- 1Key Laboratory of Zoonosis Research of the Ministry of Education, The Institute of Zoonosis, and the College of Veterinary Medicine, Jilin University, Changchun, China
- 2Electron Microscopy Core Facility, The Institute of Zoonosis, Jilin University, Changchun, China
Apicomplexan parasites possess several unique secretory organelles, including rhoptries, micronemes, and dense granules, which play critical roles in the invasion of host cells. The molecular content of these organelles and their biological roles have been well-studied in Toxoplasma and Plasmodium, but are underappreciated in Cryptosporidium, which contains many parasites of medical and veterinary importance. Only four proteins have previously been identified or proposed to be located in micronemes, one of which, GP900, was confirmed using immunogold electron microscopy (IEM) to be present in the micronemes of intracellular merozoites. Here, we report on the discovery of four new microneme proteins (MICs) in the sporozoites of the zoonotic species C. parvum, identified using immunofluorescence assay (IFA). These proteins are encoded by cgd3_980, cgd1_3550, cgd1_3680, and cgd2_1590. The presence of the protein encoded by cgd3_980 in sporozoite micronemes was further confirmed using IEM. Cgd3_980 encodes one of the three C. parvum rhomboid peptidases (ROMs) and is, thus, designated CpROM1. IEM also confirmed the presence of CpROM1 in the micronemes of intracellular merozoites, parasitophorous vacuole membranes (PVM), and feeder organelles (FO). CpROM1 was enriched in the pellicles and concentrated at the host cell–parasite interface during the invasion of sporozoites and its subsequent transformation into trophozoites. CpROM1 transcript levels were also higher in oocysts and excysted sporozoites than in the intracellular parasite stages. These observations indicate that CpROM1, an intramembrane peptidase with membrane proteolytic activity, is involved in host–parasite interactions, including invasion and proteostasis of PVM and FO.
Introduction
The apicomplexan Cryptosporidium parvum is a zoonotic protozoan parasite that infects humans and several farm animals (1). Cryptosporidium species are monoxenous parasites transmitted via fecal–oral contamination, with sporozoite-containing oocysts representing the environmental stage parasite responsible for transmission between hosts. Following ingestion by a host, C. parvum oocysts release sporozoites which then invade intestinal epithelial cells. Intracellular C. parvum is contained within a parasitophorous vacuole membrane (PVM) and undergoes two or more rounds of asexual development (i.e., merogony to form merozoites) and sexual development (i.e., gametogenesis to form macro- and micro-gametes that then fused to form zygotes), eventually forming new oocysts that are excreted in feces (2).
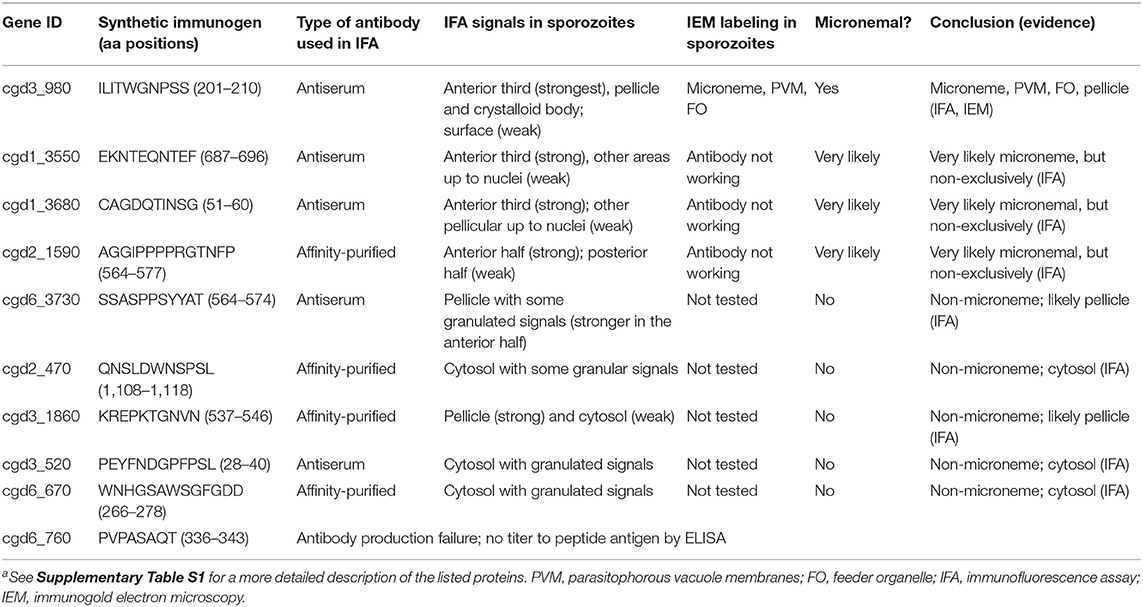
Like in other apicomplexans, such as Plasmodium spp. and Toxoplasma gondii, sporozoites and merozoites are the invasive stages of C. parvum and possess a number of specialized subcellular structures, including rhoptries, micronemes, and dense granules (3–6). Cryptosporidium parasites possess a secretory machinery composed of a single rhoptry, numerous micronemes, and a number of dense granules that are discharged during invasion (3, 4, 7–9). The morphologies of the three secretory organelles in Cryptosporidium sporozoites and merozoites have been studied using transmission electron microscopy (TEM) (3, 7, 9–12). The rhoptry is a single flask-shaped organelle whose tip is attached to the apical end of the parasite, with its end extending to slightly less than a third of the zoite. The micronemes are small oval/rode-shaped organelles clustered in the anterior third section of the zoite. A small number of micronemes are also distributed up to approximately two-thirds of the parasite. Several large dense granules are present near the center of the zoites (Figure 1A).
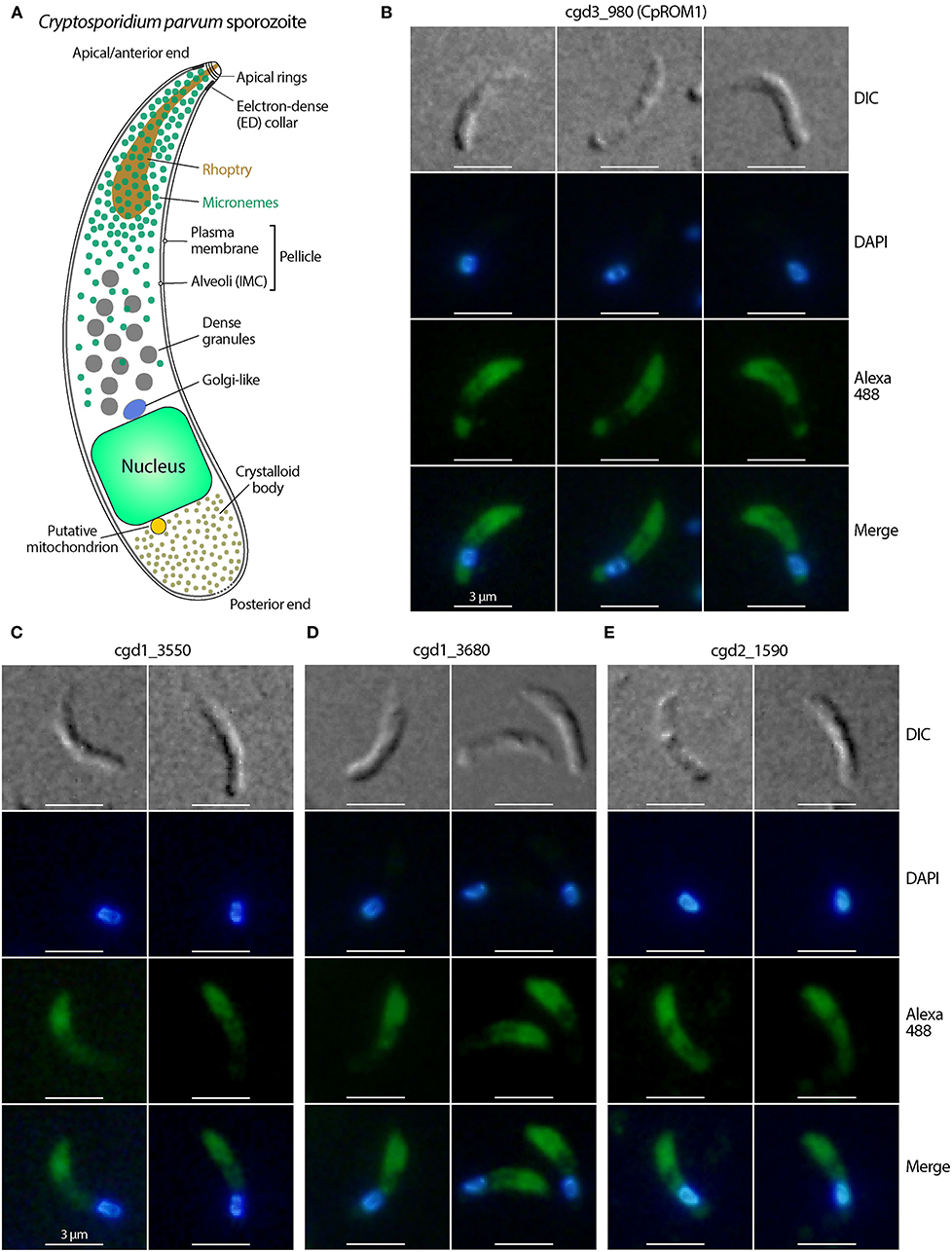
Figure 1. Immunofluorescence micrographs of the four Cryptosporidium parvum proteins likely located in sporozoite micronemes. (A) Structural illustration of C. parvum sporozoites showing the distributions of secretory organelles (micronemes, rhoptry, and dense granules). (B–E) Immunofluorescence assays (IFA) of C. parvum proteins cgd3_980 (CpROM1; Peptidase S54 rhomboid domain containing protein), cgd1_3550 (Apple domain-containing protein), cgd1_3680 (EGF-like domain containing protein), and cgd2_1590 (Apple/EGF-like/Apple domain-containing extracellular protein) in sporozoites. All four proteins showed fluorescence distribution in the anterior third of the sporozoites corresponding to the distribution of micronemes, but varied in signal distribution in other areas of the sporozoites. Proteins were labeled with Alexa Fluor 488 (green) and the nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (blue). The morphology of sporozoites was determined using differential interference contrast microscopy (DIC).
Despite their essential roles in parasite invasion, the molecular contents of these secretory organelles remain poorly understood in Cryptosporidium parasites. This is in contrast to the related T. gondii and Plasmodium species, for which numerous rhoptry, microneme, and dense granule proteins (commonly named ROP, MIC, or DG, followed by a number) have been discovered and biological roles revealed using molecular, biochemical, and/or genetic approaches (13–18). In C. parvum, no dense granule-specific proteins have been reported, although a C-type lectin (CpClec) located in the anterior half of the sporozoite pellicle has also been found in dense granules of merozoites (19). CpPRP1 (encoded by cgd3_440) was previously identified on the tips of rhoptries (20). Six new rhoptry bulb proteins have recently been identified, and the interaction between ROP1 (encoded by cgd3_1771) and the host protein LMO7 during sporozoite invasion has been validated (8).
