
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 22 November 2021
Sec. Parasitology
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.764763
This article is part of the Research TopicCurrent Knowledge on Pathogenic and Endosymbiotic Tick-Borne BacteriaView all 13 articles
 Ratree Takhampunya1*
Ratree Takhampunya1* Jira Sakolvaree1
Jira Sakolvaree1 Nitima Chanarat1
Nitima Chanarat1 Nittayaphon Youngdech1
Nittayaphon Youngdech1 Kritsawan Phonjatturas1
Kritsawan Phonjatturas1 Sommai Promsathaporn1
Sommai Promsathaporn1 Bousaraporn Tippayachai1
Bousaraporn Tippayachai1 Wirunya Tachavarong2
Wirunya Tachavarong2 Kanchit Srinoppawan3
Kanchit Srinoppawan3 Betty K. Poole-Smith1
Betty K. Poole-Smith1 P. Wesley McCardle1
P. Wesley McCardle1 Suwanna Chaorattanakawee2
Suwanna Chaorattanakawee2Ticks are known vectors for a variety of pathogens including bacteria, viruses, fungi, and parasites. In this study, bacterial communities were investigated in active life stages of three tick genera (Haemaphysalis, Dermacentor, and Amblyomma) collected from Khao Yai National Park in Thailand. Four hundred and thirty-three questing ticks were selected for pathogen detection individually using real-time PCR assays, and 58 of these were subjected to further metagenomics analysis. A total of 62 ticks were found to be infected with pathogenic bacteria, for a 14.3% prevalence rate, with Amblyomma spp. exhibiting the highest infection rate (20.5%), followed by Haemaphysalis spp. (14.5%) and Dermacentor spp. (8.6%). Rickettsia spp. were the most prevalent bacteria (7.9%) found, followed by Ehrlichia spp. (3.2%), and Anaplasma spp. and Borrelia spp. each with a similar prevalence of 1.6%. Co-infection between pathogenic bacteria was only detected in three Haemaphysalis females, and all co-infections were between Rickettsia spp. and Anaplasmataceae (Ehrlichia spp. or Anaplasma spp.), accounting for 4.6% of infected ticks or 0.7% of all examined questing ticks. The prevalence of the Coxiella-like endosymbiont was also investigated. Of ticks tested, 65.8% were positive for the Coxiella-like endosymbiont, with the highest infection rate in nymphs (86.7%), followed by females (83.4%). Among tick genera, Haemaphysalis exhibited the highest prevalence of infection with the Coxiella-like endosymbiont. Ticks harboring the Coxiella-like endosymbiont were more likely to be infected with Ehrlichia spp. or Rickettsia spp. than those without, with statistical significance for Ehrlichia spp. infection in particular (p-values = 0.003 and 0.917 for Ehrlichia spp. and Rickettsia spp., respectively). Profiling the bacterial community in ticks using metagenomics revealed distinct, predominant bacterial taxa in tick genera. Alpha and beta diversities analyses showed that the bacterial community diversity and composition in Haemaphysalis spp. was significantly different from Amblyomma spp. However, when examining bacterial diversity among tick life stages (larva, nymph, and adult) in Haemaphysalis spp., no significant difference among life stages was detected. These results provide valuable information on the bacterial community composition and co-infection rates in questing ticks in Thailand, with implications for animal and human health.
Ticks are recognized as a medically important group of arthropods that transmit a number of diseases to humans (1). Different tick species favor different habitats, which ultimately defines their geographical distribution and thus the risk areas for human or animal infections. Several pathogens are known to be carried by either hard ticks (Ixodidae) or soft ticks (Argasidae), including a wide range of viruses, bacteria, fungi, and protozoa (2–6).
In Southeast Asia, 97 tick species (5 in Argasidae and 93 in Ixodidae) have been described (7). In Thailand, four species within Argasidae (2 Argas and 2 Ornithodoros) and 58 species within Ixodidae have been recorded. Of the hard ticks in Thailand, there are 25 species within genus Haemaphysalis, 10 Ixodes, 13 Amblyomma, 5 Dermacentor, 4 Rhipicephalus, and Nosomma monstrosum. Among tick species found in Thailand, Amblyomma spp. and Dermacentor spp. were associated with human otoacariasis, especially Amblyomma testudinarium (8) and Dermacentor steini (9). However, Haemaphysalis spp. and other tick genera are occasionally found on humans as well. Rickettsia spp. are the main tick-borne pathogens causing human infections in Thailand (10–12) with scattered reports of other tick-borne disease (TBD) in human and animals such as Q fever (13, 14) and anaplasmosis in companion pets and animals (15–17).
Ticks also harbor many non-pathogenic organisms. Bacterial endosymbionts have been recognized as important microorganisms required for tick fitness, especially with regard to regulating host reproduction and immunity (18, 19). Some studies suggest that these symbionts have a potential role in providing key vitamins absent in the bloodmeal (20, 21). Others reported that they might have an important role in facilitating pathogen colonization in the gut as in the case in Ixodes scapularis ticks, in which alteration of the symbiont abundance resulted in decreased Borrelia burgdorferi colonization (22). Likewise, the level of Anaplasma marginale acquisition was lower in Dermacentor andersoni when their microbiome was altered by antibiotic treatment leading to an increase in proportion and quantity of Rickettsia bellii in the microbiome (23). Moreover, the prevalence and transovarial transmission of bacterial endosymbionts occurs at a high rate, suggesting that they might have an obligate relationship with the host (24, 25). In addition to the bacterial microbiota in ticks, other microorganisms are as important and abundant. For example, several studies reported that two groups of bunyaviruses (South Bay virus and Phlebovirus) were commonly associated with Ixodes spp. ticks in America and Europe (26–28) and were more abundant than bacteria or eukaryote counterparts (29).
It is thought that because ticks harbor a diverse range of microorganisms, co-infection or co-occurrence of bacteria, parasites, and viruses in ticks is possible (30). Co-infection may lead to increased disease severity as it complicates disease diagnosis and treatment (31–34). Co-infections between B. burgdorferi and Anaplasma phagocytophilum are widely recognized and reported (35). Other co-infections among different types of microorganisms have also been reported, such as between B. burgdorferi and South Bay virus (SBV), Babesia microti, and B. burgdorferi, as well as between a novel filarial worm (Onchocercidae sp. ex. Ixodes scapularis) and Wolbachia spp (29).
In this study, we used both conventional and high-throughput sequencing methods to study bacterial pathogen co-infections and pathogen association with bacterial endosymbionts in questing ticks collected in Khao Yai National Park. Metagenomics were used to determine the bacterial communities in individual ticks and conventional methods (real-time PCR, PCR, and Sanger sequencing) were used to detect pathogenic bacteria: Rickettsia spp., Anaplasma spp., Ehrlichia spp., Borrelia spp., Coxiella burnetii, and pathogenic Francisella spp. in Amblyomma spp., Dermacentor spp., and Haemaphysalis spp. ticks individually. All positive samples underwent DNA sequencing to identify pathogens to the species level. Co-infections between bacteria and the association between pathogenic bacteria and the Coxiella-like endosymbiont were investigated.
Questing ticks were collected by dragging a 1-m2 cotton cloth over vegetation in four tourist attraction sites in Khoa Yai National Park (14°26'19.5“N 101°22'20.1”E). Sampling was conducted by six people at each site for 1 h. Ticks were collected in one trip in November 2020 and immediately preserved in 90% ethanol before transporting to the laboratory for further processing. All sampling procedures and experimental manipulations were reviewed and approved as part of the animal collection protocol entitled “Surveillance of Tick- and Flea-Borne Diseases of Public Health Importance” (PN# 21-01). The project was also approved by Mahidol University – Institute animal care and use committee (MU-IACUC 2019/3). Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations related to animals and experiments involving animals, and adhered to principles outlined in the “Guide for the Care and Use of Laboratory Animals,” NRC Publication, 2011 edition.
Questing ticks (unfed ticks) were morphologically identified using taxonomic keys (36, 37). Each tick was vortexed for 1 min in 3% sodium hypochlorite and then transferred to a new tube and vortexed for 1 min in 70% alcohol followed by three washes in sterile PBS in the same manner. Ticks were air dried for 10 min on Whatman® filter paper (Sigma-Aldrich, Saint Louis, MO, USA) before DNA extraction. Whole ticks in 250 μl of ATL buffer (lysis buffer, component of the extraction kit) were punctured with a fine tip under a stereomicroscope to release the tissue from the hard chitin exoskeleton prior to adding 2 mg/ml of Proteinase K solution. Samples were then incubated at 55°C overnight. A total volume of 250 μl of homogenized solution was then used for DNA extraction on the QIAsymphony® SP instrument with QIAsymphony® DSP DNA Mini Kit using Tissue LC 200 DSP protocol (Qiagen, Hombrechtikon, Switzerland). The DNA was eluted in 50 μl of ATE buffer (elution buffer, component of extraction kit) and stored at−20°C until use. Ultrapure DNA/RNA-free distilled water as well as PBS buffer used for tick surface sterilization were also included as an extraction control.
