- 1Inmunology and Genomics, Marine Research Institute (IIM-CSIC), Vigo, Spain
- 2Department of Life Sciences, University of Trieste, Trieste, Italy
- 3Division of Oceanography, National Institute of Oceanography and Applied Geophysics, Trieste, Italy
The marine environment includes diverse microeukaryotic organisms that play important functional roles in the ecosystem. With molecular approaches, eukaryotic taxonomy has been improved, complementing classical analysis. In this study, DNA metabarcoding was performed to describe putative pathogenic eukaryotic microorganisms in sediment and marine water fractions collected in Galicia (NW Spain) from 2016 to 2018. The composition of eukaryotic communities was distinct between sediment and water fractions. Protists were the most diverse group, with the clade TSAR (Stramenopiles, Alveolata, Rhizaria, and Telonemida) as the primary representative organisms in the environment. Harmful algae and invasive species were frequently detected. Potential pathogens, invasive pathogenic organisms as well as the causative agents of harmful phytoplanktonic blooms were identified in this marine ecosystem. Most of the identified pathogens have a crucial impact on the aquacultural sector or affect to relevant species in the marine ecosystem, such as diatoms. Moreover, pathogens with medical and veterinary importance worldwide were also found, as well as pathogens that affect diatoms. The evaluation of the health of a marine ecosystem that directly affects the aquacultural sector with a zoonotic concern was performed with the metabarcoding assay.
Introduction
Marine ecosystems harbor highly diverse eukaryotic microorganisms, many of which are unknown or ignored due to difficulties in culturing and identifying them. Biodiversity in coastal marine environments is endangered due to climate change and human activity, and many species could disappear without being identified (1). Many marine eukaryotic species remain to be described, with the proportion of unknown diversity ranging from 24 to 98% depending on the taxonomic group (2). For example, the small eukaryotes (<1 mm) that play important ecological roles in aquatic ecosystems remain poorly described (3). In this context, the study of the taxonomic and functional diversity of small eukaryotic organisms in marine ecosystems is being enriched by incorporating novel molecular tools (2, 4) that complement conventional methods based on the morphological and nutritional characterization of specimens in vivo, light and fluorescence microscopy, electron microscopy, or liquid chromatography (HPLC) (5, 6), improving our knowledge on their ecological role in ecosystem functioning (7).
DNA metabarcoding technology allows the identification of thousands of species at the same time by sequencing the DNA extracted from organisms or environmental DNA (eDNA) in water or sediments on a high-throughput sequencing (HTS) platform (8, 9). Initially, this technology was used for the characterization of marine prokaryotes (10, 11). Subsequently, studies were also focused on eukaryotic diversity characterization, including metazoans, plants, fungi, and protists. In the marine environment, benthic and pelagic organisms have been described from the sediment (12–14) and water column (14–17), permitting the detection of new or rare taxa in the ecosystem (3, 18). DNA metabarcoding has also been used to detect exotic marine species (16), helping to establish early biosecurity alerts (19).
Protists constitute the bulk of eukaryotic diversity in marine communities (20) and play a variety of crucial roles in aquatic ecosystems, acting as photosynthesizers (primary producers), heterotrophs (predators and parasites) and mixotrophs (3, 6). Moreover, many protists, together with fungi or even metazoans, can act as potential pathogens affecting organisms and ecosystems and have been implicated in global-scale declines in a wide range of marine and terrestrial species (21–25).
Total protist diversity is usually analyzed by using the V9 and V4 regions of the 18S rRNA gene (26, 27). The V9 region is widely used in HTS platforms due to its short length (180 bp), although it provides lower phylogenetic information than the V4 region (400–500 bp length). In contrast, V4 is not adequate for Illumina platforms, and amplicon length variability could produce PCR or sequencing biases. Organisms belonging to the Alveolata group have usually remained unexplored using V4 but have been detected using V9. Thus, it seems that V9 reveals much better resolution in most protistan supergroups than V4 (28).
Abiotic and biotic factors (solar irradiation, nutrients, temperature, or predation) influence the dynamics of the eukaryote community (29). Moreover, wind-driven upwelling and downwelling events that cause alternation between water column stratification and mixing (30) induce seasonal succession of small planktonic eukaryotes in surface waters (31). In particular, the Ría de Vigo (NW, Spain) is highly influenced by the upwelling dynamics which occurs mainly in summer periods, when oceanic currents from the depths enter in zone (32). However, high-throughput sequencing technology only has been used to study seasonal succession of small-eukaryote community in surface waters (31) and co-ocurrence networks based on eukaryotic and prokaryotic associations were performed (33).
The area of study has been the scenario of the detection of Toxic Harmful Algae Blooms (THAB), invasive species and pathogens that affects the aquaculture sector. Toxic algae generate toxins that causes severe impact in the aquaculture sector and human heath by the consumption of contaminated bivalves. The importance of fish and shellfish aquaculture in the area makes that the control of pathogens is a priority. Several parasites associated with bivalve mortalities such as Perkinsus olseni (34) or different species of Marteilia (35, 36) have been detected near of Ría de Vigo (37, 38). Mortalities of turbot Scophthalmus maximus, one of the most fish farmed species have been registered due to the parasite Philasterides dicentrarchi in farms of Galicia (39, 40). These studies on the detection of pathogens have been made by histopathology, morphological studies and real-time PCR assay and, until now, metabarcoding and high-throughput sequencing have not been used before in Ría de Vigo.
Human sewage and run-off from farms are released through sewage disposal systems, reaching the marine environment (41). In this context, DNA metabarcoding technology can also detect the presence of these pathogens and could be used to predict the ecological conditions of the marine ecosystem (42). However, this technology requires the construction of a robust reference database and the use of highly efficient primers for the correct amplification of the target gene without missing any taxon information (4, 8).
In this study, we evaluated the use of DNA metabarcoding technology to describe eukaryote biodiversity in a highly productive coastal marine ecosystem. In addition to identifying non-indigenous species and harmful algae, our objective was to detect potential pathogens for both marine cultured species and for relevant species in the ecosystem.
Materials and Methods
Sampling
Eukaryotic diversity was evaluated in water and sediment from a bivalve production area located in the Ría de Vigo, NW Spain (Meira: 42°17′6.72″ N/8°43′18.80″ W) (Figure 1A). The sampling period was from summer 2016 to summer 2018. Samples were collected every 3 months (Figure 1B). Superficial sediment was collected and kept at −20°C. Seawater was successively filtered by different pore sizes. A volume of 75 m3 of seawater was filtered with a 200 μm plankton net (mesoplankton). A volume of 40 L was filtered again through a 65 μm pore size net (microplankton), and finally, 2 L of seawater was filtered by using a 0.22 μm pore size (nanoplankton-picoplankton). The nomenclature of the water fractions was adapted from Sieburth et al. (43). Filters were also kept at −20°C until use.
Figure 1. (A) Location of the bivalve production area in the Ría of Vigo (Meira, NW, Spain). (B) The sampling period extended from 2016 to 2018. Environmental samples were collected for analysis.
DNA Extraction, Amplification, and Sequencing
A total of 24 samples of sediment and water fractions were processed. DNA from the sediment and water was isolated using the PowerSoil DNA isolation Kit (Qiagen, Düsseldorf, Germany) following the manufacturer's protocol.
The V9 region of the 18S rDNA (180 bp long) was amplified using the universal eukaryotic-specific primers 1380F/1510R described by Amaral-Zettler et al. (26). Amplicons were purified using the Qiaquick PCR purification kit (Qiagen) and quantified. A library for 18S metagenomic sequencing was prepared using the Herculase II Fusion DNA Polymerase Nextera XT Index Kit V2 (Illumina), and paired-end sequencing (2 x 300) was performed on an Illumina MiSeq platform (Macrogen, Korea). The raw read sequences obtained were deposited in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under the BioProject accession number PRJNA761019.
Bioinformatic Analysis
A custom 18S eukaryotic reference database was previously constructed to classify metagenomic reads assigning an operational taxonomic unit (OTU) to the sequences obtained after the Illumina assay. Briefly, a total of 726,478 sequences ranging from 100 to 10,000 bp in length were downloaded from the NCBI database. Sequences from bacteria, unverified organisms, environmental samples, and predicted sequences were excluded. Their taxonomic information was assigned using the Python script “entrez_qiime.py” (44), and the non-annotated sequences showing 99% coverage and 100% similarity were subjected to additional BLAST analysis against the nucleotide collection nr/rt to complete the taxonomy information. A total of 723,146 sequences (99.54%) were taxonomically classified.
The CD-HIT_Est tool from the CD-hit package 4.6.8 (45) was used to cluster the sequences with 99% coverage and 100% similarity. Briefly, sequences were first sorted in order of decreasing length. The longest sequence became the representative sequence of the first cluster, which included all sequences with 99% similarity and 90% coverage. When the similarity of the sequence with the representative sequence was below 99%, a new cluster was defined with that sequence as the representative. CLC workbench 12 was used to merge the taxonomy of the database with the representative sequence of each cluster and their accession number.
Database Annotation, Trimming, and Clustering
The Microbial Genomics Module of Qiagen CLC Workbench 12 was used for data analysis. Paired-end reads were trimmed using the sequences of the primers (1380F and 1510R). Low-quality reads were also trimmed by quality scores (limit 0.05 = minimum average quality score 20), by the number of ambiguous nucleotides (maximal 2 ambiguous nucleotides), and by the length of the primers (below 100 and above 200 bp).
Paired reads were then merged and grouped using the OTU-clustering tool. The similarity threshold was set at 94% for 18S reference-based OTU clustering and 80% for de novo OTU clustering. Finally, one representative sequence of each OTU was assigned to the best match in the reference database. Singletons and chimeras were excluded from the analysis. Pathogens sequences were further verified by a Blast confirming their identity with a 100% of similarity in the NCBI database.
Alpha and Beta Diversity
Alpha diversity evaluates how many different species can be detected in a microbial ecosystem by the number of different OTUs and how they are distributed within a community (46). Alpha diversity was estimated by constructing rarefaction curves calculated by subsampling OTU abundances in the different samples at different depths. Samples were rarefied at the minimum sample read depth for each amplicon. The alpha diversity was also evaluated by measuring the relative abundance (percentage of reads associated with each OTU or taxon) and richness (number of OTUs or taxa included in each sample) in all samples.
Beta diversity shows the differences between microbial communities from different environmental samples, focusing on the difference in their taxonomic abundance profiles (47). Beta diversity analysis was performed by calculating Bray–Curtis distances between each pair of samples and applying principal coordinate analysis (PCoA) on the distance matrices. Permutational multivariate analysis of variance (PERMANOVA) based on the Bray-Curtis dissimilarity index was also estimated.
Results
Sequencing Data
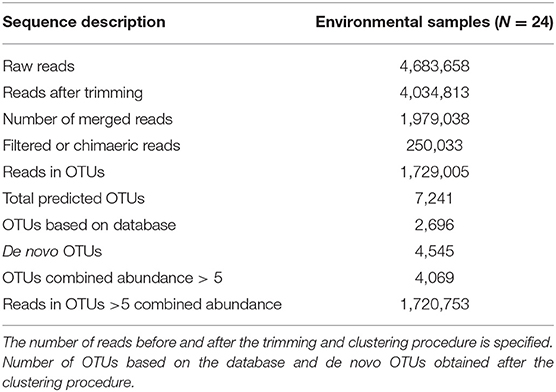
After sequencing and trimming, a total of 4,034,813 reads were obtained from environmental samples (sediment and water) (Table 1) with an average length of 146 nucleotides. A detailed description of the number of sequences obtained in each step of the analysis is specified in Table 1 and Supplementary Tables 1–3. The OTU-clustering process generated 7,241 OTUs using a total of 1,729,005 filtered reads. To decrease the number of non-representative taxa, OTUs with a combined abundance in all samples ≤ 5 were excluded from the analysis. A total of 4,069 OTUs were finally selected and used for the following analyses.
Overview of Eukaryotic Diversity
The number of different taxonomic groups and their distribution within the ecosystem were evaluated by alpha diversity. Rarefaction curves were constructed with a subsampling depth of 46,108 reads, corresponding to the sample with the lowest number of reads. The curves show that the subsampling sequencing effort successfully represented a correct total diversity in the environmental samples (Figure 2A). Seasonal variations in alpha diversity were observed. In addition, the diversity of eukaryotes in sediment was almost constant during the sampling period, and significant seasonal variations were observed in the water. In the meso- and microplankton, the number of OTUs was low during summer and increased in winter. Opposite kinetics were observed in the nanoplankton, where the highest diversity was detected during summer and decreased in winter (Figure 2B).
