, which make the dairy cattle more vulnerable to the mastitis development.
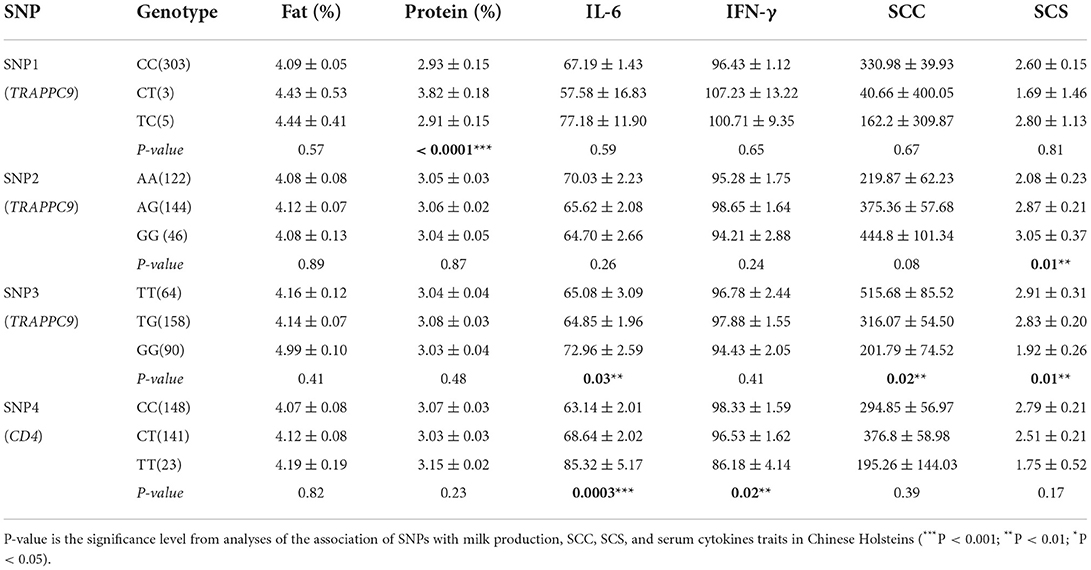
Table 3. Association of SNPs in TRAPPC9 and CD4 genes on SCC, SCS, milk production, and serum cytokines traits in Chinese Holsteins.
The additive and dominant effect of SNPs in CD4 and TRAPPC9 genes are summarized in Table 4. The current association analysis for the additive and dominant effect of polymorphisms (SNP1, SNP2, SNP3and SNP4) revealed that dominant effect of SNP1 is significantly (P < 0.05) associated with milk protein, whereas SNP3 showed a significant additive effect on SCC, SCS and IL-6 (P < 0.05). Similarly, the additive effect of SNP2 was significantly associated with SCS and the SNP4 in CD4 was correlated with IFN-γ and IL-6 (P < 0.05) (Table 4).
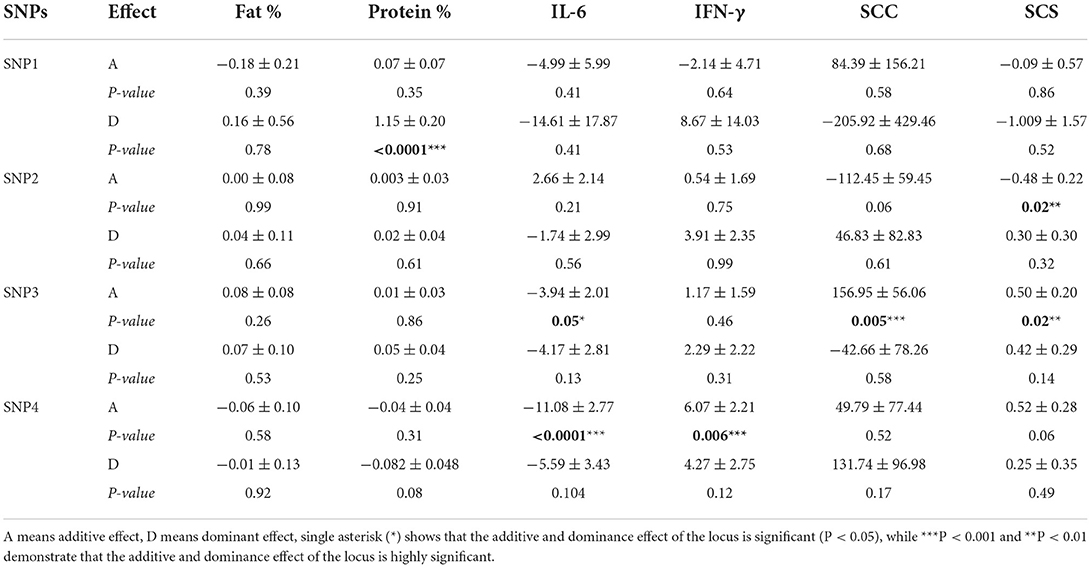
Table 4. Genetic effect of SNPs in TRAPPC9 and CD4 genes on milk production, mastitis resistance and serum cytokine traits in Chinese Holstein.
