
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 07 December 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.771755
This article is part of the Research TopicEmerging/Re-emerging Poultry VirusesView all 9 articles
 Jun Zhang1,2,3,4
Jun Zhang1,2,3,4 Tuofan Li1,2,3,4
Tuofan Li1,2,3,4 Weikang Wang1,2,3,4
Weikang Wang1,2,3,4 Quan Xie1,2,3,4
Quan Xie1,2,3,4 Zhimin Wan1,2,3,4
Zhimin Wan1,2,3,4 Aijian Qin1,2,3,4
Aijian Qin1,2,3,4 Jianqiang Ye1,2,3,4
Jianqiang Ye1,2,3,4 Hongxia Shao1,2,3,4*
Hongxia Shao1,2,3,4*In recent years, the emergence of avian orthoreovirus (ARV) has caused significant losses to the poultry industry worldwide. In this study, a novel ARV isolate, designated as AHZJ19, was isolated and identified from domestic chicken with viral arthritis syndrome in China. AHZJ19 can cause typical syncytial cytopathic effect in the chicken hepatocellular carcinoma cell line, LMH. High-throughput sequencing using Illumina technology revealed that the genome size of AHZJ19 is about 23,230 bp, which codes 12 major proteins. Phylogenetic tree analysis found that AHZJ19 was possibly originated from a recombination among Hungarian strains, North American strains, and Chinese strains based on the sequences of the 12 proteins. Notably, the σC protein of AHZJ19 shared only about 50% homology with that of the vaccine strains S1133 and 1733, which also significantly differed from other reported Chinese ARV strains. The isolation and molecular characteristics of AHZJ19 provided novel insights into the molecular epidemiology of ARV and laid the foundation for developing efficient strategies for control of ARV in China.
Avian orthoreovirus (ARV) belongs to the Orthoreovirus genus (Reoviridae family) with a double-stranded and segmented RNA genome (1, 2). According to electrophoretic mobility, ARV has 10 genomic segments including the large genomic segments (L1 to L3), the medium genomic segments (M1 to M3), and the small genomic segments (S1 to S4), which encode 12 major proteins (1–4). Among them, σC protein, a component of the outer capsid of the virion, encoded by S1 segment, is the main immunogenic protein and the viral cell attachment protein that induces specific neutralizing antibodies (3, 5, 6). Based on the genetic characteristics of σC protein, ARV isolates are divided into four to six clusters (7–11). ARV was initially discovered as the causative agents of tenosynovitis in chicks (12), and subsequently spread globally to multiple hosts including turkeys, pheasants, Muscovy ducks, geese, and other wild birds (13–15). ARV infection mainly causes arthritis syndrome and runting–stunting syndrome, and is also related to hepatitis, myocarditis, osteoporosis, respiratory diseases, and even central nervous system (16–18). However, only the direct link of ARV infection with viral arthritis syndrome has been conclusively confirmed (19). Notably, ARV infection results in immunosuppression, poor production performance, poor feed utilization, and conversion rate. ARV infection also increases the susceptibility to other pathogens (20–22).
Although vaccines have been used to prevent and control the diseases caused by ARV, the frequent emergence of novel ARV strains with variant antigenicity is challenging the current vaccine strategy (7, 8, 23–25). Thus, it is critical to clarify the prevalence and variant characteristics of ARV strains. In this study, a novel ARV strain, designated as AHZJ19, was isolated from a chicken flock with viral arthritis syndrome using LMH cells, and its complete genome sequence was determined. Sequence analysis revealed that AHZJ19 was significantly different from the ARVs previously isolated in China, and is a novel recombinant ARV originated from Hungarian strains, North American strains, and Chinese strains.
In 2019, two liver tissue samples were collected from chickens on a farm in Anhui, China. The native chickens were all 90-day-old and showed lameness, viral arthritis, and poor production performance. The liver tissue samples were homogenized with phosphate-buffered saline (PBS) and supernatants were collected by centrifugation at 12,000 rpm for 30 min at 4°C. Then, the collected supernatants were filtered through 0.22-μm filters and used for virus isolation.
The total RNA was extracted from the liver tissue sample suspension using the AxyPrep Multisource RNA Miniprep Kit (Axygen, USA) and cDNA was synthesized using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, China). For detection of ARV, the PCR was performed with specific primers of S2 gene listed in Table 1 (26). The PCR reaction volume was 25 μl containing 12.5 μl of 2× Taq PCR Master Mix (Vazyme, China), 1 μl of each primer, 9.5 μl of double-distilled water (ddH2O), and 1 μl of the cDNA template. The PCR cycling conditions for the S2 gene amplifications were as follows: 1 cycle of 94°C for 5 min, 35 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 1 min, followed by a final extension step of 72°C for 10 min. In addition, PCR detection for chicken infectious anemia virus (CIAV), reticuloendotheliosis virus (REV), A subgroup of avian leukosis virus (ALV-A), B subgroup of avian leukosis virus (ALV-B), J subgroup of avian leukosis virus (ALV-J), and K subgroup of avian leukosis virus (ALV-K) was performed. The primers used are listed in Table 2.
