- 1Departamento de Medicina Preventiva Animal, Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, La Pintana, Chile
- 2Instituto de Patología Animal, Universidad Austral de Chile, Valdivia, Chile
- 3College of Veterinary Medicine, University of Minnesota, Saint Paul, MN, United States
- 4The Ithree Institute, University of Technology Sydney, Sydney, NSW, Australia
Porcine Astrovirus (PoAstV) causes mild diarrhea in young pigs and is considered an emerging virus in the swine industry worldwide. PoAstV has high genetic diversity and has been classified into five genetic lineages, PoAstV1–5. In Chile, only human astroviruses have been reported. This study aimed to determine the presence and genetic diversity of PoAstV circulating in intensive pig farms in Chile. Seventeen Chilean intensive swine farms from Valparaíso, Metropolitana, O'Higgins, Ñuble and Araucanía regions were sampled. A selection of oral fluid and fecal material samples from 1–80 days-old pigs were collected and analyzed using next-generation sequencing. The circulation of PoAstV was confirmed in all studied farms. We obtained complete or partial sequences of PoAstV-2 (n = 3), PoAstV-4 (n = 2), and PoAstV-5 (n = 7). In 15 out of 17 farms, we detected more than one lineage co-circulating. Phylogenetic analyses grouped the seven PoAstV-5 strains in a monophyletic cluster, closely related to the United States PoAstV-5 strains. The three PoAstV-2 were located into two separate sub-clusters. PoAstV-4 sequences are also grouped in two different clusters, all related to Japanese strains. Thus, our results indicate that PoAstV circulates in Chile with high frequency and diversity. However, the lack of reference sequences impairs local evolution patterns establishment and regional comparisons. This is the first contribution of PoAstV genomes in Latin America; more studies are needed to understand the diversity and impact of PoAstV on swine health.
Introduction
Astroviruses (AstVs) are emerging pathogens, belonging to members of the family Astroviridae. These viruses are divided into two genera: Mamastrovirus and Avastrovirus, which infect mammals and birds, respectively (1). AstV infections cause a wide range of clinical signs from gastroenteric (e.g., human, turkey, sheep, and pig) to neurologic (e.g., human, mink, cattle, sheep, and pig) disease (2–10). They are non-enveloped small viruses (30 nm) with a positive-sense single-stranded RNA genome of 6.4–7.9 kb (11). The genome contains three open reading frames (ORFs): the ORF1a and ORF1b, encoding non-structural proteins, and the ORF2, which encodes the capsid (11, 12).
In humans, AstV is the third most common cause of viral diarrhea in young children worldwide, with high seroprevalences as 94% in children of 6–9 years old (13, 14). Despite this high prevalence, due to the lack of cell culture systems and animal models, AstV are among the least studied enteric RNA viruses (12). However, advances in sequencing technologies have increased the availability of genome sequences and the identification of new strains. Typically, human AstV (HAstV) infections cause acute self-limiting mild diarrhea (12). Nevertheless, immunocompromised patients occasionally exhibit systemic spread, resulting in neurologic disease (15). In addition, a zoonotic potential of AstV is suspected but remains unclear (16–18). Genetic and evolutionary studies support the idea that both cross-species transmission and recombination events among AstV of human, porcine, and other species origin, may have occurred (11, 16).
Porcine astrovirus (PoAstV) has high genetic diversity, it is worldwide distributed, and it is commonly detected and shed by healthy and diarrheic swine (19). Five lineages of porcine AstV (PoAstV1–5) have been described by Laurin, Dastor (20), but only PoAstV-1 has been officially classified under the species Mamastrovirus 3 (21). In the US, pigs are commonly (13.9%) co-infected with multiple astrovirus strains (19). High prevalence and co-infections may create appropriate conditions for viral recombination and the potential emergence of viral variants that pose a higher risk of clinical disease. Recently, PoAstV has been linked to extraintestinal infections suggesting more complex pathogenesis and serious outcomes than previously thought (3, 4, 22).
In Chile, a recent study reported human AstV infections (14%) as a predominant cause of viral gastroenteritis in rural zones, in addition to norovirus (15%) and rotavirus (14%) (23). However, animal AstV has not been described in Chile. Considering the high prevalence and worldwide distribution of the AstV and PoAstV, the suspected zoonotic potential, and the lack of information regarding these viruses in a regional context, this study aims to determine the presence and genetic diversity of PoAstV circulating in Chilean intensive pig farms. These samples were taken in the context of other viral disease surveillance programs.