Following DNA extraction, the bacterial-specific 16S rDNA (V3–V4, a 550-bp fragment) was amplified as previously described (15). Negative control PCR reactions were included in every experimental run using Ultrapure DNA/RNA-free distilled water in place of DNA template. PCR reactions were also performed with eluates from mock DNA extractions as well as from PBS buffer used for tick surface sterilization. PCR product was cleaned using AMPure magnetic bead-based purification system (Beckman Coulter, UK) following the manufacturer's instructions. Purified PCR products were eluted and quantified using the Qubit dsDNA HS Assay Kit (Invitrogen Life Technologies, MA).
The library was prepared with dual indices and Illumina sequencing adapters attached to purified PCR products using the Nextera XT Index Kit following the manufacturer's protocol (Illumina Inc., San Diego, CA). For index control reaction, a combination of index primers that were not used with samples was also included with PCR grade water as template. The number of reads recovered from these particular index combinations were used to filter the cross-contaminations between indexed PCR primers and to identify errors in an Illumina sample sheet. Libraries were cleaned using Agencourt AMPure XP beads. The purity of the libraries was checked on the QIAxcel Advanced System (Qiagen) with a QIAxcel DNA High Resolution Cartridge. Purified amplicon libraries were quantified using the Qubit dsDNA HS Assay Kit (Invitrogen). DNA concentration was calculated and normalized to reach 4.0 nM for each library. Five microliters of DNA from each library was pooled for a NGS run. Pooled libraries were denatured and diluted to a final concentration of 8 pM with a 10% PhiX (Illumina) control. Sequencing was performed using the MiSeq Reagent Kit V3 on the Illumina MiSeq System.
The sequence reads generated by the 16S rRNA on MiSeq sequencers were processed on CLC Genomics workbench v 12.0.3 (Qiagen, Aarhus A/S, http://www.clcbio.com). High-throughput sequences were imported into CLC Genomics Workbench according to quality scores of Illumina pipeline 1.8. In order to achieve the highest quality sequences for clustering, paired reads were merged in CLC microbial genomics module v 4.8 using default settings (mismatch cost = 1; minimum score = 40; gap cost = 4 and maximum unaligned end mismatch = 5). Primer sequences were trimmed from merged reads using parameters (trim using quality scores = 0.01, trim ambiguous nucleotides = 2, and discard read length shorter than 150 bp). Samples were removed from analysis if the number of reads was <100 or <25% from the median (the median number of reads across all samples). Chimeric sequences were detected and removed. Only filtered and merged sequences were clustered into operational taxonomic units (OTUs) according to a threshold of 97% sequence identity. All such processes were performed using CLC microbial genomics module v 4.8. Reference OTU data used in the present study were downloaded from the Greengenes database V13.8 (38). OTUs with combined abundance <10 reads were removed from the downstream analysis. Alpha diversity estimates (Observed OTUs, Simpson's index, and Shannon entropy) were analyzed on the quality-filtered OTU table at the genus level using CLC Microbial Genomics Module v 4.8. Beta diversity analysis for microbiome compositional difference between groups was calculated using a distance-based non-parametric test, the generalized UniFrac distances (39). The statistical significance of microbiome compositional difference between groups (tick genera, tick developmental stages, Francisella persica infection, Coxiella-like endosymbiont infection, and Rickettsia spp. infection statuses) was then compared using PERMANOVA using 99,999 permutations of the distance values. All analyses mentioned here were performed with CLC Microbial Genomics Module v 4.8.
Real-time PCR and PCR assays were performed on 433 individual ticks for the detection of bacteria (Rickettsia spp., Borrelia spp., Anaplasma spp., Ehrlichia spp., and Coxiella-like endosymbiont) and the taxonomic species assignment. Other potential pathogenic bacteria (Francisella spp. and Coxiella spp.) detected by NGS analysis (read count > 1) were also confirmed by real-time PCR and PCR assays. Detailed methods for assays and target gene(s) for selected pathogens are provided as online Supplementary data (Supplementary Table 1). For all real-time PCR, the reaction consisted of 1X Platinum quantitative PCR SuperMix-UDG (Invitrogen) using standard real-time PCR conditions with primer/probe concentrations and annealing temperatures as indicated in Supplementary Table 1. For conventional PCR, the assay was carried out in a 50-μl reaction volume containing 0.5 U of iProof High-Fidelity DNA Polymerase, 200 μM dNTPs, MgCl2, and primer concentration as indicated (Supplementary Table 1). The PCR conditions consisted of 98°C for 3 min, followed by 40 cycles of 98°C for 10 s, annealing temperature for 30 s (indicated in Supplementary Table 1 for each pathogen), and 72°C for 45 s.
PCR amplicons were purified using the QIAquick® PCR Purification Kit (Qiagen) according to the manufacturer's instructions. The PCR products were cycle-sequenced using an ABI BigDye™ Terminator v3.1 Cycle Sequencing Kit, ethanol precipitated, and run on a SeqStudio Genetic Analyzer (Applied Biosystems ThermoFisher, Thailand). Sequences of each sample and pathogen were assembled using Sequencher™ ver. 5.4.6 (GeneCodes Corp., Ann Arbor, MI). The pathogen sequences were aligned with reference sequences retrieved from the GenBank database using the MUSCLE codon alignment algorithm (40). A maximum likelihood phylogenetic tree was then constructed from bacterial target genes (Supplementary Table 1) using the best fit model of nucleotide substitution with bootstrapping (1,000 replicates) in MEGA 6 (41).
Differences in alpha diversity indices of the bacterial community composition, based on metagenomics data, were determined by Mann–Whitney U test (between two groups) or Kruskal–Wallis test (across all groups) and the critical range (p < 0.05) was determined. Statistical analyses (two-way ANOVA tests and mean, 95% confidence interval) and scatter plots were performed using GraphPad Prism version 5.04 for Windows (GraphPad Software, San Diego, CA. www.graphpad.com). Some graphical illustrations presented in this study as well as the Chi-square independence test and Fisher's exact test were performed in the R environment for statistical computing (42, 43). A nucleotide distance matrix was generated using “Compute Pairwise Distance” in MEGA 6 (41).
A total of 21,229 questing ticks were collected in November 2020 in Khao Yai National Park. The majority of ticks collected were larvae (n = 20,916) accounting for 98.5%, followed by females (n = 145, 0.7%), males (n = 85, 0.4%), and nymphs (n = 83, 0.4%). Species identification was done for the adult stage only and Haemaphysalis lagrangei, Haemaphysalis obesa, and Haemaphysalis longicornis were found at the highest rate (22%−24%), followed by Haemaphysalis shimoga (15%), Dermacentor auratus (9%), Amblyomma testudinarium (4%), Haemaphysalis papuana (1%), and Dermacentor steini (1%). All adult ticks (n = 230) and nymphs (n = 83) and 0.6% of larvae (n = 120) were selected for pathogen detection individually (n = 433). Identification of larval stage was performed on 120 selected samples and Amblyomma spp. (n = 35), Dermacentor spp. (n = 35), and Haemaphysalis spp. (n = 50) were found. In total, there were Haemaphysalis spp. (331, 76.4%), Dermacentor spp. (58, 13.4%), and Amblyomma spp. (44, 10.2%) included in this study. Additionally, a subset of ticks (n = 82) were selected for studying the bacterial community profile using 16S rRNA Next-Generation Sequencing (Table 1). Five to six ticks per species and life stage were selected for NGS, with the exception of immature Haemaphysalis spp., where 10 larvae and 20 nymphs were included. The number of ticks selected for NGS and the final number of ticks that passed the quality filter are described in detail in the online Supplementary data (Supplementary Table 2).
Overall, pathogenic bacteria were detected in 62 out of the total number of examined ticks collected from Khao Yai National Park, for a 14.3% prevalence rate. Pathogens were found in 48 of the tested Haemaphysalis spp. (14.5%), nine in Amblyomma spp. (20.5%), and five in Dermacentor spp. (8.6%). Rickettsia spp. was the most common pathogenic bacteria (7.9%), followed by Ehrlichia spp. (3.2%), and Anaplasma spp. and Borrelia spp. with a similar rate of 1.6% (Table 2). Haemaphysalis spp. (n = 331) were infected by all pathogens mentioned with Rickettsia spp. at the highest rate (7.6%), followed by Ehrlichia spp. (3.2%), Anaplasma spp. (1.8%), and Borrelia spp. (0.9%). In Amblyomma spp. (n = 44) and Dermacentor spp. (n = 58), two bacterial pathogens were found in each genus. Both were infected with Rickettsia (18.2% and 1.7% prevalence rates, respectively). Anaplasma spp. was detected only in Amblyomma spp. (2.3%), while Borrelia spp. was detected only in Dermacentor spp. (6.9%). When examining pathogen prevalence in tick life stages, the infection rate was found to increase from 10.0 to 19.3% from larvae to adults, respectively (Figure 1). The trend of increasing pathogen prevalence by life stage is clearly observed for Rickettsia spp. and Ehrlichia spp.