The supernatants of the RT-PCR-positive liver tissue samples were inoculated into LMH cells for virus isolation and the cells were incubated at 37°C with 5% CO2. The supernatants were collected at 36–48 h post infection (hpi) by centrifugation at 12,000 rpm for 15 min at 4°C and passaged every 2 days. The supernatants of LMH cells after seven consecutive blind passages were all tested by RT-PCR.
The total RNA of the isolate was sequenced using the high-throughput sequencing (Illumina sequencing technology: Illumina Novaseq 6000, ABclonal Whole RNA-seq Lib Prep kit, NovaSeq S4 flowcell) by Shanghai Tanpu Biological Technology Co. (Shanghai, China), and the sequences were assembled with de novo SPAdes assembly software, LASTZ, and SAMtools commands (10, 30, 31). The GenBank accession numbers for the obtained sequences of the isolate are listed in Table 3. The sequences of 12 major proteins of the isolate were aligned with the reference strains deposited in GenBank using the ClustalW methods in Megalign program by the Lasergene 7.0 software. The phylogenetic tree was constructed by using the neighbor-joining method in MEGA6.1 software with 1,000 bootstrap replicates and the intra-segmental recombination detection was performed by Bootscan analysis within the Simplot program version 3.5.1, using the neighbor-joining method, with a Kimura 2-parameter applied and 100 replicates.
Six 3-day-old specific-pathogen-free (SPF) chicks were inoculated with 200 μl of the isolate each through unilateral food pad. Sera from the infected chickens were collected at 3 weeks post inoculation (wpi), and the antibodies against the isolate were detected by indirect immunofluorescence assay (IFA). Briefly, the infected LMH cells were fixed with pre-chilled acetone-ethanol (3:2) for 5 min. After washing once with PBS, the cells were incubated with chicken sera against the isolate (1:200) for 45 min at 37°C. Then, the cells were washed with PBS three times followed by incubation with FITC-labeled rabbit anti-chicken IgG (1:150) for 45 min at 37°C. After three washes with PBS, the cells were observed under a fluorescence microscope and the viral titer of the isolate was calculated by the Reed-Muench method (32).
The disease of ARV has become endemic in China since its first report in 1985 (33). Although the mortality caused by ARV is not high, the infection of ARV generally results in immunosuppression, affects the normal growth and production performance of broilers, and increases the susceptibility to other pathogens. Notably, the continuous emergence of ARV mutants in recent years have caused huge economic losses to the poultry industry (8, 24, 25).
In 2019, an indigenous chicken flock in Anhui in China showed clinical symptoms of viral arthritis syndrome, hepatitis, and stunting/malabsorption syndrome. To detect whether the chickens in that farm were infected with ARV, the liver tissue samples were collected from clinically ill chickens. Total RNA was extracted from the supernatant obtained from homogenized liver tissue samples. The ARV detection was performed by RT-PCR using specific primers listed in Table 1. The liver tissue was positive in the RT-PCR assay for ARV (Figure 1A). In addition, the sample was free of CIAV, REV, ALV-A, ALV-B, ALV-J, and ALV-K infection by PCR using specific primers listed in Table 2. The results of the RT-PCR detection and the sequence of the PCR product (data not shown) confirmed that the chickens were infected with ARV. To further isolate the ARV, the supernatants of the RT-PCR-positive liver tissue samples were inoculated into LMH cells and the supernatants were blindly passaged every 2 days. The infected cells showed typical characteristic of syncytial cytopathic effect at 24 hpi (Figures 1B,C) and the RT-PCR detection of the supernatant of LMH cells after 7 passages was also ARV positive. All these demonstrated that a chicken origin ARV is efficiently isolated and designated as AHZJ19. Moreover, the chicken sera specific to AHZJ19 was generated by the inoculation of AHZJ19 in 3-day-old SPF chicks through unilateral foot pad. As shown in Figure 1D, the prepared chicken sera against AHZJ19 efficiently reacted with the LMH cells infected with AHZJ19, but not with the negative control cells (Figure 1E). In addition, we found that the isolate AHZJ19 could replicate efficiently in LMH cells and the virus titer of AHZJ19 could reach 8.7 × 108 TCID50/ml.