Only four proteins have been reported to be associated with C. parvum micronemes. These include GP900, TRAP-C1, TSP8, and Cpa135 (21). Although the micronemal location of GP900 in merozoites was confirmed using immunogold electron microscopy (IEM), their micronemal location in C. parvum sporozoites was yet unconfirmed at the ultrastructural level (see the Discussion section for more detail). In addition to the serendipitous discovery of cryptosporidium microneme proteins, earlier in silico studies predicted candidate microneme proteins in C. parvum (n = 22), C. hominis (n = 16), and other apicomplexans by identifying genes encoding signatures of known apicomplexan microneme proteins in the parasite genome (22).
This study is the first to use IEM to validate the presence of a protein in the micronemes of C. parvum sporozoites. Ten C. parvum candidate microneme proteins (22) were selected and synthetic peptides designed and used to generate polyclonal antibodies in rabbits for primary screening using immunofluorescence assay (IFA) and validation using IEM. Four probable microneme proteins were identified using IFA based on their localization in the anterior third of C. parvum sporozoites. IEM was used to validate the presence of these proteins, the peptidase S54 rhomboid family (CpROM1; encoded by cgd3_980), in the micronemes of extracellular sporozoites and intracellular merozoites. CpROM1 was also found to be enriched at the host cell–parasite interface during sporozoite invasion of host cells, as well as in the PVM, suggesting its involvement in parasite invasion and PVM proteostasis.
Materials and Methods
Selection of Candidate Microneme Proteins and Generation of Polyclonal Antibodies
An earlier in silico analysis identified a large number of candidate secretory organelle proteins in 12 apicomplexan parasites, including 22 candidate microneme proteins in C. parvum, based on the presence of one or more Pfam domains found in microneme proteins (e.g., adhesive domains and rhomboid/S8 peptidase domains) (22). To identify and validate microneme proteins in C. parvum as a basis for further investigations of mechanisms underlying parasite invasion and host cell–parasite interactions, 10 proteins from the list of predicted candidate proteins were selected and used to produce polyclonal antibodies for IFA (Table 1; Supplementary Table S1). The remaining candidate proteins were evaluated in separate laboratory studies, including studies focusing on thrombospondin type-1 (TSP1) repeat-containing proteins or peptidases.
For each selected protein, short peptides with good antigenicity were predicted using BepiPred-2.0 software (http://www.cbs.dtu.dk/services/BepiPred/). A sequence unique to the protein was identified using a BLAST search across the NCBI genome database, and the identified sequence was synthesized by the China Peptides Company (Shanghai, China) (Table 1). Synthetic peptides were conjugated to keyhole limpet hemocyanin (KLH) as previously described (23). Each KLH-linked synthetic peptide was used to immunize two pathogen-free rabbits via subcutaneous injection following a standard immunization protocol (24). Rabbits were subjected to four injections at 14-day intervals, with KLH-linked peptide emulsified with Freund's complete adjuvant (300 μg) being given as the first injection, and incomplete adjuvant (150 μg) being administered for subsequent injections. Pre-immune sera and antisera were collected prior to the first injection and 14 days after the last injection. Antibody titers were measured using ELISA coated with synthetic peptide conjugated with BSA (0.25 μg/well). Antisera and pre-immune sera from rabbits showing non-specific labeling with pre-immune sera were subjected to a nitrocellulose membrane-based affinity purification, as previously described (25).
Preparation of Parasite Developmental Stages
A strain of C. parvum harboring the gp60 gene subtype IIaA17G2R1 was propagated in-house in calves. The purification of oocysts using sucrose gradient centrifugation and the preparation of free sporozoites using an in vitro excystation protocol was conducted as previously reported (26, 27). Intracellular forms of C. parvum were prepared by infecting HCT-8 cells (a human ileocecal colorectal adenocarcinoma cell line; ATCC # CCL-244) in complete culture medium [RPMI-1640 medium containing 10% fetal bovine serum (FBS)] incubated at 37°C in a 5% CO2 atmosphere in 96-well plates to generate cell lysates, or in 48-well plates containing poly-L-lysin-treated round glass coverslips to generate substrates for IFA. The invasion stage was prepared by inoculating host cell monolayers at ~60% confluence with freshly excysted sporozoites for 15–45 min, followed by fixation with 4% formaldehyde for 30 min. Other intracellular stages were prepared by infecting cell monolayers at ~80% confluence with oocysts (viability >80% based on in vitro excystation rate) at 37°C for 3 h, followed by medium exchange to remove uninvaded parasites and continuous cultivation for 24 h. Samples were then fixed with 4% formaldehyde for 30 min for IFA (48-well plates) or lysed with iScript qRT-PCR sample preparation reagent (lysis buffer) (50 μl/well) in 96-well plates (Bio-Rad Labs, Hercules, CA, USA) to prepare cell lysates.
Immunofluorescence Microscopy Assay
In this assay, C. parvum sporozoites were prepared by excystation, fixed in suspension with 4% formaldehyde for 30 min, washed with PBS, and applied onto glass slides coated with poly-L-lysine. After air drying for 1 h, samples were permeabilized with 0.2% Triton X-100 for 5 min and exposed to a blocking solution containing 3% FBS in PBS for 1 h. Invading sporozoites and intracellular meronts on host cell monolayers were prepared and fixed as described above, and then permeabilized and blocked. Samples were incubated with individual antisera or affinity-purified antibodies and their corresponding pre-immune sera (original/neat or following subjection to affinity purification) at a 1:200 dilution for antiserum/pre-immune serum pairs or undiluted for affinity-purified pairs. The samples were then incubated for 1 h. Alexa Fluor 488-labeled goat anti-rabbit antibody (Thermo Scientific, West Palm Beach, FL, USA), diluted 1:2,000 in PBS, was used as the secondary antibody and was incubated with the substrate for 1 h. Finally, cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (1 μg/ml) and mounted with antifade mounting medium (Beyotime Biotechnology, Shanghai, China). Three 5-min PBS washes were undertaken between the steps described. All procedures were performed at room temperature. Slides were examined under an Olympus BX53 research microscope equipped with appropriate filter sets. Images were captured with an Olympus DP72 camera in the TIFF format. Signal levels of the images were linearly adjusted in Adobe Photoshop 2021 without local manipulations and presented using Adobe Illustrator 2021.
Colloidal Gold Immuno-Electron Microscopy
Four of the nine antibodies were subjected to further validation using immunoelectron microscopy (IEM). The four antibodies showed labeling in the anterior third of the sporozoites based on IFA, suggesting that their antigens were likely micronemal proteins. C. parvum sporozoites were prepared by excystation and fixed in 4% paraformaldehyde mixed with 0.1% glutaraldehyde in PBS for 2 h at room temperature. After three washes with PBS, fixed sporozoites were embedded in melted 2% agar in 50 mM maleate buffer at 50°C. After cooling to room temperature, the pellets were cut into small blocks (~0.5 mm3), washed with maleate buffer containing 0.5 mM CaCl2 and 2% sucrose, incubated for 30 min at room temperature in 0.5% uranyl acetate in maleate buffer, and washed again in maleate buffer. For intracellular stages, we used mouse intestinal samples infected with a local isolate of C. tyzzeri that had been propagated in the laboratory. This parasite was chosen simply due to convenience. The C. tyzzeri genome has been sequenced (28) and contains orthologs of the four candidate micronemal proteins, with sequences matching those of the synthesized antigen peptides. To prepare the intracellular parasite stages, the terminal ileum from a C. tyzzeri-infected mouse was collected, cut into small sections (~0.5 mm3), and fixed for 2 h in a PBS solution containing paraformaldehyde (2%) and glutaraldehyde (0.1%).
All fixed samples were dehydrated in increasing concentrations of ethanol (30, 50, 70, 80, 90, and 100%; 1 h each), infiltrated in LR White (Sigma-Aldrich, St. Louis, MO, USA) at −20°C for 48 h, and polymerized under UV at −15°C for 24 h. Ultrathin sections were prepared using a diamond knife on a Leica EM UC6 ultramicrotome and mounted on film-coated grids. Grids were blocked with PBS containing 2.5% non-fat milk and 0.01% Tween-20 and incubated with primary antibodies at 4°C overnight. After three washes, samples were labeled with goat anti-rabbit IgG secondary antibodies conjugated with 10 nm colloidal gold (Sigma-Aldrich) for 1 h at 37°C, washed, and stained with 2% uranyl acetate (Electron Microscopy Sciences, Hatfield, PA, USA). Colloidal gold-labeled thin sections were examined using a Hitachi H7650 transmission electron microscope. Images were captured using an AMT XR40B CCD and saved in TIFF format. Signal levels of the images were linearly adjusted using Adobe Photoshop 2021 without local manipulations and presented using Adobe Illustrator 2021.
Western Blot and qRT-PCR Analyses
Native CpROM1 protein was detected by Western blot analysis using a previously described procedure (29), but with some modifications. Briefly, C. parvum oocysts were suspended in reducing sample buffer containing a protease inhibitor cocktail (107 per lane), disrupted using six freeze/thaw cycles, incubated overnight on ice and then for 1 h at room temperature. After centrifugation, supernatants were separated by SDS-PAGE (10% gel) and proteins were transferred onto a nitrocellulose membrane in a transfer buffer containing 48 mM Tris (pH 10.1), 39 mM glycine, 0.1% SDS, and 20% methanol using a semidry transfer apparatus (Bio-Rad Laboratories). The transfer buffer was adjusted to a higher basicity (pH 10.1) than that used in a standard buffer (pH 9.2) to facilitate the transfer of the highly basic CpROM1 protein (pI = 9.7). The blots were probed with anti-CpROM1 antiserum and horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Immunoway, Plano, TX, USA) and then developed in an enhanced chemiluminescence reagent and visualized using UVP Chemstudio analyzer (Analytik Jena, Upland, CA, USA).
The CpROM1 transcript was detected by qRT-PCR as described previously (29), using a pair of previously reported primers (5′-CTG CGT TGT AGC AGT TGG TG-3′ and 5′-CAA TAG CTG ATG ATG GGT TTC C-3′) (30). The well-studied CpLDH gene (cgd7_480) was included as a reference. C. parvum 18S rRNA (Cp18S) was measured using 5′-TGG TGG CCA TGG CGA TGG TAT G-3′ and 5′-AGC AGC GGC TGG TGC AAA GT-3′ primers and used for normalization as previously described (29). Total RNA was extracted from oocysts, sporozoites, and intracellular parasites infecting HCT-8 cells at various time points using iScript qRT-PCR sample preparation reagent (Bio-Rad Laboratories, Hercules, CA, USA). qRT-PCR was carried out using HiScript II One-Step qRT-PCR SYBR Green Kit (Vazyme Biotech, Nanjing, China) and diluted cell lysates as templates, as described previously (29, 31). The relative levels of gene transcripts were calculated using the 2−ΔΔCT formula, as previously described (29, 32).
Effect of Anti-Cryptosporidium parvum Rhomboid Peptidase 1 Polyclonal Antibody on Cryptosporidium parvum Infection
The effect of anti-CpROM1 polyclonal antibody on C. parvum infection was evaluated using a qRT-PCR assay. HCT-8 cells were seeded in a 96-well plate for overnight growth until they reached ~80% confluence. Freshly excysted C. parvum sporozoites (8 × 104 per well) were suspended in FBS-free RPMI-1640 medium containing pre-immune rabbit serum and antiserum against CpROM1 (1:50 and 1:100 dilutions) and added to the plate via medium exchange to allow invasion of host cells at 37°C for 2 h. After washing with culture medium to remove non-invading sporozoites, infected cell monolayers were allowed to grow for 16 h (total 18 h infection time) in RPMI-1640 medium containing 10% FBS and corresponding pre-immune serum and antiserum (1:50 and 1:100 dilutions). The assay also included blank controls containing no serum. Cell lysates were then prepared using iScript qRT-PCR sample preparation reagent (50 μl/well) (Bio-Rad Laboratories) to determine parasite loads by qRT-PCR detection of parasite 18S (Cp18S) and host cell 18S (Hs18S) rRNA transcripts, as described previously (33, 34). Relative parasite loads were calculated using an empirical formula based on ΔΔCT values of Cp18S and Hs18S in the serum-containing groups and blank controls. That is, × 100(%) (34).
