
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 24 December 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.806653
This article is part of the Research Topic Molecular Mechanisms of Bacterial Disease in Cultured Fishes View all 10 articles
Antimicrobial resistance (AMR) has become a major concern worldwide. To evaluate the AMR of Escherichia coli in aquaculture farms of Zhanjiang, China, a total of 90 samples from the water, soil, and sediment of three aquaculture farms (farms I, II, and III) in Zhanjiang were collected, and 90 strains of E. coli were isolated for drug resistance analysis and AMR gene detection. The results indicated that the isolated 90 strains of E. coli have high resistance rates to penicillin, amoxicillin, ampicillin, tetracycline, compound sulfamethoxazole, sulfisoxazole, chloramphenicol, florfenicol, and rifampin (≥70%). Among these antimicrobial drugs, the resistance rate to rifampicin is as high as 100%. Among the isolated 90 strains of E. coli, all of them were resistant to more than two kinds of antimicrobial drugs, the number of strains resistant to nine kinds of drugs was the largest (19 strains), and the most resistant strain showed resistance to 16 kinds of antibacterial drugs. Regarding the AMR genes, among the three aquaculture farms, the most resistance genes were detected in farm II (28 species). The detection rate of blaTEM, blaCIT, blaNDM, floR, OptrA, cmlA, aphA1, Sul2, oqxA, and qnrS in 90 isolates of E. coli was high (≥50%). The detection rate of carbapenem-resistant genes, such as blaKPC, blaIMP, and cfr, was relatively lower ( ≤ 30%), and the detection rate of mcr2 was the lowest (0). At least four AMR genes were detected for each strain, and 15 AMR genes were detected at most. Among them, the number of strains that carried 10 AMR genes was the largest (15 strains). Finally, a correlation analysis found that the AMR genes including blaTEM, blaCIT, floR, OptrA, cmlA, aac(3)-II, Sul2, ereA, ermB, oqxB, qnrA, mcr1, and mcr2 had a high correlation rate with drug resistance (≥50%). To summarize, the 90 strains of E. coli isolated from water, surrounding soil, and sediment samples showed resistance to multi-antimicrobial drugs and carried various antimicrobial resistance genes. Thus, it is essential to strengthen the rational use of antimicrobial drugs, especially the amide alcohol drugs, and control the AMR in the aquaculture industry of Zhanjiang, China.
Currently, due to human demand for a variety of animal proteins, the aquaculture industry is developing rapidly (1). China has been one of the largest producers of aquatic products, and the coastal area is the main aquaculture base, including the coastal city of Zhanjiang (2). Because of poor resistance and susceptibility to disease of aquatic animals, antibiotics are widely used in aquaculture to prevent and treat bacterial diseases (1). As a result, the residual antibiotics in aquatic products pose a potential risk to food safety, and antibiotics excreted from aquatic animals are dispersed in the water and sediments, thus leading to the emergence of drug-resistant bacteria and antimicrobial resistance (AMR) genes in aquaculture farms and the surrounding environment (3, 4). Under the pressure of excessive use of antibiotics, the drug-resistant bacteria even appeared to demonstrate multi-drug resistance, which causes serious negative impacts on public health (5). Besides this, according to the report of Pruden et al. (6), the AMR genes are novel environmental pollutants. The existence of drug resistance genes is the key to the development of drug resistance in bacteria. The AMR genes not only spread vertically but also spread to other bacteria horizontally through genetic elements, increasing the number of drug-resistant strains and causing difficulties in the treatment of clinical diseases and infection in both humans and animals (7, 8).
Escherichia coli is a gram-negative and common conditional pathogenic bacteria in the intestines and environment of humans and animals (9). Pathogenic Escherichia coli results in intestinal diseases and infections of farm animals (10, 11). Escherichia coli also induces diseases in various aquatic animals, thereby causing serious economic losses for the aquaculture industry. In this context, the excessive use of antibacterial drugs in aquaculture has become inevitable, and aquaculture has become an important source of antibiotic-resistant strains and AMR genes of E. coli (1). However, the terrible thing is that the antibiotic-resistant bacteria can infect humans through the food chain or transfer AMR genes to human pathogens, leading to serious diseases such as meningitis, sepsis, and enterotoxemia in humans (12). Furthermore, the frequent application of similar antimicrobial drugs in the treatment of E. coli induced the diseases of animals (including aquatic animals) and humans nowadays, which makes it difficult to find an effective antimicrobial agent in case of bacterial infections in humans (13). However, little is known about the drug resistance and the distribution of AMR genes of E. coli isolates from aquaculture farms and their surroundings in Zhanjiang, China. Therefore, to provide basic data for a better understanding of AMR in aquaculture, the present study was conducted to evaluate the distribution of AMR genes and analyze the drug resistance of E. coli in aquaculture farms of Zhanjiang, China.
The water, surrounding soil, and sediment samples were randomly collected from three aquaculture farms in Zhanjiang, China (farms I, II, and III). The sampling locations are shown in Figure 1. The water samples were collected at 1–10 cm in the water surface, the soil samples were collected at 1–3 cm on the surface of the surrounding aquaculture farms, and the sediment samples were collected at 1–5 cm from the surface of the sediment. There were 10 samples of water, soil, and sediment from each aquaculture farm, respectively, and a total of 90 samples were collected in this study.
MacConkey agar medium, Eosin Meilan agar medium, and ordinary nutrient broth were purchased from Beijing Luqiao Technology Co., Ltd. (Beijing, China); 5 × TBE buffers were obtained from Shenggong Bioengineering (Shanghai) Co., Ltd. (Shanghai, China); Premix EX TaqTM version 2.0 and DL1000 DNA Marker were both purchased from TaKaRa Bioengineering (Dalian) Co., Ltd. (Dalian, China); agarose (produced in Spain) was purchased from Beijing Kaioudi Biotechnology Co., Ltd. (Beijing, China); and the Goldview nucleic acid stains were from Vazyme Nanjing Biotech Co., Ltd. (Nanjing, China).
There was a total of 23 kinds of antibacterial drug sensitivity reagent tablets, including penicillin, amoxicillin, cefotaxime, ceftriaxone, aztreonam, gentamicin, amikacin, azithromycin, tetracycline, doxycycline, ciprofloxacin, ofloxacin, compound sulfamethoxazole, chloramphenicol, polymyxin B, nitrofurantoin, rifampicin, florfenicol, ampicillin, streptomycin, lomefloxacin, sulfisoxazole, and fosfomycin, which were from Chicheng Pharmaceutical Technology Co., Ltd. (Hangzhou, China). The quality control bacterial strain is E. coli ATCC 25922 (from the Laboratory of Basic Veterinary Medicine, Guangdong Ocean University, Zhanjiang, China).