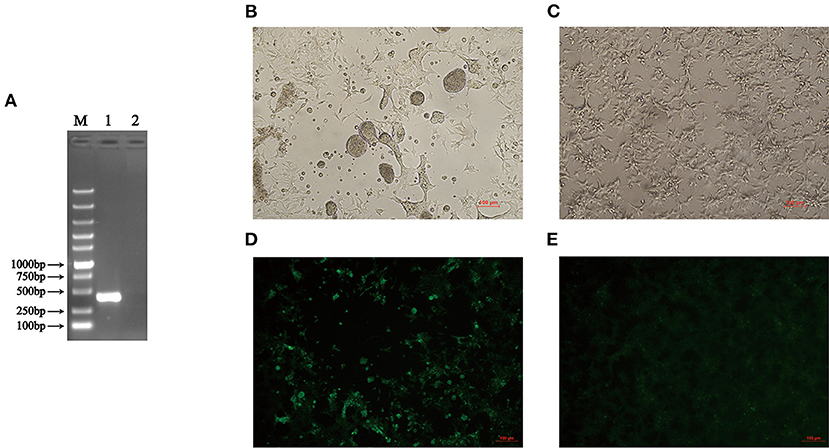
Figure 1. Isolation and identification of AHZJ19. (A) RT-PCR detection of ARV. The total RNA extracted from the liver tissue samples was tested by RT-PCR using primers specific to ARV. Lane M was Super DNA Marker; Lane 1 was the liver tissue of chickens from that farm; Lane 2 was negative control. (B,C) The CPE in LMH cells at 24 hpi after inoculation with supernatants of the RT-PCR-positive liver tissue sample. (B) The supernatants of the RT-PCR-positive liver tissue sample were inoculated into LMH cells and the cells showed the typical characteristic syncytial lesions at 24 hpi. (C) The negative LMH cells were considered as control. (D,E) IFA for detecting antibodies against ARV in chicken serum from chicks inoculated with AHZJ19. The chicken serum collected and prepared at 3 wpi was used for detected the LMH cells by IFA. (D) LMH cells infected with AHZJ19 at 24 hpi. (E) Negative LMH cells was considered as control.
To investigate the genome characteristics of AHZJ19, the high-throughput sequencing by Illumina sequencing technology was performed. The sequencing data revealed that the genome size of AHZJ19 is about 23,230 bp with the standard genome structure of ARV, which codes 12 major proteins. To elucidate the molecular characteristics of AHZJ19, the aa sequences of the 12 major proteins of the isolate were compared with the aa sequences of the reference ARV strains. As shown in Table 4, according to the aa BLAST of the 12 proteins, the σC protein of AHZJ19 had the highest homology (95.0%) with that of the American strain 99847 and only 53.2% homology with that of the vaccine strain S1133 and 1733 (data not shown). Notably, the highest homology of the σC protein between AHZJ19 and Chinese strains previously reported was only 82.8% (strain SD18). Except for σC protein, as described in Table 4, the highest homology strain with AHZJ19 for λA, λB, λC, μA, μB, μNS, P10, P17, σA, σB, and σNS was the China 919, Canada 138, China LY383, Hungary 4599-V-04, Hungary 924-Bi-05, Hungary 924-Bi-05, Hungary T1781, Hungary T1781, USA 05682/12, China LY383 and Hungary T1781, respectively. Notably, for λA, λB, λC, μA, μB, μNS, P10, P17, σA, σB, and σNS, AHZJ19 shared the highest homology with the Chinese strain 919, strain LY383, strain LY383, strain 918, strain 918, strain LY383, strain HB10-1, strain GX/2010/1, strain LY383, strain LY383, and strain 1017-1, respectively (Table 4). These data allow us to speculate about genetic reassortment of the AHZJ19 isolate and the relatively high genetic distance between the recombinant and parental strains was potentially due to the unsequenced intermediate strains. The phylogenetic tree analysis for these proteins further revealed that AHZJ19 strain might have originated from a recombination of Hungarian strains, North American strains, and Chinese strains (Figures 2–4). Moreover, according to the Bootscan analysis within Simplot program, recombination detection further highlighted the possibility of intra-segmental recombination of the λA, λB, μA, and σA genes of AHZJ19 with reference strains. The λA gene of AHZJ19 may have recombined with USA/AVS-B (GenBank accession number: FR694191.1), Reo/PA/Layer/01224B/14 (KT428308.1), and China/LY383 (MF183221.1) strains (Figure 5A) and the λB gene may have recombined with Reo/PA/Broiler/05682/12 (KM877326.1), Reo/PA/Broiler/15511/13 (KP731612.1), and China/LY383 (MF183212.1) strains (Figure 5B). Similarly, there is evidence that the μA gene may have recombined with Hungary/924-Bi-05 (KX398265.1), Hungary/4599-V-04 (KX398295.1), and Hungary/T1781 (KC865789.1) strains (Figure 5C); besides, it appears that the σA gene recombined with Canada/138 (AF059717.1), Reo/PA/Broiler/15511/13 (KP731618.1), and Reo/PA/Layer/01224B/14 (KT428315.1) strains (Figure 5D). Taken together, the generation of AHZJ19 reflected the genomic reassortment and intra-segmental recombination between previously identified ARV strains throughout the world.