Results
Primary Screening Using Immunofluorescence Assay Identified Four Proteins With Possible Distribution in Cryptosporidium parvum Sporozoite Micronemes
Ultrastructural studies of C. parvum sporozoites (3, 11) show that micronemes are mainly distributed in the anterior third of a sporozoite, with their presence reducing gradually up to the central region where dense granules are present (Figure 1A). In this study, nine of the 10 synthetic peptides produced antisera in rabbits with satisfactory ELISA titers. Antisera were used if the corresponding pre-immune sera showed no IFA signals in sporozoites; otherwise, it was subjected to affinity purification (Supplementary Figure S4). Of the nine antibodies (antisera or affinity-purified antibodies), four labeled mostly the anterior third of C. parvum sporozoites, as determined using IFA (Figures 1B–E and Table 1). These four antigens are encoded by cgd3_980 (CryptoDB description: Peptidase S54 rhomboid domain-containing protein), cgd1_3550 (Apple domain containing protein), cgd1_3680 (EGF-like domain containing protein), and cgd2_1590 (Apple/EGF-like/Apple domain containing extracellular protein) (Table 1). In addition to significant labeling in the anterior third of sporozoites, the four antibodies also produced some signals in other areas, including along the pellicles in the central region before the nuclei and crystalloid bodies after the nuclei (cgd3_980) (Figure 1B). There were also weak signals in other areas before the nuclei (cgd1_3550) (Figure 1C), slightly weaker signals along the pellicles in the central region before the nuclei with no signals after the nuclei (cgd1_3680) (Figure 1D), and slightly weaker signals in most areas except for a spot just before the nuclei (cgd2_1590) (Figure 1E). These observations suggest that these four antigens are very likely micronemal proteins, but with some other subcellular distributions.
The other five antibodies produced immunofluorescence signals in locations other than the anterior third of the sporozoites, indicating that their antigens were unlikely to be micronemal proteins (Figure 2). These antigens included proteins cgd6_3730 (signal peptide, peptidase S8/S53 domain-containing protein) (Figure 2A), cgd2_470 (uncharacterized protein) (Figure 2B), cgd3_1860 (EGF-like domain containing extracellular protein) (Figure 2C), cgd3_520 (PAN/Apple domain containing protein) (Figure 2D), and cgd6_670 (aspartic acid and asparagine hydroxylation/peptidase C11, clostripain domain-containing protein) (Figure 2E). Because this study was focused on the discovery of micronemal proteins, the five apparent non-micronemal proteins were excluded from subsequent experiments.
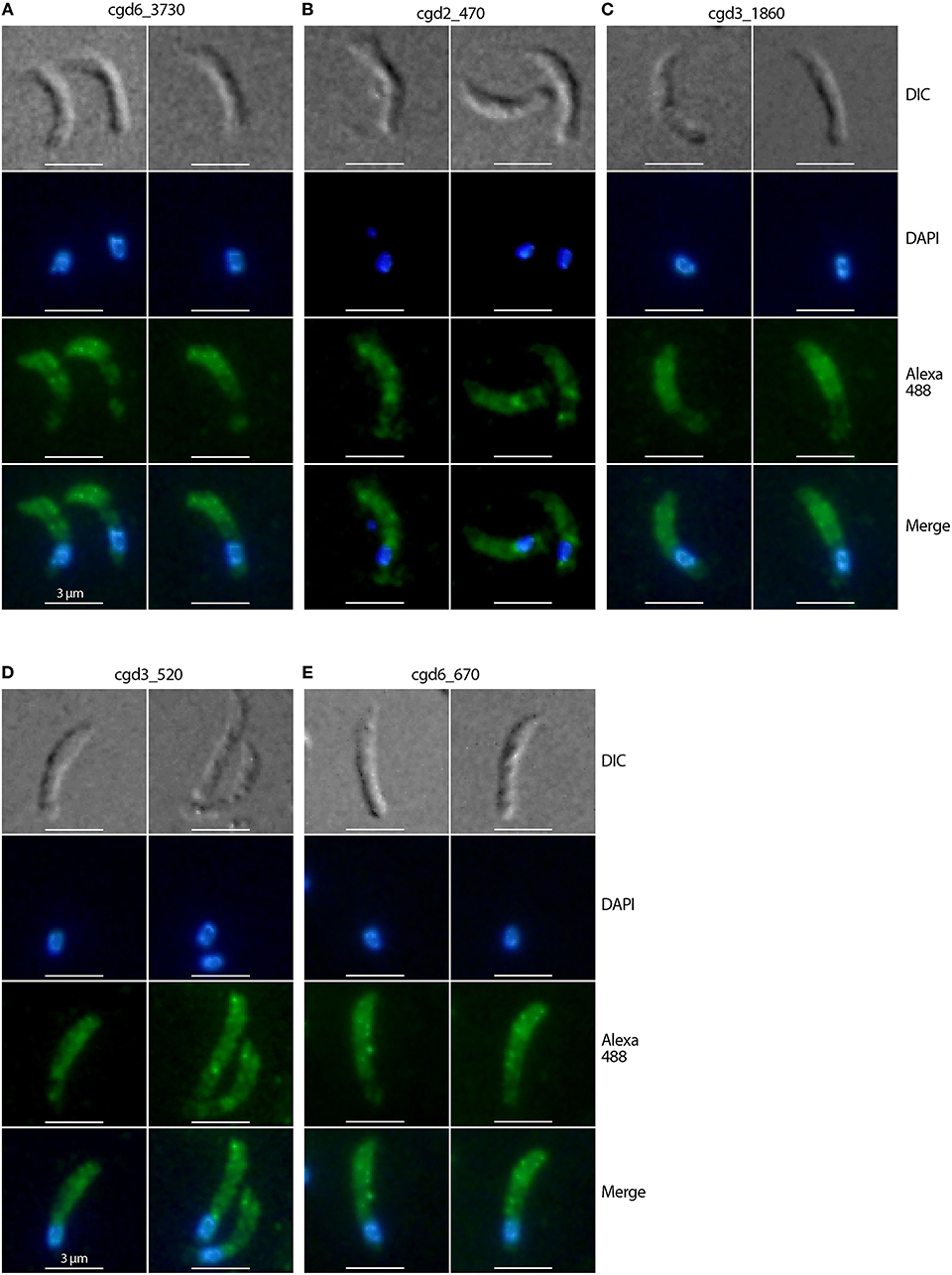
Figure 2. Immunofluorescence micrographs of five Cryptosporidium parvum proteins showing non-micronemal distribution. The proteins are cgd6_3730 (signal peptide, peptidase S8/S53 domain containing protein) (A), cgd2_470 (uncharacterized protein) (B), cgd3_1860 (EGF-like domain containing extracellular protein) (C), cgd3_520 (PAN/Apple domain containing protein) (D), and cgd6_670 (aspartic acid and asparagine hydroxylation/peptidase C11) (E). Proteins were labeled with Alexa Fluor 488 (green) and the nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (blue). The morphology of sporozoites was determined using differential interference contrast microscopy (DIC).
The Distribution of Crytosporidium parvum Rhomboid Peptidase 1 in Sporozoite Micronemes Was Confirmed Using Immuno-Electron Microscopy
The four “very likely” micronemal proteins identified using IFA were subjected to additional validation using IEM, and only antiserum to cgd3_980 produced positive results. Colloidal gold particles with anti-cgd3_980 antibodies were distributed inside or on the edges of rod- or oval-shaped micronemes or microneme clusters in the sporozoites, but not across the entire microneme (Figure 3, marked with “m”). No gold particles were present in the rhoptries, which were clearly visible as flask-shaped rods in longitudinal sections or oval-shaped rods in cross sections (Figure 3, marked with “r”). Some gold particles were visible on the “neck” of the rhoptry, but this was later identified as part of the micronemes that extended over the rhoptry based on morphology and lighter electron density [Figure 3, left panel marked with “m (over r)”]. There were particles between the pellicle and micronemes, and a few particles on the pellicle (e.g., Figure 3, right upper panel marked with “p”).
Figure 3. Immunogold electron microscopy (IEM) labeling of CpROM1 protein (cgd3_980) in free Cryptosporidium parvum sporozoites showing major distribution of gold particles in micronemes (m) and minor distribution on the pellicle (p) as exemplified by arrows. There were no gold particles on other organelles such as rhoptry (r) and dense granules (dg).
However, no gold particles were distributed over crystalloid bodies, as seen using IFA (data not shown). Antibodies against cgd1_3550, cgd1_3680, and cgd2_1590 produced no IEM signals, despite repeated attempts using different antibody concentrations and fixation conditions (e.g., 4% paraformaldehyde with varied concentrations of glutaraldehyde). It is not uncommon for an antibody to give positive IFA results and negative IEM results. Nonetheless, the IEM experiments in this study successfully validated one of the four proteins as a micronemal protein in C. parvum sporozoites, representing a 25% success rate for our approach.
Cryptosporidium parvum Rhomboid Peptidase 1 and Its Counterparts Are One of the Three S54 Rhomboid-Type Membrane Peptidases Present in Cryptosporidium Parasites
Following the confirmation by IFA and IEM that the antigen encoded by cgd3_980 is a micronemal protein, we investigated its molecular features and distribution in the intracellular parasite stage. Based on genome annotations and protein homolog searches, the cgd3_980 gene was shown to encode a 282-amino acid-long protein belonging to the S54 rhomboid peptidase and has seven transmembrane domains (TMDs) (Figure 4A). Rhomboid/rhomboid-like peptidases and rhomboid domain-containing proteins are present in all known apicomplexan genomes and are typically referred to as ROMs (e.g., TgROM1 and PfROM2 for rhomboid peptidases 1 and 2 in Toxoplasma gondii and Plasmodium falciparum, respectively) (35, 36).