The collected 90 samples (water, soil, and sediment) were diluted with sterile water to a suitable quantity. Then, 100 μl was taken and spread evenly on MacConkey agar medium and incubated at 37°C for 18–24 h. A single colony with a smooth, moist, pink surface was selected and placed on eosin. Streak purification was done on blue agar medium, and the sample was cultured at 37°C for 18–24 h. A single colony with metallic luster, black or brown-green, was selected and inoculated on eosin melan agar medium for streak purification again; finally, the suspected E. coli was removed. The strains were inoculated into nutrient broth medium, cultivated at 37°C for 24 h, and then stored at −20°C with 30% glycerol for later analysis.
PCR method was used for the identification of isolated E. coli strains from the samples. The PCR primers were designed based on the specific phoA gene sequence of E. coli—upstream primer: 5′-TACAGGTGACTGCGGGCTTATC-3′, downstream primer: 5′-CTTACCGGGCAATACACTCACTA-3′–and the primers were synthesized and produced by Shenggong Bioengineering Co., Ltd. (Shanghai, China). The length of the product is 622 bp. The DNA of the isolated strains was extracted by using DNA extraction kits (Tiangen Biochemical Technology Co., Ltd., Beijing, China). The PCR reaction system is 15 μl: DNA template 1 μl, each 0.5 μl of upstream and downstream primers (25 μmol/L), and Taq 13 μl. The PCR reaction conditions were as follows: pre-denaturation at 94°C for 7 min, denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s—for a total of 30 cycles, and final extension at 72°C for 5 min. The PCR products were electrophoresed on 1% agarose gel, and E. coli ATCC 25922 was used as standard bacteria and positive control; ddH2O was used as the blank negative control.
The drug susceptibility reagent tablet (test strips) method, which was recommended by the Clinical and Laboratory Standards Institute, was performed to detect and analyze the drug resistance of the isolated E. coli strains. Briefly, 90 strains of E. coli isolated from 90 samples were resuscitated, and 100 μl bacterial liquid was evenly spread on a common nutrient agar medium, and 23 kinds of antimicrobial drug-sensitive reagent tablets were used on the surface of the medium and then incubated at 37°C for 18–24 h. The diameter of the inhibition zone was observed and recorded, and the results were determined for drug resistance (14). There were three replicates for each antibacterial drug resistance analysis. Escherichia coli ATCC 25922 was used as the quality control bacteria. Finally, the resistance to each antimicrobial drug and the multi-drug resistance of the isolated E. coli strains from water, soil, and sediment were statistically analyzed by using Excel 2012.
The PCR method was used to detect 29 kinds of AMR genes carried in the 90 isolated strains of E. coli from the water, soil, and sediment samples of the aquaculture farms. The primers are presented in Table 1 and designed according to previous studies (the reference details are described in Table 1). The PCR reaction system is 15 μl, which contained 1 μl of DNA template, 0.5 μl of upstream and downstream primers, and 13 μl of Premix EX TaqTM. ddH2O was used as blank negative control. The PCR reaction procedure was as follows: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 s, annealing for 30 s at melting temperature (Tm, °C) (the Tm of each AMR gene is listed in Table 1), extension at 72°C for 1 min for 30 cycles, and a final extension at 72°C for 10 min. The PCR products were stored at 4°C. The PCR products were analyzed by 1% agarose gel electrophoresis, and the results were recorded.
Positive fragments of the target gene were randomly picked and recovered using Poly-Gel DNA Extraction Kit (OMEGE) and cloned according to vector ligation (pMD19-T vector, TaKaRa) and competent cells (DH-5α, TaKaRa). The plasmid (Plasmid DNA Kit, OMEGE) was extracted and then sent to Shanghai Biotech, China for sequencing. The sequences were compared using BLAST (http://www.ncbi.nlm.nih.gov/blast) on the National Center for Biotechnology Information (NCBI) website. The homology between the obtained sequences and the corresponding target genes on NCBI was very high, and the matching degree was more than 98%. Ultimately, the AMR gene detection rates of the isolated E. coli strains from water, soil, and sediment were statistically analyzed by using Excel 2012.
A correlation analysis between the results of AMR genes and drug resistance was performed, and the correlation rate of AMR genes and drug resistance was calculated. The correlation rate of AMR genes and drug resistance was calculated as to the following formula: (number of resistant strains with positive genes + number of sensitive strains with negative genes) / total number of strains × 100% (40).
One E. coli strain was isolated from each sample of water, soil, and sediment, totaling to 90 strains. The colonies of the isolated strains grown on Macconkey agar medium have a smooth, moist, and pink surface, and the colonies grown on eosin melan agar medium have a metallic luster, either black or brown-green. Besides this, the PCR products of E. coli-specific phoA gene can be observed by gel electrophoresis and showed that 90 strains of bacteria have an amplified 622-bp band, and it is determined that the isolated and cultured 90 strains are E. coli.
The results of antimicrobial drug resistance of the isolated E. coli strains are presented in Table 2. Overall, the isolated E. coli strains from the three aquaculture farms all have high antimicrobial drug resistance rates to penicillin and rifampicin, and the resistance rate to rifampicin reaches 100%. The E. coli isolates of the water, soil, and sediment samples of the three aquaculture farms all have relatively low resistance to β-lactam aztreonam, aminoglycosides gentamicin, amikacin, streptomycin, macrolide azithromycin, doxycycline, ciprofloxacin, ofloxacin, lomefloxacin, polymyxin B, nitrofurantoin, and fosfomycin (≤50%). Specifically, the E. coli isolates from the water samples of aquaculture farm I have a low resistance rate to amoxicillin, ampicillin, tetracyclines, sulfonamides, and amide alcohol drugs (≤30%), but the E. coli isolates of the soil and sediment samples from aquaculture farms I, II, and III have high resistance rates to the above-mentioned drugs (≥60%). In addition, there is high resistance to cefotaxime and ceftriaxone for the E. coli isolates of the water and soil samples from aquaculture farm II (≥70%).