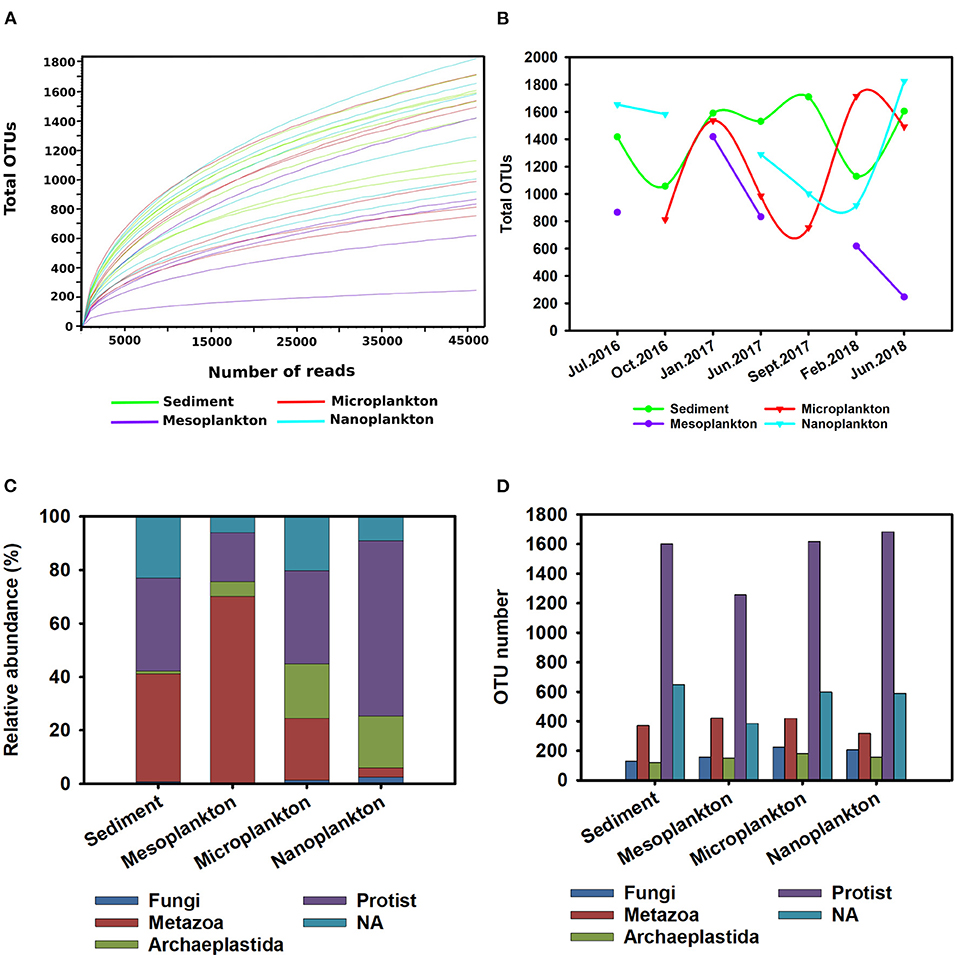
Figure 2. Overview of eukaryotic diversity. (A) Rarefaction curves constructed to analyse the alpha diversity in the environmental samples. A subsampling depth of 46,108 reads was used. (B) Seasonal variations in the alpha diversity registered during the sampling period. (C,D) General description of eukaryotic diversity [protist, metazoan, Archaeplastida, Fungi, and non-classified eukaryotes (NA group)] presented in the environment. The relative abundance (percentage) of the different taxonomic groups in environmental samples (C) is shown. The richness of the different taxonomic groups based on the number of OTUs included in each group was analyzed (D).
The most abundant taxonomic groups were metazoans and protists. The metazoan group was the largest (34%) composing 40 and 70% of the total abundance of sediment and mesoplankton, respectively. In contrast, the protist group was predominant in the nanoplankton (65%) (Figure 2C). The protists represented 38.56% abundance in the environment and 52% of environmental richness with the highest number of OTUs, followed by the unclassified eukaryotes (NA group) (20.54% of total OTUs). Thus, the high number of unknown eukaryotes present in the marine environment was confirmed. The metazoan group included 300–400 OTUs, representing 15.61% of the total OTUs, while Archaeplastida and Fungi included the lowest number of OTUs (264 and 206, respectively), which barely represented 5% of the total OTUs (Figure 2D).
Significant differences in the composition of eukaryotic communities among environmental compartments were clearly observed by beta diversity analysis. Although no significant differences were observed between the water fractions in pairwise comparisons, the sediment was clearly clustered and separated from the meso-, micro-, and nano-plankton fractions (Figure 3A). Exclusive OTUs were obtained from each environmental compartment. Sediment showed 2,296 exclusive OTUs (5,644% total OTUs) but also in nanoplankton (317), microplankton (258) and mesoplankton (204). Only 204 OTUs where shared among all environmental compartments (5% total OTUs) (Figure 3B).
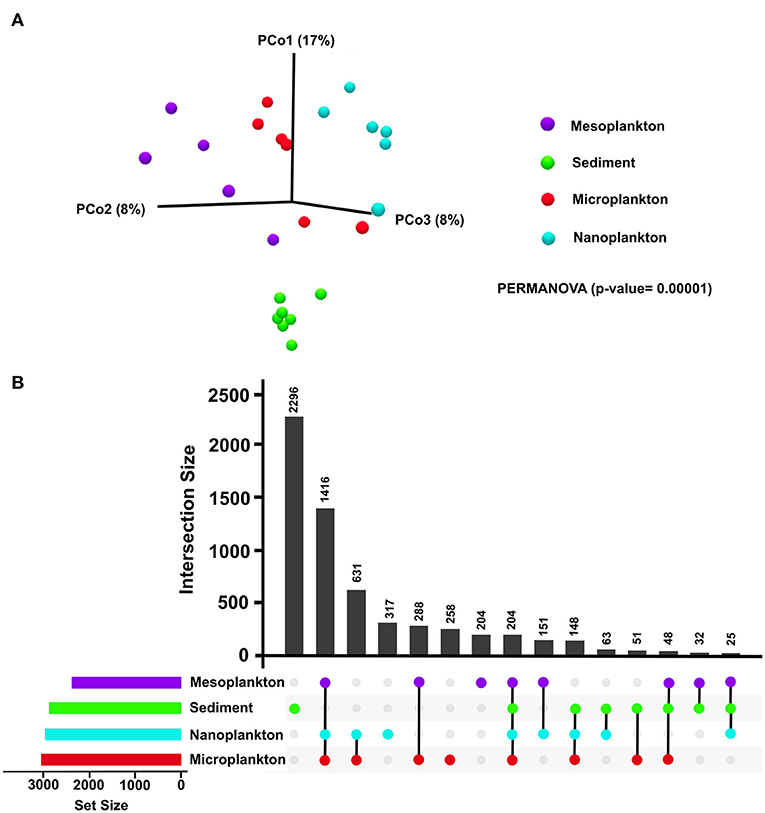
Figure 3. (A) Beta diversity analysis of each environmental compartment (sediment, mesoplankton, microplankton, and nanoplankton). Statistical differences were calculated using the pseudo f statistic (2.51) and a p = 0.00001. (B) Representation of the number of common and exclusive OTUs detected in sediment and water. Horizontal bars represent the total number of input OTUs in each environmental compartment.
Diversity of Eukaryotes in the Marine Ecosystem
A detailed description of the different eukaryotic groups present in water and sediment is described below. Special attention was given to the protist group since it is the most diverse group in the marine environment.
Archaeplastida
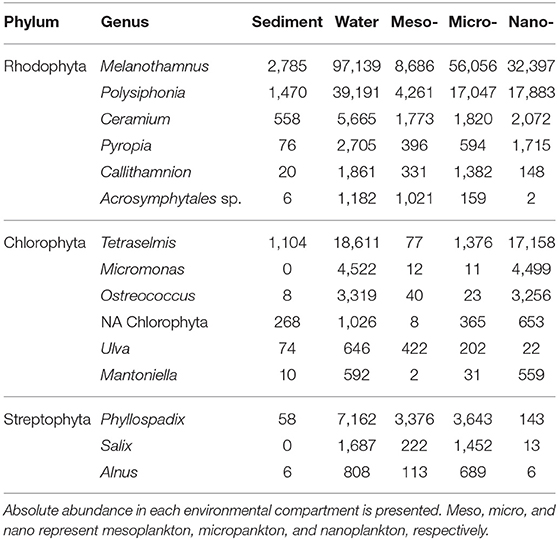
The Archeplastida group comprises red and green macroalgae, land plants, and some groups of small unicellular algae. This kingdom represented 20% of the total abundance in micro- and nanoplankton. At the phylum level, Rhodophyta and Chlorophyta were the dominant groups among the micro- and nanoplankton. The 15 most abundant genera are represented in Table 2.
Fungi
Fungi was the least abundant group (<2% of the total abundance in the environment). The phylum Ascomycota (42%) and the phylum Chrytridiomycota (46%) were the most abundant phyla in the water and sediment, respectively. The phylum Basidiomycota was present in both water and sediment at high abundance (32%) (Figure 4A).
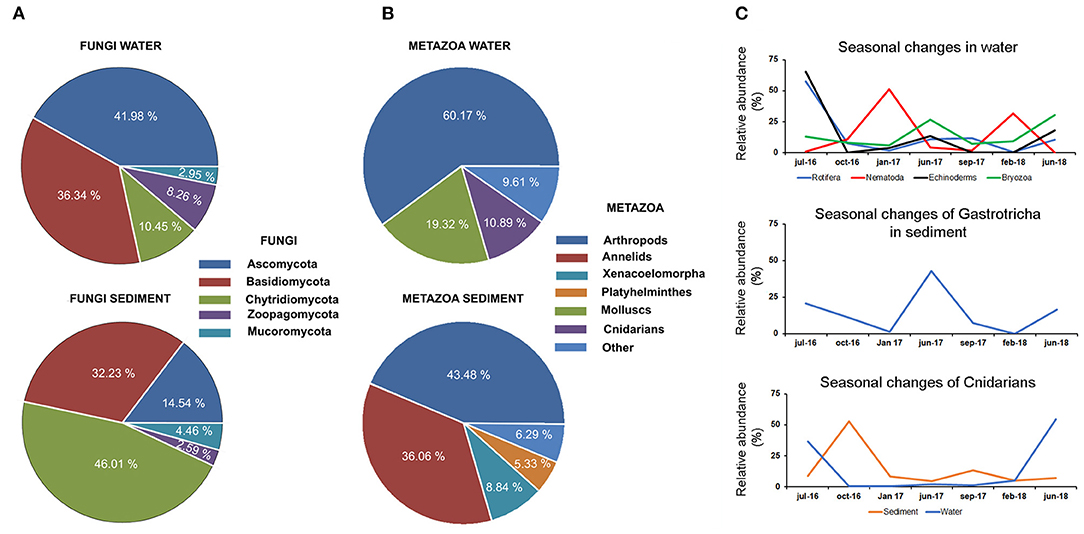
Figure 4. Distribution of fungi (A) and metazoan (B) in the water and sediment. (C) Temporal data allowed us to describe seasonal changes in the abundance of several taxonomic groups in water and sediment. Seasonal changes were also described for cnidarians in water and sediment.
Metazoans
The abundance of metazoans in the water was ~60% higher than that observed in the sediment (Figure 4B). The eukaryotic communities found in the water were dominated by arthropods and molluscs, which represented 60 and 20%, respectively, of total metazoans. Planktonic forms of cnidarians constituted 10% of the total abundance. Chordates were mainly represented by fish species (clupeids), which were only present in the water fraction (2.5% of total abundance) (Figure 4B). The distribution of metazoans in the sediment was completely different, being mainly composed of arthropods (43%) and other kinds of worms, such as annelids and platyhelminths (41%) (Figure 4B). Temporal data allowed the description of seasonal changes in the abundance of several metazoan groups (Figure 4C). A high abundance of several taxonomic groups was observed during the summer period, followed by a decrease in autumn and winter. This pattern was observed in cnidarians, rotifers, echinoderms, and bryozoans detected in the water and in the Gastrotricha group in the sediment. In contrast, the nematodes increased in the sediment during winter. In the cnidarian group, the decrease in swimming forms after summer was followed by an increase in benthonic stages in the sediment (Figure 4C).