Materials and Methods
Sample Collection
During influenza virus and rotavirus surveillance and diagnosis programs, we collected oral fluids and fecal samples from 1–80 days-old pigs in 17 intensive pig farms from mainland Chile in 2015 and 2017. The sampled farms are located in an area that concentrates 95% of the national intensive pig production (Valparaíso, Metropolitana, Libertador General Bernardo O'Higgins, Maule, Ñuble, and Araucanía Regions), which represents approximately 50% of the pig inventory in Chile (24, 25). Each fecal sample corresponds to a pool of 5 diarrheic feces, which were collected using nylon gloves, deposited in sterile 50 mL tubes with 20 mL of viral transport media (Minimum Essential Medium, 1X Trypsin TPCK, 2% bovine serum albumin, and 1% antifungal antibiotic solution), and then centrifuged at 7,000 rpm for 5 min. Oral fluids were collected by groups of 20–30 healthy pigs kept in pens. Briefly, a 16 mm braided cotton rope was hung in each pen for about 30 min. The ropes were deposited inside plastic ziplock bags and squeezed to obtain the oral fluid and deposited into 50 mL tubes. All samples were kept at −20°C until processing. One sample per farm (n = 17) was selected for next-generation sequencing. The criteria to select the samples included the location, geographic distance between farms and detection of other pathogens such as rotavirus and influenza.
Viral RNA Extraction and Whole-Genome Sequencing
The RNA extraction was carried out using the Chomczynski-phenol solution (Winkler, BM-1,755, Chile) following the manufacturer's recommendations. The Next-generation sequencing (NGS) was performed at the Molecular Diagnostic Development Laboratory at the Veterinary Diagnostic Laboratory of the University of Minnesota (MVDL, UMN), USA, using the Illumina MiSeq platform. Library pre-paration was performed using the SMARTer Stranded Total RNA-Seq Kit v2–Pico Input Mammalian (Takara bio, USA). De novo assembly of the reads was carried out using an automated pipeline that identifies viral reads using DIAMOND protein alignment and the Swissprot Uniref90 database. The viral reads are then grouped by the lowest common ancestor and assembled using SPAdes and subsequently, the contigs are joined using an Advanced Genome Aligner (http://www.genomedetective.com/app/typingtool/virus/). Complementary, the assembly using PoAstV reference sequences was performed using Geneious Prime® 2021.2.2.
Phylogeny
Complete or near to complete Chilean PoAstV genomes, with >77% of coverage, were used for phylogenetic analysis (Table 1). These sequences were compared with all complete or near complete PoAstV genome available in GenBank database. We used the lineage classification described by Lee et al. (26). The final data set comprised 93 Astrovirus genome sequences that were aligned using MUSCLE (27). The phylogeny was constructed using RAxML with the GTR+G+I substitution model and 1,000 bootstrap replications in Geneious Prime® 2021.2.2. Additionally, the phylogeny was constructed for the ORF2 region with the same methodology, and p-distances at the nucleotide and amino acid level of the ORF2 sequences were estimated using MEGA X (28).