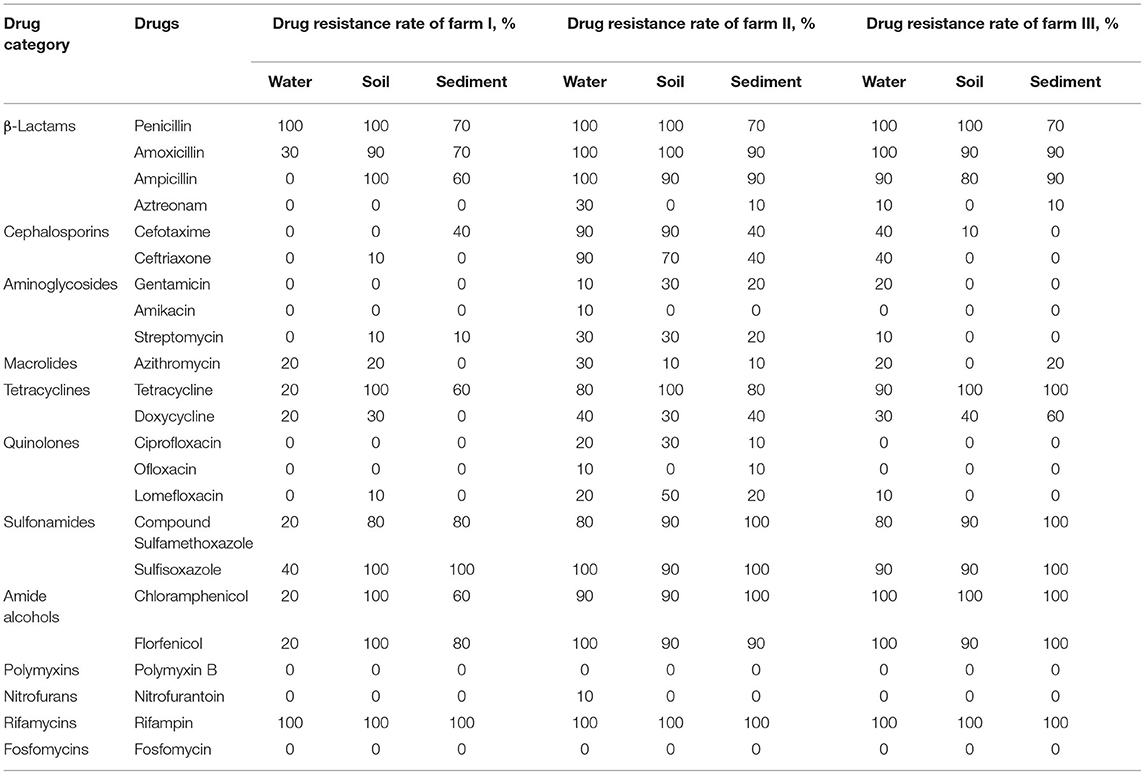
Table 2. Drug resistance rate of isolated strains of Escherichia coli from three aquaculture farms in Zhanjiang, China.
The resistance of the 90 isolates of E. coli to 23 antibacterial drugs is shown in Table 3. The resistance rate of the 90 isolates of E. coli to penicillin, amoxicillin, ampicillin, tetracycline, compound sulfamethoxazole, sulfisoxazole, chloramphenicol, florfenicol, and rifampin is high (≥70%), and the resistance rate to rifampicin is 100%. The resistance rates to aztreonam, gentamicin, amikacin, streptomycin, azithromycin, ciprofloxacin, ofloxacin, lomefloxacin, polymyxin B, nitrofurantoin, and fosfomycin were low (≤30%), and the resistance rate to polymyxin B and fosfomycin was 0.
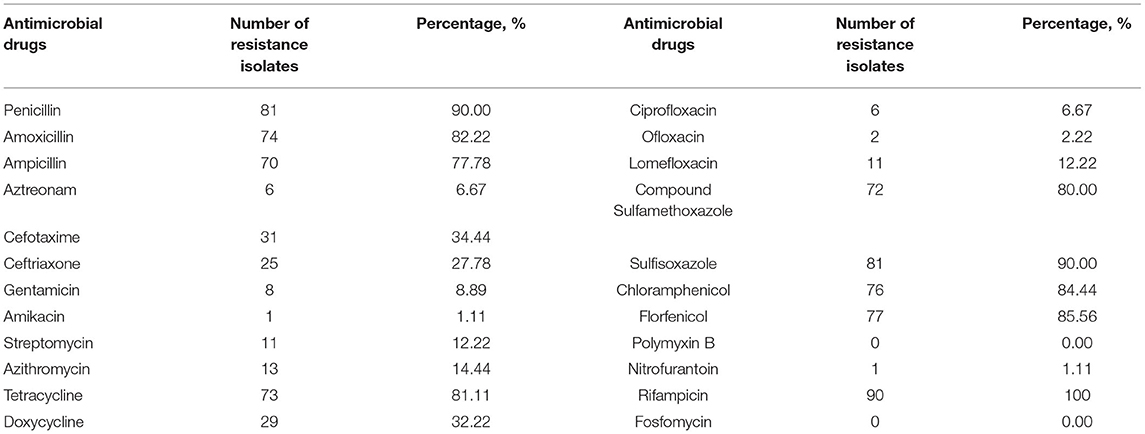
Table 3. Resistance of 90 isolated strains of Escherichia coli from three aquaculture farms in Zhanjiang, China, to 23 kinds of antimicrobial drugs.
As shown in Figure 2, all 90 isolated E. coli strains exhibited varying degrees of multi-drug resistance, with resistance to at least two drugs and resistance to at most 16 kinds of antibacterial drugs. Meanwhile, 60% or more isolated E. coli strains are resistant to more than nine drugs; especially the number of strains resistant to nine kinds of antibacterial drugs is the largest (19 strains, 21.11%), and the number of strains resistant to four, five, and 16 kinds of antibacterial drugs is the least (one strain, 1.11%).
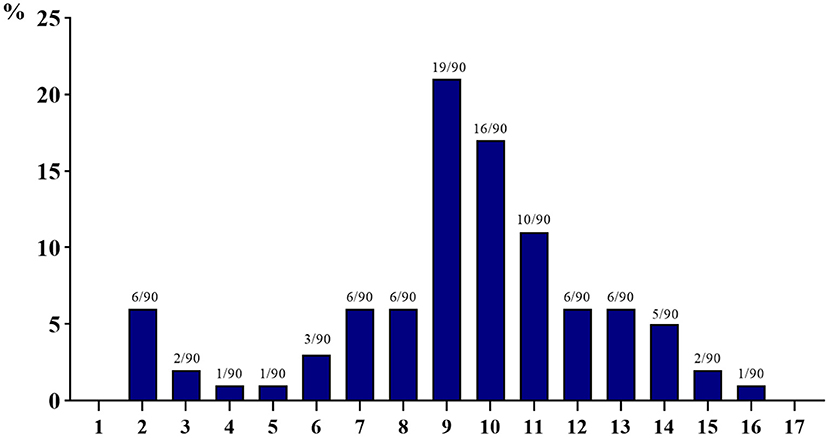
Figure 2. Multi-drug resistance of 90 isolated strains of Escherichia coli from aquaculture farms in Zhanjiang, China (the X-axis shows the number of drug resistance, while the Y-axis shows the percentage of multi-drug-resistant strains among 90 isolates).
The detection results of 90 strains of E. coli isolated from the water, soil, and sediment samples of aquaculture farms for carrying 29 kinds of AMR genes are shown in Supplementary Table 1. Regarding aquaculture farm I, the detection rate of the following AMR genes of the E. coli isolates from the water samples was high: blaCTX−M (80%), blaCIT (100%), blaDHA (80%), fexA (70%), OptrA (90%), aphA1 (90%) and mcr1 (70%). The AMR genes with a high detection rate of E. coli isolates in soil samples include blaCIT (100%), blaDHA (70%), floR (100%), fexB (60%), OptrA (70%), Sul2 (60%), tetM (90%), oqxA (60%), qnrS (100%), and mcr1 (60%). The high detection rates of the AMR genes of E. coli isolates in the sediment samples are as follows: blaTEM(60%), blaNDM (80%), fexB (60%), OptrA (90%), cmlA (70%), aphA1 (70%), Sul2 (60%), oqxA (90%), and qnrS (80%).