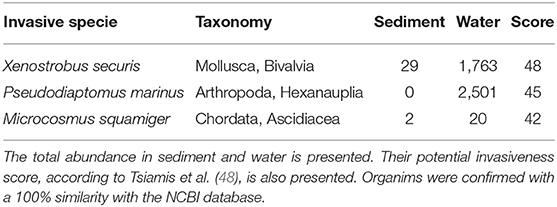
The analysis of the metazoan group also revealed the presence of four alien or invasive species in the environment (Table 3). Their potential invasiveness was scored following Tsiamis et al. (48) based on the likelihood of arrival, establishment, spread, and potential impact with a maximum value of 48. The bivalve mollusc Xenostrobus securis, which had the highest score (48), was detected in all environmental compartments at high abundance. The invasive arthropod Pseudodiaptomus marinus was only present in water fractions. The ascidian Microcosmus squamiger was also detected in all environmental samples (Table 3).
Protists
The majority of eukaryotes found in the environment belonged to the protist group. A total of 663,556 reads were clustered in 2,127 OTUs. Protists were classified according to Burki et al. (49) into different supergroups, including Amebozoa, Excavata, TSAR (Telonemia, Rhizaria, Alveolata, and Stramenopila) and a fourth group including other minor organisms. In general, the supergroup TSAR was dominant in the environment, representing more than 90% of all protists. Amoebozoa and Excavata constituted <5% of the protist abundance (Figures 5A–C).
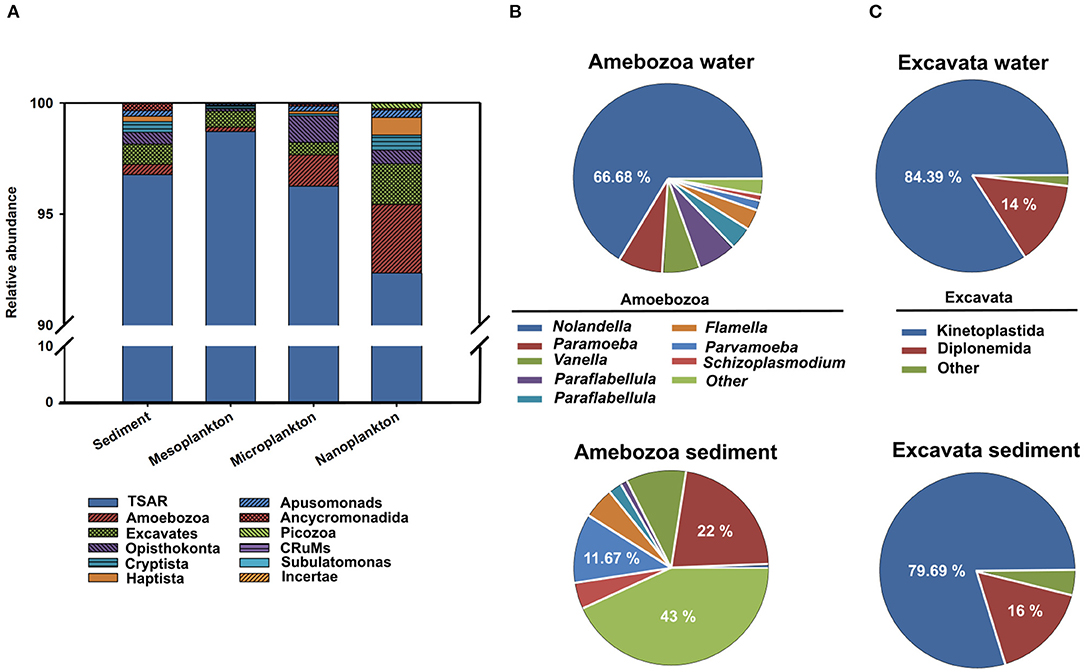
Figure 5. The presence of protists classified according to Burki et al. (49) into four supergroups (Amebozoa, Excavata, TSAR, and a fourth group including other minor organisms) is represented. (A) Relative abundance of the different supergroups in sediment, meso-, micro-, and nanoplankton. Relative abundances of Amebozoa (B) and Excavata (C) in sediment and water.
TSAR supergroup was the most abundant group of protists in all environmental compartments. Stramenopiles were more abundant in the sediment and represented 70% of the TSAR abundance. Alveolates and Rhizaria were more abundant in water and represented 28 and 24% of the total quantity, respectively (Figure 6A). The Stramenopiles were basically composed of diatoms (Bacillariophyceae) and other minor groups (Figure 6B), more abundant in sediment. The most abundant genera were Navicula, Amphora, Talaroneis, Pseudo-nitzschia, Sellaphora, Thalassiosira, Thalassionema, and Alaucoseira (Figure 7A). Brown algae (Phaeophyceae) of the genera Ectocarpus, Myrionema, Battersia, and Sargassum were abundant in the mesoplankton (Figure 7B).
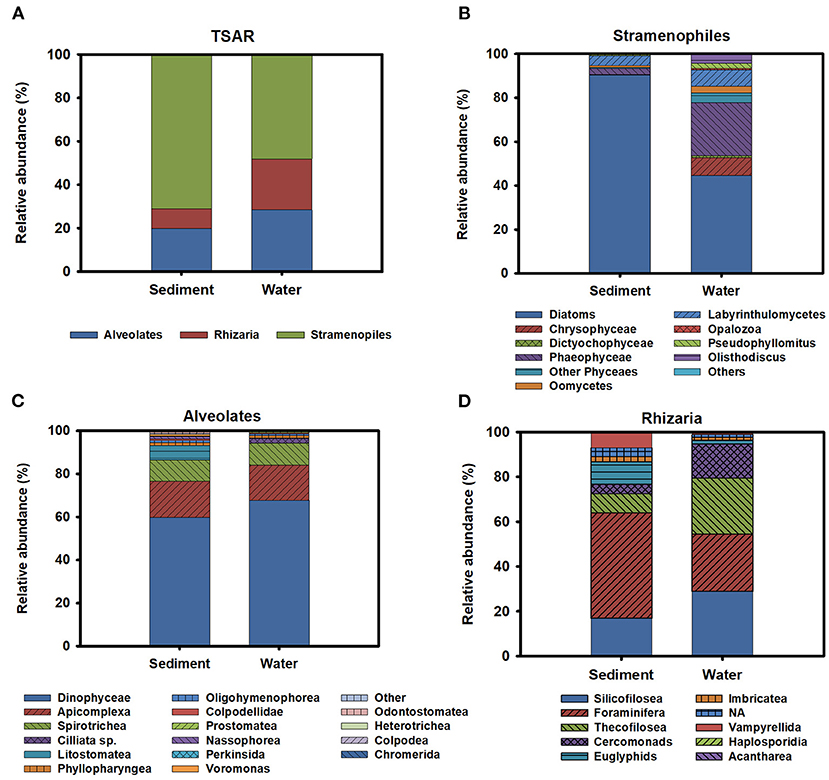
Figure 6. (A) General description of the diversity observed inside the TSAR supergroup. Relative abundance of the different members of Stramenopiles (B), Alveolates (C), and rhizarians (D) in sediment and water.
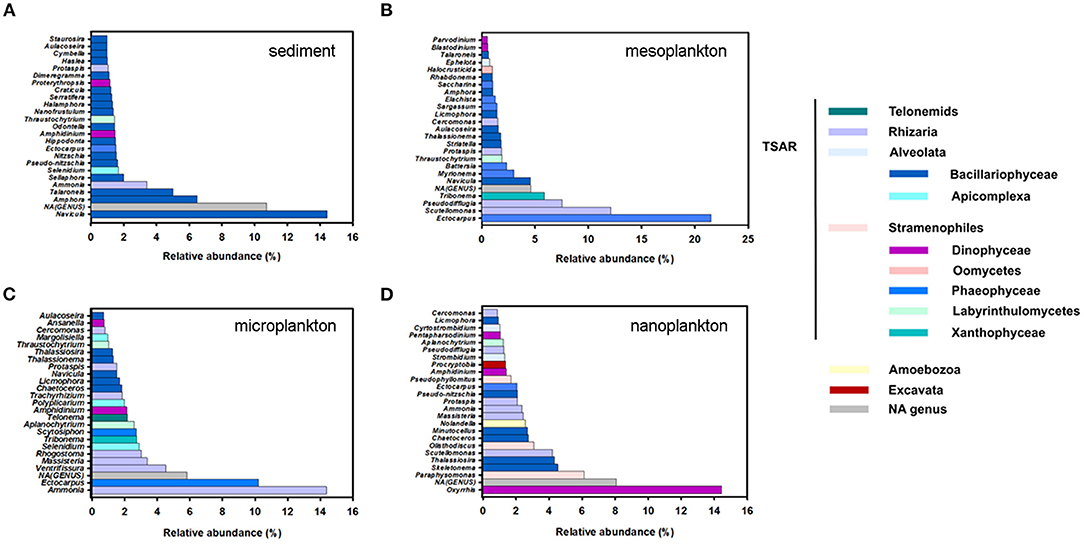
Figure 7. Top 25 most abundant genera of protists in sediment (A), mesoplankton (B), microplankton (C), and nanoplankton (D). Genera were classified in the legend according to their pertinence of higher taxonomic groups, mainly TSAR, Amoebozoa, and Excavata.
The Alveolata group was mainly composed of dinoflagellates (Dinophyceae), followed by apicomplexan organisms and ciliates (Figure 6C). Apicomplexans of the genera Selenidium and similar to Margolisiella were also found in the top 25 most abundant protist groups, especially in sediment and microplankton (Figures 7A,C). Dinoflagellates were abundant in the environment, although the genus Oxyrrhis was the largest taxon in the nanoplankton. Other abundant genera were Pentapharsodium, Ansanella, Blastodinium, Parvodinium, and Proterythropsis (Figure 7D). Dinoflagellates that cause toxic blooms, such as Amphidinium, were detected in the sediment and nanoplankton. Other harmful dinoflagellates, such as Alexandrium, Gymnodinium, and Prorocentrum, were also detected, although they were not included in the top 25 most abundant genera.
Rhizaria was mainly composed of benthonic Foraminifera (Ammonia), Silicofilosea, Thecofilosea, and other minor organisms, such as cercomonads, Haplosporidia, and Vampyrellida (Figure 6D). The genera Massisteria and Cercomonas (Cercozoan), Scutellomonas and Protapsis (Thaumatomonadida), and Rhogostoma (Thecofilosea) were detected in water and sediment, but they were predominant in the microplankton (Figure 7C).
Diversity of Potential Pathogens Found in the Ecosystem
A high number of potential pathogens belonging to several taxonomic groups were found in the ecosystem. Although some pathogens were included in the metazoan and fungi groups, the wide majority belonged to the protist group.
Pathogenic metazoans included several species of cnidarians, platyhelminths and arthropods that were mainly associated with water (Table 4). Platyhelminthes within the genus Parvatrema, as well as Gymnophalloides seoi and Bucephalus minimus, were the most abundant. The cnidarians (Kudoa, Myxobolus, and Parvicapsula) and the arthropod Demodex folliculorum were also detected (Table 4). The pathogenic fungi Malassezia and Chrytridiomycota were found in water and sediment (Table 4).
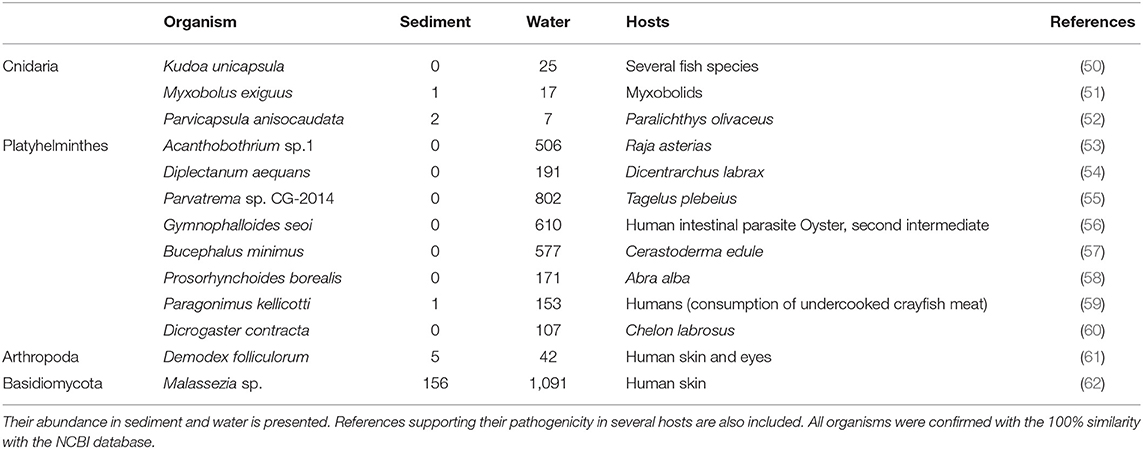
Table 4. Diversity of potential pathogens included in the metazoan and fungi taxonomic groups detected in the environment.