The expression level of mutations in the TRAPPC9 and CD4 genes was measured by real-time quantitative PCR. The analysis showed that the relative mRNA expression of AA genotype in SNP2 had higher expression compared to GG and AG genotypes in the TRAPPC9 gene. Similarly, the GG genotype in SNP3 in TRAPPC9 demonstrated significantly higher expression than TG genotype in the given Chinese Holsteins dairy cattle population (Figure 1A, P < 0.05). Moreover, the genotype TT's mRNA expression level was comparatively higher than CC and CT genotypes in the SNP4 of CD4 gene (Figure 1B).
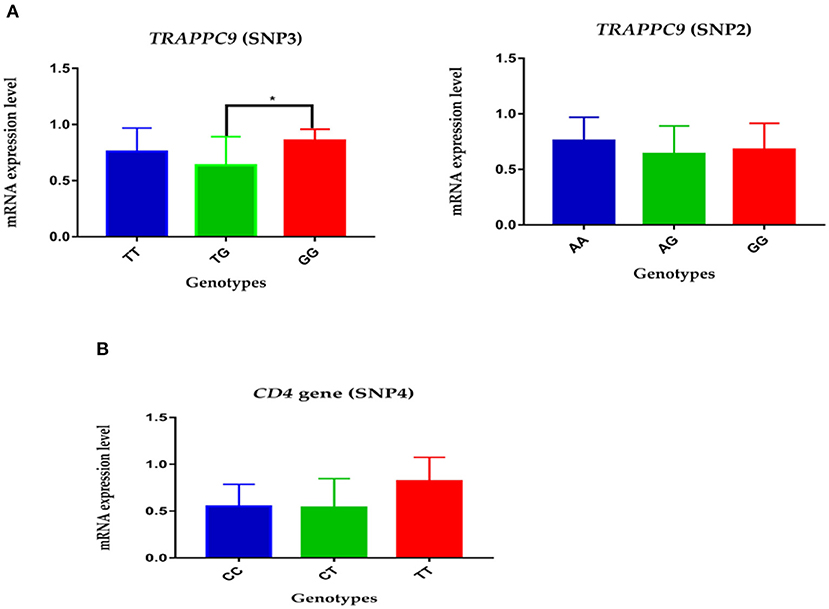
Figure 1. The relative mRNA expression level of polymorphisms in TRAPPC9 and CD4 gene: (A) The expression level of genotypes in SNP2 and SNP3 of TRAPPC9: the GG genotype show significantly higher relative mRNA expression than TG in SNP3; however, no significant difference was reported among the three genotypes of SNP2. (B) The relative genotypes expression of SNP4 in CD4 gene: the TT genotype showed comparatively higher relative mRNA expression than CT and CC.
Recently, single nucleotide polymorphisms in many genes have been found to be associated with milk production and mastitis resistance traits suggesting that these variants could be used as potential genetic markers in modern breeding schemes for the improvement of production and increasing resistance to mastitis. In the present study, the polymorphisms in TRAPPC9 and CD4 genes that cause variation in the economic and health traits (milk production and mastitis resistance phenotypic traits) were selected from our previous studies and analyzed for validation in a new and larger population by using CCSC-I. To our knowledge, this is the first study in which we practically applied CCSC-I in mastitis resistance research. Bovine SNP Chip's application was previously used by Mullen and his co-workers in 2013 for dairy and beef production research (32). Similarly, a research study has also reported that SNP Chip is a cost and time-effective approach for implementing genomic selection in livestock (33). Keeping in view the importance of the SNP Chip from various published studies we used this technique in the present study to validate the role of the significant variants from our previous studies in CD4 and TRAPPC9 genes for production and mastitis resistance traits. Our research team reported in previous studies that SNPs at position 2484891 C/T, 2525852 T/G and 2607583 A/G in TRAPPC9 were associated with milk protein and fat percentages (34). In contrast, polymorphism at position 2525852 T/G (TRAPPC9) did not show any link with milk contents (35). Furthermore, the mutation (2607583 A/G) was noticed to be associated with SCS, while the SNPs at position 2484891 C/T, and 2525852 T/G were linked to IL-6 and IFN-γ, respectively, however no correlation of 2607583 A/G in TRAPPC9 gene was found with SCC, SCS and serum cytokines (IL-6 and IFN-γ) (36). In comparison, we found that the SNP (2484891 C/T) was linked to protein percentage, whereas the polymorphisms 2525852 T/G and 2607583 A/G were associated with IL-6, SCS, SCC, and SCS, respectively. Moreover, the mutation at point C104010752T was significantly correlated to milk SCC in Chinese Holsteins (26) and mutation at point g.13598C>T was linked to milk yield, protein and SCS (27). Importantly, in the new dairy population, our results further revealed the noteworthy correlation of SNP (C104010752T) with IL-6 and IFN-γ instead of SCC and SCS (P < 0.05). Based on our current findings, we reported that the SNPs in TRAPPC9 and CD4 genes show pleiotropic ability; however, it is also possible that the documented polymorphisms in the current study and our previous research might be influenced by population size and environmental factors.