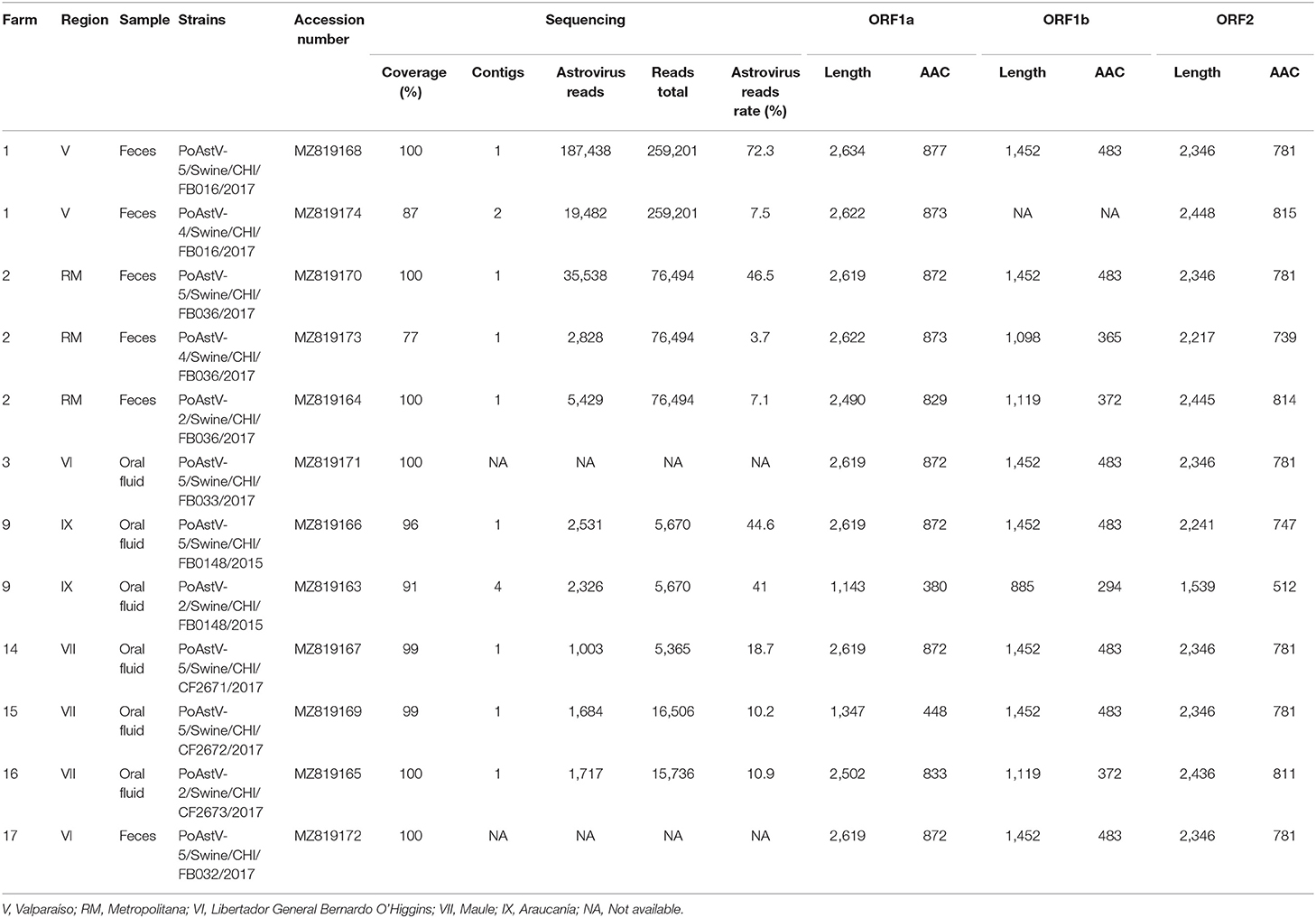
Table 1. Summary of porcine astroviruses whole-genome sequencing results obtained from intensive farms in Chile.
Results and Discussion
All samples were successfully sequenced by Illumina. The most consistent virus family found in the samples was Astroviridae. Additionally, reads that belong to other families, such as Caliciviridae, Parvoviridae, and Reoviridae, were found at lower rates and were not considered for further analysis as they are beyond the aim of this study. Porcine astrovirus reads were observed in all samples/farms included, confirming its ubiquity in Chilean swine intensive production. Overall results of PoAstV identified 229,496 reads for PoAstV-5, 33,917 for PoAstV-4 and 31,751 for PoAstV-2. Only 10 reads were classified as PoAstV3, however, the limited reads do not provide sufficient evidence to confirm the presence of this lineage.
PoAstV-4 was identified in 16 out of 17 farms, PoAstV-2 in 15 and PoAstV-5 in 13 farms (Supplementary Table 1). These results are in agreement with estimations made in the US and some European and Asian countries, where the most prevalent lineage is PoAstV-4, mainly followed by PoAstV-2 (19, 29–33). Contrary to the situation in China, where the most widely distributed strain is presumably PoAstV-2 (34).