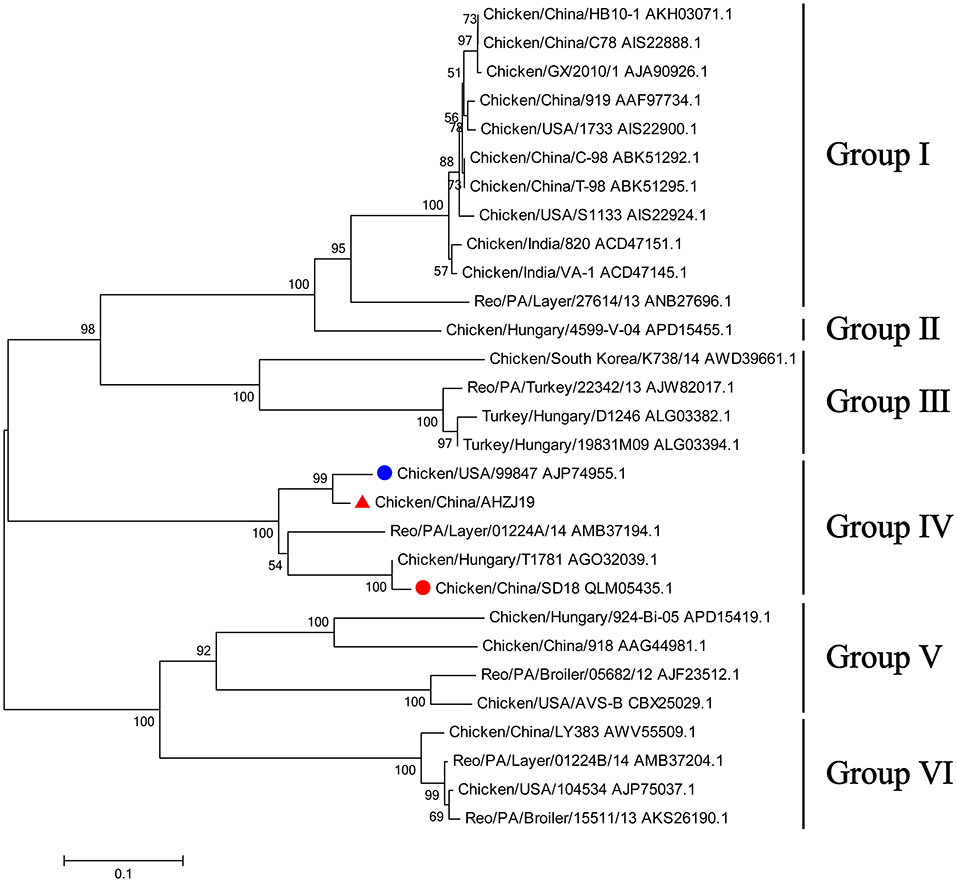
Figure 2. Phylogenetic tree of AHZJ19 and reference strains based on aa sequences of σC protein. The phylogenetic tree was constructed using the neighbor-joining method (1,000 bootstraps) with MEGA6. The AHZJ19 strain isolated in Anhui in 2019 is indicated by the red triangle. The strain sharing the highest homology with AHZJ19 in the world and in China is indicated by the blue and red circle, respectively.