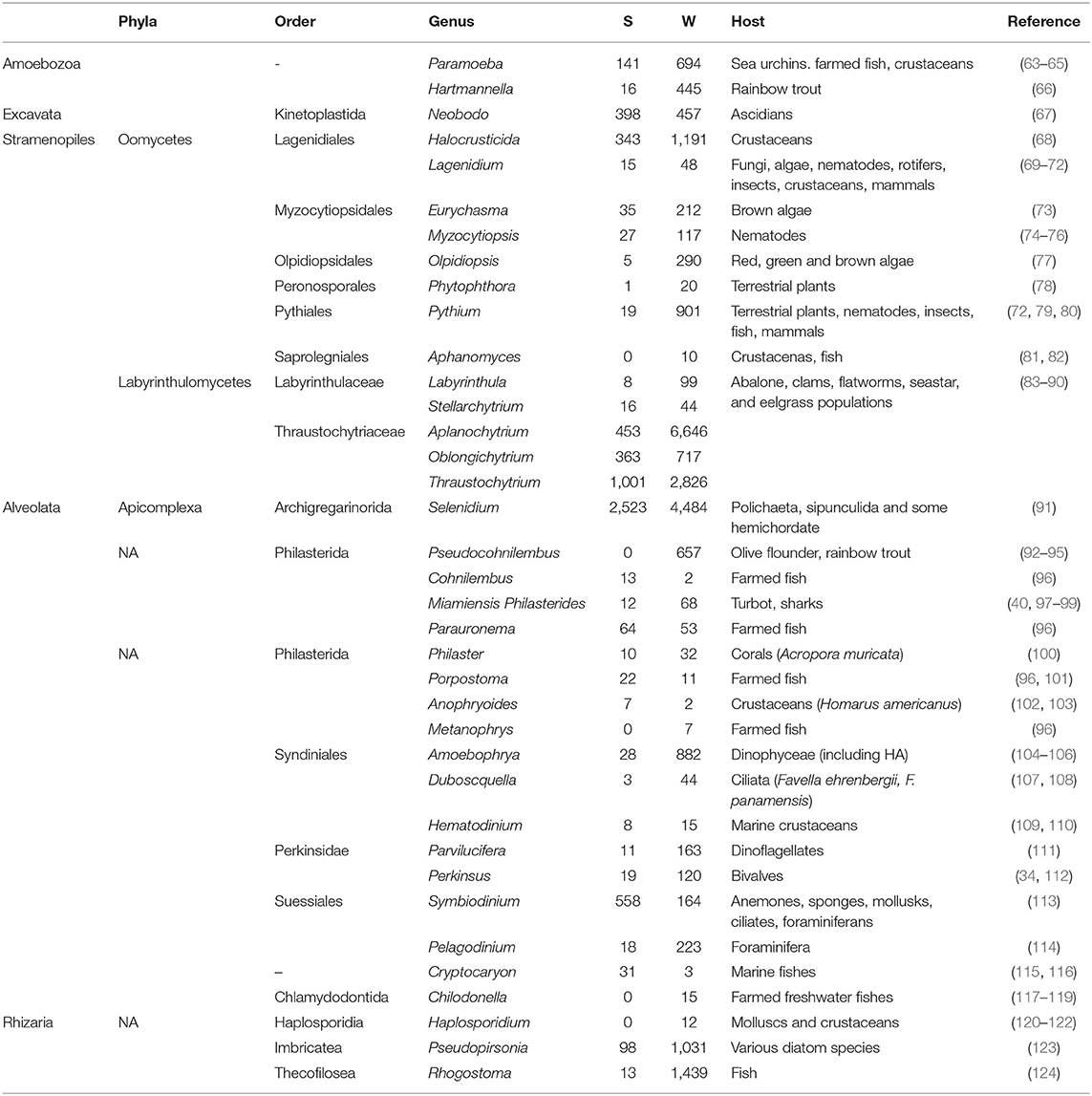
The protists included the highest number of potential pathogens in a wide variety of marine organisms. Recognized pathogens are members of the Amoebozoa, Excavata, and TSAR supergroups, as detailed in Table 5. Pathogenic Amoebozoa of farmed fish and marine invertebrates, such as the genera Paramoeba and Hartmannella, were detected in water. Pathogens belonging to the Excavata group included the genera Neobodo, which are responsible for diseases in a wide range ascidian.
The highest number of protists considered potential pathogens were included inside the different TSAR supergroups: Stramenopiles, Alveolata, and Rhizaria.
Pathogenic Stramenopiles were detected in sediment and water, and they include several members of the phyla Labyrinthulomycetes (Aplanochytrium and Thraustochytrium) and Oomycetes (Halocrusticida), which infect marine invertebrates. Aplanochytrium, Thraustochytrium, and Halocrusticida were in the top 25 most abundant protists in the marine environment (Figure 7). Low abundant genera of Oomycetes described as marine and terrestrial pathogens, such as Lagenidiales, Pythiales, Myzocytiopsidales, Peronosporales, Olpidiopsidales, and Saprolegniales, were also detected. The designated pathogen declared by the World Organization for Animal Health (O.I.E.), Aphanomyces (Saprolegniales), which is responsible for fish and crustacean diseases, was detected in the environment.
Pathogens included in the alveolates were mainly grouped in the orders Apicomplexa, Philasterida Syndinial, Perkinsidae, and Suessiales. Apicomplexans are parasites of marine organisms such as molluscs, polychaetes, fishes, and vertebrates, including domestic animals and even humans. Among all the apicomplexans, the genus Selenidium (Archigregarinorida) was the most abundant in sediment and microplankton (Figures 7A,C, Table 5) but other non-classified apicomplexans were also detected. The Philasterida order detected in the water was mainly associated with parasites of farmed fish. They included the genera Pseudocohnilembus, Parauronema, Philasterides/Miamiensis, Cohnilembus, Anophyoides, Porpostoma, Phylaster, and Metanophrys (Table 5). The genus Amoebophrya (Syndiniales), described as a pathogen in dinoflagellates, was dominant in water. Hematodinium and Duboscquella were also found, although in low abundance. Perkinsus (Perkinsidae), a O.I.E listed pathogen of mollusc was mainly detected in the water, but also in sediment. Cryptocarion, a well-known pathogen in marine fishes, was also present in sediment (Table 5).
Pathogens belonging to the rhizarians included the genus Rhogostoma, primarily associated with microplankton (Figure 7C). Moreover, the genus Pseudopirsonia, which affect diatoms and algae, was abundant. In lower abundance the genus Haplosporidium was also detected.
Discussion
The “One Health” approach recognizes that human and animal health are interconnected within the ecosystem, especially in coastal marine environments, where human activities can dramatically alter microbial biodiversity (125). Marine infections have been reported in relation to cultured species or after massive mortalities of different species. As an example, coral diseases have received increasing attention because of the recent decline in coral reefs (126). However, although mass mortalities and disease outbreaks have been reported, it is difficult to understand the extent of these events. Moreover, we do not always know the causes or even if these diseases can be affected by anthropogenic effects. In recent years, concerns have risen regarding how human-mediated climate change can alter the composition of ecological communities and therefore modify pathogen abundance and patterns of disease transmission (127). Although many studies have been conducted in different parts of the world on marine biodiversity, the absence of an efficient and coordinated monitoring system could be a significant obstacle to our understanding of the pathogen distribution in the marine environment and the possible effects of anthropogenic activities, such as pollution and fishing. Global trade and travel can introduce new species, particularly in estuarine habitats, and successfully introduced pathogens could have broad host specificity and be pathogenic to new species.
One interesting result of our study was the identification of invasive species. Early detection of invasive species is essential for their management and to avoid the displacement of autochthonous species producing variations in the ecosystem (19). The European Union has ranked 267 marine species based on their likelihood of arrival, establishment, spread, and impact in EU waters (128). We detected four invasive species on the Galician coast, with the bivalve Xenostrobus securis being one of the most harmful invasive molluscs. The presence of X. securis on the Galician coast has been described (129), and its invasiveness and potential impact on autochthonous communities have already been evaluated (130, 131). In contrast, the microscopic arthropod Pseudodiaptomus marinus has not been previously reported, and its high abundance could suggest the presence of a stable population of this species in northwestern Spain between the previously recorded distribution areas of northern Europe and Mediterranean Sea (132) using a comparable metabarcoding approach (133). The appearance of previously non-reported organisms, could indicate that maritime transport (fouling and ballast water) plays a fundamental role in expanding the geographic distribution of exotic species.
Although our aim was mainly to obtain an overview of invasive and putative pathogenic species in the Ría de Vigo, and the study of V9 amplicons provides good resolution of protist diversity, our study also offers information on the presence and variety of fish species (134, 135). Metagenomics is being used as a less costly and faster monitoring tool of stocks based on the apparent good correlation between eDNA and fish biomass (136). Our results allowed the detection of 41 fish species grouped into 22 different orders, representing ~60% of the total orders previously described on the Galician coast (137). These results highlight the potential of eDNA metabarcoding to conduct a correct estimation of fish biodiversity, management of fisheries, including the detection of spawning seasons, regulation of protected spawning areas, etc., although this methodology requires a proper sampling and further validation (138). In this context, we detected a high prevalence of clupeids (Clupea harengus) and moronids (Dicentrarchus punctatus) during autumn and winter, corresponding with their spawning season (139).
The impact of harmful algal blooms (HABs) has increased in recent decades, and several management strategies have been adopted by countries to mitigate, prevent, and control HABs in marine waters (140). Their uncontrolled proliferation produces toxic and harmful effects on fish, shellfish, marine mammals, birds, and humans, affecting the aquacultural sector and human public health (141, 142). In this study, organisms included in the Taxonomic Reference List of Harmful Microalgae (IOC-UNESCO) (143) were very abundant, and some of them are responsible for producing toxins, such as the diatom Pseudonitzschia [that produces domoic acid neurotoxin (144, 145) and generates Amnesic Shelfish Poisining (ASP) outbreaks in Galician waters (146)] and the dinoflagellate Amphidinium, that contains the species Amphidinium carterae recognized to produce haemolytic substances and ichthyotoxins with harmful effects in invertebrates (147, 148).
Our work was conducted in an important area bivalve's production, therefore, the detection of potential pathogens of these animals could have a great impact on bivalve culture. This is the case of Perkinsus olseni, detected in sediment and planktonic fractions, which has been cataloged by the O.I.E. as a notifiable parasite that causes perkinsosis disease in bivalves, mainly in clams. Haplosporidium sp. was also found in our samplings, although at low abundance. This genus infects a wide range of marine invertebrates like oysters or mussels (122).
Our study also revealed pathogens that produce important diseases in fish, with a strong economic impact on aquaculture. The order Philasterida includes several pathogens that infect cultured flatfish, causing scuticociliatosis, one of the most critical parasitological diseases in marine aquaculture worldwide. This disease has led to severe economic losses, particularly in olive flounder and turbot aquaculture (40, 92, 98). Philasterida genera were detected in sediment and water in our study, and the genus Pseudocohnilembus was the most abundant in the water column. This species has been responsible for scuticociliatosis in the olive flounder Paralichthys olivaceus in Korea (93) and the rainbow trout Oncorhynchus mykiss in the United States (95). However, in Galicia, scuticociliatosis has been mainly attributed to Philasterides dicentrarchi or Miamiensis avidus (40, 98), which were also detected in sediment and water fractions but in lower abundance than Pseudocohnilembus.
Another important group of fish pathogens that were identified in our work were those causing Nodular Gill Disease (NGD) in salmonids in Europe (66, 124, 149). The protist Rhogostoma, highly detected in water, has been associated with NGD, in particular, the species Rhogostoma minus, which induces hyperplasia of gill lamellae and, as a consequence, is one of the most critical fish disorders (149). Other abundant pathogenic genera, such as Paramoeba and Hartmanella, including harmful amoeba species, were detected in the sediment and water column. These species have been associated with NGD in freshwater fish and with Amoebic Gill Disease (AGD) in other species, such as Atlantic salmon (Salmo salar), rainbow trout (Oncorhynchus mykiss), brown trout (Salmo trutta) or turbot (Scophthalmus maximus) (150). Moreover, although Nolandella, the most abundant amoeba genus in our study, does not seem to be pathogenic, it has been reported to colonize the gills of farmed Atlantic salmon with AGD (151).