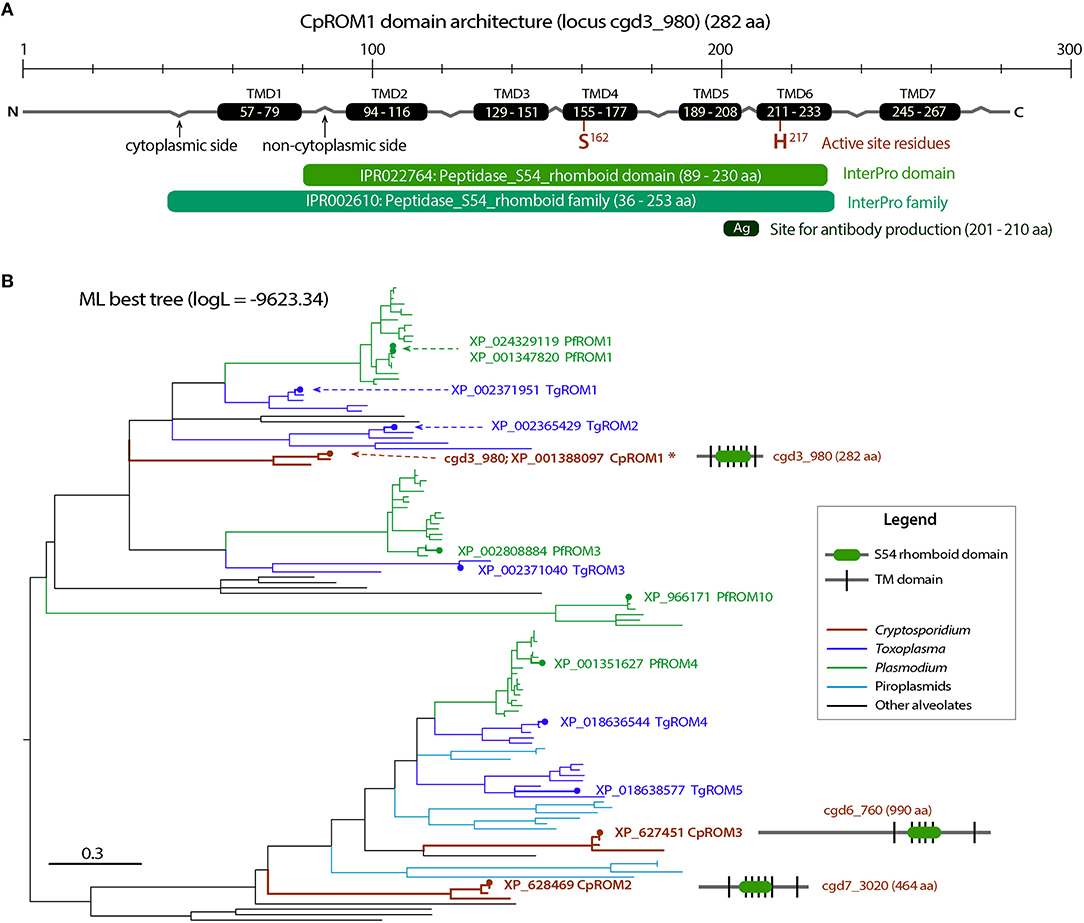
Figure 4. Molecular and phylogenetic features of CpROM1 (cgd3_980). (A) Architecture of CpROM1 (282 aa), showing seven transmembrane domains (TMDs), two active site residues (S162 and H217) in TMD4 and TMD6, regions homologous to specified InterPro domain and family, and the site which was used to design the short peptide used for antibody production. Small zigzags indicate the positions of inter-TMD regions (cytoplasmic vs. non-cytoplasmic sides of the membrane). The lengths of domains are scaled. (B) A maximum likelihood (ML) tree (logL = −9,623.34) showing the relationship between the three CpROM proteins and orthologs in other major apicomplexan groups. Branches representing CpROM1 (cgd3_980; marked with an asterisk), CpROM2 (cgd7_3020) and CpROM3 (cgd6_760) are annotated with gene names, GenBank accession numbers, and domain organizations. Branches representing orthologs from Toxoplasma gondii (TgROMs) and Plasmodium falciparum (PfROMs) are indicated with GenBank accession numbers and names for reference.
Cryptosporidium parasites possess three ROM peptidases encoded by cgd3_980, cgd7_3020, and cgd6_760, and their orthologs are present in other species (Figure 4B and Supplementary Figure S1). This number is less than that of ROMs in T. gondii and P. falciparum, which have at least 11 and 8 ROMs, respectively (35). The three C. parvum ROMs (CpROMs) differ in size but contain a single rhomboid domain and six or seven TMDs (Figure 4B). The rhomboid domain belongs to the S54 family of membrane-bound serine endopeptidases, as indicated in the MEROPS peptidase database (https://www.ebi.ac.uk/merops) (37). All three CpROMs contain serine (Ser) and histidine (His) residues at their active sites, for example, Ser162 and His217 in cgd3_980 (Figure 4A), indicating that they are biochemically functional peptidases.
Maximum likelihood-based phylogenetic analysis placed cgd3_980 at the base of the cluster containing TgROM1/TgROM2 and PfROM1, and cgd7_3020 and cgd6_760 at the base of the cluster containing TgROM4/TgROM5 and PfROM4 (Figure 4B), suggesting that the three CpROM proteins cannot simply be numbered based on their phylogenetic clustering with known TgROMs as proposed previously (35). For simplicity and clarity, we assigned numbers to the three CpROM peptidases based on their sizes: CpROM1 for cgd3_980 (282 aa), CpROM2 for cgd7_3020 (464 aa), and CpROM3 for cgd6_760 (990 aa) (Figure 4B).
The CpROM1 (cgd3_980) transcript was detected by qRT-PCR throughout the life cycle of the in vitro-cultured parasites (Figure 5A). Transcript levels were substantially higher in oocysts and sporozoites than in the intracellular stages, for example, transcript levels were 24.3-fold higher in sporozoites than in intracellular parasites 6 h post-infection (hpi) and 4.8-to 7.5-fold higher than the levels at 3, 12, 24, 48, and 72 hpi. The qRT-PCR assay was reliable because the transcriptomic profiles detected in parallel for three reference genes (CpEF1α, CpLDH, and CpTIPH) followed the same patterns as those previously reported (29, 38, 39). The native protein in C. parvum oocysts was also detected using Western blot analysis (Figure 5B). These observations imply that CpROM1, as a membrane peptidase involved in intramembrane cleavage, might play a role in sporozoite invasion.
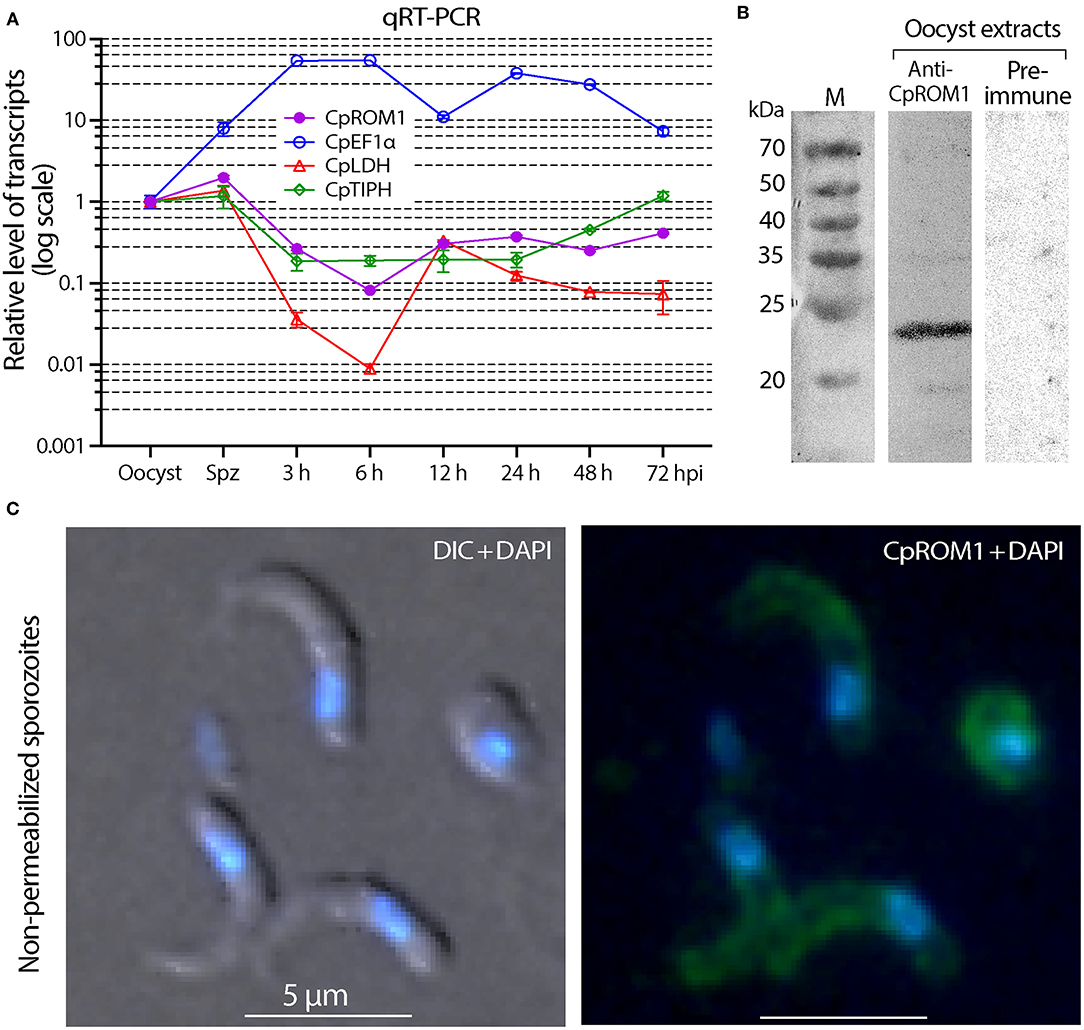
Figure 5. Detection of CpROM1 gene transcript in various life cycle stages and identification of CpROM1 protein in oocyst extracts using Western blot and on sporozoite surfaces using Immunofluorescence assay (IFA). (A) Relative levels of CpROM1 transcript in oocysts, excysted sporozoites, and intracellular parasite stages at different hours post-infection (hpi). Three previously identified genes were detected in parallel and used as references and for quality control. (B) Western blot analysis of CpROM1 using an optimized protocol (described in Materials and Methods). Note that the band recognized by the anti-CpROM1 antibody has a smaller molecular weight (~23 kDa) than predicted (31.3 kDa) due to the high number of hydrophobic amino acids which have high affinity for the SDS molecules. (C) IFA of CpROM1 in free sporozoites fixed with paraformaldehyde but not permeabilized used to show the presence of the protein on the surface of sporozoites. This assay confirmed the presence of some CpROM1 protein on the sporozoite surface (green), although the fluorescence signals were weak compared with signals of fixed and permeabilized samples. The signal levels were adjusted in Photoshop for better visualization. The nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (blue); DIC, differential interference contrast.
As CpROM1 is a membrane protein with seven TMDs and IFA showed some pellicular distributions, we analyzed whether it was also present on the surface of C. parvum sporozoites. This was done using the same antibody recognizing an epitope of CpROM1 between TMD5 and TMD6, which was predicted to be extracellular. In this IFA experiment, sporozoites were fixed but unpermeabilized, and fluorescence signals were observed on the sporozoite surface (Figure 5C). However, the signals were much weaker than those from fixed and permeabilized sporozoites, and were only visible after overexposure to capture fluorescent signals. Thus, IFA confirmed the presence of some CpROM1 on the outer layer of the sporozoite pellicle.
Cryptosporidium Rhomboid Peptidase 1 Is Present in Parasitophorous Vacuole Membranes and Feeder Organelles During the Intracellular Development of Parasites
The presence of CpROM1 in intracellular stages of C. parvum was also observed using IFA and IEM. In IFA, CpROM1 signals exhibited different patterns at different focal points, that is, either as relatively homogenous signals at one focal point (Figure 6A, labeled as CpROM1:focal point 1; Figure 6B) or as non-homologous signals corresponding to intracellular merozoites (Figure 6A, labeled as CpROM1:focal point 2; Figure 6C). These IFA patterns differ from those of a protein present mainly in the parasitophorous vacuole membrane (PVM), for example, lactate dehydrogenase (CpLDH) (38), or a protein present mainly in intracellular merozoites, for example, phosphopantetheinyl transferase (CpPPTase) (40). Therefore, the IFA signal patterns for CpROM1 indicated that this membrane peptidase is present in both the PVM and intracellular merozoites. While the subcellular locations of CpROM1 in intracellular merozoites could not be resolved using IFA, we observed the same pattern of IFA signal distribution of CpROM1 in free merozoites, that is, strong signals in the anterior third of the merozoites and weaker signals in other areas of the merozoites (Figure 6D).