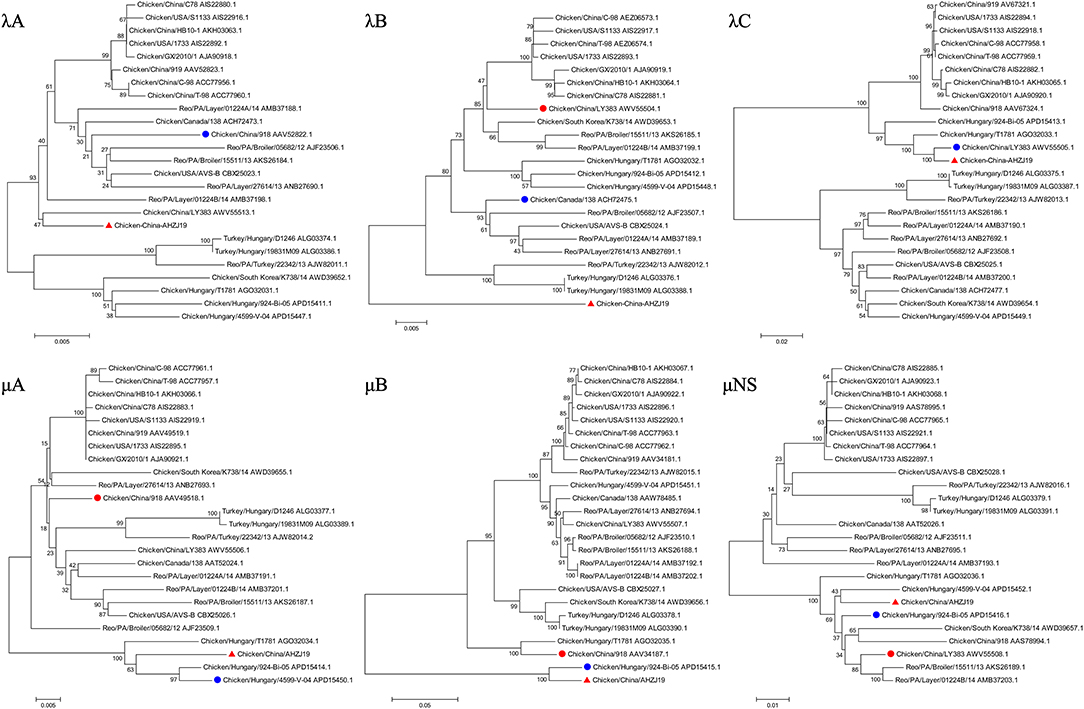
Figure 3. Phylogenetic trees of AHZJ19 and reference strains based on aa sequences of λA, λB, λC, μA, μB, and μNS proteins. The phylogenetic tree was constructed using the neighbor-joining method (1,000 bootstraps) with MEGA6. The AHZJ19 strain isolated in Anhui in 2019 is indicated by the red triangle. The strain sharing the highest homology with AHZJ19 in the world and in China is indicated by the blue and red circle, respectively.
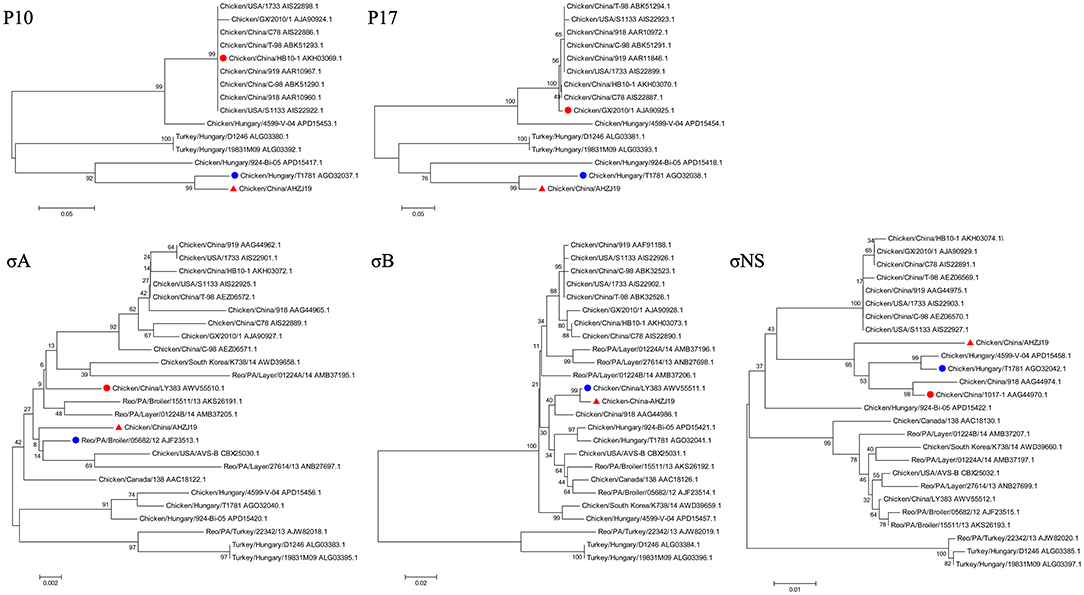
Figure 4. Phylogenetic trees of AHZJ19 and reference strains based on aa sequences of P10, P17, σA, σB, and σNS proteins. The phylogenetic tree was constructed using the neighbor-joining method (1,000 bootstraps) with MEGA6. The AHZJ19 strain isolated in Anhui in 2019 is indicated by the red triangle. The strain sharing the highest homology with AHZJ19 in the world and in China is indicated by the blue and red circle, respectively.