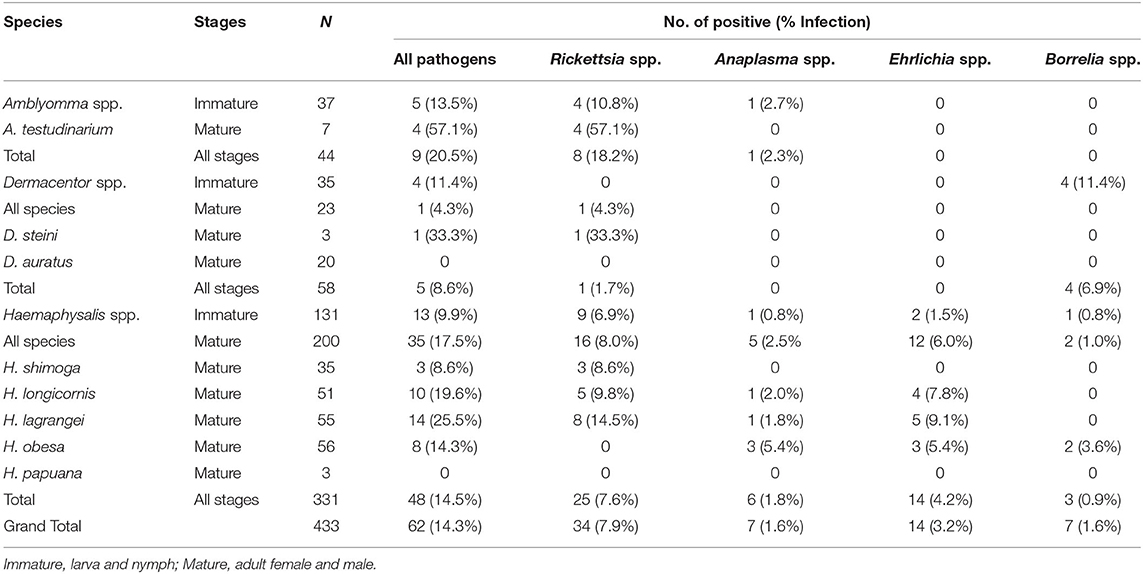
Table 2. Prevalence of pathogenic bacteria (Rickettsia spp., Anaplasma spp., Ehrlichia spp., and Borrelia spp.) in questing ticks from Khao Yai National Park, by species and stages.
Sequence and phylogenetic analyses showed that among Rickettsia species detected in ticks, R. montana (n = 21) was the predominant species found in Haemaphysalis spp., while R. raoultii (n = 6) was detected mostly in Amblyomma spp. (n = 5), and three other Rickettsia species (2 = R. heilongjiangensis, 3 = R. monacensis, and 2 = Rickettsia sp.) were detected in Haemaphysalis spp. and Amblyomma spp. (Table 3). Borrelia theileri (n = 7) was the only Borrelia species found, and was found in nearly equal numbers in Dermacentor spp. and Haemaphysalis spp. ticks. The majority of Anaplasmataceae bacteria (7 = Anaplasma spp. and 14 = Ehrlichia spp.) were mostly detected in Haemaphysalis spp. with E. ewingii (n = 9) being the dominant species found. Phylogenetic analyses for all bacteria can be found in online Supplementary Data (Supplementary Figure 1).
Co-infection between pathogenic bacteria examined in this study only occurred in a small number of ticks, primarily female Haemaphysalis spp. (n = 3). All co-infections occurred with Rickettsia spp. and Anaplasmataceae (Ehrlichia spp. or Anaplasma spp.) (Figures 2A,B) and accounted for 4.8% of infected ticks and 0.7% of all ticks examined. All three co-infected ticks were also positive for the Coxiella-like endosymbiont.
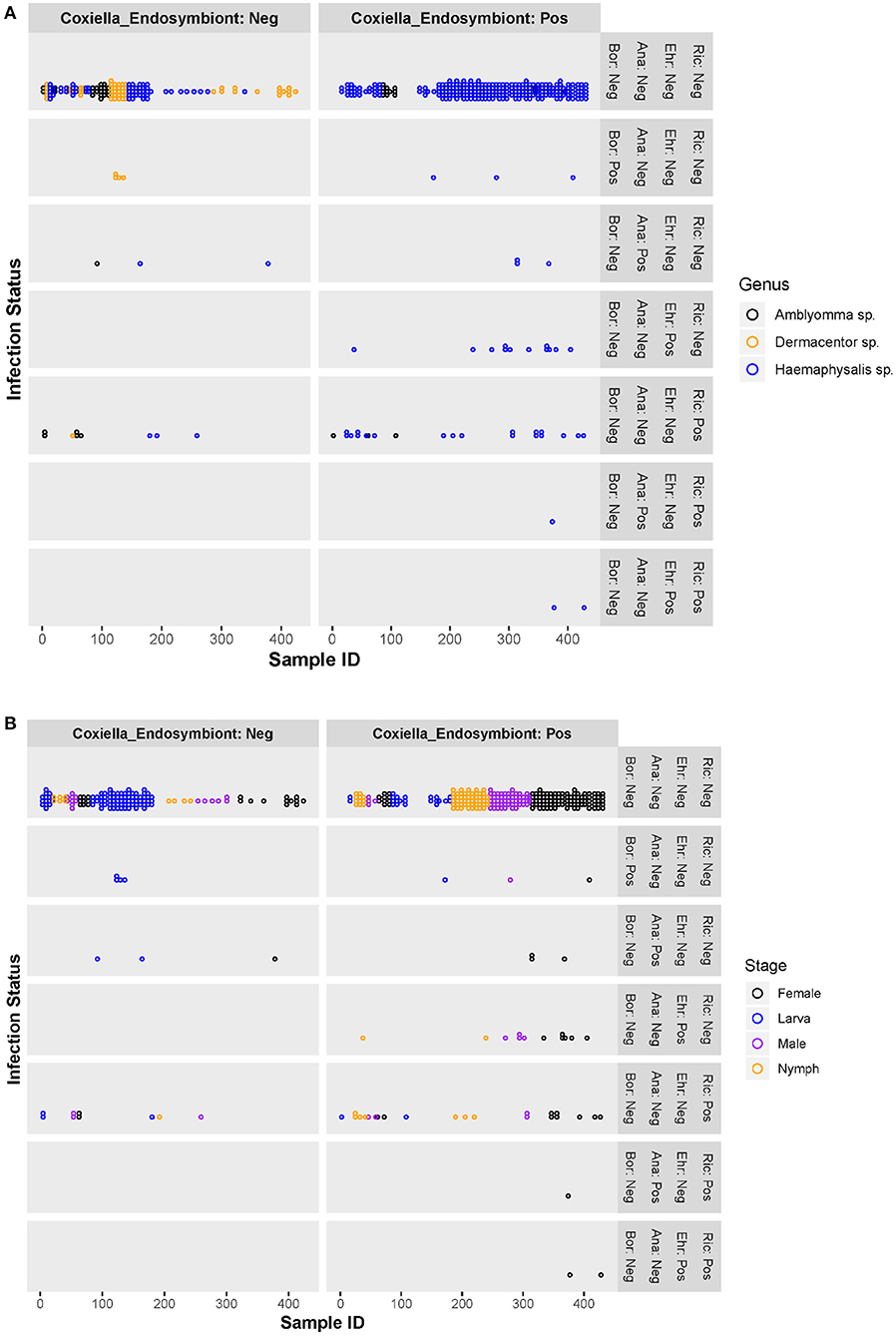
Figure 2. Scatter plots of individual ticks (with or without Coxiella-like endosymbiont) and their pathogenic bacterial infection status, by tick genera (A) and tick stages (B). Ric, Rickettsia spp.; Ehr, Ehrlichia spp.; Ana, Anaplasma spp.; Bor, Borrelia spp.; Coxiella_Endosymbiont, Coxiella-like endosymbiont; Pos, positive; Neg, negative.