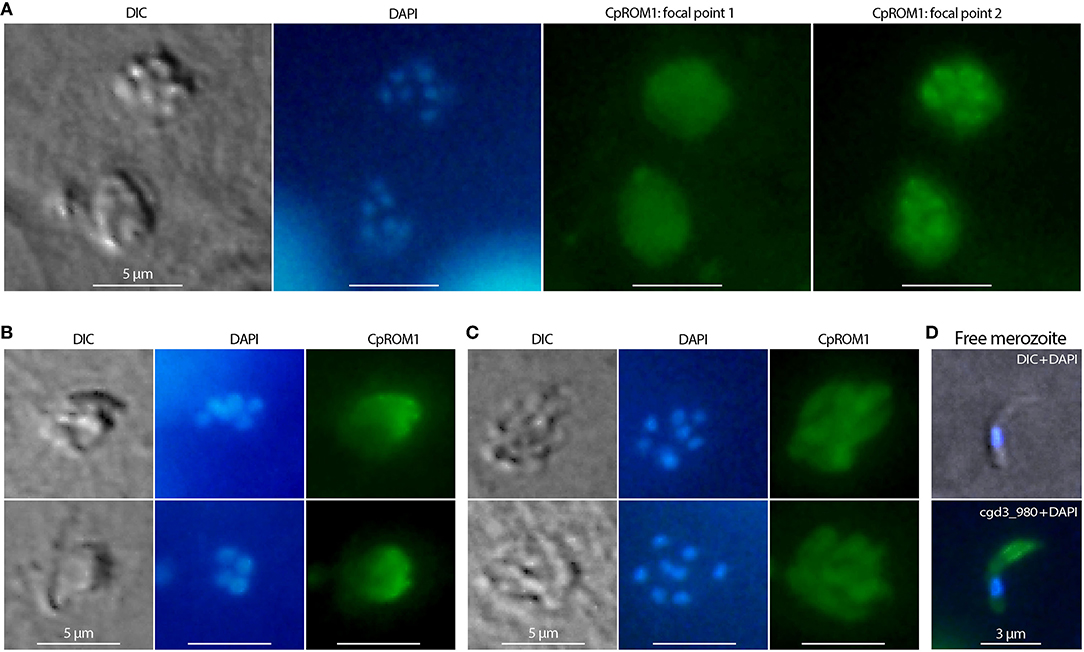
Figure 6. Immunofluorescence assay (IFA) detection of CpROM1 in intracellular Cryptosporidium parvum. Intracellular type I or II meronts containing up to eight nuclei counterstained with 4,6-diamidino-2-phenylindole (DAPI) (blue) and labeled with anti-CpROM1 antibody (green). Cell morphology was illustrated using differential interference contrast (DIC) microscopy. (A) IFA of CpROM1 in two type I meronts at two different focal points showing relatively homogenous distribution of fluorescent signals on parasitophorous vacuole membrane (PVM) (focal point 1) and major signal distribution in the intracellular parasites (focal point 2). (B) IFA of CpROM1 in type I and type II meronts at a focal point showing relatively homogenous signal distribution on a portion of PVM. (C) IFA of CpROM1 in two type I meronts showing signals in intracellular merozoites. (D) IFA of CpROM1 in a free merozoite.
We also performed IEM to detect ROM1 in intracellular parasites in a C. tyzzeri-infected mouse. The mouse parasite C. tyzzeri was selected purely for convenience, as it was available in the laboratory. CpROM1 and CtROM1, its counterpart in C. tyzzeri, share the same epitope as the antibody used for labeling, and C. tyzzeri is also evolutionarily the closest species to C. parvum and C. hominis (28). In this experiment, gold particles were observed in or on the edge of the microneme clusters and in some parts of the PVM, but not in the dense granules or other organelles (Figures 7A,B). Additionally, gold particles were also present in the feeder organelles (FO) (Figure 7C, labeled as “fo”). Gold particles were absent in specimens labeled with pre-immune serum, which was used as a negative control (Figure 7D).
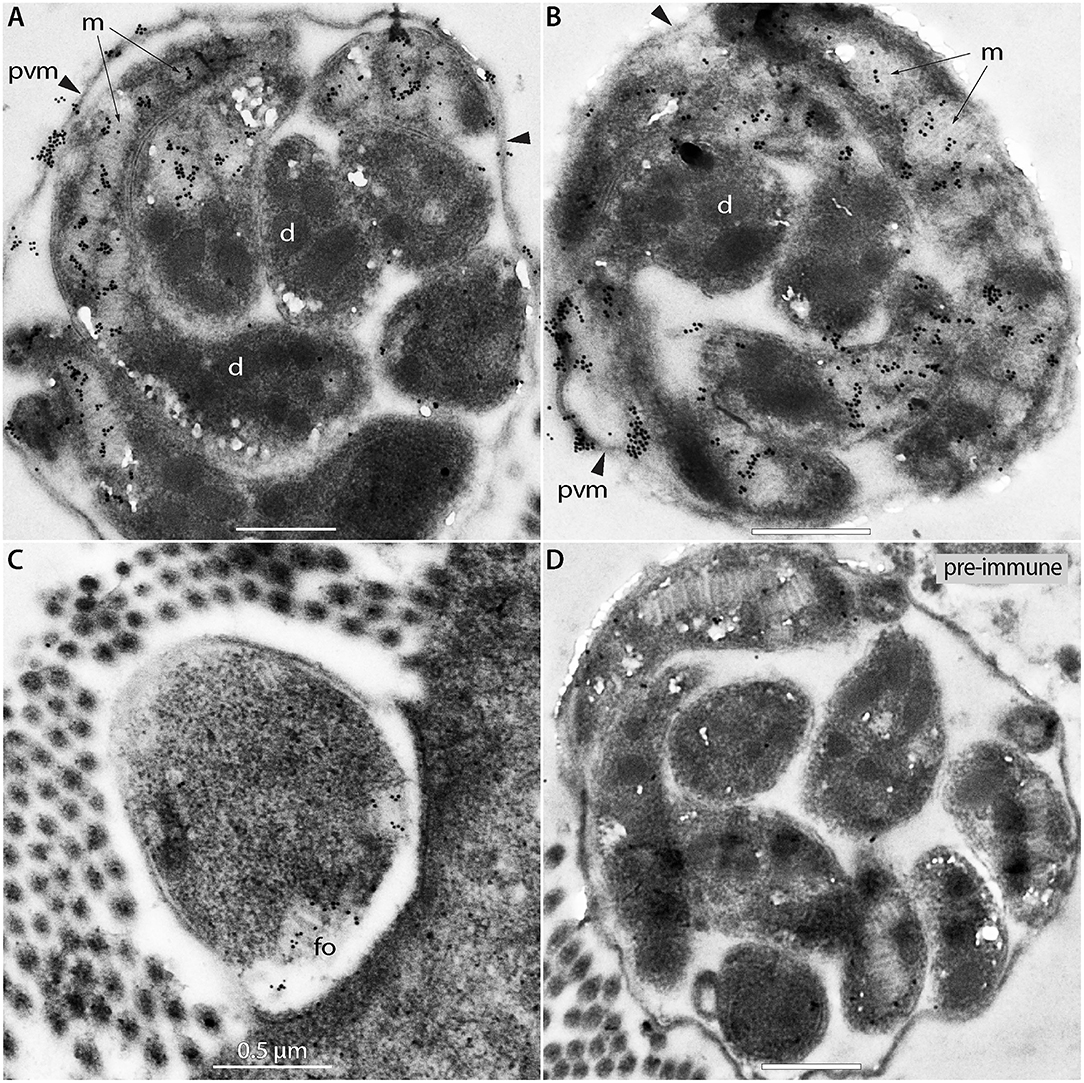
Figure 7. Immunogold electron micrographs of intracellular meronts of Cryptosporidium tyzzeri labeled with anti-CpROM1 antibody (A–C) or pre-immune serum (D). Specimens were obtained from a mouse ileum infected with C. tyzzeri. Gold particles are present over the micronemes (m; arrows) of intracellular merozoites, parasitophorous vacuole membrane (pvm; arrowheads) and a feeder organelle (fo). No gold particles were observed on dense granules (d).
We can, therefore, conclude that CpROM1 is present in various Cryptosporidium membrane structures, including sporozoite and merozoite micronemes, PVM, FO, and, to a lesser degree, sporozoite cell surfaces.
Cryptosporidium parvum Rhomboid Peptidase 1 Concentrates at the Host Cell–Parasite Interface During Sporozoite Invasion
Micronemes are part of the secretory machinery involved in the invasion of Cryptosporidium and other apicomplexans. The IFA showed that CpROM1 was concentrated in the anterior region of C. parvum sporozoites upon their attachment to host cells (Figure 8A) and were then distributed to the pellicles or surfaces of sporozoites during invasion and transformation of sporozoites into trophozoites (Figures 8B–F). CpROM1 was further enriched at the host cell–parasite interface, which was more apparent in the later stages of invasion (Figures 8D–F). Finally, CpROM1 signals were observed around the nuclei when trophozoites were formed (Figures 8G,H). These observations suggest that CpROM1 is likely discharged into the host cell–sporozoite interface during the parasite invasion process.