In aquaculture farm II, 28 kinds of AMR genes were detected, which was the largest number of detected AMR genes among the three farms. Specifically, the high detection rates of the AMR genes of E. coli isolates from water samples are blaCIT (70%), blaDHA (60%), floR (80%), OptrA (70%), cmlA (80%), aphA1 (80%), Sul2 (80%), tetA (70%), qnrS (90%), and mcr1 (70%). The high detection rates of AMR genes of E. coli isolates from soil samples include blaCTX−M (70%), blaTEM (60%), blaCIT (90%), blaNDM (90%), blaIMP (70%), floR (90%), fexB (60%), OptrA (80%), cmlA (90%), aphA1 (60%), Sul2 (70%), tetC (60%), oqxA (100%), qnrS (70%), and mcr1 (70%). High detection rates of E. coli isolates in sediment samples are demonstrated for the following AMR genes: blaTEM (90%), blaCIT (60%), blaNDM (70%), floR (90%), OptrA (70%), Sul2 (70%), oqxA (80%), and qnrS (80%).
Regarding aquaculture farm III, high detection rates of E. coli isolates in water samples are observed for the following AMR genes: blaDHA (70%), floR (80%), fexB (60%), OptrA (80%), cmlA (70%), Sul2 (90%), and qnrS (70%). The high detection rates of AMR genes of E. coli isolates from soil samples include blaCTX−M (80%), blaTEM (80%), blaCIT (60%), blaNDM (100%), floR (90%), fexA (80%), cmlA (70%), oqxA (90%), and qnrS (80%). High detection rates of AMR genes of E. coli isolates from sediment samples are observed as follows: blaTEM (70%), blaNDM (100%), floR (100%), OptrA (100%), Sul2 (90%), oqxA (90%), and qnrS (80%).
As shown in Table 4, on the whole, among the carried AMR genes in 90 strains of E. coli isolates, β-lactams (blaTEM and blaCIT), amide alcohols (floR, OptrA, and cmlA), aminoglycosides (aphA1), sulfonamides (Sul2), and quinolones (oqxA and qnrS) have a high detection rate (≥50%). The detection rate of AMR genes of carbapenems (blaKPC and blaIMP), amide alcohols (cfr and cat1), aminoglycosides (aac(3)-II), sulfonamides (Sul1), tetracyclines (tetM and tetA), macrolides (ereA and ermB), quinolones (oqxA and qnrB), and colistins (mcr2) is relatively low (≤30%), and the detection rate of mcr2 is 0.
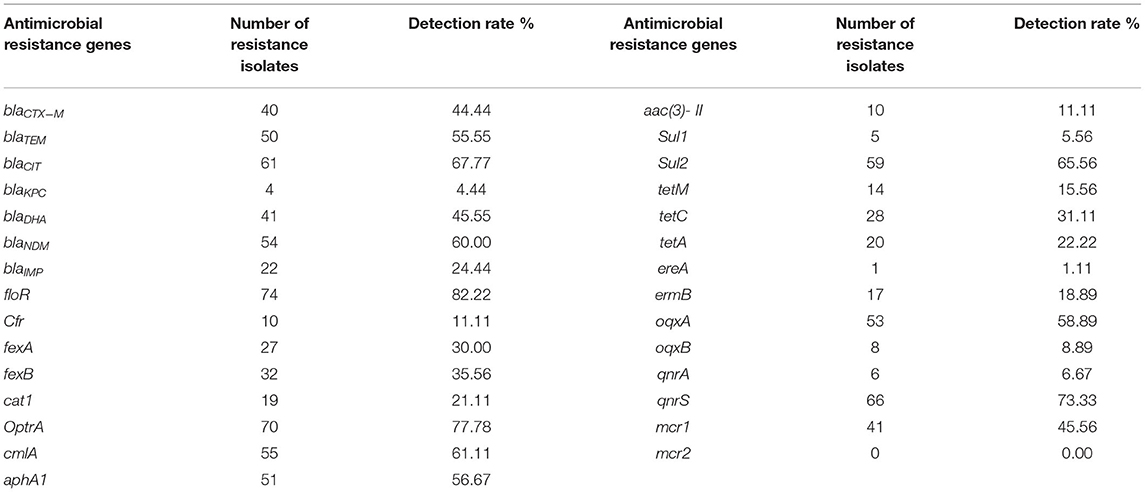
Table 4. Detection of antimicrobial resistance genes in a total of 90 isolated strains of Escherichia coli from aquaculture farms in Zhanjiang, China.
The number and detection rate of 29 AMR genes in 90 isolated E. coli strains are shown in Figure 3. AMR genes of at least four species were detected, and a maximum of 15 AMR genes were detected in each strain of isolated E. coli. More than 60% of the isolated E. coli strains carried more than 10 kinds of AMR genes, of which the number of isolated E. coli strains carrying 10 kinds of AMR genes is the largest (15 strains, 15.56%), and the number of isolated E. coli strains carrying four kinds of AMR genes is the least (one strain, 1.11%).
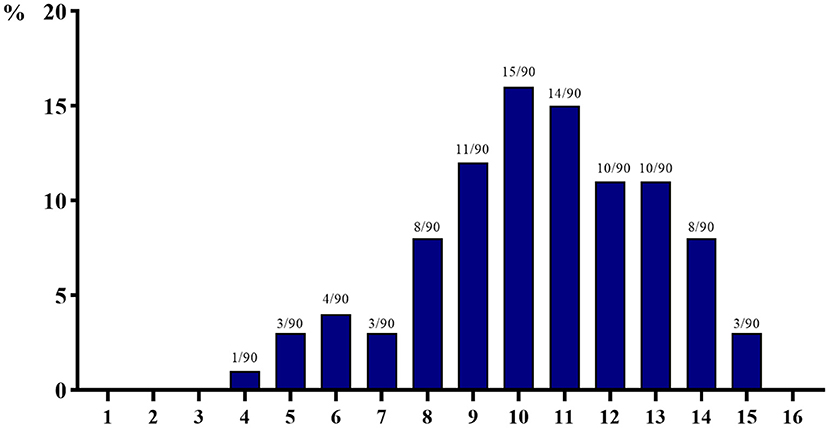
Figure 3. The number of 29 antimicrobial resistance genes detected in 90 isolated strains of Escherichia coli from aquaculture farms in Zhanjiang, China (the X-axis indicates the number of resistance genes, while the Y-axis indicates the percentage of bacteria carrying multi-drug resistance genes among 90 isolates).