The Coxiella-like endosymbiont was detected in 65.8% of all ticks examined, with the highest infection rate in nymphs (86.7%) and females (83.4%) (Table 4). Of all genera, Haemaphysalis spp. exhibited the highest rate of infection with the Coxiella-like endosymbiont, with an 81.0% infection rate. The majority of Haemaphysalis spp. infected were females (95.1%), followed by nymphs (88.9%), males (87.0%), and, to a lesser extent, larvae (24.0%). The second highest infection rate belonged to Amblyomma spp. with a 38.6% infection rate and only detected in females (66.7%) and larvae (37.1%). Surprisingly, no Dermacentor spp. were positive for the Coxiella-like endosymbiont.
Figures 2A,B are scatter plots of individual ticks depicting the pathogenic bacterial infection status compared between two tick populations—those harboring the Coxiella-like endosymbiont and those without. There was no significant difference in the number of ticks infected with Borrelia spp. or Anaplasma spp. between the Coxiella-like endosymbiont-positive and -negative groups. However, ticks harboring the Coxiella-like endosymbiont had greater rates of Ehrlichia spp. or Rickettsia spp. infection than those without the Coxiella-like endosymbiont. Chi-square and Fisher's exact tests were used to test for significant differences of Rickettsia spp. or Ehrlichia spp. infection between endosymbiont-positive and -negative ticks. Results show that the proportion of Rickettsia spp. infection in ticks harboring the Coxiella-like endosymbiont [8.8%, CI (5.5, 12.1%)] was not significantly different from the proportion in ticks without the Coxiella-like endosymbiont [6.1%%, CI (2.2, 9.9%)] with statistical values; Chi-square = 0.638, df = 1, p-value = 0.424. Likewise, the proportion of all pathogenic bacterial infection between ticks harboring the Coxiella-like endosymbiont [16.1%, CI (11.9, 20.4%)] and ticks without Coxiella-like endosymbiont [10.8%, CI (5.8, 15.8%)] was not significantly different (Chi-square = 0.7374, df = 1, p-value = 0.3905). Ehrlichia infection was observed only in ticks harboring Coxiella-like endosymbiont with the proportion of 4.9%, CI [2.4%, 7.4%]. Fisher's Exact test was used to determine the association between Ehrlichia infection and infection with the Coxiella-like endosymbiont. Results indicate that infection by these two bacteria in ticks is dependent (alpha = 0.05, p-value = 0.003), suggesting that the Ehrlichia infection is significantly associated with ticks harboring the Coxiella-like endosymbiont.
Of 433 samples tested in this study, 82 samples were selected for metagenomics NGS analysis (Table 1). After performing sequence quality filters and removing samples with low reads, only 58 samples (range: 1,085–472,236, mean number of reads ± SD = 37,780 ± 98,225) passed the criteria and were subjected to further OTU clustering and alpha and beta diversity analyses. A total of 2,222,970 reads passed quality filters and 268 OTUs were found across all samples. Comparison of the number of passed-filter reads being used for OTU clustering between tick stages showed that larvae (n = 19; 96,713 ± 157,880) generated more reads than nymphs (n = 17; 10,161 ± 9,845) and adults (n = 22; 8,226 ± 8,603). Eight Amblyomma spp., 10 Dermacentor spp., and 40 Haemaphysalis spp. were included in metagenomics analysis (Table 1).
The classification of OTUs from each sample was made against the Greengenes reference database and the similarity threshold was set at 0.97 in the CLC microbial genomics module. There were nine recorded phyla found among all tick samples studied: Proteobacteria (91%), Bacteroidetes (4%), Firmicutes (3%), Cyanobacteria (1%), and Actinobacteria (1%) (Figure 3A). There were five major phyla found in controls for DNA extraction, PCR, and Indexing; however, the majority of Proteobacteria phylum (73%) detected was genus Enterobacteria (89%), and very small amount of genus Rickettsia (0.0092%) (Supplementary Figure 2). Only 1% of reads could not be classified using Greengenes reference and were removed before performing downstream process of abundance analysis. Figure 3B shows the abundance of bacterial taxa in tick genera at the family level. There were one or two predominant bacterial taxa in each tick genus. Rickettsiaceae (83%) was the predominant bacterial taxon in Amblyomma spp., while Francisellaceae (50%) and Methylobacteriaceae (26%) were the major taxa found in Dermacentor spp. Haemaphysalis spp. harbored a more diverse bacterial spectrum than the other two tick genera. Coxiellaceae (43%) and Methylobacteriaceae (24%) were the predominant bacterial taxa found, with a lesser abundance of Rickettsiaceae (4.6%). Beta analysis using distance-based non-parametric test indicated that microbiome composition differed significantly across tick genera by the generalized UniFrac distance (df = 2, pseudo-F = 5.812, p-value = 0.00001).
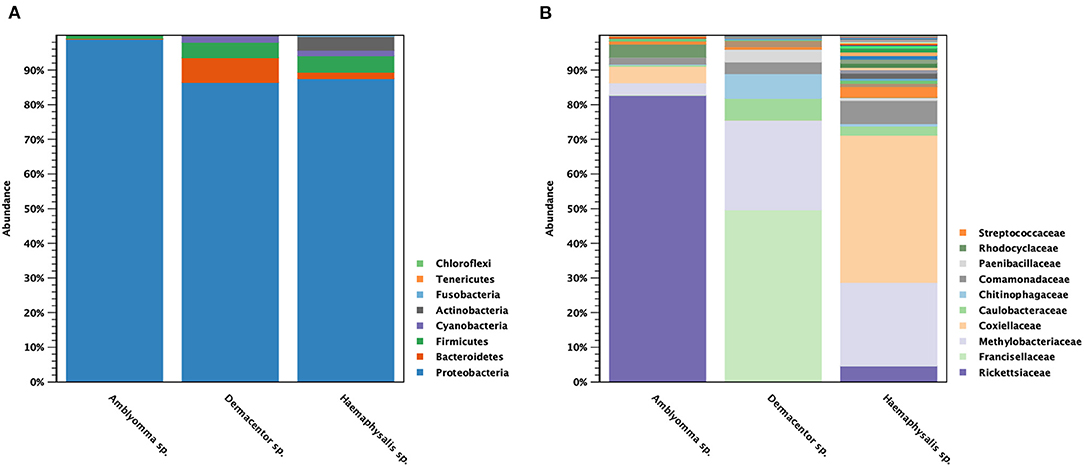
Figure 3. Taxonomic diversity and relative abundance at the phylum (A) and family (B) level of bacterial community in questing ticks. The percent relative abundances are of the total number of OTUs. Color legend for each phylum (A) or family (B) was indicated next to the bar graph.
Alpha diversity estimates of bacterial communities in ticks and statistical significance (p-value) for group comparison are summarized in Table 5. The alpha diversity analyses showed that the bacterial community of Haemaphysalis spp. (n = 40) was significantly different from Amblyomma spp. (n = 8) with p-value < 0.05 for all tests (Observed OTUs, Simpson's index, Shannon entropy) when measured at the genus level as shown in Figure 4A. While there were few ticks positive for Francisella persica (n = 4), alpha diversity estimates indicated that their bacterial profiles were quite different from ticks that were not infected with F. persica with statistical significance (p-value < 0.001) (Figure 4B).
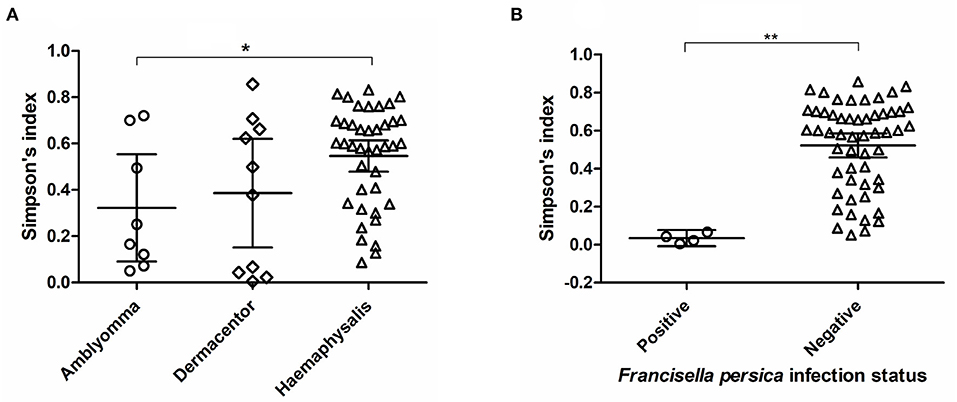
Figure 4. Alpha diversity measures (Simpson's index) based on 16S rRNA gene database for each tick genus (A) and Francisella persica infection status (B). The statistically significant differences between groups are indicated (*p-value < 0.05, **p-value < 0.001). The solid lines show mean and 95% confidence interval (CI) of alpha diversity for each group. Scatter plots and Mann–Whitney U test were created and tested using GraphPad Prism version 5.04.