Putative pathogens can play essential functions in the ecosystem or affect other species without commercial interest but can also be valuable for the health of the ecosystem. Apicomplexans such as gregarines are still poorly studied and restricted to invertebrate hosts (152). Nevertheless, Archigregarine Selenidium was one of the most abundant protists in sediment and microplankton. This parasite infects the intestine of marine invertebrates, such as Polichaeta, Sipunculida, and some Hemichordata, explaining their high abundance in water but also in sediment (91, 153). Other gregarines from the families Lecudinidae and Eugregaronida were detected in the environment. These organisms infect invertebrates, including polychaetes, crustaceans or bivalves (152, 154); however, available information on gregarines in Spain is limited (155), therefore an accurate classification of organisms from this group at genus level would be necessary with the combination of other molecular techniques. These parasites have an important impact on invertebrates, such as crustaceans disrupting the ecosystem, and have been described as affecting shrimp aquaculture (156).
The oomycete Aphanomyces astaci, an organism native to North America, is also cataloged as a notifiable pathogen by the O.I.E. This organism infects aquatic decapods, and has been responsible for severe mortalities of native crayfish species in Europe in which their populations have declined (82). Although their abundance was very low in the environment, future surveillance of this pathogen should be performed in the study area to avoid possible propagation of this pathogen.
Oomycetes and Labyrithulomycetes infect many marine invertebrates (68, 157) and microalgae, even those involved in harmful algae blooms (73, 158). Our results showed a high abundance of Lagenidiales, which are parasites in several crustaceans and microalgal genera (69, 73, 157). Labyrinthulomycetes are opportunistic pathogens in bivalves, clams, and flatworms, and they cause severe diseases in eelgrass populations (83).
Planktonic diatoms, one of the main dominant groups in our metagenomic study, are susceptible to infections caused by parasitoids such as the rhizarian Pseudopirsonia, which it is very abundant in the environment (123, 159, 160). They are also affected by fungal organisms belonging to the Chitridiomycota group (161), detected mainly in sediment, and previously described as dominant in the nearshore and sediment samples (162).
Our metabarcoding study also allowed us to detect pathogens in mammals, confirming that human and animal health are interconnected within the marine ecosystem. The phylum Apicomplexa, as in previous studies such as the Tara Ocean expedition (163), was highly represented in the Ría de Vigo. Apicomplexan parasites are known to infect a wide range of animals, including humans and domestic animals with medical and veterinary importance worldwide, and include Toxoplasma (164), Cryptosporidium (165) and Babesia (166). In our study, pathogens similar to Cryptosporidium were amply detected in sediment and planktonic fractions, however, further analysis should be done to clearly identify these organisms because these parasites, with a waterborne transmission, have been associated with economic losses in animals such as ruminants (167). Transmissible stages are ubiquitous in aquatic habitats such as irrigation water, recreational areas, and wastewater and drinking water treatment plants. This spread could explain the high abundance obtained in planktonic fractions, according to its previous detection in Galicia (167, 168).
In conclusion, the metagenomic assay of the V9 region of the 18S SSU rRNA gene has provided valuable information on the eukaryotic composition in Ría de Vigo, and the identification of several pathogenic organisms included protists, fungi, and metazoans. The presence of specific genera and species should be validated in the future by using other techniques and taking into account the gene copy number normalization (169) and even the bias produced by the amplicon PCR but the metabarcoding method allowed us to establish a baseline of putative invasive and pathogenic organisms in this marine ecosystem.
Ría de Vigo is known for its high fish and shellfish productivity and diversity dynamics, especially during upwelling periods. The release of pollutants to the marine ecosystem, the movement of species from other areas or the introduction of foreign species by ballast water, could be factors that could change the composition of the ecosystem in Ría de Vigo in the future and benefit the uncontrolled proliferation of the known eukaryotic pathogens, causing severe mortalities to species with a high importance in the aquaculture sector. For these reasons, the use of metabarcoding assays for the monitoring of potential pathogens, invasive species, and the causative agents of harmful phytoplanktonic blooms could be important to evaluate the health of a marine ecosystem that directly affects the aquacultural sector and even human and veterinary health.
Data Availability Statement
The data presented in the study are deposited in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra), accession number PRJNA761019.
Author Contributions
BN and AF conceived and designed the project. RR-C, AR, and RA conducted the analyses. AP, AF, RR-C, and EB performed the data analysis and the bioinformatic studies. RR-C wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was conducted with the support of the projects AGL2015-65705-R (Ministerio de Economía y Competitividad, Spain), IN607B 2019/01 (Consellería de Economía, Emprego e Industria–GAIN, Xunta de Galicia), 0474_BLUEBIOLAB Fondo Europeo de Desarrollo Regional FEDER en el marco del programa Interreg V A España – Portugal (POCTEP) 2014-2020, and VIVALDI (678589) (EU H2O20). RR-C wishes to thank the Axencia Galega de Innovación (GAIN, Xunta de Galicia) for her predoctoral contract IN606A-2018/020.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We want to thank Begoña Villaverde for her technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.765606/full#supplementary-material
References
1. Pan J, Marcoval MA, Bazzini SM, Vallina VM, De-Marco SG. Coastal marine biodiversity challenges and threats. In: Arias AH, Menéndez MC, editors. Marine Ecology in a Changing World. Boca Raton, FL: CRC Press (2013). p. 43–67.
2. Leray M, Knowlton N. Censusing marine eukaryotic diversity in the twenty-first century. Philos Trans R Soc Lond B Biol Sci. (2016) 371:20150331. doi: 10.1098/rstb.2015.0331
3. Simon M, Jardillier L, Deschamps P, Moreira D, Restoux G, Bertolino P, et al. Complex communities of small protists and unexpected occurrence of typical marine lineages in shallow freshwater systems. Environ Microbiol. (2015) 17:3610–27. doi: 10.1111/1462-2920.12591
4. Pavan-Kumar A, Gireesh-Babu P, Lakra WS. DNA metabarcoding: a new approach for rapid biodiversity assessment. J Cell Sci Mol Biol. (2015) 2:111.
6. Massana R. Eukaryotic picoplankton in surface oceans. Annu Rev Microbiol. (2011) 65:91–110. doi: 10.1146/annurev-micro-090110-102903
7. Bakker J, Wangensteen OS, Baillie C, Buddo D, Chapman DD, Gallagher AJ, et al. Biodiversity assessment of tropical shelf eukaryotic communities via pelagic eDNA metabarcoding. Ecol Evol. (2019) 9:14341–55. doi: 10.1002/ece3.5871
8. Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol. (2012) 21:2045–50. doi: 10.1111/j.1365-294X.2012.05470.x
9. Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, et al. Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol Ecol. (2017) 26:5872–95. doi: 10.1111/mec.14350
10. Sogin ML, Morrison HG, Huber JA, Mark WD, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. (2006) 103:12115–20. doi: 10.1073/pnas.0605127103
11. Louca S, Mazel F, Doebeli M, Parfrey LW (2019). A census-based estimate of Earth's bacterial and archaeal diversity. PLoS Biol 17:e3000106. doi: 10.1371/journal.pbio.3000106
12. Fonseca VG, Carvalho GR, Sung W, Johnson HF, Power DM, Neill SP, et al. Second-generation environmental sequencing unmasks marine metazoan biodiversity. Nat Commun. (2010) 1:98. doi: 10.1038/ncomms1095
13. Cowart DA, Pinheiro M, Mouchel O, Maguer M, Grall J, Miné J, Arnaud-Haond S. Metabarcoding is powerful yet still blind: a comparative analysis of morphological and molecular surveys of seagrass communities. PLoS ONE. (2015) 10:e0117562. doi: 10.1371/journal.pone.0117562
14. Polinski JM, Bucci JP, Gasser M, Bodnar AG. Metabarcoding assessment of prokaryotic and eukaryotic taxa in sediments from Stellwagen Bank National Marine Sanctuary. Sci Rep. (2019) 9:14820. doi: 10.1038/s41598-019-51341-3
15. Cowart DA, Murphy KR, Cheng CC. Metagenomic sequencing of environmental DNA reveals marine faunal assemblages from the West Antarctic Peninsula. Mar Genomics. (2018) 37:148–60. doi: 10.1016/j.margen.2017.11.003
16. Holman LE, de Bruyn M, Creer S, Carvalho G, Robidart J, Rius M. Detection of introduced and resident marine species using environmental DNA metabarcoding of sediment and water. Sci Rep. 9:11559. Erratum in: Sci Rep. (2020) 10:2457. doi: 10.1038/s41598-019-47899-7
17. Schroeder A, Stanković D, Pallavicini A, Gionechetti F, Pansera M, Camatti E. DNA metabarcoding and morphological analysis - assessment of zooplankton biodiversity in transitional waters. Mar Environ Res. (2020) 160:104946. doi: 10.1016/j.marenvres.2020.104946
18. Medinger R, Nolte V, Pandey RV, Jost S, Ottenwälder B, Schlötterer C, et al. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol Suppl. (2010) 1(Suppl 1):32–40. doi: 10.1111/j.1365-294X.2009.04478.x
19. Borrell YJ, Miralles L, Do Huu H, Mohammed-Geba K, Garcia-Vazquez E. DNA in a bottle-Rapid metabarcoding survey for early alerts of invasive species in ports. PLoS ONE. (2017) 12:e0183347. doi: 10.1371/journal.pone.0183347
20. Sherr BF, Sherr EB, Caron DA, Vaulot D, Worden AZ. Oceanic protists. Oceanography. (2007) 20:130–4. doi 10.5670/oceanog.2007.57
21. Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, et al. Climate warming and disease risks for terrestrial and marine biota. Science. (2002) 296:2158–62. doi: 10.1126/science.1063699
22. Granek EF, Brumbaugh DR, Heppell SA, Heppell SS, Secord D. A blueprint for the oceans: implications of two national commission reports for conservation practitioners. Conserv Biol. (2005) 19:1008–18. doi: 10.1111/j.1523-1739.2005.00221.x
23. Panek FM. Epizootics and disease of coral reef fish in the Tropical Western Atlantic and Gulf of Mexico. Rev Fish Sci. (2005) 13:1–21. doi: 10.1080/10641260590885852
24. King DA, Peckham C, Waage JK, Brownlie J, Woolhouse ME. Epidemiology. Infectious diseases: preparing for the future. Science. (2006) 313:1392–3. doi: 10.1126/science.1129134
25. Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci USA. (2006) 103:3165–70. doi: 10.1073/pnas.0506889103
26. Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE. 4:e6372. Erratum in: PLoS ONE. (2009) 4:e6372. doi: 10.1371/annotation/50c43133-0df5-4b8b-8975-8cc37d4f2f26
27. Geisen S, Lara E, Mitchell EAD, Völcker E, Krashevska V. Soil protist life matters! Soil Org. (2020) 92:189–96. doi: 10.25674/so92iss3pp189
28. Choi J, Park JS. Comparative analyses of the V4 and V9 regions of 18S rDNA for the extant eukaryotic community using the Illumina platform. Sci Rep. (2020) 10:6519. doi: 10.1038/s41598-020-63561-z
29. Simon M, López-García P, Deschamps P, Moreira D, Restoux G, Bertolino P, et al. Marked seasonality and high spatial variability of protist communities in shallow freshwater systems. ISME J. (2015) 9:1941–53. doi: 10.1038/ismej.2015.6
30. Casas B, Varela M, Canle M, González N, Bode A. Seasonal variations of nutrients, Seston and phytoplankton, and upwelling intensity off La Coruña (NW Spain). Estuar Coast Shelf Sci. (1997) 44:767–78.
31. Hernández-Ruiz M, Barber-Lluch E, Prieto A, Álvarez-Salgado XA, Logares R, Teira E. Seasonal succession of small planktonic eukaryotes inhabiting surface waters of a coastal upwelling system. Environ Microbiol. (2018) 20:2955–73. doi: 10.1111/1462-2920.14313
32. Figueiras FG, Abarta U, Fernández Reiriz MJ. Coastal upwelling, primary production and mussel growth in the Rías Baixas of Galicia. Hydrobiologia. (2002) 484:121–31. doi: 10.1023/A:1021309222459
33. Joglar V, Álvarez-salgado XA, Gago-Martínez A, Leao JM, Pérez- Martínez C, Pontiller B, et al. Cobalamin and microbial plankton dynamics along a coastal to offshore transect in the Eastern North Atlantic Ocean. Environ Microbiol. (2020) 2:1559–83. doi: 10.1111/1462-2920.15367
34. Villalba A, Reece KS, Ordas MC, Casas SM, Figueras A. Perkinsosis in molluscs: a review. Aquat Living Res. (2004) 17:411–32. doi: 10.1051/alr:2004050
35. Robledo JAF, Cáceres-Martínez J, Figueras A. Marteilia refringens in mussel (Mytilus galloprovincialis LMK.) beds in Spain. Bull. Eur. Ass. Fish Pathol. (1994) 14:61.