As described in Table 5, the following AMR genes have a high correlation rate with the drug resistance of the isolated E. coli strains: blaTEM (55.56%), blaCIT (67.78%), floR (85.56%), OptrA (71.11%), cmlA (64.44%), aac(3)-II (81.11%), Sul2 (70 %), ereA (84.44%), ermB (73.33%), oqxB (85.56%), qnrA (83.33%), mcr1 (56.67%), and mcr2 (95.56%). The correlation rate of other AMR genes with drug resistance is relatively low (<50%).
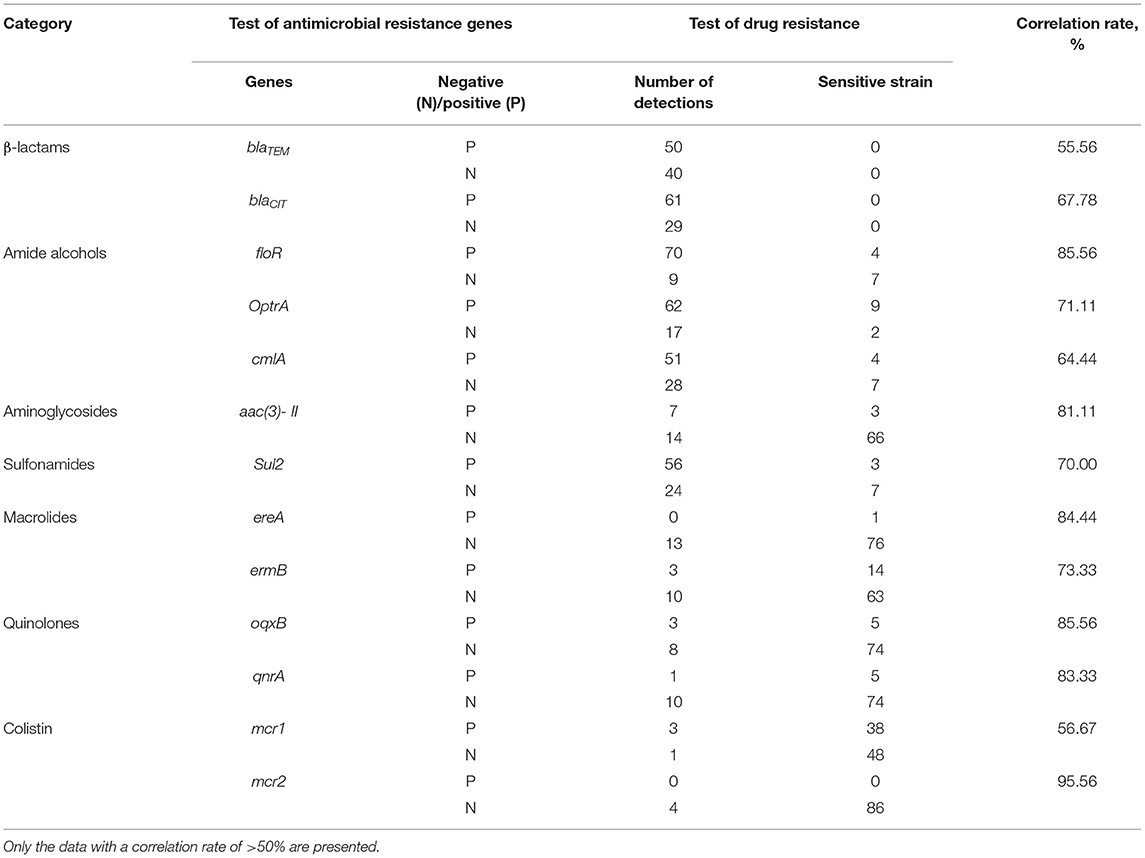
Table 5. Correlation analysis between antimicrobial resistance genes and drug resistance of Escherichia coli isolates from aquaculture farms in Zhanjiang, China.
China is a huge market for animal product consumption. With the increase in consumer demand for meat products other than livestock products, the scale of aquaculture has significantly increased in recent years, and Zhanjiang is an important place for aquatic product production in China (2). However, in order to improve the economic benefits of aquaculture, antimicrobial drugs (including preventive and therapeutic drugs) are widely used in the aquaculture industry to prevent and treat bacterial diseases (1). The overuse and/or abuse of antimicrobial drugs in animal production leads to the generation of AMR genes and drug-resistant bacteria, thus threatening to complicate the treatment of bacterial infections in humans (8, 9). Due to the fact that E. coli is a common pathogen in both humans and animals, the AMR issue of E. coli has been a prominent concern in public health (10, 11). Accordingly, the application of antibiotics for growth promotion in farm animals was banned entirely in the European Union since 2000 and in South Korea and Japan after 2011 (41). Recently, China also completely banned the preventive antibiotics for promoting the growth performance of animals to mitigate the deleterious consequences of AMR to public health and encouraged research on alternatives to preventive antibiotics (42–45). Nevertheless, in large-scale aquaculture, the extensive use of therapeutic antibacterial drugs induced the AMR issues and was still a major concern. In this context, the present study investigated the distribution of AMR genes and analyzed the drug resistance of E. coli in aquaculture farms of Zhanjiang and their surrounding environments, and the findings have positive implications for public health.
Through the drug resistance analysis of the present study, it was found that the E. coli strains isolated from three aquaculture farms in Zhanjiang, China, have different levels of resistance to various antimicrobial drugs. The E. coli isolates from aquaculture environments are highly resistant to β-lactams (except aztreonam), tetracycline, sulfonamides, amide alcohols, and rifamycin but have a relatively low resistance rate to aztreonam, aminoglycosides, macrolides, doxycycline, quinolones, polymyxins, nitrofurans, and fosfomycins. It is worth noting that the 90 isolated E. coli strains in this study were resistant to at least two drugs and at most 16 drugs, indicating that the multi-drug resistance of E. coli was a serious concern in the aquaculture farms of Zhanjiang, China. Similarly, Saharan et al. (46) reported that the majority of the E. coli isolates from a fish farm in India were found to be resistant to more than three antibiotics (>89%), suggesting that there was multi-drug resistance for E. coli in the fish farm. According to the study of Ng et al. (47), it was found that 33 multi-drug-resistant E. coli were isolated from surface waters in aquaculture sites of Singapore. Lihan et al. (48) investigated the antibiotic-resistant E. coli from Sarawak rivers and aquaculture farms in northwest of Borneo. They found that resistance to piperacillin (100%) was observed in all E. coli isolates, resistance to amoxicillin (100%) and ampicillin (100%) was observed in E. coli isolated from aquaculture farms, and resistance to streptomycin (93%) was observed in E. coli isolated from rivers. The rivers and aquaculture farms showed similar partial drug resistance, indicating that the drug-resistant E. coli produced by aquaculture can be transferred to surrounding waters. On the other hand, compared to these previous reports, different aquaculture farms showed different degrees of resistance to various antibacterial drugs. This may be due to a variety of feed formulas and various medication regimes of the aquaculture farms in different regions.