As three Haemaphysalis spp. life stages (Larva = 9, Nymph = 15, Male = 6, Female = 10) were included in NGS, this genus was selected for testing the bacterial community difference among tick life stages (Figures 5A,B). Alpha diversity estimates (Simpson's index and Shannon entropy) showed that while the overall bacterial profiles among life stages were not significantly different with regard to bacterial taxa present, the number of observed OTUs among life stages was significantly different (Table 5; Figure 5B). Likewise, the beta diversity analysis using distance-based non-parametric test showed no significant difference across Haemaphysalis spp. life stages by the generalized UniFrac distance (df = 3, pseudo-F = 0.905, p-value = 0.5638). Comparing Coxiellaceae abundance across life stages, this group comprised 57–59% abundance of the total bacteria community in male and female ticks, 40% in nymphs, and 28% in larvae (Figure 5A).
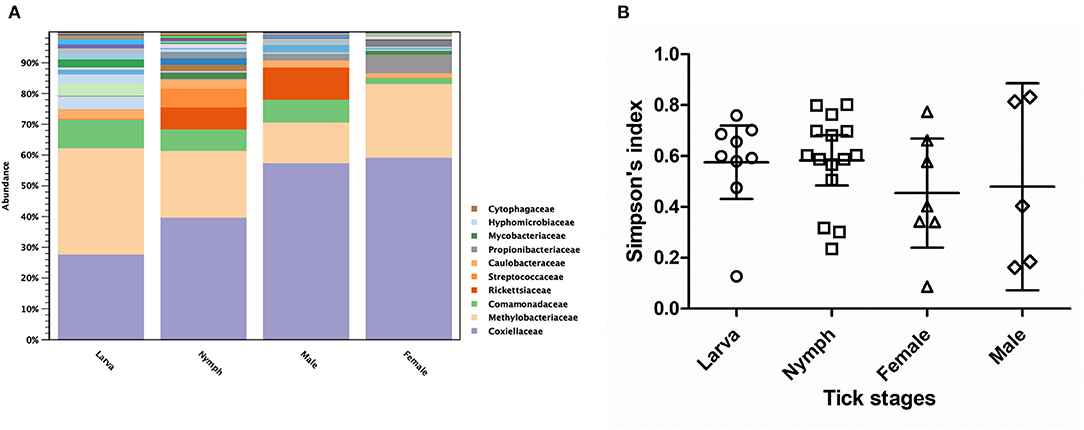
Figure 5. Taxonomic diversity and relative abundance at the family level of bacterial community (A) and scatter plots of alpha diversity measure (Simpson's index) (B) in three developmental stages of Haemaphysalis spp. ticks. The solid lines show mean and 95% Confidence Interval of alpha diversity for each group. Color legend for each family was indicated next to the bar graph.
In this study, we examined the bacterial community profile and co-infection of questing ticks collected by dragging from Khao Yai National Park using metagenomics. The majority of ticks collected were in the larval stage; however, adults and nymphs were also collected and identified as Haemaphysalis spp., Amblyomma spp., and Dermacentor spp. The greater number of Haemaphysalis spp. found in comparison to other genera may be due to the collection method used (dragging). The prevalence of Rickettsia spp. in ticks collected in Khao Yai National Park in this study was lower than in a previous report that found 30% of Amblyomma testudinarium positive for Rickettsia spp. and approximately 17% positive in Haemaphysalis spp. (44). However, they did not detect other bacteria such as Borrelia spp., Francisella spp., or the common symbiont Wolbachia spp. In this study, we detected Rickettsia DNA in 18% and 8% in Amblyomma spp. (n = 44) and Haemaphysalis spp. (n = 331) respectively, along with other bacteria such as Anaplasma spp., Ehrlichia spp., and Borrelia spp. at lower prevalence rates. We did not detect Wolbachia spp. in any of the ticks tested but found other bacterial endosymbionts, namely Rickettsia spp., Francisella spp., and Coxiella spp., using metagenomics and real-time PCR assay. Sequence analyses of Coxiella spp. and Francisella spp. detected by NGS confirmed the detection of these bacterial endosymbionts in examined ticks (Supplementary Figure 1). Rickettsia spp. that were detected by NGS but were not confirmed by subsequent qPCR and PCR assays are suspected to be a Rickettsia endosymbiont.
Co-infection of pathogenic bacteria occurred in a small number of ticks (n = 3), with the majority of co-infections between Rickettsia spp. and the Anaplasmataceae family. High prevalence of Rickettsia spp. in domestic animals and their ectoparasites, especially fleas, in Thailand has been recognized (15, 16, 45). Less than 10% of Rickettsia spp. was found in ticks collected from animals (45). However, some studies found infection rates as high as 24% for Rickettsia spp. and 32% for Anaplasma spp. in adult ticks collected from under leaves along animal trails across the country (46). The finding of co-infections between Rickettsia spp. and other bacteria in female ticks in this study was likely due to the ability of the bacteria to be maintained in ticks through transovarial and transstadial transmission (47–49). As females have already taken two bloodmeals during their development from larva to adult, the probability for Rickettsia-infected or Anaplasmataceae-infected ticks acquiring additional bacteria during feeding would be high (50–52). Our study also revealed the increased infection rate in adult ticks compared with larvae or nymphs. Similar findings were reported for an increasing A. phagocytophilum infection in adult ticks, especially in females, in Hanover, Germany (53). The study also reported that the co-infection between A. phagocytophilum and Rickettsia spp. was higher in females (5.2%) than in males (2.4%) or nymphs (1.6%).
A similar study was conducted by Nooroong et al. (46), in which ticks were collected under leaves along animal trails across Thailand were screened for bacterial pathogens. They found co-infections between Rickettsia spp. and Anaplasma spp. This co-infection was also observed in adult A. testudinarium ticks carrying the Coxiella-like endosymbiont in Nakhon Nayok, the same province where Khao Yai National Park is located. In Nooroong et al. (46), only adult ticks were collected and a 5.97% co-infection rate between Rickettsia spp. and Anaplasma spp., as well as a total infection rate of 35.8% for Rickettsia, was reported in the location near our study. This higher infection rate is likely due to the fact that Nooroong et al. (46) focused on the adult stage and did not include data from immature ticks. While all tick life stages collected by dragging were included in our study, the majority of ticks collected were larvae. Several previous studies in Thailand have primarily focused on ticks collected from animals; therefore, true co-infection status is inconclusive as pathogens may have been from the host animal. Our study focuses on co-infections in questing ticks actively seeking hosts, which directly represents the human risk of encountering ticks that can transmit multiple pathogens simultaneously. The co-infections investigated in previous studies were mostly among pathogenic bacteria and/or with bacterial endosymbionts (54), or among bacteria and protozoa such as Coxiella and Babesia spp. in H. bispinosa (55), or even co-infection with two to four pathogens of Anaplasmataceae, Babesia spp., and Hepatozoon spp. in Rhipicephalus sanguineus sensu lato collected from dogs in Bangkok, Thailand (56). Co-infection with more than three microorganisms was also reported in questing Ixodes scapularis ticks in Wisconsin, USA (29). Additionally, another study reported by Moutailler et al. (57) found up to 45% of questing I. ricinus ticks were co-infected with five to eight different pathogens including bacteria (Borrelia spp.), parasites, viruses, and endosymbionts. No significant interactions between endosymbionts and pathogens were found but there was a significant association between two Borrelia species: B. garinii and B. afzelii. Our results show that Ehrlichia infection in ticks harboring Coxiella-like endosymbiont was significantly higher than those without, suggesting that there might be some kind of relationship between Ehrlichia spp. and the Coxiella-like endosymbiont. However, we found no other association between Anaplasmataceae (Ehrlichia spp. and Anaplasma spp.) or Rickettsia spp. infection with the Coxiella-like endosymbiont. The data observed in this study implied that there was no negative or competitive effect of the Coxiella-like endosymbiont on the maintenance or existence of other bacteria in tick hosts. Endosymbionts relate to their host and pathogenic bacteria in many ways, such as providing nutrition lacking in blood meal, and sometimes possess an obligate relationship with the host (58, 59). However, other endosymbionts may interfere with the transmission to vertebrate hosts as was shown in R. rickettsii and R. peacockii in D. andersoni (60, 61). Another example of transmission interference can be seen in the salivary glands of Amblyomma spp., where a Coxiella-related symbiont impairs the transmission of E. chaffeensis (62). Alteration of the bacterial microbiome can also interfere with the colonization of pathogenic bacteria as seen in B. burgdorferi by modulation of the host immune response (63), which indicates that gut microbiota in ticks also plays a role in pathogen colonization in the gut lumen (22, 63).