36. Villalba A, Mourelle SG, Carballal MJ. Symbionts and diseases of farmed mussels Mytilus galloprovincialis throughout the culture process in the Rías of Galicia (NW Spain). Dis. Aquat. Organ. (1997) 31:127–39.
37. Balseiro P, Montes J, Fernández-Conchas R, Novoa B, Figueras A. Comparison of diagnostic techniques to detect the clam pathogen Perkinsus olseni. Dis Aquat Organ. (2010) 90:143–51. doi: 10.3354/dao02194
38. Ríos R, Aranguren R, Gastaldelli M, Arcangeli G, Novoa B, Figueras A. Development and validation of a specific real-time PCR assay for the detection of the parasite Perkinsus olseni. J Invertebr Pathol. (2020) 169:107301. doi: 10.1016/j.jip.2019.107301
39. Dyková I, Figueras A. Histopathological changes in turbot Scophthalmus maximus due to histophagous ciliate. Dis Aquat Organ. (1994) 18:5–9.
40. Iglesias R, Paramá A, Alvarez MF, Leiro J, Fernández J, Sanmartín ML. Philasterides dicentrarchi (Ciliophora, Scuticociliatida) as the causative agent of scuticociliatosis in farmed turbot Scophthalmus maximus in Galicia (NW Spain). Dis Aquat Organ. (2001) 46:47–55. doi: 10.3354/dao046047
41. Girones R, Ferrús MA, Alonso JL, Rodriguez-Manzano J, Calgua B, Corrêa A, et al. Molecular detection of pathogens in water–the pros and cons of molecular techniques. Water Res. (2010) 44:4325–39. doi: 10.1016/j.watres.2010.06.030
42. Chariton AA, Stephenson S, Morgan MJ, Steven ADL, Colloff MJ, Court LN, et al. Metabarcoding of benthic eukaryote communities predicts the ecological condition of estuaries. Environ Pollut. (2015) 203:165–74. doi: 10.1016/j.envpol.2015.03.047
43. Sieburth J, Smetacek V, Lenz J. Pelagic ecosystem structure: Heterotrophic compartments of the plankton and their relationship to plankton size fractions. Limnol Oceanogr. (1978) 23:1256–63.
44. Baker CCM. Entrez Qiime: A Utility for Generating QIIME Input Files From the NCBI Databases. (2016). Available online at: https://github.com/bakerccm/entrez_qiime/ (accessed October 15, 2021).
45. Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. (2006) 22:1658–9. doi: 10.1093/bioinformatics/btl158
46. Thukral AK. A review on measurement of alpha diversity in biology. Agric Res J. (2017) 54:1–10. doi: 10.5958/2395-146X.2017.00001.1
47. McKnight MW, White PS, McDonald RI, Lamoreux JF, Sechrest W, Ridgely RS, et al. (2007). Putting beta-diversity on the map: broad-scale congruence and coincidence in the extremes. PLoS Biol. (2007) 5:e272. doi: 10.1371/journal.pbio.0050272
48. Tsiamis K, Palialexis A, Stefanova K, Gladan ŽN, Skejić S, Despalatović M, et al. Non-indigenous species refined national baseline inventories: a synthesis in the context of the European Union's Marine Strategy Framework Directive. Mar Pollut Bull. (2019) 145:429–35. doi: 10.1016/j.marpolbul.2019.06.012
49. Burki F, Roger AJ, Brown MW, Simpson AGB. The new tree of eukaryotes. Trends Ecol Evol. (2020) 35:43–55. doi: 10.1016/j.tree.2019.08.008
50. Li YC, Inoue K, Tanaka S, Zhang JY, Sato H. Identification of four new Kudoa spp. (Myxozoa: Myxosporea: Multivalvulida) in commercial fishes collected from South China Sea, Atlantic Ocean, and Bering Sea by integrated taxonomic approach. Parasitol Res. (2020) 119:2113–28. doi: 10.1007/s00436-020-06707-2
51. Bahri S, Andree KB, Hedrick RP. Morphological and phylogenetic studies of marine Myxobolus spp. from mullet in Ichkeul Lake, Tunisia. J Eukaryot Microbiol. (2003) 50:463–70. doi: 10.1111/j.1550-7408.2003.tb00272.x
52. Cho JB, Lee MK, Huh Min-Do, Kim Ki-Hong. Co-infection of two myxosporean parasites parvicapsula anisocaudata and an unidentified myxosporean - in the kidney of cultured olive flounder, paralichthys olivaceus. J Fish Pathol. (2005) 18:119–24. doi: 10.7847/jfp.2018.31.2.123
53. Sanmartín ML, Alvarez MF, Peris D, Iglesias R, Leiro J. Parasite community study of the undulate ray Raja undulata in the Ria of Muros (Galicia, northwest Spain). Aquaculture. (2000) 184:189–201. doi: 10.1016/S0044-8486(99)00332-4
54. Ozer A, Okkay S, Öztürk T, Baki B, Acar G, Güven A, et al. Comparative Diplectanum aequans (Monogenea) infestations in cultured European seabass (Dicentrarchus labrax) in the Black Sea and the Aegean Sea. EgeJFAS. (2019) 36:163–9. doi: 10.12714/egejfas.2019.36.2.08
55. Da Silva PM, Cremonte F, Sabry RC, Rosa RD, Cantelli L, Barracco MA. Presence and histopathological effects of the Parvatrema sp. (Digenea, Gymnophallidae) in the stout razor clam Tagelus plebeius (Bivalvia, Psammobiidae). J Invertebr Pathol. (2009) 102:14–20. doi: 10.1016/j.jip.2009.05.014
56. Lee SH, Chai JY A. review of Gymnophalloides seoi (Digenea: Gymnophallidae) and human infections in the Republic of Korea. Korean J Parasitol. (2001) 39:85–118. doi: 10.3347/kjp.2001.39.2.85
57. Magalhães L, Freitas R, De-Montaudouin X. Review: Bucephalus minimus, a deleterious trematode parasite of cockles Cerastoderma spp. Parasitol Res. (2015) 114:1263–78. doi: 10.1007/s00436-015-4374-6
58. Eydal M, Freeman MA, Kristmundsson Á, Bambir SH, Jónsson PM, Helgason S. Prosorhynchoides borealis Bartoli, Gibson & Bray, 2006 (Digenea: Bucephalidae) cercariae from Abra prismatica (Mollusca: Bivalvia) in Icelandic waters. J Helminthol. (2013) 87:34–41. doi: 10.1017/S0022149X11000782
59. Procop GW. North American paragonimiasis (Caused by Paragonimus kellicotti) in the context of global paragonimiasis. Clin Microbiol Rev. (2009) 22:415–46. doi: 10.1128/CMR.00005-08
60. Sarabeev VL, Balbuena JA. Morphological variability of Dicrogaster contracta Looss, 1902 (Digenea: Haploporidae) and its proposed synonymy with D. perpusilla Looss, 1902. Syst Parasitol. (2003) 55:25–31. doi: 10.1023/A:1023937309228
61. Aktaş Karabay E, Aksu Çerman A. Demodex folliculorum infestations in common facial dermatoses: acne vulgaris, rosacea, seborrheic dermatitis. An Bras Dermatol. (2020) 95:187–93. doi: 10.1016/j.abd.2019.08.023
62. Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev. (2012) 25:106–41. doi: 10.1128/CMR.00021-11
63. Feehan CJ, Johnson-Mackinnon J, Scheibling RE, Lauzon-Guay JS, Simpson AG. Validating the identity of Paramoeba invadens, the causative agent of recurrent mass mortality of sea urchins in Nova Scotia, Canada. Dis Aquat Organ. (2013) 103:209–27. doi: 10.3354/dao02577
64. Bloecher N, Powell M, Hytterød S, Gjessing M, Wiik-Nielsen J, Mohammad SN, et al. Effects of cnidarian biofouling on salmon gill health and development of amoebic gill disease. PLOS ONE. (2018) 13:e0199842. doi: 10.1371/journal.pone.0199842
65. Nowak BF, Archibald JM. Opportunistic but lethal: the mystery of Paramoebae. Trends Parasitol. (2018) 34:4. doi: 10.1016/j.pt.2018.01.004
66. Dyková I, Kostka M, Wortberg F, Nardy E, Pecková H. New data on aetiology of nodular gill disease in rainbow trout, Oncorhynchus mykiss. Folia Parasitol. (2010) 57:157–63. doi: 10.14411/fp.2010.021
67. Jang HB, Kim YK, Del-Castillo CS, Nho SW, Cha IS, Park SB. RNA-Seq- Based metatranscriptomic and microscopic investigation reveals novel metalloproteases of Neobodo sp as potential virulence factors for Soft Tunic Syndrome in Halocynthia roretzi. PLoS ONE. (2012) 7:e52379. doi: 10.1371/journal.pone.0052379
68. Yasunobu H. Prevention of Lagenidiales infections in Artemia salina larvae by high pH of rearing water. Fish Pathol. (2001) 36:79–82. doi: 10.3147/jsfp.36.79
69. Mendoza L, Vilela R. The mammalian pathogenic oomycetes. Curr Fungal Infect Rep. (2013) 7:198–208. doi: 10.1007/s12281-013-0144-z
70. Barron GL. Fungal parasites and predators of rotifers, nematodes and other invertebrates. In: Muller GM, Bills G, Foster MS, editors. Biodiversity of Fungi, Inventory and Monitoring Methods. Amsterdam: Elsevier Academic Press (2004). p. 435–50. doi: 10.1016/B978-012509551-8/50022-2
71. Mendoza L, Taylor JW, Walker ED, Vilela R. Description of three novel Lagenidium (Oomycota) species causing infection in mammals. Rev Iberoam Micol. (2016) 33:83–91. doi: 10.1016/j.riam.2015.07.005
72. Saunders WB. Fungal skin diseases. In: Hnilica KA, Patterson AP, editors. Small Animal Dermatology (Fourth Edition). (2017). p. 94–131. doi: 10.1016/B978-0-323-37651-8.00004-3
73. Strittmatter M, Grenville-Briggs LJ, Breithut L, Van WP, Gachon CM, Küpper FC. Infection of the brown alga Ectocarpus siliculosus by the oomycete Eurychasma dicksonii induces oxidative stress and halogen metabolism. Plant Cell Environ. (2016) 39:259–71. doi: 10.1111/pce.12533
74. Glockling SL, Beakes G A. review of the taxonomy, biology and infection strategies of “biflagellate holocarpic” parasites of nematodes. Fungal Divers. (2000) 4:1–20.
75. Jaffee BA, Bastow J, Strong DR. Suppression of nematodes in a coastal grassland soil. Biol Fertil Soils. (2007) 44:19–26. doi: 10.1007/s00374-007-0174-1
76. Osman GA, Fasseas MK, Koneru SL, Essmann CL, Kyrou K, Srinivasan MA, et al. Natural infection of C. elegans by an oomycete reveals a new pathogen-specific immune response. Curr Biol. (2018) 28:640–8.e5. doi: 10.1016/j.cub.2018.01.029
77. Klochkova T, Shin YJ, Moon KH, Motomura T, Kim GH. New species of unicellular obligate parasite, Olpidiopsis pyropiae sp. nov, that plagues Pyropia sea farms in Korea. J Appl Phycol. (2016) 28:73–83. doi: 10.1007/s10811-015-0595-4
78. Erwin DC, Bartnicki-Garcia S, Tsao PH. Phytophthora: Its Biology, Taxonomy, Ecology and Pathology. St. Paul, MN: American Phytopathological Society (1983).