We identified at least two different PoAstV lineages co-circulating in 15 out of 17 farms (Supplementary Table 1). Thus, the Chilean swine exhibits conditions for PoAstV recombination events. The circulation of multiple PoAstV strains in the same farm has been reported previously in China, Denmark, Slovakia, Thailand, and the USA (31, 35–37). Co-infection of different lineages in the same individual has also been reported (19, 38).
Eleven complete or near-to-complete genomes of PoAstV were obtained, which were used for the phylogenetic analysis (Table 1). The genomes were obtained from both fecal and oral fluids samples and were recovered from 8 different farms. Figure 1 shows the geographic distribution of the sequences obtained (Figure 1). Interestingly, from farm two, it was possible to obtain the genome of PoAst-2, 4, and 5.
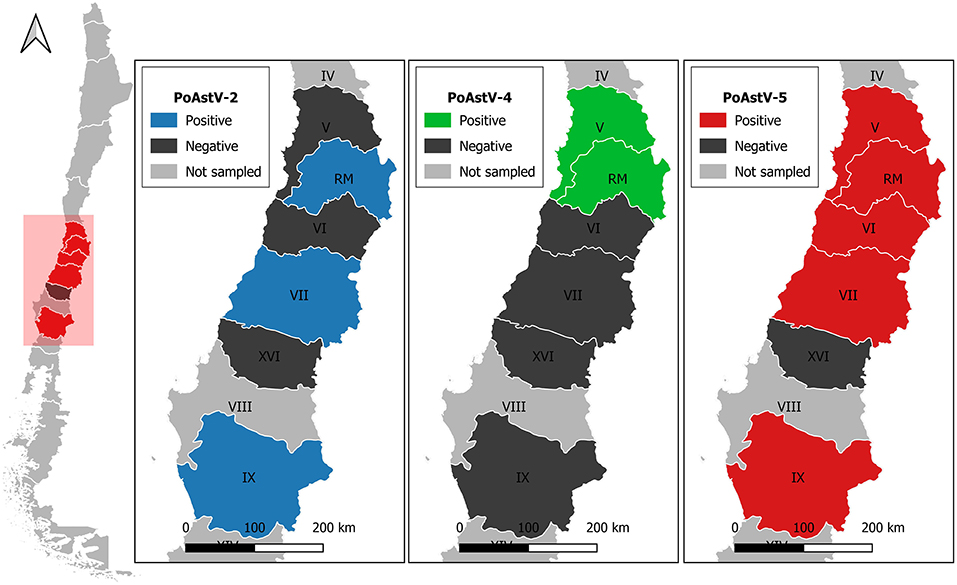
Figure 1. PoAstV strains with >77% of sequencing coverage were detected in Porcine intensive farms from different regions of Chile. Regions are labeled as V, Valparaíso; RM, Metropolitana; VI, Libertador General Bernardo O'Higgins; VII, Maule; XVI, Ñuble; IX, Araucanía.
The phylogenetic analysis grouped the Chilean strains into 5 (Figure 2 and Supplementary Figure 1). The seven PoAstV-5 strains are grouped into one monophyletic cluster (97.4% pairwise identity), closely related to strains detected in the USA. Interestingly, the PoAstV-5 were obtained from seven different farms distributed in five geographic regions. The three PoAstV-2 genomes were detected from three different farms in different regions and grouped into two separate sub-clusters. The PoAstV-2/Swine/CHI/FB036/2017 (Metropolitan region) and PoAstV-2/Swine/CHI /CF2673/2017 (Maule region) genomes formed one sub-cluster, while PoAstV-2 /Swine/CHI/FB0148/2017 (Araucania region) grouped with sequences from USA and Japan. Finally, the two PoAstV-4 from two different farms and regions (Metropolitan and Valparaiso region) were phylogenetically distant also related to Japanese strains. The phylogeny demonstrates a genetic relationship between Chilean PoAstV-5 strains, suggesting the same origin for those strains. On the contrary, PoAstV-2 and PoAstV-4 results indicate more diversity even with fewer sequences (For details on genetic distances see Supplementary Tables 2, 3).