Within an SNP, allele combinations and genotypes exert a critical role in the regulation of any traits. In the current study, we found that the homozygous GG genotype in SNP3 (TRAPPC9) and TT genotype in SNP4 (CD4) were associated with a higher level of IL-6 and a low level of SCC. These findings suggested that the mentioned genotypes (GG and TT) are linked with mastitis resistance and should be considered as potential markers while selecting genetically mastitis resistance cattle. Finally, the primary functional validation of SNPs in TRAPPC9 and CD4 genes were verified through RT-qPCR. Similar trends for all the genotypes in SNPs (TRAPPC9 and CD4) that were found for their association with serum cytokines and mastitis resistance phenotypic traits were also reported in expression analysis. In relation to the association of the genetic variants to the specific mastitis resistance phenotypic trait and the level of significance documented in our previous studies, there were some differences in the present study's findings and in the previous studies, which still need further validation in large and different population of Chinese Holsteins.
In general, the present study using a newly designed CCSC-I for genotyping to validate the association of SNPs in TRAPPC9 and CD4 genes with milk production and mastitis resistance traits. Although we reported the significant link of the selected SNPs in TRAPPC9 and CD4 genes milk production and mastitis resistance phenotypic traits, however, we recommend further in-depth studies to test the documented reported SNPs of TRAPPC9 and CD4 in a large Chinese Holstein population as well as in other different dairy breeds from various regions of the world by using our newly designed CCSC-I for the validation of its capability to improve milk production and mastitis resistance in dairy cattle.
Overall, the present study validated the three SNPs of TRAPPC9 and one SNP of CD4 in a large and different Chinese Holstein population by using our newly designed CCSC-I. The results verify that the documented SNPs in both genes (TRAPPC9 and CD4) could be considered as powerful genetic markers against bovine mastitis resistance. The targeted SNPs in the TRAPPC9 gene might be used as a marker for improved milk protein percentage as well. The study proposed that the CCSC-I could also be validated in more large dairy cattle population not only in China but also in other countries across the globe. Additionally, the upshot of a study infers that not only SCS and SCC but IL-6 and IFN-γ association can be establish with genetic markers while selecting genetically mastitis resistance dairy cattle.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Ethical Committee of the College of Animal Science and Technology, China Agriculture University, Beijing, PR China [Permission number: DK996].
Conceptualization and methodology: MK and YY. Validation: MK. Resources: MK and GD. Writing—original draft preparation: MK. Writing—review and editing: MK, AK, YY, and GD. Supervision: YY. All authors have read and agreed to the published version of the manuscript.