79. Tzean SS, Estey RH. Species of Phytophthora and Pythium as Nematode-destroying Fungi. J Nematol. (1981) 13:160–3.
80. Mendoza L, Vilela R. Anomalous fungal and fungal-like infections: lacaziosis, pythiosis, and rhinosporidiosis. Clin Mycol. (2009) 2:403–15. doi: 10.1016/B978-1-4160-5680-5.00018-9
81. Boddy L. Interactions with humans and other animals. In: Watkinson SC, Boddy L, Money, editors. The Fungi. Amsterdam; Boston, MA: Elsevier Academic Press (2016). p. 293–336. doi: 10.1016/B978-0-12-382034-1.00009-8
82. Martín-Torrijos L, Campos LM, Pou-Rovira Q, Diéguez-Uribeondo J. Resistance to the crayfish plague, Aphanomyces astaci (Oomycota) in the endangered freshwater crayfish species, Austropotamobius pallipes. PLoS ONE. (2017) 12:e0181226. doi: 10.1371/journal.pone.0181226
83. Bower SM. Pathogenicity and host specificity of Labyrinthuloides haliotidis (Protozoa: Labyrinthomorpha), a parasite of juvenile abalone. Can J Zool. (1987) 65:2008–12. doi: 10.1139/z87-305
84. Ragan MA, MacCallum GS, Murphy CA, Cannone JJ, Gutell RR, McGladdery SE. Protistan parasite QPX of hard-shell clam Mercenaria mercenaria is a member of Labyrinthulomycota. Dis Aquat Organ. (2000) 42:185–90. doi: 10.3354/dao042185
85. Raghukumar S. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur J Protistol. (2002) 38:127–45. doi: 10.1078/0932-4739-00832
86. Anderson RS, Kraus BS, McGladdery SE, Reece KS, Stokes NA. A thraustochytrid protist isolated from Mercenaria mercenaria: molecular characterization and host defense responses. Fish Shellfish Immunol. (2003) 15:183–94. doi: 10.1016/S1050-4648(02)00157-2
87. Schärer L, Knoflach D, Vizoso DB, Rieger G, Peintner U. Thraustochytrids as novel parasitic protists of marine free-living flatworms: Thraustochytrium caudivorum sp. nov parasitized Macrostomum lignano. Mar Biol. (2007) 152:1095–104. doi: 10.1007/s00227-007-0755-4
88. Sullivana BK, Sherman TD, Damare VS, Lilje O, Gleason FH. Potential roles of Labyrinthula spp. in global seagrass population declines. Fungal Ecol. (2013) 6:328–38. doi: 10.1016/j.funeco.2013.06.004
89. FioRito R, Leander C, Leander B. Characterization of three novel species of Labyrinthulomycota isolated from ochre sea stars (Pisaster ochraceus). Marine Biol. (2016) 163:170. doi: 10.1007/s00227-016-2944-5
90. Fossier ML, Chang KJL, Nichols PD, Mitchell WJ, Polglase JL, Gutierrez T. Taxonomy, ecology and biotechnological applications of thraustochytrids: a review. Biotechnol Adv. (2018) 36:26–46. doi: 10.1016/j.biotechadv.2017.09.003
91. Schrével J, Valigurová A, Prensier G, Chambouvet A, Florent I, Guillou L. Ultrastructure of Selenidium pendula, the type species of archigregarines, and phylogenetic relations to other marine apicomplexa. Protist. (2016) 167:339–68. doi: 10.1016/j.protis.2016.06.001
92. Kim SM, Cho JB, Kim SK, Nam YK, Kim KH. Occurrence of scuticociliatosis in olive flounder Paralichthys olivaceus by Philasterides dicentrarchi (Ciliophora: Scuticociliatida). Dis Aquat Organ. (2004) 62:233–8. doi: 10.3354/dao062233
93. Kim SM, Cho JB, Lee EH, Kwon SR, Kim SK, Nam YK, et al. Pseudocohnilembus persalinus (Ciliophora: Scuticociitida) is an additional species causing scuticociliatosis in olive flounder 1Paralichthys olivaceus. Dis Aquat Organ. (2004) 62:239–44. doi: 10.3354/dao062239
94. Song JY, Kitamura S, Oh MJ, Kang HS, Lee JH, Tanaka SJ, et al. Pathogenicity of Miamiensis avidus (syn. Philasterides dicentrarchi), Pseudocohnilembus persalinus, Pseudocohnilembus hargisi and Uronema marinum (Ciliophora, Scuticociliatida). Dis Aquat Organ. (2009) 83:133–43. doi: 10.3354/dao02017
95. Jones SR, Prosperi-Porta G, LaPatra SE. First isolation of Pseudocohnilembus persalinus (Ciliophora: Scuticociliatida) from freshwater-reared rainbow trout, Oncorhynchus mykiss. J Parasitol. (2010) 96:1014–6. doi: 10.1645/GE-2500.1
96. Gao F, Strüder-Kypke M, Yi Z, Miao M, Al-Farraj SA, Song W. Phylogenetic analysis and taxonomic distinction of six genera of pathogenic scuticociliates (Protozoa, Ciliophora) inferred from small-subunit rRNA gene sequences. Int J Syst Evol Microbiol. (2012) 62:246–56. doi: 10.1099/ijs.0.028464-0
97. Stidworthy MF, Garner MM, Bradway DS, Westfall BD, Joseph B, Repetto S, et al. Systemic Scuticociliatosis (Philasterides dicentrarchi) in sharks. Vet Pathol. (2014) 51:628–32. doi: 10.1177/0300985813492800
98. De Felipe AP, Lamas J, Sueiro RA, Folgueira I, Leiro JM. New data on flatfish scuticociliatosis reveal that Miamiensis avidus and Philasterides dicentrarchi are different species. Parasitology. (2017) 29:1–18. doi: 10.1017/S0031182017000749
99. Lama R, Pereiro P, Costa MM, Encinar JA, Medina-Gali RM, Pérez L, et al. Turbot (Scophthalmus maximus) Nk-lysin induces protection against the pathogenic parasite Philasterides dicentrarchi via membrane disruption. Fish Shellfish Immunol. (2018) 82:190–9. doi: 10.1016/j.fsi.2018.08.004
100. Sweet M, Bythell J. White syndrome in Acropora muricata: nonspecific bacterial infection and ciliate histophagy. Mol Ecol. (2015) 24:1150–9. doi: 10.1111/mec.13097
101. Ofelio C, Blanco A, Roura Á, Pintado J, Pascual S, Planas M. Isolation and molecular identification of the scuticociliate Porpostoma notata Moebius, 1888 from moribund reared Hippocampus hippocampus (L.) seahorses, by amplification of the SSU rRNA gene sequences. J Fish Dis. (2014) 37:1061–5. doi: 10.1111/jfd.12207
102. Lavallée J, Hammell KL, Spangler ES, Cawthorn RJ. Estimated prevalence of Aerococcus viridans and Anophryoides haemophila in American lobsters Homarus americanus freshly captured in the waters of Prince Edward Island, Canada. Dis Aquat Organ. (2001) 46:231–6. doi: 10.3354/dao046231
103. Clark KF, Acorn AR, Greenwood SJ. A transcriptomic analysis of American lobster (Homarus americanus) immune response during infection with the bumper car parasite Anophryoides haemophila. Dev Comp Immunol. (2013) 40:112–22. doi: 10.1016/j.dci.2013.02.009
104. Yih W, Coats DW. Infection of Gymnodinium sanguineum by the dinoflagellate Amoebophrya sp.: effect of nutrient environment on parasite generation time, reproduction, and infectivity. J Eukaryot Microbiol. (2020) 47:504–10. doi: 10.1111/j.1550-7408.2000.tb00082.x
105. Lu Y, Wohlrab S, Glöckner G, Guillou L, John U. Genomic insights into processes driving the infection of Alexandrium tamarense by the Parasitoid Amoebophrya sp. Eukaryot Cell. (2014) 13:1439–49. doi: 10.1128/EC.00139-14
106. Chen T, Liu Y, Xu S, Song S, Li C. Variation of Amoebophrya community during bloom of Prorocentrum donghaiense Lu in coastal waters of the East China Sea Estuarine, Coastal and Shelf. Science. (2020) 243:106887. doi: 10.1016/j.ecss.2020.106887
107. Harada A, Ohtsuka S, Horiguchi T. Species of the parasitic genus Duboscquella are members of the enigmatic Marine Alveolate Group I. Protist. (2007) 158:337–47. doi: 10.1016/j.protis.2007.03.005
108. Coats DW, Bachvaroff TR, Delwiche CF. Revision of the family Duboscquellidae with description of Euduboscquella crenulata n. gen., n. sp. Dinoflagellata, Syndinea), an intracellular parasite of the ciliate Favella panamenensis Kofoid & Campbell, 1929. J Eukaryotic Microbiol. (2012) 59:1–11. doi: 10.1111/j.1550-7408.2011.00588.x
109. Small HJ. Advances in our understanding of the global diversity and distribution of Hematodinium spp. - significant pathogens of commercially exploited crustaceans. J Invertebr Pathol. (2012) 110:234–46. doi: 10.1016/j.jip.2012.03.012
110. Ryazanova TV, Eliseikina MG, Semenchenko AA A. new host for Hematodinium infection among lithodid crabs from the Sea of Okhotsk. J Invertebr Pathol. (2018) 153:12–9. doi: 10.1016/j.jip.2018.02.002
111. Alacid E, Reñé A, Gallisai R, Paloheimo A, Garcés E, Kremp A. Description of two new coexisting parasitoids of blooming dinoflagellates in the Baltic sea: Parvilucifera catillosa sp. nov and Parvilucifera sp. (Perkinsea, Alveolata). Harmful Algae. (2020) 100:101944. doi: 10.1016/j.hal.2020.101944
112. Perkins FO, Menzel RW. Ultrastructure of sporulation in the oyster pathogen Dermocystidium marinum. J Invertebr Pathol. (1967) 9:205–29. doi: 10.1016/0022-2011(67)90009-2
113. Mordret S, Romac S, Henry N, Colin S, Carmichael M, Berney C, et al. The symbiotic life of Symbiodinium in the open ocean within a new species of calcifying ciliate (Tiarina sp.). ISME J. (2016) 10:1424–36. doi: 10.1038/ismej.2015.211
114. Siano R, Montresor M, Probert I, Not F, De-Vargas C. Pelagodinium gen. nov and P béii comb nov, a dinoflagellate symbiont of planktonic foraminifera. Protist. (2010) 161:385–99. doi: 10.1016/j.protis.2010.01.002
115. Cardoso PHM, Soares HS, Martins ML, Balian SC. Cryptocaryon irritans, a ciliate parasite of an ornamental reef fish yellowtail tang Zebrasoma xanthurum. Rev Bras de Parasitol Vet. (2019) 28:750–3. doi: 10.1590/s1984-29612019033
116. Chi H, Goldstein M, Pichardo A, Wei ZH, Chang WJ, Gong H. Infectivity and genes differentially expressed between young and aging theront cells of the marine fish parasite Cryptocaryon irritans. PLoS ONE. (2020) 15:e0238167. doi: 10.1371/journal.pone.0238167
117. Rintamäki P, Torpström H, Bloigu A. Chilodonella spp. at four fish farms in Northern Finland. Eukaryotic Microbiol. (1994) 41:602–7. doi: 10.1111/j.1550-7408.1994.tb01522.x
118. Mitra AK, Haldar DP. First record of Chilodonella hexasticha (Kiernik, 1909) Kahl, 1931 (Ciliophora: Chilodonellidae) infesting a freshwater fish Nandus nandus (Hamilton) from Gangetic West Bengal, India. Anim Biol. (2004) 54:111–8. doi: 10.1163/1570756041445182
119. Bastos-Gomes G, Miller TL, Vaughan DB, Jerry DR, McCowan C, Bradley TL, et al. Evidence of multiple species of Chilodonella (Protozoa, Ciliophora) infecting Australian farmed freshwater fishes. Vet Parasitol. (2017) 237:8–16. doi: 10.1016/j.vetpar.2017.03.004
120. Urrutia A, Bass D, Ward G, Ross S, Bojko J, Marigomez I, et al. Ultrastructure, phylogeny and histopathology of two novel haplosporidians parasitising amphipods, and importance of crustaceans as hosts. Dis Aquat Organ. (2019) 136:89–105. doi: 10.3354/dao03417
121. Davies CE, Bass D, Ward GM, Batista FM, Malkin SH, Thomas JE, et al. Diagnosis and prevalence of two new species of haplosporidians infecting shore crabs Carcinus maenas: Haplosporidium carcini n. sp., and H. cranc n. sp. Parasitol. (2020) 147:1229–37. doi: 10.1017/S0031182020000980
122. Lynch SA, Lepée-Rivero S, Kelly R, Quinn E, Coghlan A, Bookelaar B, et al. Detection of haplosporidian protistan parasites supports an increase to their known diversity, geographic range and bivalve host specificity. Parasitology. (2020) 147:584–92. doi: 10.1017/S0031182019001628