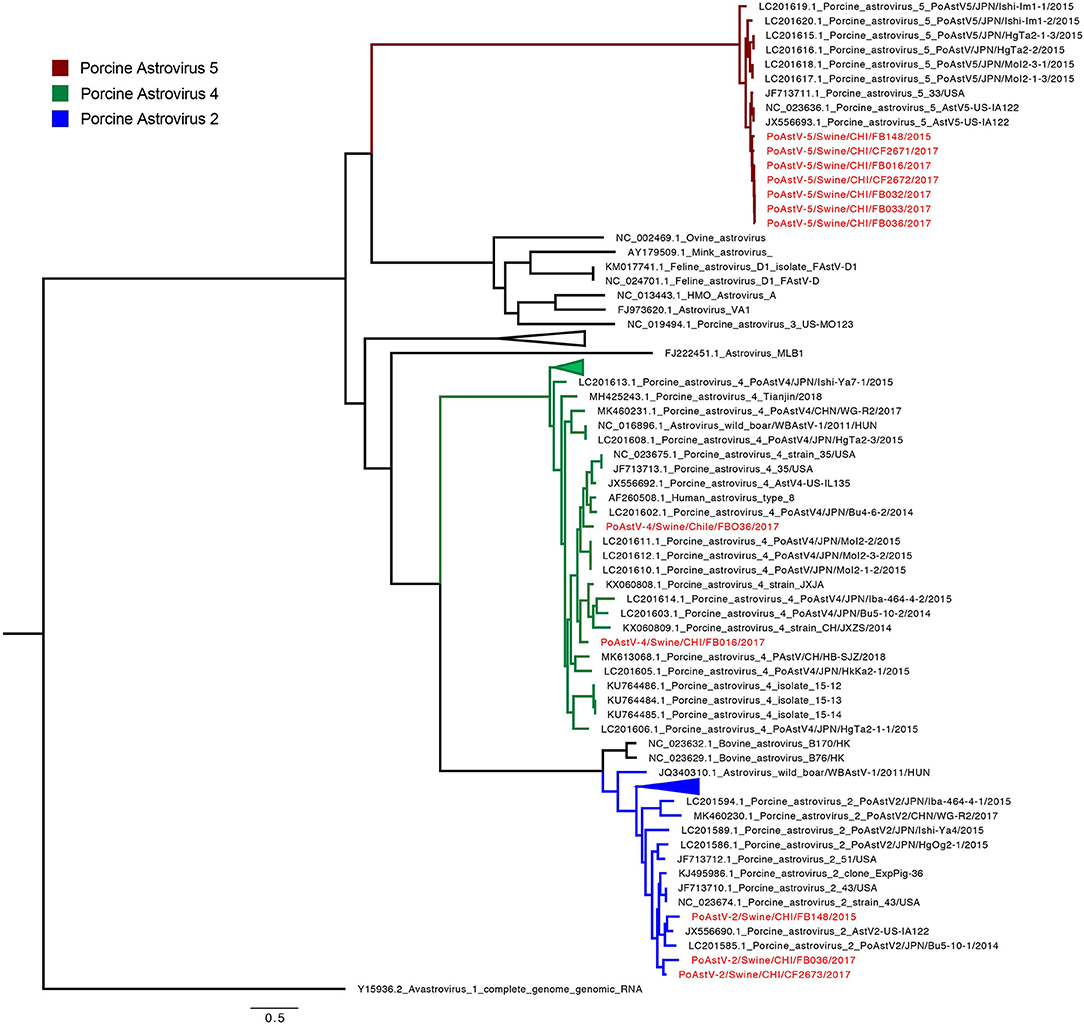
Figure 2. Phylogenetic tree of PoAstV by using the complete genome. The final dataset included 93 genomes. Chilean sequences are highlighted in red. Clusters by species are highlighted in colors: PoAstV-5 (Red), PoAstV-4 (Green) and PoAstV-2 (Blue).
Due to the limited number of sequences obtained in this study and the scarce of sequences in GenBank database, conclusions about the origin of the viral strains cannot be elucidated. Indeed, most of the available sequences in GenBank database are from the USA, Japan, and China. Other limitations of the phylogenetic analysis are the incomplete genome coverage in several samples (Table 1) and the potential errors derived from sequencing methods, as these may alter the phylogenetic tree estimation.
This is the first report characterizing the PoAstV sequences circulating in Chile.