Nowadays, the AMR genes are considered as new environmental pollutants; the AMR genes not only spread to surrounding environments (such as water) and the food chain but also spread to other bacteria horizontally through genetic elements, leading to antibiotic-resistant bacteria and causing difficulties in therapy. Therefore, the AMR in bacteria associated with food and water has been a global concern (7, 8). Previously, it has been reported that multiple AMR genes (mainly sul1, sul2, and tetM) were detected in E. coli isolates from surface water samples of aquaculture farms in America (49). Hassan et al. (50) also found that around five strains of E. coli isolated from a rainbow trout farm carried the mcr-1 gene. In this study, the 90 strains of E. coli isolates from three aquaculture farms in Zhanjiang, China, mainly carried the AMR genes of β-lactams (blaTEM and blaCIT), carbapenems (blaNDM), amide alcohols (floR, OptrA, and cmlA), aminoglycosides (aphA1), sulfonamides (sul2), and quinolones (oqxA and qnrS), among which floR has the highest carrying rate (82.2%). It was consistently found that the floR gene detection rate in the isolates from aquaculture farms in coastal areas of Jiangsu, China, was 76.67% (51). By contrast, Ng et al. (47) demonstrated that there was high relative abundance of sul1, qnrA, and intI1 genes in the sediments of aquaculture farms in Singapore. Changkaew et al. (52) suggested that the tet(A) gene was the most common AMR gene (69.1%) in the 55 E. coli isolates from shrimp farms and their surrounding environment in Thailand. Ng et al. (53) isolated 19 strains of E. coli from 10 aquaculture farms and their environment in Malaysia, and they found that the AMR gene of Cat I has the highest detection rate (100%, 19 of 19). On the basis of these reports, we can clearly realize that there were various AMR genes carried by E. coli in aquaculture farms of different regions. This may be because of the different epidemic diseases of aquatic animals and the variety of medication regimens that were executed in these regions, thus resulting in the generation of inconsistent AMR genes. Therefore, the contamination of AMR genes in the aquaculture industry of different regions requires investigation case by case. Most notably, the floR gene had the highest detection rate in this experiment, which has been confirmed to be the main AMR gene that mediates drug resistance in E. coli (54). Thus, the aquaculture farms in Zhanjiang area should avoid the overuse of amide alcohol drugs. Besides this, the 90 strains of E. coli isolates in this study have a low carrying rate of tetracycline (tetM, tetC, and tetA), macrolide (ereA and ermB), and colistin (mcr1 and mcr2), and none of the AMR genes was detected in the 90 E. coli isolates at the same time. Bacteria have different resistance mechanisms to different antimicrobial drugs, for instance, tetracycline-resistant bacteria usually produce a protein that interacts with ribosomes; thereby, protein synthesis is not affected by the antimicrobial drugs, which is called ribosome protection (36). Escherichia coli can produce macrolide phosphotransferase, which destroys the lactone ring of macrolide antibiotics, resulting in a high degree of resistance to macrolide drugs (27). Moreover, the detection rate of mcr1 was not high (45.56%), and the mcr2 gene was not detected among the 90 E. coli isolates from aquaculture farms. The possible reasons are as follows: (1) mcr1 is a relatively universal AMR gene in animals, but the mcr2 gene was carried by a rare plasmid (IncX4 type), and (2) the colistin drugs are one of the “last resorts” for the treatment of multi-drug resistant gram-negative bacterial infections; therefore, the aquaculture farms use this type of drug less.
In this study, the AMR genes of β-lactams (blaTEM and blaCIT), amide alcohols (floR, OptrA, and cmlA), aminoglycosides [aac(3)-II], sulfonamides (sul2), macrolides (ereA and ermB), quinolones (oqxB and qnrA), and colistin (mcr1 and mcr2) are highly correlated with their drug resistance (>55%), while the other AMR genes are relatively less related to their drug resistance. E. coli has a strong ability to accumulate AMR genes. Previous studies have shown that E. coli from animals often carry multiple AMR genes, but not every AMR gene exhibits corresponding resistance (34, 55). There are many reasons for the development of drug resistance in bacteria, which are related to the characteristics of the bacteria, the spread of AMR genes, and the use of drugs (1, 3). Among them, although the most important factor of the AMR gene, the expression of AMR genes was associated with the complex regulation by multiple substances (5–7). Therefore, the AMR genes are detected in some bacteria, but the related drug resistance phenotypes may not be shown because the AMR genes are not expressed or are expressed in low abundance (7, 55). Exploring the intrinsic relationship between AMR genes and drug resistance and reducing the spread of AMR genes from the inner roots to curb the trend of drug resistance still require further in-depth research.
Collectively, the E. coli isolated from the aquaculture farms and their environment in Zhanjiang, China, showed multi-drug resistance and carried a large number of AMR genes. Among them, the floR gene had the highest detection rate. Meanwhile, there was a high correlation between some of the AMR genes and the drug resistance phenotypes. Therefore, in order to reduce the spread of AMR genes and the production of drug-resistant bacteria, the present study suggests for aquaculture practitioners in Zhanjiang area to decrease the use of amide alcohol drugs and regulate the use of various antibacterial drugs. The current findings persuade the prudent use of antimicrobial agents in aquatic animal farming and also provide basic information for better understanding of AMR in aquaculture farms in Zhanjiang, China, thus being beneficial to public health.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
W-CL, YM, and BB: conceptualization, writing—review, and editing. C-YL and J-JP: methodology and data curation. C-YL, J-JP, and S-RT: analysis. W-CL, C-YL, and BB: writing—original draft preparation. YM: supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