Bacterial profiles using metagenomics used in this study showed a few dominant bacterial taxa harbored by questing ticks collected in Khao Yai National Park. Three main bacteria taxa were found in each tick genus—Coxiella spp. in Haemaphysalis spp., Rickettsia spp. in Amblyomma spp., and Francisella spp. in Dermacentor spp., making up the majority of bacterial taxa in ticks with relative abundance ranged from 40 to 80%. Our findings were consistent with other studies reporting that hard tick microbiomes are dominated by a small number of bacterial species, most of which are endosymbionts (20, 64). For example, Coxiella spp. was the main taxon found with a relative prevalence of 89.5% in Rhipicephalus turanicus (65), 89–100% for A. americanum (66–68), 98.2% in the female ovaries of Rhipicephalus (Boophilus) microplus (69), and 39.2% in Haemaphysalis spp. collected from animals in Malaysia (64). Coxiella spp. is a ubiquitous bacterium found in many tick species and maintained through transovarial transmission, as it was shown to pass on to their eggs and larvae from adult laboratory-reared R. sanguineus ticks (65, 70). Other bacterial endosymbionts detected in our study were Rickettsia spp., and depending on the tick species, Rickettsia spp. could represent up to 83% of the bacterial community, as observed in Amblyomma ticks in our study, while it was found to be less prevalent in the other two genera (Haemaphysalis and Dermacentor). A Francisella-like endosymbiont was another taxon we detected in high prevalence along with Methylobacteria in Dermacentor spp. The same finding was previously reported by several groups in which two Dermacentor species were dominated by three core bacterial taxa: Francisella, Sphinogomonas, and Methylobacterium (61, 71, 72). In addition to Dermacentor spp., A. maculatum was reported to harbor a Francisella-like endosymbiont at high abundance as well (73, 74). Our study also found that bacterial community composition varied significantly among tick genera, especially between Amblyomma and Haemaphysalis ticks. However, the species richness and diversity slightly decreased during development in Haemaphysalis spp. Other studies found that microbiome richness and diversity significantly decreased during development and varied greatly among species (72, 75). In one study, the obvious difference was on core OTUs of endosymbiont bacteria among tick species where Dermacentor spp. was dominated by Francisella spp., while H. leporispalustris and I. pacificus had Coxiella spp. and Rickettsia spp. as dominant bacterial species, respectively (72). Differences in reports might be from the techniques used to study the microbiome, geography, and natural vs. laboratory-reared tick populations (22, 76–78). However, the striking similarity among these studies is the difference in bacterial endosymbionts among tick genera, suggesting that there could be a competition among symbionts within tick genera. As it was reported in R. turanicus, Coxiella was the primary symbiont and Rickettsia was the secondary, having lesser relative abundance (65). Other studies discovered competition between Rickettsia species in Dermacentor spp. in which one species prevented another from transovarial transmission (79, 80).
Understanding the microbiome composition in ticks of different species, their vector capacities, as well as the role of bacterial endosymbionts on tick physiology, including their influence on pathogen transmission, may provide insight into vector control to prevent human infection and the emergence of tick-borne diseases in the future. This study provides important information on bacterial community composition and co-infection rates in questing ticks in Thailand with implications for animal and human health.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank accession number(s) OK161258-61, OK161265-68, OK161272-79, OK161279-84, and OK180561-611. The SRA BioProject ID PRJNA766341.
The animal study was reviewed and approved by AFRIMS Institutional Animal Care and Use Committee (IACUC), U.S. Army Medical Directorate-Armed Forces Research Institute of Medical Sciences.
RT and SC conceived and designed the experiments. JS, NC, NY, SP, and BT performed the experiments. SP, BT, NC, WT, KS, and SC conducted field works. RT and JS analyzed the data. RT wrote/revised the manuscript. BP-S and PM reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the Armed Forces Health Surveillance Division (AFHSD) Global Emerging Infections Surveillance Branch (GEIS), ProMIS ID # P0128_20_AF_04.05.
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. Research was conducted under an approved animal use protocol as part of an AAALAC International-accredited facility in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank the staff at Khao Yai National Park for their excellent collaboration and assistance during this study. The authors are grateful to Dr. Erica J. Lindroth for providing very valuable comments, review, and assisting with the editing of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.764763/full#supplementary-material
1. Dantas-Torres F, Chomel BB, Otranto D. Ticks and tick-borne diseases: a One Health perspective. Trends Parasitol. (2012) 28:437–46. doi: 10.1016/j.pt.2012.07.003
2. Ebel GD, Campbell EN, Goethert HK, Spielman A, Telford SR. Enzootic transmission of deer tick virus in New England and Wisconsin sites. Am J Trop Med Hyg. (2000) 63:36–42. doi: 10.4269/ajtmh.2000.63.36
3. Scoles GA, Papero M, Beati L, Fish D. A relapsing fever group spirochete transmitted by Ixodes scapularis ticks. Vector Borne Zoonotic Dis. (2001) 1:21–34. doi: 10.1089/153036601750137624
4. Dolan MC, Hojgaard A, Hoxmeier JC, Replogle AJ, Respicio-Kingry LB, Sexton C, et al. Vector competence of the blacklegged tick, Ixodes scapularis, for the recently recognized Lyme borreliosis spirochete candidatus borrelia mayonii. Ticks Tick Borne Dis. (2016) 7:665–9. doi: 10.1016/j.ttbdis.2016.02.012
5. Nelder MP, Russell CB, Sheehan NJ, Sander B, Moore S, Li Y, et al. Human pathogens associated with the blacklegged tick ixodes scapularis: a systematic review. Parasit Vectors. (2016) 9:265. doi: 10.1186/s13071-016-1529-y
6. Zimmermann DE, Penzhorn BL, Vorster I, Troskie M, Oosthuizen MC. Babesia bicornis, theileria bicornis and theileria equi in metapopulations of two black rhinoceros (Diceros bicornis) subspecies in South Africa and their potential impact on conservation. Ticks Tick Borne Dis. (2021) 12:101635. doi: 10.1016/j.ttbdis.2020.101635
7. Petney TN, Saijuntha W, Boulanger N, Chitimia-Dobler L, Pfeffer M, Eamudomkarn C, et al. Ticks (argasidae, ixodidae) and tick-borne diseases of continental Southeast Asia. Zootaxa. (2019) 4558:1–89. doi: 10.11646/zootaxa0.4558.1.1
8. Nakao Y, Tanigawa T, Shibata R. Human otoacariasis caused by Amblyomma testudinarium: diagnosis and management: case report. Medicine. (2017) 96:e7394. doi: 10.1097/MD.0000000000007394
9. Mariana A, Srinovianti N, Ho TM, Halimaton I, Hatikah A, Shaharudin MH, et al. Intra-aural ticks (metatigmata: ixodidae) from human otoacariasis cases in Pahang, Malaysia. Asian Pac J Trop Med. (2008) 1:20–4. https://www.cabdirect.org/cabdirect/abstract/20093152429
10. Kollars TM, Tippayachai B, Bodhidatta D. Short report: Thai tick typhus, Rickettsia honei, and a unique rickettsia detected in Ixodes granulatus (ixodidae: acari) from Thailand. Am J Trop Med Hyg. (2001) 65:535–7. doi: 10.4269/ajtmh.2001.65.535
11. Lewin MR, Musher D. Spotted-fever-group rickettsioses in north Asia. Lancet. (2004) 363:1076. doi: 10.1016/S0140-6736(04)15853-4
12. Gaywee J, Sunyakumthorn P, Rodkvamtook W, Ruang-Areerate T, Mason CJ, Sirisopana N. Human infection with Rickettsia sp. related to R japonica, Thailand. Emerg Infect Dis. (2007) 13:657–9. doi: 10.3201/eid1304.060585
13. Pachirat O, Fournier PE, Pussadhamma B, Taksinachanekij S, Lulitanond V, Baggett HC, et al. The first reported cases of Q fever endocarditis in Thailand. Infect Dis Rep. (2012) 4:e7. doi: 10.4081/idr.2012.3910
14. Yingst SL, Opaschaitat P, Kanitpun R, Thammasart S, Ekgatat M, Jirathanawat V, et al. Q Fever surveillance in ruminants, Thailand, 2012. Emerg Infect Dis. (2013) 19:2056–8. doi: 10.3201/eid1912.130624
15. Takhampunya R, Korkusol A, Pongpichit C, Yodin K, Rungrojn A, Chanarat N, et al. Metagenomic approach to characterizing disease epidemiology in a disease-endemic environment in northern Thailand. Front Microbiol. (2019) 10:319. doi: 10.3389/fmicb.2019.00319
16. Foongladda S, Inthawong D, Kositanont U, Gaywee J. Rickettsia, ehrlichia, anaplasma, and bartonella in ticks and fleas from dogs and cats in bangkok. Vector Borne Zoonotic Dis. (2011) 11:1335–41. doi: 10.1089/vbz.2010.0174
17. Ahantarig A, Trinachartvanit W, Milne JR. Tick-borne pathogens and diseases of animals and humans in Thailand. Southeast Asian J Trop Med Public Health. (2008) 39:1015–32.
18. Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. (2008) 6:741–51. doi: 10.1038/nrmicro1969
19. Rudolf I, Mendel J, Sikutova S, Svec P, Masarikova J, Novakova D, et al. 16S rRNA gene-based identification of cultured bacterial flora from host-seeking Ixodes ricinus, dermacentor reticulatus and haemaphysalis concinna ticks, vectors of vertebrate pathogens. Folia Microbiol. (2009) 54:419–28. doi: 10.1007/s12223-009-0059-9
20. Hunter DJ, Torkelson JL, Bodnar J, Mortazavi B, Laurent T, Deason J, et al. The Rickettsia endosymbiont of Ixodes pacificus contains all the genes of de novo folate biosynthesis. PLoS ONE. (2015) 10:e0144552. doi: 10.1371/journal.pone.0144552
21. Bonnet SI, Binetruy F, Hernandez-Jarguin AM, Duron O. The tick microbiome: why non-pathogenic microorganisms matter in tick biology and pathogen transmission. Front Cell Infect Microbiol. (2017) 7:236. doi: 10.3389/fcimb.2017.00236
22. Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the lyme disease spirochete. Cell Host Microbe. (2014) 15:58–71. doi: 10.1016/j.chom.2013.12.001
23. Gall CA, Reif KE, Scoles GA, Mason KL, Mousel M, Noh SM, et al. The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility. ISME J. (2016) 10:1846–55. doi: 10.1038/ismej.2015.266
24. Lo N, Beninati T, Sassera D, Bouman EA, Santagati S, Gern L, et al. Widespread distribution and high prevalence of an alpha-proteobacterial symbiont in the tick Ixodes ricinus. Environ Microbiol. (2006) 8:1280–7. doi: 10.1111/j.1462-2920.2006.01024.x
25. Sassera D, Beninati T, Bandi C, Bouman EaP, Sacchi L, Fabbi M, et al. 'Candidatus Midichloria mitochondrii', an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int J Syst Evol Microbiol. (2006) 56:2535–40. doi: 10.1099/ijs.0.64386-0
26. Tokarz R, Williams SH, Sameroff S, Sanchez Leon M, Jain K, Lipkin WI. Virome analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. J Virol. (2014) 88:11480–92. doi: 10.1128/JVI.01858-14
27. Sakamoto JM, Ng TFF, Suzuki Y, Tsujimoto H, Deng X, Delwart E, et al. Bunyaviruses are common in male and female Ixodes scapularis ticks in central Pennsylvania. PeerJ. (2016) 4:e2324. doi: 10.7717/peerj.2324
28. Vanmechelen B, Laenen L, Vergote V, Maes P. Grotenhout virus, a novel nairovirus found in ixodes ricinus in Belgium. Genome Announc. (2017) 5. doi: 10.1128/genomeA.00288-17
29. Cross ST, Kapuscinski ML, Perino J, Maertens BL, Weger-Lucarelli J, Ebel GD, et al. Co-infection patterns in individual Ixodes scapularis ticks reveal associations between viral, eukaryotic and bacterial microorganisms. Viruses. (2018) 10:1–19. doi: 10.3390/v10070388
30. Steiner FE, Pinger RR, Vann CN, Grindle N, Civitello D, Clay K, et al. Infection and co-infection rates of anaplasma phagocytophilum variants. babesia spp, borrelia burgdorferi, and the rickettsial endosymbiont in ixodes scapularis (acari: ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J Med Entomol. (2008) 45:289–97. doi: 10.1093/jmedent/45.2.289
31. Grunwaldt E, Barbour AG, Benach JL. Simultaneous occurrence of babesiosis and Lyme disease. N Engl J Med. (1983) 308:1166. doi: 10.1056/NEJM198305123081918
32. Golightly LM, Hirschhorn LR, Weller PF. Fever and headache in a splenectomized woman. Rev Infect Dis. (1989) 11:629–37. doi: 10.1093/clinids/11.4.629
33. Thomas V, Anguita J, Barthold SW, Fikrig E. Coinfection with Borrelia burgdorferi and the agent of human granulocytic ehrlichiosis alters murine immune responses, pathogen burden, and severity of Lyme arthritis. Infect Immun. (2001) 69:3359–71. doi: 10.1128/IAI.69.5.3359-3371.2001
34. Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, et al. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. JAMA. (2004) 292:716–20. doi: 10.1001/jama.292.6.716
35. Nieto NC, Foley JE. Meta-analysis of coinfection and coexposure with Borrelia burgdorferi and Anaplasma phagocytophilum in humans, domestic animals, wildlife, and Ixodes ricinus-complex ticks. Vector Borne Zoonotic Dis. (2009) 9:93–102. doi: 10.1089/vbz.2008.0072
36. Hoogstraal H, Kohls GM. Southeast Asian Haemaphysalis ticks (Ixodoidea, Ixodidae). H bandicota sp n from bandicoot rats in Taiwan, Thailand, and Burma. J Parasitol. (1965) 51:460–6. doi: 10.2307/3275973
37. Tanskull P, Inlao I. Keys to the adult ticks of Haemaphysalis Koch, 1844, in Thailand with notes on changes in taxonomy (acari: ixodoidea: ixodidae). J Med Entomol. (1989) 26:573–601. doi: 10.1093/jmedent/26.6.573
38. Desantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. (2006) 72:5069–72. doi: 10.1128/AEM.03006-05
39. Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. (2012) 28:2106–13. doi: 10.1093/bioinformatics/bts342
40. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7. doi: 10.1093/nar/gkh340
41. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30:2725–9. doi: 10.1093/molbev/mst197
42. Chang W. R Graphics Cookbook. Gravenstein Highway North Sebastopol, CA: O'Reilly Media, Inc (2012).
43. R Core Team. A language and environment for statistical computing. R foundation for statistical computing. Vienna, Austria (2018). Available online at: URL https://www.R-project.org/
44. Hirunkanokpun S, Kittayapong P, Cornet JP, Gonzalez JP. Molecular evidence for novel tick-associated spotted fever group rickettsiae from Thailand. J Med Entomol. (2003) 40:230–7. doi: 10.1603/0022-2585-40.2.230
45. Chaorattanakawee S, Korkusol A, Tippayachai B, Promsathaporn S, Poole-Smith BK, Takhampunya R. Amplicon-based next generation sequencing for rapid identification of Rickettsia and ectoparasite species from entomological surveillance in Thailand. Pathogens. (2021) 10. doi: 10.3390/pathogens10020215
46. Nooroong P, Trinachartvanit W, Baimai V, Ahantarig A. Phylogenetic studies of bacteria (rickettsia, coxiella, and anaplasma) in amblyomma and dermacentor ticks in Thailand and their co-infection. Ticks Tick Borne Dis. (2018) 9:963–71. doi: 10.1016/j.ttbdis.2018.03.027
47. Kelly PJ, Mason PR. Transmission of a spotted fever group rickettsia by amblyomma hebraeum (acari: ixodidae). J Med Entomol. (1991) 28:598–600. doi: 10.1093/jmedent/28.5.598
48. Socolovschi C, Huynh TP, Davoust B, Gomez J, Raoult D, Parola P. Transovarial and trans-stadial transmission of rickettsiae africae in amblyomma variegatum ticks. Clin Microbiol Infect. (2009) 15 Suppl 2:317–8. doi: 10.1111/j.1469-0691.2008.02278.x
49. Hauck D, Jordan D, Springer A, Schunack B, Pachnicke S, Fingerle V, et al. Transovarial transmission of borrelia spp. rickettsia spp and anaplasma phagocytophilum in ixodes ricinus under field conditions extrapolated from DNA detection in questing larvae. Parasit Vectors. (2020) 13:176. doi: 10.1186/s13071-020-04049-7
50. Halos L, Bord S, Cotte V, Gasqui P, Abrial D, Barnouin J, et al. Ecological factors characterizing the prevalence of bacterial tick-borne pathogens in Ixodes ricinus ticks in pastures and woodlands. Appl Environ Microbiol. (2010) 76:4413–20. doi: 10.1128/AEM.00610-10
51. Otranto D, Dantas-Torres F, Giannelli A, Latrofa MS, Cascio A, Cazzin S, et al. Ticks infesting humans in Italy and associated pathogens. Parasit Vectors. (2014) 7:328. doi: 10.1186/1756-3305-7-328
52. Stromdahl E, Hamer S, Jenkins S, Sloan L, Williamson P, Foster E, et al. Comparison of phenology and pathogen prevalence, including infection with the Ehrlichia muris-like (EML) agent, of Ixodes scapularis removed from soldiers in the midwestern and the northeastern United States over a 15 year period (1997-2012). Parasit Vectors. (2014) 7:553. doi: 10.1186/s13071-014-0553-z
53. Blazejak K, Janecek E, Strube C. A 10-year surveillance of rickettsiales (rickettsia spp. and anaplasma phagocytophilum) in the city of hanover, germany, reveals rickettsia spp as emerging pathogens in ticks. Parasit Vectors. (2017) 10:588. doi: 10.1186/s13071-017-2537-2
54. Sumrandee C, Baimai V, Trinachartvanit W, Ahantarig A. Molecular detection of rickettsia, anaplasma, coxiella and francisella bacteria in ticks collected from artiodactyla in Thailand. Ticks Tick Borne Dis. (2016) 7:678–89. doi: 10.1016/j.ttbdis.2016.02.015
55. Trinachartvanit W, Kaenkan W, Chelong I-A, Bahakheeree M, Baimai V, Ahantarig A. Co-Infection with Coxiella-like bacteria and Babesia in goat ticks from Southern Thailand. Southeast Asian J Trop Med Public Health. (2019) 50:643–50.