This result represents, in turn, the first PoAstV genomes from swine in Latin America. PoAstV studies in Latin America are very scarce, and only two have been published. One study identified PoAstV in healthy pigs from a farm in Brazil (39), and another study conducted in Colombia, which obtained partial PoAstV sequences from diarrheic piglets and humans (40). To date, most of the PoAstV sequences available in GenBank database were obtained in the Northern hemisphere.
Our results support the detection of PoAstV in the Chilean swine population, similar to other observations worldwide. Further studies are needed to understand the relevance of PoAstV to swine health and the evolution and spread of PoAstV locally and globally.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committees of the Universidad de Chile, protocol number 02–2016. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
NA, GR-T, and VN: study design and conceptualization. SM and VN: funding and resources. CF and JM: samples collection and processing. CF, JM, and SM: performed the assays. CF, NA, CV, BB, and VN: data analysis. CF, NA, GR-T, and VN: wrote the paper. All authors critically evaluated the paper. All authors contributed to the article and approved the submitted version.
Funding
Animal Virology Laboratory, Faculty of Veterinary and Animal Sciences, Universidad de Chile and Programa Fondecyt 11170877 and 1211517.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the staff of Animal Virology Lab, Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile for all their support in sampling processing, specially to Dr. Felipe Berrios and Valentina Valdes. This study was partially funded by the Animal Virology Laboratory, Faculty of Veterinary and Animal Sciences, Universidad de Chile, and Programa Fondecyt N° 11170877 and N° 1211517 to VN.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.764837/full#supplementary-material
References
1. Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. (2017) 46:D708–D17. doi: 10.1093/nar/gkx932
2. Opriessnig T, Xiao C-T, Halbur PG. Porcine astrovirus type 5-associated enteritis in pigs. J Comp Pathol. (2020) 181:38–46. doi: 10.1016/j.jcpa.2020.09.014
3. Ulloa JC, Olarte-Aponte AM, Ospina JC, Rincon MA. Experimental infection of conventional newly-weaned piglets with porcine astrovirus. Acta Virol. (2019) 63:96–102. doi: 10.4149/av_2019_112
4. Boros Á, Albert M, Pankovics P, Bíró H, Pesavento P, Phan TG, et al. Outbreaks of neuroinvasive astrovirus associated with encephalomyelitis, weakness, and paralysis among weaned pigs, hungary. Emerg Infect Dis J. (2017) 23:1982. doi: 10.3201/eid2312.170804
5. Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, et al. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis. (2015) 60:881–8. doi: 10.1093/cid/ciu940
6. Koci MD, Moser LA, Kelley LA, Larsen D, Brown CC, Schultz-Cherry S. Astrovirus induces diarrhea in the absence of inflammation and cell death. J Virol. (2003) 77:11798–808. doi: 10.1128/JVI.77.21.11798-11808.2003
7. Pfaff F, Schlottau K, Scholes S, Courtenay A, Hoffmann B, Höper D, et al. A novel astrovirus associated with encephalitis and ganglionitis in domestic sheep. Transbound Emerg Dis. (2017) 64:677–82. doi: 10.1111/tbed.12623
8. Snodgrass DR, Angus KW, Gray EW, Menzies JD, Paul G. Pathogenesis of diarrhoea caused by astrovirus infections in lambs. Arch Virol. (1979) 60:217–26. doi: 10.1007/BF01317493
9. Englund L, Chriél M, Dietz HH, Hedlund KO. Astrovirus epidemiologically linked to pre-weaning diarrhoea in mink. Vet Microbiol. (2002) 85:1–11. doi: 10.1016/S0378-1135(01)00472-2
10. Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, et al. Divergent astrovirus associated with neurologic disease in cattle. Emerg Infect Dis. (2013) 19:1385–92. doi: 10.3201/eid1909.130682