123. Kim S, Jeon CB, Park MG. Morphological observations and phylogenetic position of the parasitoid nanoflagellate Pseudopirsonia sp. (Cercozoa) infecting the marine diatom Coscinodiscus wailesii (Bacillariophyta). Algae. (2017) 32:181–7. doi: 10.4490/algae.2017.32.7.28
124. Dyková I, Tyml T. Testate amoeba Rhogostoma minus Belar, 1921, associated with nodular gill disease of rainbow trout, Oncorhynchus mykiss (Walbaum). J Fish Dis. (2016) 39:539–46. doi: 10.1111/jfd.12384
125. Mackenzie JS, Jeggo M. The one health approach-why is it so important? Trop Med Infect Dis. (2019) 4:88. doi: 10.3390/tropicalmed4020088
126. Fontoura L, Zawada KJA, D'agata S, Álvarez-Noriega M, Baird AH, Boutros N, et al. Climate-driven shift in coral morphological structure predicts decline of juvenile reef fishes. Glob Chang Biol. (2020) 26:557–67. doi: 10.1111/gcb.14911
127. Pecl GT, Araújo MB, Bell J.D, Blanchard J, Bonebrake TC, Chen IC, et al. Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science. (2017) 355:eaai9214. doi: 10.1126/science.aai9214
128. Tsiamis K, Azzurro E, Bariche M, Cinar M, Crocetta F, De-Clerck O, et al. Prioritizing marine invasive alien species in the European Union through horizon scanning. Aquat Conserv. (2020) 30:794–845. doi: 10.1002/aqc.3267
129. Garcí ME, Trigo JE, Pascual AF, Rocha F, Guerra A. Xenostrobus securis (Lamarck, 1819) (Mollusca: Bivalvia): first report of an introduced species in Galician waters. Aquacult Int. (2007) 15:19–24. doi: 10.1007/s10499-006-9062-1
130. Romero A, Aranguren R, Moreira R, Novoa B, Figueras A. Integrated transcriptomic and functional immunological approach for assessing the invasiveness of bivalve alien species. Sci Rep. (2019) 9:19879. doi: 10.1038/s41598-019-56421-y
131. Montes A, Olabarria C, Vázquez E. Reproductive plasticity in the invasive -Xenostrobus securis (Bivalvia: Mytiloidea) in northwestern Spain. J Sea Res. (2020) 159:101893. doi: 10.1016/j.seares.2020.101893
132. Reyes-Martínez M, González-Gordillo JI. New record of the non-indigenous copepod Pseudodiaptomus marinus Sato, 1913 (Calanoida, pseudodiaptomidae) from the Guadalquivir estuary (Gulf of Cádiz, SW Spain). Crustaceana. (2019) 92:675–83. doi: 10.1163/15685403-00003903
133. Stefanni S, Stanković D, Borme D, De-Olazabal A, Juretić T, Pallavicini A, et al. Multi-marker metabarcoding approach to study mesozooplankton at basin scale. Sci Rep. (2018) 8:12085. doi: 10.1038/s41598-018-30157-7
134. Fujii K, Doi H, Matsuoka S, Nagano M, Sato H, Yamanaka H. Environmental DNA metabarcoding for fish community analysis in backwater lakes: a comparison of capture methods. PLoS ONE. (2019) 14:e0210357. doi: 10.1371/journal.pone.0210357
135. Morey KC, Bartley T, Hanner R. Validating environmental DNA metabarcoding for marine fishes in diverse ecosystems using a public aquarium. Environ DNA. (2020) 2:76. doi: 10.1002/edn3.76
136. Wang S, Yan Z, Hänfling B, Zheng X, Wang P, Fan J, et al. Methodology of fish eDNA and its applications in ecology and environment. Sci Total Environ. (2021) 755(Pt 2):142622. doi: 10.1016/j.scitotenv.2020.142622
137. Bañon R, Villegas-Ríos D, Serrano A, Mucientes G, Arronte JC. Marine fishes from Galicia (NW Spain): an updated checklist. Zootaxa. (2010) 2667:1–27. doi: 10.11646/zootaxa.2667.1.1
138. Evans NT, Li Y, Renshaw MA, Olds BP, Deiner K, Turner CR, et al. Fish community assessment with eDNA metabarcoding: effects of sampling design and bioinformatic filtering. Can J Fish Aquat Sci. (2017) 74:1362–74. doi: 10.1139/cjfas-2016-0306
139. Carrera P, Porteiro C. Stock dynamic of the Iberian sardine (Sardina pilchardus, W.) and its implication on the fishery off Galicia (NW Spain). Sci Mar. (2003) 67:245–58. doi: 10.3989/scimar.2003.67s1245
140. Anderson DM. Approaches to monitoring, control and management of harmful algal blooms (HABs). Ocean Coast Manag. (2009) 52:342. doi: 10.1016/j.ocecoaman.2009.04.006
141. Grattan LM, Holobaugh S, Morris JG Jr. Harmful algal blooms and public health. Harmful Algae. (2016) 57(Pt B):2–8. doi: 10.1016/j.hal.2016.05.003
142. Gallardo-Rodríguez JJ, Astuya-Villalón A, Llanos-Rivera A, Avello-Fontalba V, Ulloa-Jofré V A. critical review on control methods for harmful algal blooms. Rev Aquacult. (2019) 11:661–84. doi: 10.1111/raq.12251
143. Moestrup Ø, Akselmann-Cardella R, Churro C, Fraga S, Hoppenrath M, Iwataki M, et al. (editors). IOC-UNESCO Taxonomic Reference List of Harmful Micro Algae. (2009). Available online at: www.marinespecies.org/hab (accessed October 15, 2021).
144. Bates SS, Hubbard KA, Lundholm N, Montresor M, Leaw CP. Pseudo-nitzschia, Nitzschia, and domoic acid: New research since 2011. Harmful Algae. (2018) 79:3–43. doi: 10.1016/j.hal.2018.06.001
145. Broullón E, López-Mozos M, Reguera B, Chouciño P, Doval MD, Fernández-Castro B, et al. Thin layers of phytoplankton and harmful algae events in a coastal upwelling system. Prog Oceanogr. (2020) 189:102449. doi: 10.1016/j.pocean.2020.102449
146. Trainer VL, Pitcher GC, Reguera B, Smayda TJ. The distribution and impacts of harmful algal bloom species in eastern boundary upwelling systems. Prog Oceanogr. (2010) 85:79–6611. doi: 10.1016/j.pocean.2010.02.003
147. Yasumoto T, Seino NY, Murakami Y. Toxins produced by benthic dinoflagellates. Biol Bull. (1987) 172:128–31. doi: 10.2307/1541612
148. Echigoya R, Rhodes L, Oshima Y, Satake M. The structures of five new antifungal and hemolytic amphidinol analogs from Amphidinium carterae collected in New Zealand. Harmful Algae. (2005) 4:383–9. doi: 10.1016/j.hal.2004.07.004
149. Quaglio F, Perolo A, Bronzatti P, Gustinelli A, Menconi V, Cavazza G, et al. Nodular gill disease in farmed rainbow trout (Oncorhynchus mykiss) in Italy. J Fish Dis. (2016) 39:1139–42. doi: 10.1111/jfd.12446
150. Padrós F, Constenla M. Diseases caused by amoebae in fish: an overview. Animals. (2021) 11:991. doi: 10.3390/ani11040991
151. English CJ, Tyml T, Botwright NA, Barnes AC, Wynne JW, Lima PC, et al. A diversity of amoebae colonise the gills of farmed Atlantic salmon (Salmo salar) with amoebic gill disease (AGD). Eur J Protistol. (2019) 67:27–45. doi: 10.1016/j.ejop.2018.10.003
152. Schrével J, Desportes I. Gregarines. Encyclopedia of Parasitology. Heidelberg: Springer-Verlag Berlín (2015). doi: 10.1007/978-3-642-27769-6_1335-2
153. Rueckert S, Horák A. Archigregarines of the English Channel revisited: new molecular data on Selenidium species including early described and new species and the uncertainties of phylogenetic relationships. PLoS ONE. (2017) 12:e0187430. doi: 10.1371/journal.pone.0187430
154. Silva TJ, Soares EC, Casal G, Rocha S, Santos EL, Nascimento R, et al. Ultrastructure of phagocytes and oocysts of Nematopsis sp. (Apicomplexa, Porosporidae) infecting Crassostrea rhizophorae in Northeastern Brazil. Rev Bras Parasitol Vet. (2019) 28:97–104. doi: 10.1590/s1984-29612019010
155. Criado-Fornelio A, Verdú-Expósito C, Martin-Pérez T, Heredero-Bermejo I, Pérez-Serrano J, Guàrdia-Valle L, et al. A survey for gregarines (Protozoa: Apicomplexa) in arthropods in Spain. Parasitol Res. (2017) 116:99–110. doi: 10.1007/s00436-016-5266-0
156. Jiménez R, de Barniol L, Machuca M. Nematopsis marinus n. sp. a new septate gregarine from cultured penaeoid shrimp Litopenaeus vannamei (Boone), in Ecuador. Aquac Res. (2002) 33:231–40. doi: 10.1046/j.1355-557x.2002.00647.x
157. Bower SM, McGladdery SE, Price IM. Synopsis of infectious diseases and parasites of commercially exploited shellfish: Haliphthoros sp. (Fungus Disease) of Lobsters. Ann Rev Fish Dis. (1994) 4:1–3. doi: 10.1016/0959-8030(94)90028-0
158. Garvetto A, Nézan E, Badis Y, Bilien G, Arce P, Bresnan E, et al. Novel widespread marine oomycetes parasitising diatoms, including the toxic genus pseudo-nitzschia: genetic, morphological, and ecological characterisation. Front Microbiol. (2018) 9:2918. doi: 10.3389/fmicb.2018.02918
159. Tillmann U, Hesse KJ, Tillman A. Large-scale parasitic infection of diatoms in the Northfrisian Wadden Sea. J Sea Res. (1999) 42:255–61. doi: 10.1016/S1385-1101(99)00029-5
160. Gong Y, Patterson DJ Li Y, Hu Z, Sommerfeld M, Chen Y, et al. Vernalophrys algivore gen. nov, sp nov (Rhizaria: Cercozoa: Vampyrellida), a new algal predator isolated from outdoor mass culture of Scenedesmus dimorphus. Appl Environ Microbiol. (2015) 81:3900–13. doi: 10.1128/AEM.00160-15
161. Gutierrez MH, Jara AM, Pantoja S. Fungal parasites infect marine diatoms in the upwelling ecosystem of the Humboldt current system off central Chile. Environ Microbiol. (2016) 18:1646–53. doi: 10.1111/1462-2920.13257
162. Richards TA. Molecular diversity and distribution of marine fungi across 130 European environmental samples. Proc Biol Sci. (2015) 282:20152243. doi: 10.1098/rspb.2015.2243
163. De Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, et al. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science. (2015) 348:1261605. doi: 10.1126/science.1261605
164. Saadatnia G, Golkar M A. review on human toxoplasmosis. Scand J Infect Dis. (2012) 44:805–14. doi: 10.3109/00365548.2012.693197
165. Bouzid M, Hunter PR, Chalmers RM, Tyler KM. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev. (2013) 26:115–34. doi: 10.1128/CMR.00076-12
166. Schnittger L, Rodriguez AE, Florin-Christensen M, Morrison DA. Babesia: a world emerging. Infect Genet Evol. (2012) 12:1788–809. doi: 10.1016/j.meegid.2012.07.004
167. Castro-Hermida JA, García-Presedo I, González-Warleta M, Mezo M. Cryptosporidium and Giardia detection in water bodies of Galicia, Spain. Water Res. (2010) 44:5887–96. doi: 10.1016/j.watres.2010.07.010
168. Castro-Hermida JA, García-Presedo I, Almeida A, González-Warleta M, Correia Da Costa JM, Mezo M. Cryptosporidium spp. and Giardia duodenalis in two areas of Galicia (NW Spain). Sci Total Environ. (2011) 409:2451–9. doi: 10.1016/j.scitotenv.2011.03.010
Keywords: metabarcoding, eukaryote, high-throughput sequencing, eDNA, environment, pathogens
Citation: Ríos-Castro R, Romero A, Aranguren R, Pallavicini A, Banchi E, Novoa B and Figueras A (2021) High-Throughput Sequencing of Environmental DNA as a Tool for Monitoring Eukaryotic Communities and Potential Pathogens in a Coastal Upwelling Ecosystem. Front. Vet. Sci. 8:765606. doi: 10.3389/fvets.2021.765606
Received: 27 August 2021; Accepted: 08 October 2021;
Published: 03 November 2021.
Edited by:
Fabian Horn, Federal Office of Consumer Protection and Food Safety (BVL), GermanyCopyright © 2021 Ríos-Castro, Romero, Aranguren, Pallavicini, Banchi, Novoa and Figueras. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio Figueras, YW50b25pb2ZpZ3VlcmFzQGlpbS5jc2ljLmVz
 Raquel Ríos-Castro
Raquel Ríos-Castro Alejandro Romero
Alejandro Romero Raquel Aranguren
Raquel Aranguren Alberto Pallavicini
Alberto Pallavicini Elisa Banchi
Elisa Banchi Beatriz Novoa
Beatriz Novoa Antonio Figueras
Antonio Figueras