56. Do T, Phoosangwalthong P, Kamyingkird K, Kengradomkij C, Chimnoi W, Inpankaew T. Molecular detection of tick-borne pathogens in stray dogs and rhipicephalus sanguineus sensu lato ticks from Bangkok, Thailand. Pathogens. (2021) 10:1–12. doi: 10.3390/pathogens10050561
57. Moutailler S, Valiente Moro C, Vaumourin E, Michelet L, Tran FH, Devillers E, et al. Co-infection of ticks: the rule rather than the exception. PLoS Negl Trop Dis. (2016) 10:e0004539. doi: 10.1371/journal.pntd.0004539
58. Guizzo MG, Parizi LF, Nunes RD, Schama R, Albano RM, Tirloni L, et al. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci Rep. (2017) 7:17554. doi: 10.1038/s41598-017-17309-x
59. Ben-Yosef M, Rot A, Mahagna M, Kapri E, Behar A, Gottlieb Y. Coxiella-like endosymbiont of Rhipicephalus sanguineus is required for physiological processes during ontogeny. Front Microbiol. (2020) 11:493. doi: 10.3389/fmicb.2020.00493
60. Childs JE, Paddock CD. Passive surveillance as an instrument to identify risk factors for fatal rocky mountain spotted fever: is there more to learn? Am J Trop Med Hyg. (2002) 66:450–7. doi: 10.4269/ajtmh.2002.66.450
61. Ahantarig A, Trinachartvanit W, Baimai V, Grubhoffer L. Hard ticks and their bacterial endosymbionts (or would be pathogens). Folia Microbiol. (2013) 58:419–28. doi: 10.1007/s12223-013-0222-1
62. Klyachko O, Stein BD, Grindle N, Clay K, Fuqua C. Localization and visualization of a coxiella-type symbiont within the lone star tick, Amblyomma americanum. Appl Environ Microbiol. (2007) 73:6584–94. doi: 10.1128/AEM.00537-07
63. Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. (2011) 332:855–8. doi: 10.1126/science.1201618
64. Khoo JJ, Chen F, Kho KL, Ahmad Shanizza AI, Lim FS, Tan KK, et al. Bacterial community in Haemaphysalis ticks of domesticated animals from the Orang Asli communities in Malaysia. Ticks Tick Borne Dis. (2016) 7:929–37. doi: 10.1016/j.ttbdis.2016.04.013
65. Lalzar I, Harrus S, Mumcuoglu KY, Gottlieb Y. Composition and seasonal variation of rhipicephalus turanicus and rhipicephalus sanguineus bacterial communities. Appl Environ Microbiol. (2012) 78:4110–6. doi: 10.1128/AEM.00323-12
66. Jasinskas A, Zhong J, Barbour AG. Highly prevalent Coxiella sp. bacterium in the tick vector Amblyomma americanum. Appl Environ Microbiol. (2007) 73:334–6. doi: 10.1128/AEM.02009-06
67. Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, Fuqua C. Microbial communities and interactions in the lone star tick, amblyomma americanum. Mol Ecol. (2008) 17:4371–81. doi: 10.1111/j.1365-294X.2008.03914.x
68. Heise SR, Elshahed MS, Little SE. Bacterial diversity in amblyomma americanum (acari: ixodidae) with a focus on members of the genus rickettsia. J Med Entomol. (2010) 47:258–68. doi: 10.1093/jmedent/47.2.258
69. Andreotti R, Perez De Leon AA, Dowd SE, Guerrero FD, Bendele KG, Scoles GA. Assessment of bacterial diversity in the cattle tick rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. (2011) 11:6. doi: 10.1186/1471-2180-11-6
70. Noda H, Munderloh UG, Kurtti TJ. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. (1997) 63:3926–32. doi: 10.1128/aem.63.10.3926-3932.1997
71. Niebylski ML, Peacock MG, Fischer ER, Porcella SF, Schwan TG. Characterization of an endosymbiont infecting wood ticks, dermacentor andersoni, as a member of the genus Francisella. Appl Environ Microbiol. (1997) 63:3933–40. doi: 10.1128/aem.63.10.3933-3940.1997
72. Chicana B, Couper LI, Kwan JY, Tahiraj E, Swei A. Comparative microbiome profiles of sympatric tick species from the Far-Western United States. Insects. (2019) 10. doi: 10.3390/insects10100353
73. Scoles GA. Phylogenetic analysis of the francisella-like endosymbionts of dermacentor ticks. J Med Entomol. (2004) 41:277–86. doi: 10.1603/0022-2585-41.3.277
74. Varela-Stokes AS, Park SH, Stokes JV, Gavron NA, Lee SI, Moraru GM, et al. Tick microbial communities within enriched extracts of amblyomma maculatum. Ticks Tick Borne Dis. (2018) 9:798–805. doi: 10.1016/j.ttbdis.2018.02.022
75. Ruiling Z, Zhendong H, Guangfu Y, Zhong Z. Characterization of the bacterial community in haemaphysalis longicornis (acari: ixodidae) throughout developmental stages. Exp Appl Acarol. (2019) 77:173–86. doi: 10.1007/s10493-019-00339-7
76. Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC. Molecular analysis of microbial communities identified in different developmental stages of ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol. (2006) 8:761–72. doi: 10.1111/j.1462-2920.2005.00955.x
77. Landesman WJ, Mulder K, Fredericks LP, Allan BF. Cross-kingdom analysis of nymphal-stage Ixodes scapularis microbial communities in relation to borrelia burgdorferi infection and load. FEMS Microbiol Ecol. (2019) 95:1–14. doi: 10.1093/femsec/fiz167
78. Tokarz R, Tagliafierro T, Sameroff S, Cucura DM, Oleynik A, Che X, et al. Microbiome analysis of ixodes scapularis ticks from New York and Connecticut. Ticks Tick Borne Dis. (2019) 10:894–900. doi: 10.1016/j.ttbdis.2019.04.011
79. Niebylski ML, Schrumpf ME, Burgdorfer W, Fischer ER, Gage KL, Schwan TG. Rickettsia peacockii sp. nov, a new species infecting wood ticks, dermacentor andersoni, in western Montana. Int J Syst Bacteriol. (1997) 47:446–52. doi: 10.1099/00207713-47-2-446
Keywords: co-infection, metagenomics in ticks, Amblyomma spp., Haemaphysalis spp., Dermacentor spp., questing ticks in Thailand
Citation: Takhampunya R, Sakolvaree J, Chanarat N, Youngdech N, Phonjatturas K, Promsathaporn S, Tippayachai B, Tachavarong W, Srinoppawan K, Poole-Smith BK, McCardle PW and Chaorattanakawee S (2021) The Bacterial Community in Questing Ticks From Khao Yai National Park in Thailand. Front. Vet. Sci. 8:764763. doi: 10.3389/fvets.2021.764763
Received: 25 August 2021; Accepted: 15 October 2021;
Published: 22 November 2021.
Edited by:
Cornelia Silaghi, Friedrich-Loeffler-Institute, GermanyReviewed by:
Donato Antonio Raele, Istituto Zooprofilattico Sperimentale di Puglia e Basilicata (IZSPB), ItalyCopyright © 2021 Takhampunya, Sakolvaree, Chanarat, Youngdech, Phonjatturas, Promsathaporn, Tippayachai, Tachavarong, Srinoppawan, Poole-Smith, McCardle and Chaorattanakawee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ratree Takhampunya, cmF0cmVldC5mc25AYWZyaW1zLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.