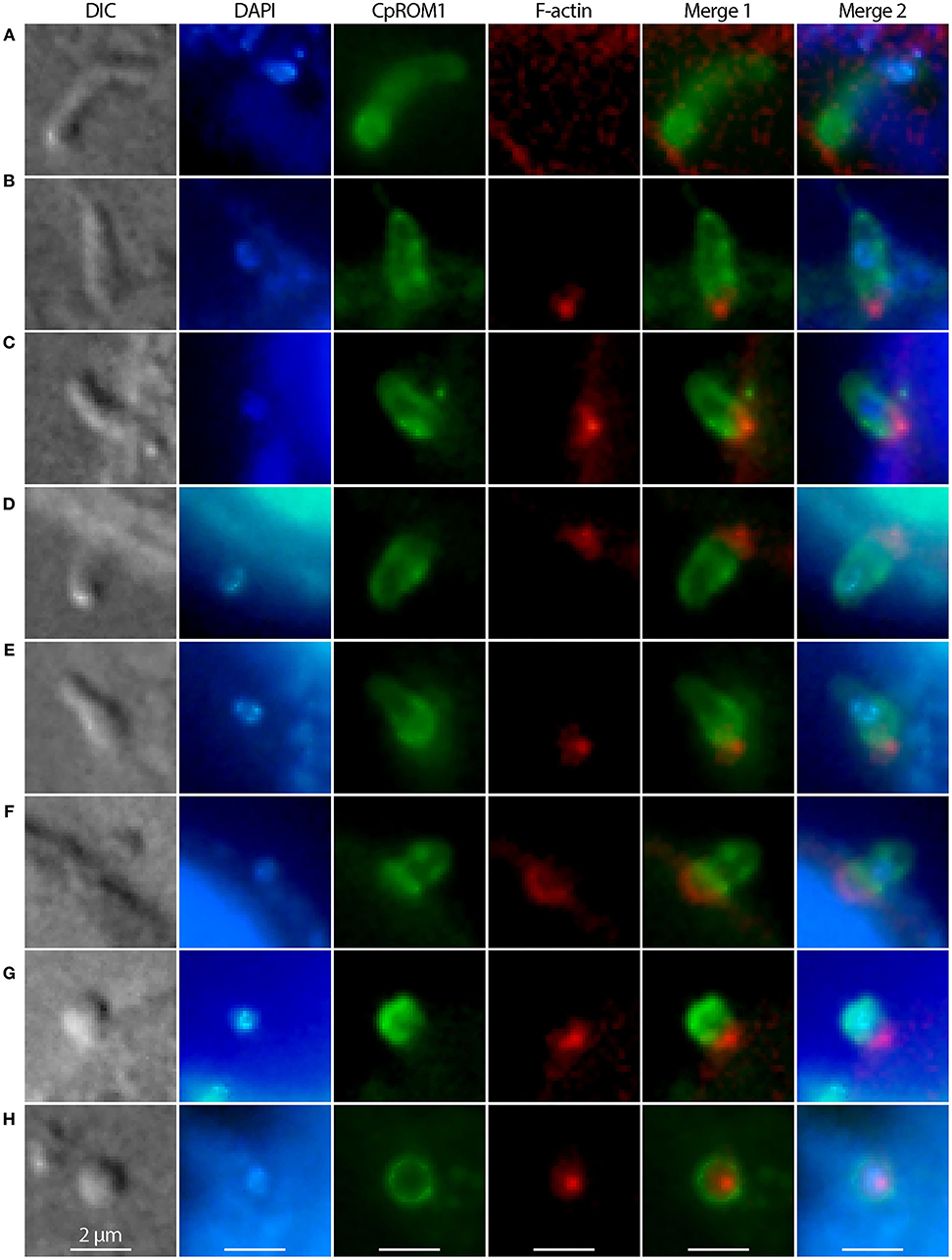
Figure 8. Distribution of CpROM1 (green) in sporozoites during its invasion and transformation into trophozoites, identified using immunofluorescence assays. CpROM1 accumulated in the apex of the sporozoite upon its attachment to the host cell (A), redistributed to pellicles or subpellicles (B–D), enriched at the host cell–parasite interface (D–G), and eventually became relatively evenly distributed on the plasma membrane of the round trophozoite (H). The morphologies of invading and transforming sporozoites were determined using differential interference contrast (DIC) microscopy. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (blue). Phalloidin–rhodamine (non-reactive to the parasite F-actin) was used to stain host cell F-actin aggregated and accumulated beneath the infection sites (red).
Anti-Cryptosporidium parvum Rhomboid Peptidase 1 Antibody Had No Effect on Cryptosporidium parvum Infection
We tested the effect of an anti-CpROM1 antibody that recognizes a non-cytoplasmic epitope located between TMD5 and TMD6 on host cell infection by C. parvum sporozoites using an 18-h infection assay. No significant differences were observed between groups receiving pre-immune and antiserum treatments (Figure 9). These results show that this anti-CpROM1 antibody has no effect on both invasion and intracellular parasite development. However, this observation does not imply that CpROM1 plays no role in parasite invasion or intracellular development. The ineffectiveness of the anti-CpROM1 antibody on infection was likely a result of the inaccessibility of the CpROM1 protein located inside the parasite. Although some CpROM1 was distributed on the surface of sporozoites, the quantity was determined to be minute, based on IFA staining of fixed but unpermeabilized sporozoites (Figure 5C). Additionally, the epitope recognized by this antibody was mostly embedded in the membrane, and only three non-cytoplasmic residues were exposed (Figure 4A and Supplementary Figure S2), making it inaccessible to the antibody when the parasite was live and unpermeabilized. Additionally, antibody binding may not fully inactivate the intramembrane peptidase. Thus, more effective tools and approaches are needed to elucidate the functional roles of CpROM1 and the other two Cryptosporidium ROM peptidases, as exemplified by functional investigations of Toxoplasma and Plasmodium ROM peptidases (36, 41–44).
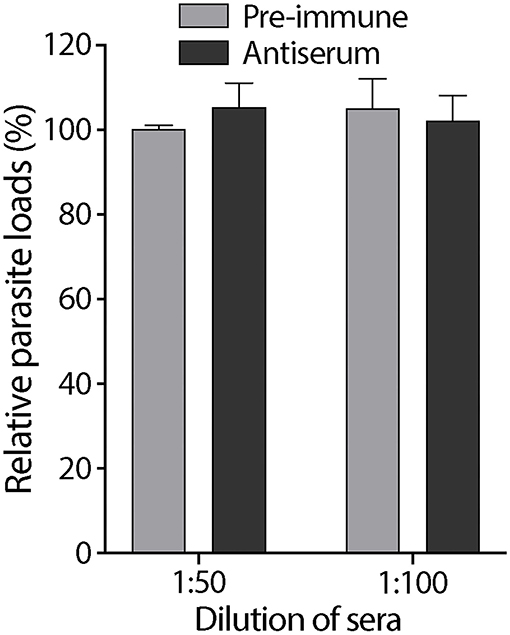
Figure 9. Effect of rabbit antiserum against CpROM1 and pre-immune serum at 1:50 and 1:100 dilutions on C. parvum sporozoite infection of HCT-8 cells in vitro. HCT-8 cell monolayers were inoculated with excysted sporozoites for 2 h to allow invasion, washed to remove non-invading sporozoites, and allowed to grow for an additional 16 h. Pre-immune serum and antiserum were included in the invasion and intracellular growth stages. Blank control was subjected to the same infection procedure but with no serum treatment. Relative parasite loads were determined using qRT-PCR, with the blank control as the baseline.
Discussion
Although micronemes are one of the three secretory machineries essential for invasion by apicomplexan parasites, only four microneme proteins have previously been identified in Cryptosporidium, with their micronemal localization corroborated using mainly IFA. Of these, the mucin-like glycoprotein, GP900, was identified in micronemes of intracellular merozoites using IEM (45), and in the pellicles or surfaces of sporozoites using IFA (45–48). GP900 is located either on the surface and micronemes of zoites or are stored in micronemes prior to relocation to the surface of invasive forms of the parasite (21, 49). Micronemal localization of the other three proteins was not validated using IEM, but was based on IFA only. TRAP-C1 (TSP-related adhesive protein of Cryptosporidium-1) is likely located in the micronemes, based on labeling of the apical pole of sporozoites (50). TSP8 (TSP-related adhesive protein 8), also known as CpMIC1, appeared to be apically located on the zoite surface and was colocalized with a monoclonal antibody (mAb) TOU (51). IEM showed that mAb TOU specifically labeled sporozoite micronemes, but its antigen is unknown (52). Cpa135 (a modular protein with ricin B and LCCL domains) is localized in the anterior pole of sporozoites and is also thought to be a micronemal protein (53).
This study has expanded the number of confirmed (or potential) microneme proteins from four to eight, with CpROM1 being confirmed using IEM and the other three being confirmed using IFA. CpROM1 is the first protein whose micronemal localization in C. parvum sporozoites has been observed using IEM. The 10 candidate microneme proteins investigated in this study were selected based on previous in silico prediction studies (22), with polyclonal antibodies being successfully raised against nine of these proteins. IFA showed probable micronemal localization for four of these proteins, indicating overall the success and power of the in silico prediction. This study also suggests that it is worth following up on the remaining in silico-predicted microneme proteins, in addition to the single protein against which we were unable to raise antibodies (cgd6_760), to confirm whether they are localized in the micronemes.
CpROM1 was the smallest of the three C. parvum rhomboid peptidases (282 aa; Figure 4A). Its seven TMDs occupy a total of 158 amino acids (56% of the protein), making it a highly hydrophobic membrane protein. CpROM1 is highly basic (pI = 9.7). Therefore, experimental conditions for Western blot analysis of CpROM1, including SDS-PAGE and blotting procedures, have to be optimized, otherwise the protein would aggregate on top of the SDS-PAGE separation gel. In this study, Western blot analysis was used to successfully detect CpROM1 after the following modifications were made: Incubation of protein extracts in sample buffer prior to SDS-PAGE was performed at 37°C for 10 min instead of at 95°C for 5–10 min as is specified in conventional protocols. The basicity of the transfer buffer was also increased to pH 10.1 (vs. pH 9.2 in a standard transfer buffer). In addition, due to the high basicity of protein (hence, higher positive charge) and hydrophobicity (more SDS molecules attached to the hydrophobic amino acids), CpROM1 moved faster than an average protein on SDS-PAGE and appeared as an ~23-kDa protein band (instead of its 31.3-kDa predicted molecular weight) (Figure 5B). Such anomalous gel mobility or “gel shifting” in SDS-PAGE is common among membrane proteins (54, 55).
CpROM1 has syntenic orthologs that are highly conserved across all sequenced Cryptosporidium genomes (56–58). Protein similarities were much higher within intestinal parasites, such as C. parvum, C. hominis, C. tyzzeri, C. meleagridis, and C. ubiquitum and gastric species such as C. muris and C. andersoni, than between intestinal and gastric parasite species (Supplementary Figure S2). For example, there is only one amino acid difference between CpROM1 and ChROM1 (i.e., P to I at position 31) in C. parvum and C. hominis, the two major species that infect humans (Supplementary Figure S2). This high sequence conservation is indicative of the conserved function of ROM1 peptidases within the genus Cryptosporidium.
As a family of widely distributed intramembrane serine peptidases responsible for the cleavage of membrane proteins near the TM domain, rhomboid peptidase was first identified in Drosophila (DmROM1) and shown to participate in in vivo Spitz processing during epidermal growth factor receptor (EGFR) signaling (59). ROMs have different substrate specificities in Toxoplasma and Plasmodium and are responsible for the shedding of various adhesins, including apical membrane adhesin/antigen 1 (AMA1), microneme proteins (MICs), and thrombospondin-related adhesive proteins (TRAPs) (41, 60). Cryptosporidium possesses only three ROM or ROM domain-containing proteins (Figure 4B) in contrast to Toxoplasma and Plasmodium, which possess 11 and 8 ROMs, respectively (35). This study revealed the presence of CpROM1 not only in the micronemes of sporozoites and merozoites but also in PVM and feeder organelles. CpROM1 also accumulated on the sporozoite surface and the host cell–parasite interface (Figure 8). These observations imply that CpROM1 participates in the cleavage of certain micronemal proteins in the zoites during invasion, as well as in the proteostasis of membrane proteins in the Cryptosporidium-unique PVM and feeder organelles.
The two residues in the active site of CpROM1 (Ser162 and His217) are present in TMD4 and TMD6, along the non-cytoplasmic side (Figures 4A, 10A), suggesting that CpROM1 cleaves substrates on the non-cytoplasmic side of a TMD (Figure 10B). In apicomplexans and other organisms, the majority of known ROM substrates are type I membrane proteins containing a single TMD located near the C-terminus. In the case of CpROM1, the large non-cytoplasmic domain of a type I membrane protein substrate may be shed following cleavage from the surface of zoites during gliding, attachment, and invasion, or released to the outside of the zoites or PVM; and/or to the lumens of feeder organelles during intracellular development. Cryptosporidium parasites possess a number of type I membrane proteins that are candidate ROM substrates, including the TIP homolog and some TRAPs (29, 50, 61). It is important to verify CpROM1, CpROM2, and CpROM3 substrates in order to delineate their roles in C. parvum invasion and proteostasis in the PVM and feeder organelles.