11. Wohlgemuth N, Honce R, Schultz-Cherry S. Astrovirus evolution and emergence. Infect Genet Evol. (2019) 69:30–7. doi: 10.1016/j.meegid.2019.01.009
12. Cortez V, Meliopoulos VA, Karlsson EA, Hargest V, Johnson C, Schultz-Cherry S. Astrovirus biology and pathogenesis. Annu Rev Virol. (2017) 4:327–48. doi: 10.1146/annurev-virology-101416-041742
13. Mitchell DK, Matson DO, Cubitt WD, Jackson LJ, Willcocks MM, Pickering LK, et al. Prevalence of antibodies to astrovirus types 1 and 3 in children and adolescents in Norfolk, Virginia. Pediatr Infect Dis J. (1999) 18:249–54. doi: 10.1097/00006454-199903000-00008
14. Olortegui MP, Rouhani S, Yori PP, Salas MS, Trigoso DR, Mondal D, et al. Astrovirus infection and diarrhea in 8 countries. Pediatrics. (2018) 141:e20171326. doi: 10.1542/peds.2017-1326
15. Vu D-L, Bosch A, Pintó RM, Guix S. Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses. (2017) 9:33. doi: 10.3390/v9020033
16. De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. Astrovirus infections in humans and animals - molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol. (2011) 11:1529–44. doi: 10.1016/j.meegid.2011.07.024
17. Bosch A, Pintó RM, Guix S. Human astroviruses. Clin Microbiol Rev. (2014) 27:1048–74. doi: 10.1128/CMR.00013-14
18. Japhet MO, Famurewa O, Adesina OA, Opaleye OO, Wang B, Höhne M, et al. Viral gastroenteritis among children of 0-5 years in Nigeria: characterization of the first nigerian aichivirus, recombinant noroviruses and detection of a zoonotic astrovirus. J Clin Virol. (2019) 111:4–11. doi: 10.1016/j.jcv.2018.12.004
19. Xiao CT, Giménez-Lirola LG, Gerber PF, Jiang YH, Halbur PG, Opriessnig T. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. J Gen Virol. (2013) 94:570–82. doi: 10.1099/vir.0.048744-0
20. Laurin MA, Dastor M. L'Homme Y. Detection and genetic characterization of a novel pig astrovirus: relationship to other astroviruses. Arch Virol. (2011) 156:2095–9. doi: 10.1007/s00705-011-1088-7
21. Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Dempsey DM, et al. Changes to virus taxonomy and the statutes ratified by the international committee on taxonomy of viruses. Arch Virol. (2020) 165:2737–48. doi: 10.1007/s00705-020-04752-x
22. Rawal G, Matias Ferreyra FR, Macedo NK, Bradner LM, Harmon K, Mueller A, et al. Detection and cellular tropism of porcine astrovirus type 3 on breeding farms. Viruses. (2019) 11:1051. doi: 10.3390/v11111051
23. Lucero Y, Lagomarcino AJ, Espinoza M, Kawakami N, Mamani N, Huerta N, et al. Norovirus compared to other relevant etiologies of acute gastroenteritis among families from a semirural county in Chile. Int J Infect Dis. (2020) 101:353–60. doi: 10.1016/j.ijid.2020.10.013
24. Tapia R, García V, Mena J, Bucarey S, Medina RA, Neira V. Infection of novel reassortant H1N2 and H3N2 swine influenza A viruses in the guinea pig model. Vet Res. (2018) 49:73. doi: 10.1186/s13567-018-0572-4
25. Tapia R, Torremorell M, Culhane M, Medina RA, Neira V. Antigenic characterization of novel H1 influenza A viruses in swine. Sci Rep. (2020) 10:4510. doi: 10.1038/s41598-020-61315-5
26. Lee S, Jang G, Lee C. Complete genome sequence of a porcine astrovirus from South Korea. Arch Virol. (2015) 160:1819–21. doi: 10.1007/s00705-015-2436-9
27. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7. doi: 10.1093/nar/gkh340
28. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
29. Lee M-H, Jeoung H-Y, Park H-R, Lim J-A, Song J-Y, An D-J. Phylogenetic analysis of porcine astrovirus in domestic pigs and wild boars in South Korea. Virus Genes. (2013) 46:175–81. doi: 10.1007/s11262-012-0816-8