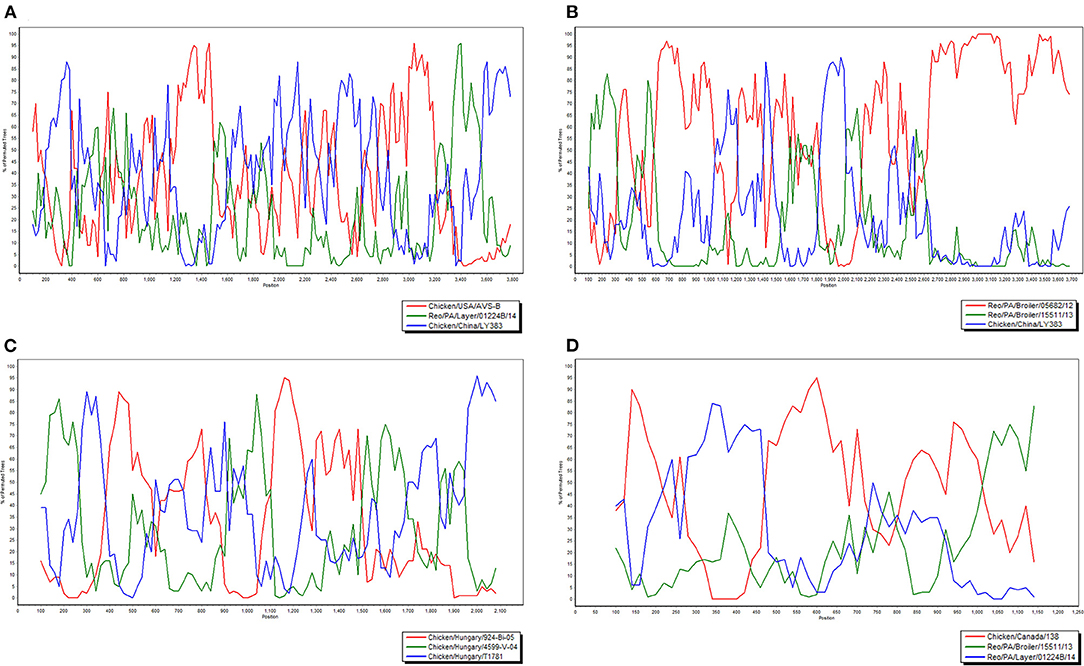
Figure 5. Bootscan analysis of AHZJ19 for detecting intra-segmental recombination. The Bootscan analysis was constructed using the neighbor-joining method (a Kimura two-parameter applied and 100 replicates) with Simplot program version 3.5.1. Query-Chicken/China/AHZJ19; (A) λA, (B) λB, (C) μA, and (D) σA.
Of note, based on the σC protein of AHZJ19 in the phylogenetic tree and a relevant study (34), AHZJ19 was clustered into Group IV (Figure 2). Although Chinese strain SD18 and Hungary strain T1781 also belonged to Group IV, the homology of the σC protein between AHZJ19 and Chinese strain SD18 or Hungary strain T1781 was only about 80%. Regardless of the genetic variability of the σC protein, from an evolution perspective, the strains had diverged and evolved independently quite a long time ago and in-depth analysis depends on more unsampled or unsequenced strains. Notably, the homology of the σC protein between Chinese strain SD18 and Hungary strain T1781 reached 98.5%, indicating that Group IV could be further divided into two subgroups, AHZJ19-like and SD18-like. As a dominant antigenic protein and the viral cell attachment protein, σC protein can efficiently induce the specific neutralizing antibodies against ARV (2, 6). However, σC protein is the highest variant protein among the 12 proteins and the hypervariable regions of σC protein are mainly located in 1–122 and 196–326 aa residues (35). In comparison with ARV reference strains, multiple mutations were identified in the σC protein between the different groups and AHZJ19 as shown in Figure 6. Moreover, six antigenic sites (74–76 aa, 78–82 aa, 86–88 aa, 93–95 aa, 109–112 aa, and 125–128 aa) of the σC protein were variant between AHZJ9 and strain SD18. As vaccine strains S1133 and 1733 have been widely used in the poultry industry, the low homology of the σC protein of AHZJ19 with that of these vaccine strains highlights that current vaccines might not provide efficient protection against the novel ARV isolate AHZJ19.

Figure 6. Alignment of AHZJ19 and eight reference strains based on σC protein. The alignment was constructed using the ClustalW methods in Megalign program included in the Lasergene 7.0 software program.