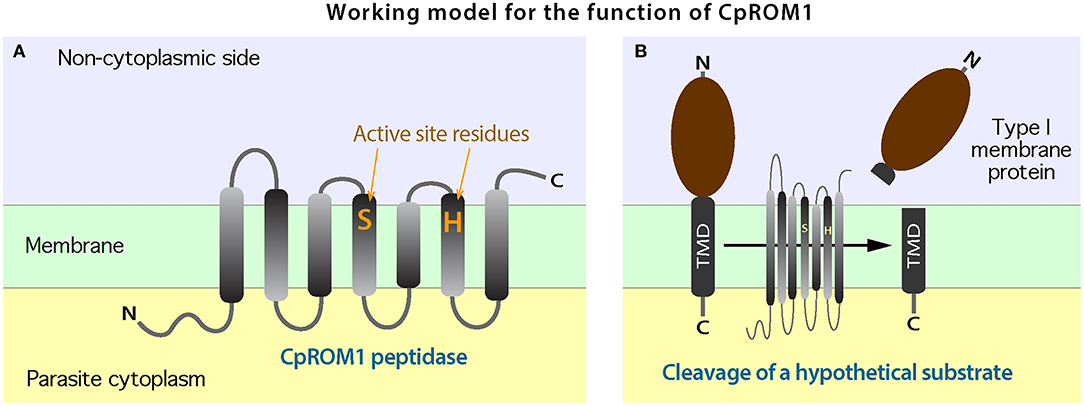
Figure 10. A working model for the biochemical function of CpROM1. (A) Arrangement of the seven transmembrane domains (TMDs) of CpROM1 across the plasma membrane. Parasite cytoplasmic and non-cytoplasmic sides are shown below and above the membrane. The two active residues at the active sites in TMD4 and TMD6 are located close to the non-cytoplasmic side. (B) Illustration of the CpROM1-mediated intramembrane cleavage of a hypothetical type I membrane protein with a single TMD at the C-terminus. After cleavage, the functional section of the substrate protein is released into the non-cytoplasmic side.
For the three additional micronemal proteins identified using IFA, it may be worth further validating their subcellular localization using IEM and new antibodies or using optimized IEM procedures. These three proteins contain no N-terminal signal peptides based on domain analysis using InterProScan (https://www.ebi.ac.uk/interpro/search/sequence/) and SignalP server (http://www.cbs.dtu.dk/services/SignalP-5.0/), but are predicted to contain non-cytoplasmic domains that function in extracellular regions (Supplementary Figure S3). The cgd1_3550-encoded product is relatively large (925 aa), contains one Apple domain (InterPro domain: IPR003609), and is predicted to be involved in protein binding in the extracellular region based on gene ontology (GO:0005576) (Supplementary Figure S3A). The cgd2_1590-encoded product (614 aa) possesses a short N-terminal cytosolic domain, a TM domain, a long non-cytoplasmic region containing an EGF-like domain (IPR000742), and an Apple domain (Supplementary Figure S3B). It is also predicted to be involved in protein binding in the extracellular region. The protein encoded by cgd1_3680 is a relatively short (263 aa), single-pass, type I membrane protein with a TM domain near the C-terminus. The N-terminal non-cytoplasmic region contains an EGF-like domain (IPR000742) (Supplementary Figure S3C).
Some microneme proteins such as AMA1 and ROM peptidases in other apicomplexans are vaccine candidates (62, 63). A previous in silico study predicted surface-exposed antigens of C. hominis as vaccine candidates based on several criteria, including the presence of glycosylphosphatidylinositol (GPI)-anchor motifs, ≥5 TMDs, or signal peptides for protein targeting to the secretory pathway (64). Of the four new microneme proteins identified in this study, only CpROM1 met one of these criteria by possessing seven TMDs. The other three proteins contained no N-terminal signal peptides, had zero or only one TMD, and lacked GPI-anchor motifs based on PredGPI analysis (http://gpcr2.biocomp.unibo.it/gpipe/). However, they were all predicted to contain non-cytoplasmic domains that function in the extracellular regions (Supplementary Figure S3). Therefore, in addition to serving as markers for probing the function of micronemes in Cryptosporidium micronemes, their potential as vaccine candidates might also be worth exploring.
Conclusions
The present study reports on the discovery of a new microneme protein (CpROM1), confirmed using IEM, and three possible microneme proteins (cgd1_3550, cgd1_3680, and cgd2_1590) identified using IFA. These observations significantly increase our understanding of the molecular composition of C. parvum micronemes. CpROM1 is also present in the PVM and feeder organelles, and is enriched on sporozoite surfaces and at the host cell–parasite interface during sporozoite invasion of host cells, implying the involvement of CpROM1 in parasite invasion as well as in membrane proteostasis of PVM and feeder organelles. Anti-CpROM1 antibody failed to block sporozoite attachment and invasion of host cells by sporozoites in vitro, presumably because of the inaccessibility of the epitope in live sporozoites.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Institute Committee for Biosafety and Ethics for Animal Use, Jilin University.
Author Contributions
GZ, JY, and XG conceived and designed the experiments. XG and DW performed the experiments. YyZ, YZ, XL, and CW provided technical assistance. GZ and XG analyzed the data. XG and JY wrote the draft manuscript. GZ edited and wrote the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by funding from the National Key R&D Program of China (2017YFD0500401) and the National Natural Science Foundation of China (31772731).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.778560/full#supplementary-material
Supplementary Figure S1. Maximum likelihood (M) tree of rhomboid peptidase orthologs in the alveolates, with detailed information on the accession numbers and species names.
Supplementary Figure S2. Multiple amino acid alignment of Cryptosporidium ROM1 orthologs available in the CryptoDB.
Supplementary Figure S3. InterProScan domain analysis of the three Cryptosporidium parvum microneme proteins encoded by cgd1_3550 (A), cgd2_1590 (B), and cgd1_3680 (C).
Supplementary Figure S4. Immunofluorescence micrographs of excysted sporozoites of Cryptosporidium parvum obtained using antibodies from specified pre-immune sera.
Supplementary Table S1. A detailed description of the candidate microneme proteins used in this study. A concise version is included in the manuscript as Table 1.
References
1. Bouzid M, Hunter PR, Chalmers RM, Tyler KM. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev. (2013) 26:115–34. doi: 10.1128/CMR.00076-12
2. Prystajecky N, Huck PM, Schreier H, Isaac-Renton JL. Assessment of Giardia and Cryptosporidium spp. as a microbial source tracking tool for surface water: application in a mixed-use watershed. Appl Environ Microbiol. (2014) 80:2328–36. doi: 10.1128/AEM.02037-13
3. Tetley L, Brown SMA, Mcdonald V, Coombs GH. Ultrastructural analysis of the sporozoite of Cryptosporidium parvum. Microbiology. (1998) 144 (Pt 12) 3249–55. doi: 10.1099/00221287-144-12-3249
4. O'hara SP, Huang BQ, Chen XM, Nelson J, Larusso NF. Distribution of Cryptosporidium parvum sporozoite apical organelles during attachment to and internalization by cultured biliary epithelial cells. J Parasitol. (2005) 91:995–9. doi: 10.1645/GE-495R.1
5. Arredondo SA, Schepis A, Reynolds L, Kappe SHI. Secretory organelle function in the Plasmodium sporozoite. Trends Parasitol. (2021) 37:651–63. doi: 10.1016/j.pt.2021.01.008
6. Sparvoli D, Lebrun M. Unraveling the elusive rhoptry exocytic mechanism of apicomplexa. Trends Parasitol. (2021) 37:622–37. doi: 10.1016/j.pt.2021.04.011
7. Petry F. Structural analysis of Cryptosporidium parvum. Microsc Microanal. (2004) 10:586–601. doi: 10.1017/S1431927604040929
8. Guerin A, Roy NH, Kugler EM, Berry L, Burkhardt JK, Shin JB, et al. Cryptosporidium rhoptry effector protein ROP1 injected during invasion targets the host cytoskeletal modulator LMO7. Cell Host Microbe. (2021) 29:1407–20. doi: 10.1016/j.chom.2021.07.002
9. Mageswaran SK, Guerin A, Theveny LM, Chen WD, Martinez M, Lebrun M, et al. In situ ultrastructures of two evolutionarily distant apicomplexan rhoptry secretion systems. Nat Commun. (2021) 12:4983. doi: 10.1038/s41467-021-25309-9
10. Uni S, Iseki M, Maekawa T, Moriya K, Takada S. Ultrastructure of Cryptosporidium muris (strain RN 66) parasitizing the murine stomach. Parasitol Res. (1987) 74:123–32. doi: 10.1007/BF00536023
11. Joung M, Yun S, Joung M, Park WY, Yu JR. Ultrastructural changes in Cryptosporidium parvum oocysts by gamma irradiation. Korean J Parasitol. (2011) 49:25–31. doi: 10.3347/kjp.2011.49.1.25
12. Aldeyarbi HM, Karanis P. The ultra-structural similarities between Cryptosporidium parvum and the Gregarines. J Eukaryot Microbiol. (2016) 63:79–85. doi: 10.1111/jeu.12250
13. Gaechter V, Hehl AB. Assembly and export of a Toxoplasma microneme complex in Giardia lamblia. Int J Parasitol. (2005) 35:1359–68. doi: 10.1016/j.ijpara.2005.06.007
14. Kats LM, Black CG, Proellocks NI, Coppel RL. Plasmodium rhoptries: how things went pear-shaped. Trends Parasitol. (2006) 22:269–76. doi: 10.1016/j.pt.2006.04.001
15. Counihan NA, Kalanon M, Coppel RL, De Koning-Ward TF. Plasmodium rhoptry proteins: why order is important. Trends Parasitol. (2013) 29:228–36. doi: 10.1016/j.pt.2013.03.003
16. Liu Q, Li FC, Zhou CX, Zhu XQ. Research advances in interactions related to Toxoplasma gondii microneme proteins. Exp Parasitol. (2017) 176:89–98. doi: 10.1016/j.exppara.2017.03.001
17. Venugopal K, Marion S. Secretory organelle trafficking in Toxoplasma gondii: a long story for a short travel. Int J Med Microbiol. (2018) 308:751–60. doi: 10.1016/j.ijmm.2018.07.007
18. Ben Chaabene R, Lentini G, Soldati-Favre D. Biogenesis and discharge of the rhoptries: key organelles for entry and hijack of host cells by the Apicomplexa. Mol Microbiol. (2021) 115:453–65. doi: 10.1111/mmi.14674
19. Bhalchandra S, Ludington J, Coppens I, Ward HD. Identification and characterization of Cryptosporidium parvum Clec, a novel C-type lectin domain-containing mucin-like glycoprotein. Infect Immun. (2013) 81:3356–65. doi: 10.1128/IAI.00436-13
20. Valentini E, Cherchi S, Possenti A, Dubremetz JF, Pozio E, Spano F. Molecular characterisation of a Cryptosporidium parvum rhoptry protein candidate related to the rhoptry neck proteins TgRON1 of Toxoplasma gondii and PfASP of Plasmodium falciparum. Mol Biochem Parasitol. (2012) 183:94–9. doi: 10.1016/j.molbiopara.2012.02.004
21. Lendner M, Daugschies A. Cryptosporidium infections: molecular advances. Parasitology. (2014) 141:1511–32. doi: 10.1017/S0031182014000237
22. Chen ZQ, Harb OS, Roos DS. In silico Identification of specialized secretory-organelle proteins in apicomplexan parasites and in vivo validation in Toxoplasma gondii. PLoS ONE. (2008) 3:e3611. doi: 10.1371/journal.pone.0003611
23. Lateef SS, Gupta S, Jayathilaka LP, Krishnanchettiar S, Huang JS, Lee BS. An improved protocol for coupling synthetic peptides to carrier proteins for antibody production using DMF to solubilize peptides. J Biomol Tech. (2007) 18:173–6.