30. Zhou W, Ullman K, Chowdry V, Reining M, Benyeda Z, Baule C, et al. Molecular investigations on the prevalence and viral load of enteric viruses in pigs from five European countries. Vet Microbiol. (2016) 182:75–81. doi: 10.1016/j.vetmic.2015.10.019
31. Kumthip K, Khamrin P, Saikruang W, Kongkaew A, Vachirachewin R, Ushijima H, et al. Detection and genetic characterization of porcine astroviruses in piglets with and without diarrhea in Thailand. Arch Virol. (2018) 163:1823–9. doi: 10.1007/s00705-018-3806-x
32. Ito M, Kuroda M, Masuda T, Akagami M, Haga K, Tsuchiaka S, et al. Whole genome analysis of porcine astroviruses detected in Japanese pigs reveals genetic diversity and possible intra-genotypic recombination. Infect Genet Evol. (2017) 50:38–48. doi: 10.1016/j.meegid.2017.02.008
33. Kattoor JJ, Malik YS, Saurabh S, Sircar S, Vinodhkumar OR, Bora DP. First report and genetic characterization of porcine astroviruses of lineage 4 and 2 in diarrhoeic pigs in India. Transbound Emerg Dis. (2019) 66:47–53. doi: 10.1111/tbed.13058
34. Su M, Qi S, Yang D, Guo D, Yin B, Sun D. Coinfection and genetic characterization of porcine astrovirus in diarrheic piglets in China From 2015 to 2018. Front Vet Sci. (2020) 7:462. doi: 10.3389/fvets.2020.00462
35. Li JS, Li MZ, Zheng LS, Liu N, Li DD, Duan ZJ. Identification and genetic characterization of two porcine astroviruses from domestic piglets in China. Arch Virol. (2015) 160:3079–84. doi: 10.1007/s00705-015-2569-x
36. Goecke NB, Hjulsager CK, Kongsted H, Boye M, Rasmussen S, Granberg F, et al. No evidence of enteric viral involvement in the new neonatal porcine diarrhoea syndrome in Danish pigs. BMC Vet Res. (2017) 13:315. doi: 10.1186/s12917-017-1239-5
37. Salamunova S, Jackova A, Mandelik R, Novotny J, Vlasakova M, Vilcek S. Molecular detection of enteric viruses and the genetic characterization of porcine astroviruses and sapoviruses in domestic pigs from Slovakian farms. BMC Vet Res. (2018) 14:313. doi: 10.1186/s12917-018-1640-8
38. Lv SL, Zhang HH, Li JY, Hu WQ, Song YT, Opriessnig T, et al. High genetic diversity and recombination events of porcine astrovirus strains identified from ill and asymptomatic pigs in 2017, Hunan Province, China. Virus Genes. (2019) 55:673–81. doi: 10.1007/s11262-019-01692-w
39. Hammerschmitt ME, de Almeida PR, de Cecco BS, Lorenzett MP, Schwertz CI, da Cruz RAS, et al. Swine polioencephalomyelitis in Brazil: identification of Teschovirus A, Sapelovirus A, and Enterovirus G in a farm from Southern Brazil. Braz J Microbiol. (2021). doi: 10.1007/s42770-021-00509-z
Keywords: porcine astrovirus, poAstV, swine intensive farm, next-generating sequencing, phylogenetic analysis
Citation: Flores C, Ariyama N, Bennett B, Mena J, Verdugo C, Mor S, Brito B, Ramírez-Toloza G and Neira V (2021) Case Report: First Report and Phylogenetic Analysis of Porcine Astroviruses in Chile. Front. Vet. Sci. 8:764837. doi: 10.3389/fvets.2021.764837
Received: 26 August 2021; Accepted: 25 October 2021;
Published: 25 November 2021.
Edited by:
Torsten Seuberlich, University of Bern, SwitzerlandReviewed by:
Shao-Lun Zhai, Guangdong Academy of Agricultural Sciences, ChinaBen Hause, South Dakota State University, United States
Bailey Arruda, Iowa State University, United States
Copyright © 2021 Flores, Ariyama, Bennett, Mena, Verdugo, Mor, Brito, Ramírez-Toloza and Neira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Victor Neira, dmljdG9ybmVpcmFAdS51Y2hpbGUuY2w=; Galia Ramírez-Toloza, Z2FsaWFyYW1AdWNoaWxlLmNs
†These authors have contributed equally to this work and share first authorship
 Carlos Flores
Carlos Flores Naomi Ariyama
Naomi Ariyama Benjamín Bennett1
Benjamín Bennett1 Juan Mena
Juan Mena Claudio Verdugo
Claudio Verdugo Sunil Mor
Sunil Mor Barbara Brito
Barbara Brito Galia Ramírez-Toloza
Galia Ramírez-Toloza Victor Neira
Victor Neira