This article was financially supported by the National Key R&D Program of China (2021YFD1200900, 2021YFD1200903), NSFC-PSF Joint Project (31961143009), Beijing Dairy Industry Innovation Team (BAIC06), China Agriculture Research System of MOF and MARA, Beijing Natural Science Foundation (6182021), and the Program for Changjiang Scholar and Innovation Research Team in University (IRT-15R62).The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gomes, F, Henriques M. Control of bovine mastitis: old and recent therapeutic approaches. Curr Microbiol. (2016) 72:377–82. doi: 10.1007/s00284-015-0958-8
2. Usman T, Ying Y, Zhai L, Chao L, Wang X, Wang Y. Association of CD4 SNPs with a fat percentage of Holstein cattle. Genet Mol Res. (2016) 15:gmr.15038697. doi: 10.4238/gmr.15038697
3. Rainard P, Cunha P, Gilbert FB. Innate and adaptive immunity synergize to trigger inflammation in the mammary gland. PLoS ONE. (2016) 11:e0154172. doi: 10.1371/journal.pone.0154172
4. Whelehan JC, Meade GK, Eckersall DP, Young, JF, O'Farrelly C. Experimental Staphylococcus aureus infection of the mammary gland induce region-specific changes in innate immune gene expression. Vet Immunol Immunopathol. (2011) 140:181–9. doi: 10.1016/j.vetimm.2010.11.013
5. Sordillo LM, Streicher KL. Mammary gland immunity and mastitis susceptibility. J Mammary Gland Biol Neoplasia. (2002) 7:135–46. doi: 10.1023/A:1020347818725
6. Lafi SQ. Use of somatic cell counts and California Mastitis Test results from udder halves milk samples to detect subclinical intramammary infection in Awassi sheep. Small Ruminant Res. (2006) 62:83–6. doi: 10.1016/j.smallrumres.2005.07.035
7. Usman, T, Yachun W, Chao, L, Wang X. Association study of single nucleotide polymorphisms in JAK2 and STAT5B genes and their differential mRNA expression with mastitis susceptibility in Chinese Holstein cattle. Animal Genet. (2015) 46:371–380. doi: 10.1111/age.12306
8. Gernand E, König S. Random regression test-day model for clinical mastitis: genetic parameters, model comparison, and correlations with indicator traits. J Dairy Sci. (2014) 97:3953–63. doi: 10.3168/jds.2013-7830
9. Wang X, Zhong J, Gao Y, Ju Z, Huang J. A SNP in intron 8 of CD46 causes a novel transcript associated with mastitis in Holsteins. BMC Genomics. (2014) 15:630. doi: 10.1186/1471-2164-15-630
10. Khan M, Wang D, Liu L, Usman T, Wen H, Zhang R, et al. Significant genetic effects of JAK2 and DGAT1 mutations on milk fat content and mastitis resistance in Holsteins. J Dairy Res. (2019) 86:388–93. doi: 10.1017/S0022029919000682
11. Hu HC, Wang HM, Li JB, Wang CF, Lai SJ, Li LQ, et al. Genetic polymorphism of the Nramp1 gene and correlation with mastitis in Holstein cattle. Yi Chuan. (2009) 31:57–62. doi: 10.3724/SP.J.1005.2009.00057
12. Rupp R, Bergonier D, Dion S, Hygonenq CM, Aurel RM, Christèle R, et al. Response to somatic cell count-based selection for mastitis resistance in a divergent selection experiment sheep. J Dairy Sci. (2009) 92:1203–19. doi: 10.3168/jds.2008-1435
13. Van G, Green LE, Guzmán D, Esparza H, Tadich N. Risk factors for bulk milk somatic cell counts and total bacterial counts in smallholder dairy farms in the 10th region of Chile. Prevent Vet Med. (2005) 67:1–17. doi: 10.1016/j.prevetmed.2004.10.002
14. Reyes J, Sanchez J, Stryhn H, Ortiz T, Olivera M, Keefe GP. Influence of milking method, disinfection and herd management practices on bulk tank milk somatic cell counts in tropical dairy herds in Colombia. Vet J. (2017) 220:34–39. doi: 10.1016/j.tvjl.2016.12.011
15. Bonifati C, Ameglio F. Cytokines in psoriasis. Int J Dermatol. (1999) 38:241–51. doi: 10.1046/j.1365-4362.1999.00622.x
16. Sakemi Y, Tamura Y, Hagiwara K. Interleukin-6 in quarter milk as a further prediction marker for bovine subclinical mastitis. J Dairy Sci. (2011) 78:118–21. doi: 10.1017/S0022029910000828
17. Fontanesi L, Calo DG, Galimberti G., Negrini R., Marino R., Nardone A., et al., Russo V. A candidate gene association study for nine economically important traits in Italian Holstein cattle. Animal Genet. (2014) 45:576–80. doi: 10.1111/age.12164
18. Hinchs D, Bennewitz J, Stamer E, Junge W, Kalm E, Thaller G. Genetic analysis of mastitis data with different models. J Dairy Sci. (2011) 94:471–8. doi: 10.3168/jds.2010-3374