24. Greenfield EA. Standard immunization of rabbits. Cold Spring Harb Protoc. (2020) 2020:100305. doi: 10.1101/pdb.prot100305
25. Kurien BT. Affinity purification of autoantibodies from an antigen strip excised from a nitrocellulose protein blot. Methods Mol Biol. (2009) 536:201–11. doi: 10.1007/978-1-59745-542-8_22
26. Truong Q, Ferrari BC. Quantitative and qualitative comparisons of Cryptosporidium faecal purification procedures for the isolation of oocysts suitable for proteomic analysis. Int J Parasitol. (2006) 36:811–9. doi: 10.1016/j.ijpara.2006.02.023
27. Zhang H, Guo F, Zhu G. Involvement of host cell integrin alpha2 in Cryptosporidium parvum infection. Infect Immun. (2012) 80:1753–8. doi: 10.1128/IAI.05862-11
28. Sateriale A, Slapeta J, Baptista R, Engiles JB, Gullicksrud JA, Herbert GT, et al. A genetically tractable, natural mouse model of cryptosporidiosis offers insights into host protective immunity. Cell Host Microbe. (2019) 26:135–46.e135. doi: 10.1016/j.chom.2019.05.006
29. Zhang T, Gao X, Wang D, Zhao J, Zhang N, Li Q, et al. A single-pass type I membrane protein from the apicomplexan parasite Cryptosporidium parvum with nanomolar binding affinity to host cell surface. Microorganisms. (2021) 9:1015. doi: 10.3390/microorganisms9051015
30. Mauzy MJ, Enomoto S, Lancto CA, Abrahamsen MS, Rutherford MS. The Cryptosporidium parvum transcriptome during in vitro development. PLoS ONE. (2012) 7:e31715. doi: 10.1371/journal.pone.0031715
31. Zhang H, Zhu G. Quantitative RT-PCR assay for high-throughput screening (HTS) of drugs against the growth of Cryptosporidium parvum in vitro. Front Microbiol. (2015) 6:991. doi: 10.3389/fmicb.2015.00991
32. Guo F, Zhang H, Payne HR, Zhu G. Differential gene expression and protein localization of Cryptosporidium parvum Fatty Acyl-CoA synthetase isoforms. J Eukaryot Microbiol. (2016) 63:233–46. doi: 10.1111/jeu.12272
33. Jin Z, Ma J, Zhu G, Zhang H. Discovery of novel anti-cryptosporidial activities from natural products by in vitro high-throughput phenotypic screening. Front Microbiol. (2019) 10:1999. doi: 10.3389/fmicb.2019.01999
34. Zhang H, Zhu G. High-throughput screening of drugs against the growth of Cryptosporidium parvum in vitro by qRT-PCR. Methods Mol Biol. (2020) 2052:319–34. doi: 10.1007/978-1-4939-9748-0_18
35. Dowse TJ, Soldati D. Rhomboid-like proteins in Apicomplexa: phylogeny and nomenclature. Trends Parasitol. (2005) 21:254–8. doi: 10.1016/j.pt.2005.04.009
36. Sibley LD. The roles of intramembrane proteases in protozoan parasites. Biochim Biophys Acta. (2013) 1828:2908–15. doi: 10.1016/j.bbamem.2013.04.017
37. Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. (2014) 42:D503–9. doi: 10.1093/nar/gkt953
38. Zhang H, Guo F, Zhu G. Cryptosporidium lactate dehydrogenase is associated with the parasitophorous vacuole membrane and is a potential target for developing therapeutics. PLoS Pathog. (2015) 11:e1005250. doi: 10.1371/journal.ppat.1005250
39. Yu X, Guo F, Mouneimne RB, Zhu G. Cryptosporidium parvum elongation factor 1alpha (CpEF1alpha) participates in the formation of base structure at the infection site during invasion. J Infect Dis. (2019) 221:1816–25. doi: 10.1093/infdis/jiz684
40. Cai X, Herschap D, Zhu G. Functional characterization of an evolutionarily distinct phosphopantetheinyl transferase in the apicomplexan Cryptosporidium parvum. Eukaryot Cell. (2005) 4:1211–20. doi: 10.1128/EC.4.7.1211-1220.2005
41. Brossier F, Jewett TJ, Sibley LD, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc Natl Acad Sci USA. (2005) 102:4146–51. doi: 10.1073/pnas.0407918102
42. Dowse TJ, Koussis K, Blackman MJ, Soldati-Favre D. Roles of proteases during invasion and egress by Plasmodium and Toxoplasma. Subcell Biochem. (2008) 47:121–39. doi: 10.1007/978-0-387-78267-6_10
43. Ejigiri I, Ragheb DR, Pino P, Coppi A, Bennett BL, Soldati-Favre D, et al. Shedding of TRAP by a rhomboid protease from the malaria sporozoite surface is essential for gliding motility and sporozoite infectivity. PLoS Pathog. (2012) 8:e1002725. doi: 10.1371/journal.ppat.1002725
44. Shen B, Buguliskis JS, Lee TD, Sibley LD. Functional analysis of rhomboid proteases during Toxoplasma invasion. MBio. (2014) 5:e01795–e01714. doi: 10.1128/mBio.01795-14
45. Barnes DA, Bonnin A, Huang JX, Gousset L, Wu J, Gut J, et al. A novel multi-domain mucin-like glycoprotein of Cryptosporidium parvum mediates invasion. Mol Biochem Parasitol. (1998) 96:93–110. doi: 10.1016/S0166-6851(98)00119-4
46. Petersen C, Gut J, Doyle PS, Crabb JH, Nelson RG, Leech JH. Characterization of a > 900,000-M(r) Cryptosporidium parvum sporozoite glycoprotein recognized by protective hyperimmune bovine colostral immunoglobulin. Infect Immun. (1992) 60:5132–8. doi: 10.1128/iai.60.12.5132-5138.1992
47. Petersen C, Gut J, Leech JH, Nelson RG. Identification and initial characterization of five Cryptosporidium parvum sporozoite antigen genes. Infect Immun. (1992) 60:2343–8. doi: 10.1128/iai.60.6.2343-2348.1992
48. Chatterjee A, Banerjee S, Steffen M, O'connor RM, Ward HD, Robbins PW, et al. Evidence for mucin-like glycoproteins that tether sporozoites of Cryptosporidium parvum to the inner surface of the oocyst wall. Eukaryot Cell. (2010) 9:84–96. doi: 10.1128/EC.00288-09
49. Wanyiri J, Ward H. Molecular basis of Cryptosporidium-host cell interactions: recent advances and future prospects. Future Microbiol. (2006) 1:201–8. doi: 10.2217/17460913.1.2.201
50. Spano F, Putignani L, Naitza S, Puri C, Wright S, Crisanti A. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Mol Biochem Parasitol. (1998) 92:147–62. doi: 10.1016/S0166-6851(97)00243-0
51. Putignani L, Possenti A, Cherchi S, Pozio E, Crisanti A, Spano F. The thrombospondin-related protein CpMIC1 (CpTSP8) belongs to the repertoire of micronemal proteins of Cryptosporidium parvum. Mol Biochem Parasitol. (2008) 157:98–101. doi: 10.1016/j.molbiopara.2007.09.004
52. Bonnin A, Dubremetz JF, Camerlynck P. Characterization of microneme antigens of Cryptosporidium parvum (Protozoa, Apicomplexa). Infect Immun. (1991) 59:1703–8. doi: 10.1128/iai.59.5.1703-1708.1991
53. Tosini F, Agnoli A, Mele R, Gomez Morales MA, Pozio E. A new modular protein of Cryptosporidium parvum, with ricin B and LCCL domains, expressed in the sporozoite invasive stage. Mol Biochem Parasitol. (2004) 134:137–47. doi: 10.1016/j.molbiopara.2003.11.014
54. Kaur J, Bachhawat AK. A modified Western blot protocol for enhanced sensitivity in the detection of a membrane protein. Anal Biochem. (2009) 384:348–9. doi: 10.1016/j.ab.2008.10.005
55. Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci USA. (2009) 106:1760–5. doi: 10.1073/pnas.0813167106
56. Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. (2004) 304:441–5. doi: 10.1126/science.1094786
57. Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, et al. The genome of Cryptosporidium hominis. Nature. (2004) 431:1107–12. doi: 10.1038/nature02977
58. Ifeonu OO, Chibucos MC, Orvis J, Su Q, Elwin K, Guo F, et al. Annotated draft genome sequences of three species of Cryptosporidium: Cryptosporidium meleagridis isolate UKMEL1, C. baileyi isolate TAMU-09Q1 and C. hominis isolates TU502_2012 and UKH1. Pathog Dis. (2016) 74:ftw080. doi: 10.1093/femspd/ftw080
59. Urban S, Dickey SW. The rhomboid protease family: a decade of progress on function and mechanism. Genome Biol. (2011) 12:231. doi: 10.1186/gb-2011-12-10-231
60. Baker RP, Wijetilaka R, Urban S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog. (2006) 2:e113. doi: 10.1371/journal.ppat.0020113
61. Deng M, Templeton TJ, London NR, Bauer C, Schroeder AA, Abrahamsen MS. Cryptosporidium parvum genes containing thrombospondin type 1 domains. Infect Immun. (2002) 70:6987–95. doi: 10.1128/IAI.70.12.6987-6995.2002
62. Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. (2008) 24:74–84. doi: 10.1016/j.pt.2007.12.002
63. Foroutan M, Zaki L, Tavakoli S, Soltani S, Taghipour A, Ghaffarifar F. Rhomboid antigens are promising targets in the vaccine development against Toxoplasma gondii. EXCLI J. (2019) 18:259–72. doi: 10.17179/excli2018-1993
Keywords: apicomplexan, Cryptosporidium parvum, microneme proteins (MICs), rhomboid peptidase (ROM), sporozoite invasion, parasitophorous vacuole membrane (PVM), feeder organelle (FO)
Citation: Gao X, Yin J, Wang D, Li X, Zhang Y, Wang C, Zhang Y and Zhu G (2021) Discovery of New Microneme Proteins in Cryptosporidium parvum and Implication of the Roles of a Rhomboid Membrane Protein (CpROM1) in Host–Parasite Interaction. Front. Vet. Sci. 8:778560. doi: 10.3389/fvets.2021.778560
Received: 17 September 2021; Accepted: 12 November 2021;
Published: 13 December 2021.
Edited by:
Valeria Analia Sander, CONICET Institute of Biotechnological Research (IIB-INTECH), ArgentinaCopyright © 2021 Gao, Yin, Wang, Li, Zhang, Wang, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guan Zhu, Y3J5cHRvc3BvcmlkYUBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Xin Gao
Xin Gao Jigang Yin
Jigang Yin Dongqiang Wang1
Dongqiang Wang1 Guan Zhu
Guan Zhu