This research was supported by the 2019 Guangdong University Features Innovation Project by the Department of Education in Guangdong Province, China (2019KTSCX057), the Project of Innovation and Strengthening of Guangdong Ocean University (Q18290), and Guangdong Provincial Department of Education 2021 Special Project for Key Fields of Ordinary Colleges and Universities (2021ZDZX4003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.806653/full#supplementary-material
1. Shao Y, Wang Y, Yuan Y, Xie Y. A systematic review on antibiotics misuse in livestock and aquaculture and regulation implications in China. Sci Total Environ. (2021) 798:149205. doi: 10.1016/j.scitotenv.2021.149205
2. Liu WC, Zhou SH, Balasubramanian B, Zeng FY, Sun CB, Pang HY. Dietary seaweed (Enteromorpha) polysaccharides improves growth performance involved in regulation of immune responses, intestinal morphology and microbial community in banana shrimp Fenneropenaeus merguiensis. Fish Shellfish Immun. (2020) 104:202–12. doi: 10.1016/j.fsi.2020.05.079
3. Jindal P, Bedi J, Singh R, Aulakh R, Gill J. Phenotypic and genotypic antimicrobial resistance patterns of Escherichia coli and Klebsiella isolated from dairy farm milk, farm slurry and water in Punjab, India. Environ Sci Pollut Res. (2021) 28:28556–70. doi: 10.1007/s11356-021-12514-8
4. Samuel O, Olalekan F, Beatrice O, Olatunda O. Veterinary pharmaceuticals in aqueous systems and associated effects: an update. Environ Sci Pollut Res Int. (2017) 24:3274–97. doi: 10.1007/s11356-016-7757-z
5. Zhang T, Ling B. Antibiotic resistance in water environment: Frontiers of fundamental research, risk assessment and control strategies. Chin Sci Bull. (2020) 65:2543–54. doi: 10.1360/TB-2020-0110
6. Pruden A, Pei R, Storteboom H, Carlson KH. Antibiotic resistance genes as emerging contaminants: Studies in Northern Colorado. Environ Sci Technol. (2006) 40:7445–50. doi: 10.1021/es060413l
7. Scott AB, Mark JW. Tracking antibiotic resistance. Science. (2014) 345:1454–5. doi: 10.1126/science.1260471
8. Al-Tawfiq JA, Rabaan AA, Saunar JV, Bazzi AM. Antimicrobial resistance of gram-negative bacteria: a six-year longitudinal study in a hospital in Saudi Arabia. J Infect Public Health. (2020) 13:737–45. doi: 10.1016/j.jiph.2020.01.004
9. Mariani M, Medici C, Ugolotti E, Losurdo G, Mesini A, Castagnola E. Colonization by Escherichia coli strains with increased minimal inhibitory concentration for cefiderocol: when resistance anticipates drug use. J Infect Public Health. (2021) 14:749–50. doi: 10.1016/j.jiph.2021.03.002
10. Lippolis JD, Holman DB, Brunelle BW, Thacker TC, Bearson BL, Reinhardt TA, et al. Genomic and transcriptomic analysis of Escherichia coli strains associated with persistent and transient bovine mastitis and the role of colanic acid. Infect Immun. (2017) 86:e00566. doi: 10.1128/IAI.00566-17
11. Sarwar A, Butt MA, Hafeez S, Danish MZ. Rapid emergence of antibacterial resistance by bacterial isolates from patients of gynecological infections in Punjab, Pakistan. J Infect Public Health. (2020) 13:1972–80. doi: 10.1016/j.jiph.2020.06.011
12. Mujeeb UR, Zhang H, Wang YJ, Khalid M, Huang SC, Muhammad KI, et al. Experimental mouse lethality of Escherichia coli strains isolated from free ranging Tibetan yaks. Microb Pathog. (2017) 109:15–9. doi: 10.1016/j.micpath.2017.05.020
13. McNally A, Cheng L, Harris SR, Corander J. The evolutionary path to extraintestinal pathogenic, drug-resistant Escherichia coli is marked by drastic reduction in detectable recombination within the core genome. Genome Biol Evol. (2013) 5:699–710. doi: 10.1093/gbe/evt038
14. CLSI. Performance Standards For Antimicrobial Susceptibility Testing. Clinical and laboratory standards institute. (2018). Available online at https://www.academia.edu/41587236/M100_Performance_Standards_for_Antimicrobial_Susceptibility_Testing_A_CLSI_supplement_for_global_application_28th_Edition
15. Sun YJ, Wu YJ, Liu ZY, Hu M, Zhao YC, Huang K, et al. Antibacterial Effect of aqueous extract of Chrysanthemum indicum L. combined with antibiotics on extended spectrump β-lactamases-producing Escherichia coli. Chin Anim Hus Vet Med. (2016) 43:2170–5. doi: 10.16431/j.cnki.1671-7236.2016.08.033
16. Monstein HJ, Ostholm BA, Nilsson MV, Nilsson M, Dornbusch K, Nilsson LE. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS. (2007) 115:1–6. doi: 10.1111/j.1600-0463.2007.00722.x
17. Ma QL, Wei DJ, Liu M, Men K. Detection of common genotypes CIT and DHA of plasmid mediated AmpC enzyme in Escherichia coli. Tianjin Med J. (2009) 37:308–9. doi: 10.1111/CNKI:SUN:TJYZ.0.2009-04-035
18. van der Zee A, Roorda L, Bosman G, Fluit AC, Hermans M, Smits PHM, et al. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect Dis. (2014) 14:27. doi: 10.1186/1471-2334-14-27
19. Ma HY. Acquired resistance-related genes and clinical characteristic in Extensively drug-resistant Klebsella pneumoniae. (ph.D. Thesis). Suzhou: Soochow University (2017).
20. Wang SM, Zhang JD, Wang YF, Liu SY. Typing of drug resistance genes in carbapenem-resistant Enterobacteriaceae. Chin J Nosocomiol. (2019) 29:2566–70. doi: 10.11816/cn.ni.2019-182135
21. Yu XH, Li XL, Yang FL, Hai Q. Cloning and sequence analysis of floR genes of pathogenic Escherichia coli in ducks. Chin Anim Hus Vet Med. (2010) 37:84–7. doi: 10.11816/CNKI:SUN:GWXK.0.2010-03-023
22. Kehrenberg C, Schwarz S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother. (2006) 50:1156. doi: 10.1128/AAC.50.4.1156-1163.2006
23. Ying YY, Wu F, Wu CY, Jiang Y, Yin M, Zhao WX, et al. Florfenicol resistance in Enterobacteriaceae and whole-genome sequence analysis of florfenicol-resistant Leclercia adecarboxylata strain R25. Int J Genomics. (2019) 2019:9828504. doi: 10.1155/2019/9828504
24. Zhao FJ, Guan M, Li QZ, Li JC, Liang Q, Gu GB. Detection and analysis of chloramphenicols resistance genes in Escherichia coli from Swine. Prog Vet Med. (2017) 38:49–51. doi: 10.16437/j.cnki.1007-5038.2017.07.011
25. Zhou Q, Zhang XY, Ma YP, Chen DW, Tang XJ, Lu JX, et al. Changes of drug resistance and detection of florfenicol resistance genes in Escherichia coli isolated from laying hens after florfenicol administration. China Poultry. (2020) 42:40–6. doi: 10.16372/j.issn.1004-6364.2020.09.007
26. Ming YY. Study on drug resistance gene detection and FexB gene transmission in pig farm environment in Zhanjiang area. (Ph.D. thesis). Zhanjiang: Guangdong Ocean University (2020).