In summary, this is the first demonstration of a novel ARV strain AHZJ19 with the unique gene constellation possibly originated from the recombination of Hungarian strains, North American strains, and Chinese strains. Although the σC protein sequences of almost all ARV strains can be obtained from GenBank, the sequences of other proteins of most strains have not been deposited in GenBank yet. In addition, coupled with the lack of some unsampled or unsequenced strains, the above factors are the limitations of this study for recombination analysis. Thus, our study shows that the σC protein of AHZJ19 is closely related to that of the American strain 99847, but significantly different from that of the vaccine strains S1133, 1733, and Chinese strains previously reported, highlighting the inefficient protection of these vaccines against such ARV variant. Notably, lots of ARV mutants have recently emerged in broiler flocks globally, and some of them have become prevalent in different regions, such as North America, which presents a challenge to the current vaccine strategies for ARV. An in-depth recombination analysis of the novel AHZJ19 ARV isolate is necessary and will certainly be performed once the complete genome sequences of all strains involved in this study (the American strain 99847 and Chinese strain SD18) become publicly available. Therefore, it is critical to study on the molecular epidemiology for these ARV variants for better controlling of the diseases caused by ARV. Of course, the pathogenesis and the mechanism of the recombination of the novel ARV isolate AHZJ19 need to be further elucidated.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Animal Care Committee at Yangzhou University in China.
HS conceived and designed the experiments. JZ, TL, WW, and QX performed the experiments. JZ, TL, and HS analyzed the data. ZW and AQ contributed reagents, materials, and analysis tools. JZ, TL, HS, and JY contributed to the writing of the manuscript. All authors have read and approved the final manuscript.
This study was supported by the National Key Research and Development (R&D) Plan (2018YFD0500106), the Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality (26116120), the Research Foundation for Talented Scholars in Yangzhou University, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Spandidos DA, Graham AF. Physical and chemical characterization of an avian reovirus. J Virol. (1976) 19:968–76. doi: 10.1128/jvi.19.3.968-976.1976
2. Pitcovski J, Goyal SM. Avian reovirus infections. In: Swayne DE, editor. Diseases of Poultry. Hoboken, NJ: Wiley-Blackwell (2020). p. 382–400.
3. Benavente J, Martinez-Costas J. Avian reovirus: structure and biology. Virus Res. (2007) 123:105–19. doi: 10.1016/j.virusres.2006.09.005
4. Xu W, Coombs KM. Conserved structure/function of the orthoreovirus major core proteins. Virus Res. (2009) 144:44–57. doi: 10.1016/j.virusres.2009.03.020
5. Martinez-Costas J, Grande A, Varela R, Garcia-Martinez C, Benavente J. Protein architecture of avian reovirus S1133 and identification of the cell attachment protein. J Virol. (1997) 71:59–64. doi: 10.1128/jvi.71.1.59-64.1997
6. Huang WR, Wang YC, Chi PI, Wang L, Wang CY, Lin CH, et al. Cell entry of avian reovirus follows a caveolin-1-mediated and dynamin-2-dependent endocytic pathway that requires activation of p38 mitogen-activated protein kinase (MAPK) and Src signaling pathways as well as microtubules and small GTPase Rab5 protein. J Biol Chem. (2011) 286:30780–94. doi: 10.1074/jbc.M111.257154
7. Dandar E, Balint A, Kecskemeti S, Szentpali-Gavaller K, Kisfali P, Melegh B, et al. Detection and characterization of a divergent avian reovirus strain from a broiler chicken with central nervous system disease. Arch Virol. (2013) 158:2583–8. doi: 10.1007/s00705-013-1739-y
8. Lu H, Tang Y, Dunn PA, Wallner-Pendleton EA, Lin L, Knoll EA. Isolation and molecular characterization of newly emerging avian reovirus variants and novel strains in Pennsylvania, USA, 2011-2014. Sci Rep. (2015) 5:14727. doi: 10.1038/srep14727
9. Farkas SL, Marton S, Dandar E, Kugler R, Gal B, Jakab F, et al. Lineage diversification, homo- and heterologous reassortment and recombination shape the evolution of chicken orthoreoviruses. Sci Rep. (2016) 6:36960. doi: 10.1038/srep36960
10. Tang Y, Lin L, Sebastian A, Lu H. Detection and characterization of two co-infection variant strains of avian orthoreovirus (ARV) in young layer chickens using next-generation sequencing (NGS). Sci Rep. (2016) 6:24519. doi: 10.1038/srep24519
11. Noh JY, Lee DH, Lim TH, Lee JH, Day JM, Song CS. Isolation and genomic characterization of a novel avian orthoreovirus strain in Korea, 2014. Arch Virol. (2018) 163:1307–16. doi: 10.1007/s00705-017-3667-8
13. Yun T, Ye W, Ni Z, Chen L, Yu B, Hua J, et al. Complete genomic sequence of goose-origin reovirus from China. J Virol. (2012) 86:10257. doi: 10.1128/JVI.01692-12
14. Tang Y, Lu H. Whole genome alignment based one-step real-time RT-PCR for universal detection of avian orthoreoviruses of chicken, pheasant and turkey origins. Infect Genet Evol. (2016) 39:120–6. doi: 10.1016/j.meegid.2016.01.018
15. Farkas SL, Varga-Kugler R, Marton S, Lengyel G, Palya V, Banyai K. Genomic sequence and phylogenetic analyses of two novel orthoreovirus strains isolated from Pekin ducks in 2014 in Germany. Virus Res. (2018) 257:57–62. doi: 10.1016/j.virusres.2018.09.001
16. Banyai K, Dandar E, Dorsey KM, Mato T, Palya V. The genomic constellation of a novel avian orthoreovirus strain associated with runting-stunting syndrome in broilers. Virus Genes. (2011) 42:82–9. doi: 10.1007/s11262-010-0550-z