19. Wiggans GR, Vanraden PM, Cooper TA. The genomic evaluation system in the United States: past, present, future. J Dairy Sci. (2011) 94:3202–11. doi: 10.3168/jds.2010-3866
20. García-Ruiz A, Cole JB, VanRaden PM, Wiggans GR, Ruiz-López FJ, Van CP. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc Natl Acad Sci USA. (2016) 113:E3995–4004. doi: 10.1073/pnas.1519061113
21. Sender G, Agnieszka K, Adrianna P, Karima G, Jolanta O. Genetic basis of mastitis resistance in dairy cattle –a review. Annals of Animal Science. (2013) 13:663– 673. doi: 10.2478/aoas-2013-0043
22. Khan MZ, Khan A, Xiao J, Ma J, Ma Y, Chen T, et al. Z. Overview of research development on the role of NF-κB signaling in mastitis. Animals. (2020) 10:1625. doi: 10.3390/ani10091625
23. Denis-Donini S, Dellarole A, Crociara P, Maria TF, Valeria B, Giorgia Q, et al. Impaired adult neurogenesis associated with short-term memory defects in NF-kappaB p50-deficient mice. J Neurosci. (2008) 28:3911–9. doi: 10.1523/JNEUROSCI.0148-08.2008
24. Oeckinghaus A, Matthew SH, Sankar G. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. (2011) 12:695–708. doi: 10.1038/ni.2065
25. Wang X, Ma P, Liu J, Zhang Q, Zhang Y, Ding X, et al. Genome-wide association study in Chinese Holstein cows reveals two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genetics. (2015) 16:16–111. doi: 10.1186/s12863-015-0263-3
26. Usman T, Yachun W, Minyan S, Xiao W., Yichun D, Chao L, et al. Novel polymorphisms in bovine CD4 and LAG-3 genes associated with somatic cell counts of clinical mastitis cows. Genetics Mol Res. (2017) 16:gmr16039859. doi: 10.4238/gmr16039859
27. He Y, Qin C, Peipei M, Yachun W, Qin Z, Dongxiao S, et al. Association of bovine CD4 and STAT5b single nucleotide polymorphisms with somatic cell scores and milk production traits in Chinese Holsteins. J Dairy Res. (2011) 78:242–9. doi: 10.1017/S0022029911000148
28. Wang D, Liu L, Augustino S, Duan T, Hall TJ, MacHugh DE, et al. Identification of novel molecular markers of mastitis caused by Staphylococcus aureus using gene expression profiling in two consecutive generations of Chinese Holstein dairy cattle. J Animal Sci Biotechnol. (2020) 11:1–17. doi: 10.1186/s40104-020-00494-7
29. Freitas PHF, Oliveira HR, Silva FF, Fleming A, Schenkel FS, Miglior LF, et al. Time-dependent genetic parameters and single-step genome-wide association analyses for predicted milk fatty acid composition in Ayrshire and Jersey dairy cattle. J Dairy Sci. (2020) 103:5263–9. doi: 10.3168/jds.2019-17820
30. Souza FN, Blagitz MG, Batista CF, Takano PV. Immune response in nonspecific mastitis: What can it tell us? J Dairy Sci. (2020) 103:5376–86. doi: 10.3168/jds.2019-17022
32. Mullen PM, McClure CM, Kearney FJ, Waters MS, Weld R, Flynn P, et al. Development of a custom SNP chip for dairy and beef cattle breeding, parentage and research. Interbull Bulletin. (2013) 47:23–25.
33. Berry DP, McClure MC, Mullen MP. Within- and across-breed imputation of high-density genotypes in dairy and beef cattle from medium- and low-density genotypes. J Animal Breed Genet. (2013) 131:165–72. doi: 10.1111/jbg.12067
34. Feng W, Dong Y, Xiao W, Liu C, Wang X, Wang Y, et al. The genetic effect of TRAPPC9 on mastitis resistance to S. aureus in dairy cows. Acta Veterin Zootecn Sina. (2016) 47:276–83. https://www.cabdirect.org/cabdirect/abstract/20163110701
35. Dong Y, Liu C, Xiao W, Wang Y, Zhang Y, Sun D, et al. Confirming the genetic effect of Bovine TRAPPC9 on milk production traits based on Post-GWAS strategies. Acta Veterin Zootecn Sina. (2015) 46:60–8. https://www.cabdirect.org/cabdirect/abstract/20153108958
Keywords: Chinese Holstein, milk protein, mastitis, SNP, CD4, TRAPPC9
Citation: Khan MZ, Dari G, Khan A and Yu Y (2022) Genetic polymorphisms of TRAPPC9 and CD4 genes and their association with milk production and mastitis resistance phenotypic traits in Chinese Holstein. Front. Vet. Sci. 9:1008497. doi: 10.3389/fvets.2022.1008497
Received: 31 July 2022; Accepted: 26 August 2022;
Published: 23 September 2022.
Edited by:
Abdul Rasheed Baloch, University of Karachi, PakistanReviewed by:
Saqib Umer, University of Agriculture, Faisalabad, PakistanCopyright © 2022 Khan, Dari, Khan and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Yu, eXV5aW5nQGNhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.