27. Sáenz Y, Briñas L, Domínguez E, Ruiz J, Zarazaga M, Vila J, et al. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother. (2004) 48:3996–4001. doi: 10.1128/AAC.48.10.3996-4001.2004
28. Liu ZM, Li JQ, Huang DH, Hao XW, Hao PG, Wu LC, et al. Study on the drug-resistance of Escherichia coli isolated from sheep in Inner Mongolia. Chin Anim Hus Vet Med. (2017) 44:839–46. doi: 10.16431/j.cnki.1671-7236.2017.03.031
29. Boerlin P, Travis R, Gyles CL, Reid SR, Janecko N, Lim H, et al. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl Environ Microbiol. (2005) 71:6753–61. doi: 10.1128/AEM.71.11.6753-6761.2005
30. Lv WF, Jiang HQ, Yu L. Detection of sulfanilamide sensitivity and sulfonamide resistant related genes in clinical Escherichia coli isolates. J Jilin Agri Univ. (2010) 32:340–4. doi: 10.13327/j.jjlau.2010.03.026
31. Xia QQ, Wang HN, Zhang AY. Detection of tetracycline resistant genes in bacterial by using a triple-PCR method. J Sichuan Univ. (2013) 50:171–6. doi: 10.13327/CNKI:SUN:SCDX.0.2013-01-033
32. Lopardo HA, Vidal P, Jeric P, Centron D, Paganini H, Facklam RR, et al. Six-month multicenter study on invasive infections due to group B Streptococci in Argentina. J Clin Microbiol. (2003) 41:4688–94. doi: 10.1128/JCM.41.10.4688-4694.2003
33. He KL. Characteristics and fate of antibiotics and antibiotic resistance genes in manure application soil. (Ph.D. thesis). Beijing: University of Chinese Academy of Sciences (2018).
34. Feng SW, Li J, Li CT, Xu LS, Pan Y, Hu S, et al. Detection and correlation analysis on the antibiotic resistance phenotype and resistance genes of swine Escherichia coli in Guangxi province. Chin J Vet Sci. (2020) 40:1170–8. doi: 10.16303/j.cnki.1005-4545.2020.06.17
35. Hansen LH, Johannesen E, Burmølle M, Sørensen AH, Sørensen SJ. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother. (2004) 48:3332–7. doi: 10.1128/AAC.48.9.3332-3337.2004
36. Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. (2007) 60:145–7. doi: 10.1093/jac/dkm167
37. Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby G, Barrett TJ, et al. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis. (2006) 43:297–304. doi: 10.1086/505397
38. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. (2016) 16:161–8. doi: 10.1016/S1473-3099(15)00424-7
39. Basil BX, Christine L, Rohit R, Samir KS, Patrick B, Herman G, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Eurosurveillance. (2016) 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280
40. Shen XY, Li JM, Cheng X, Liu M, Dai YB. Detection of drug-resistant genes and their correlation with drug sensitivity in Salmonella pullorum isolates. China Poultry. (2018) 40:15–8. doi: 10.16372/j.issn.1004-6364.2018.03.004
41. Cully M. Public health: The politics of antibiotics. Nature. (2014) 509:S16–7. doi: 10.1038/509S16a
42. Liu WC, Guo Y, Zhao ZH, Jha R, Balasubramanian B. Algae-derived polysaccharides promote growth performance by improving antioxidant capacity and intestinal barrier function in broiler chickens. Front Vet Sci. (2020) 7:601336. doi: 10.3389/fvets.2020.601336
43. Liu WC, Ou BH, Liang ZL, Zhang R, Zhao ZH. Algae-derived polysaccharides supplementation ameliorates heat stress-induced impairment of bursa of Fabricius via modulating NF-κB signaling pathway in broilers. Poult Sci. (2021) 100:101139. doi: 10.1016/j.psj.2021.101139
44. Zhao Y, Balasubramanian B, Guo Y, Qiu SJ, Jha R, Liu WC. Dietary Enteromorpha polysaccharides supplementation improves breast muscle yield and is associated with modification of mRNA transcriptome in broiler chickens. Front Vet Sci. (2021) 8:663988. doi: 10.3389/fvets.2021.663988
45. Liu WC, Zhu YR, Zhao ZH, Jiang P, Yin FQ. Effects of dietary supplementation of algae-derived polysaccharides on morphology, tight junctions, antioxidant capacity and immune response of duodenum in broilers under heat stress. Animals. (2021) 11:2279. doi: 10.3390/ani11082279
46. Saharan VV, Verma P, Singh AP. High prevalence of antimicrobial resistance in Escherichia coli, Salmonella spp. and staphylococcus aureus isolated from fish samples in India. Aqua Res. (2020) 51:1200–10. doi: 10.1111/are.14471
47. Ng C, Chen H, Goh SG, Haller L, Wu Z, Charles FR, et al. Microbial water quality and the detection of multidrug resistant E. coli and antibiotic resistance genes in aquaculture sites of Singapore. Mar Pollut Bull. (2018) 135:475–80. doi: 10.1016/j.marpolbul.2018.07.055
48. Lihan S, Lee SY, Toh SC, Leong SS. Plasmid-mediated antibiotic resistant Escherichia coli in Sarawak rivers and aquaculture farms, Northwest of Borneo. Antibiotics. (2021) 10:776. doi: 10.3390/antibiotics10070776
49. Huang Y, Zhang L, Tiu L, Wang HH. Characterization of antibiotic resistance in commensal bacteria from an aquaculture ecosystem. Front Microbiol. (2015) 6:914. doi: 10.3389/fmicb.2015.00914
50. Hassan J, Eddine RZ, Mann D, Li S, Deng X, Saoud IP, et al. The mobile colistin resistance gene, mcr-11, is carried on IncX4 plasmids in multidrug resistant E coli isolated from Rainbow trout aquaculture. Microorganisms. (2020) 8:1636. doi: 10.3390/microorganisms8111636
51. Qiao Y. Study on the major pathogenic bacteria resistance of aquaculture in the coastal regions of Jiangsu province. (Ph.D. thesis). Shanghai: Shanghai Ocean University (2015).
52. Changkaew K, Utrarachkij F, Siripanichgon K, Nakajima C, Suthienkul O, Suzuki Y. Characterization of antibiotic resistance in Escherichia coli isolated from shrimps and their environment. J Food Prot. (2014) 77:1394–401. doi: 10.4315/0362-028X.JFP-13-510
53. Ng KH, Samuel L, Kathleen MM, Leong SS, Felecia C. Distribution and prevalence of chloramphenicol-resistance gene in Escherichia coli isolated from aquaculture and other environment. Int Food Res J. (2014) 21:1321–5.
54. Li YY. Preliminary study on the detection of florfenicol resistance gene floR in avian Escherichia coli isolates and mechanism of transmission. (Ph.D. thesis). Yangzhou: Yangzhou University (2016).
Keywords: antimicrobial resistance, aquaculture farm, Escherichia coli, multi-drug resistance, resistance genes
Citation: Liao C-Y, Balasubramanian B, Peng J-J, Tao S-R, Liu W-C and Ma Y (2021) Antimicrobial Resistance of Escherichia coli From Aquaculture Farms and Their Environment in Zhanjiang, China. Front. Vet. Sci. 8:806653. doi: 10.3389/fvets.2021.806653
Received: 01 November 2021; Accepted: 24 November 2021;
Published: 24 December 2021.
Edited by:
Lixing Huang, Jimei University, ChinaReviewed by:
Wei Xu, State Oceanic Administration, ChinaCopyright © 2021 Liao, Balasubramanian, Peng, Tao, Liu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Chao Liu, bGl1d2NAZ2RvdS5lZHUuY24=; Yi Ma, bWF5aTc2MUAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.