17. Schiff LA, Nibert ML, Tyler KL. Orthoreoviruses and their replication. Fields Virology. (2007) 5:1853–915.
18. Van de Zande S, Kuhn EM. Central nervous system signs in chickens caused by a new avian reovirus strain: a pathogenesis study. Vet Microbiol. (2007) 120:42–9. doi: 10.1016/j.vetmic.2006.10.024
19. Jones RC. Avian reovirus infections. Rev Sci Tech. (2000) 19:614–25. doi: 10.20506/rst.19.2.1237
20. Hoerr FJ. Clinical aspects of immunosuppression in poultry. Avian Dis. (2010) 54:2–15. doi: 10.1637/8909-043009-Review.1
21. van der Heide L. The history of avian reovirus. Avian Dis. (2000) 44:638–41. doi: 10.2307/1593104
23. Tang Y, Lu H. Genomic characterization of a broiler reovirus field strain detected in Pennsylvania. Infect Genet Evol. (2015) 31:177–82. doi: 10.1016/j.meegid.2015.01.029
24. Zhong L, Gao L, Liu Y, Li K, Wang M, Qi X, et al. Genetic and pathogenic characterisation of 11 avian reovirus isolates from northern China suggests continued evolution of virulence. Sci Rep. (2016) 6:35271. doi: 10.1038/srep35271
25. Troxler S, Rigomier P, Bilic I, Liebhart D, Prokofieva I, Robineau B, et al. Identification of a new reovirus causing substantial losses in broiler production in France, despite routine vaccination of breeders. Vet Rec. (2013) 172:556. doi: 10.1136/vr.101262
26. Bruhn S, Bruckner L, Ottiger HP. Application of RT-PCR for the detection of avian reovirus contamination in avian viral vaccines. J Virol Methods. (2005) 123:179–86. doi: 10.1016/j.jviromet.2004.09.019
27. Wang P, Shi J, Yuan H, Ye J, Qian K, Shao H, et al. Isolation and identification of a chicken infectious anemia virus and its whole genomic sequence analysis. China Poultry. (2019) 41:57–60 (in Chinese).
28. Davidson I, Borovskaya A, Perl S, Malkinson M. Use of the polymerase chain reaction for the diagnosis of natural infection of chickens and turkeys with Marek's disease virus and reticuloendotheliosis virus. Avian Pathol. (1995) 24:69–94. doi: 10.1080/03079459508419050
29. Li T, Xie J, Liang G, Ren D, Sun S, Lv L, et al. Co-infection of vvMDV with multiple subgroups of avian leukosis viruses in indigenous chicken flocks in China. BMC Vet Res. (2019) 15:288. doi: 10.1186/s12917-019-2041-3
30. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19:455–77. doi: 10.1089/cmb.2012.0021
31. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
32. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Trop Med Hyg. (1938) 27:619–26. doi: 10.1093/oxfordjournals.aje.a118408
33. Wang X, Tang X, Liu Q, Ju F, Wang L. Application of agar diffusion test to detect chicken viral arthritis. Chin J Prev Vet Med. (1985) 03:26–27 (in Chinese).
34. Chen H, Yan M, Tang Y, Diao Y. Sequencing and phylogenetic analysis of partial S1 genes of avian orthoreovirus isolates in Shandong province during 2015-2017. Poult Sci. (2020) 99:2416–23. doi: 10.1016/j.psj.2019.11.067
Keywords: avian orthoreovirus, isolation, genomic characteristics, phylogenetic analysis, recombination, σC protein
Citation: Zhang J, Li T, Wang W, Xie Q, Wan Z, Qin A, Ye J and Shao H (2021) Isolation and Molecular Characteristics of a Novel Recombinant Avian Orthoreovirus From Chickens in China. Front. Vet. Sci. 8:771755. doi: 10.3389/fvets.2021.771755
Received: 07 September 2021; Accepted: 31 October 2021;
Published: 07 December 2021.
Edited by:
Huanmin Zhang, Agricultural Research Service (USDA), United StatesReviewed by:
Iryna Goraichuk, Institute of Experimental and Clinical Veterinary Medicine, UkraineCopyright © 2021 Zhang, Li, Wang, Xie, Wan, Qin, Ye and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongxia Shao, aHhzaGFvQHl6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.