
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 16 November 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.753297
This article is part of the Research Topic Porcine Reproductive and Respiratory Syndrome Virus - Animal Virology, Immunology, and Pathogenesis View all 9 articles
 Ying Huang
Ying Huang Xianghong Chen
Xianghong Chen Yunzhi Long
Yunzhi Long Liu Yang
Liu Yang Wenbo Song
Wenbo Song Jinjin Liu
Jinjin Liu Qianqian Li
Qianqian Li Gong Liang
Gong Liang Daobing Yu
Daobing Yu Chao Huang*
Chao Huang* Xibiao Tang*
Xibiao Tang*Porcine circovirus type 2 (PCV2) is one of the smallest known animal viruses and is the main pathogen of PCV-associated diseases (PCVAD). Epidemiological surveillance results have shown that the PCV2 infection rate is on the rise in China, thus, PCV2 disease prevention and control has become a huge challenge for the Chinese swine industry. We collected clinical samples from multiple different provinces in China from 2018 to 2020 and found that the positive rate of PCV2 was 53% (3619/6872), identity between the cloned 62 ORF2 genes was 84.4–100% and identity between the cloned 62 ORF2 sequences and reference sequence was 72.9–99.8%. Genetic evolution analysis found that PCV2d accounted for 79% (49/62 samples), PCV2a for 12.9% (8/62 samples), PCV2b for 8% (5/62 samples), and PCV2c and PCV2e genotypes were not found. However, most commercial PCV2 subunit vaccines are based on the PCV2a genotype, and there are very few vaccines based on PCV2b or PCV2d. Therefore, the homologous and heterologous protection ability of PCV2b and PCV2d Cap proteins based on the baculovirus against the PCV2b and PCV2d infections was evaluated, which is expected to design and develop excellent PCV2 protein vaccine candidates. This study found that both PCV2b and PCV2d Cap proteins can increase the level of humoral immunity and cellular immune response in mice. Importantly, both PCV2b and PCV2d cap proteins can provide homologous and heterologous protection against the PCV2b and PCV2d viruses. Overall, this study provides a reference for the prevention and control of PCVAD in mainland China and the development of PCV2 vaccines.
Porcine circovirus (PCV) belongs to the family Circoviridae and genus Circovirus (1). There are three major genotypes of PCV: PCV PCV1, PCV2, and the newly found PCV3 (2). Since the late 1990s, PCV2 has emerged as a major pig pathogen around the globe, easily infecting pigs 4–16 weeks of age, and mainly causes typical clinical appearances, consisting of post-weaning multisystemic wasting syndrome (PMWS), porcine dermatitis nephropathy syndrome (PDNS), reproductive disorders, and respiratory diseases. PCV2 mainly destroys the lymphatic system, leading to defects in the body's immune function, which may cause susceptibility to other diseases, such as porcine parvovirus, pseudorabies virus, and porcine streptococcus (3).
The PCV2 genome is composed of a single strand of circular closed DNA with a length of approximately 1.7 kb. The virus replicates in a rolling circle, producing double-stranded DNA replicas. A study found that the whole genome of PCV2 contains 11 open reading frames (ORFs) (4). The research objects are mainly concentrated in ORF1, ORF2, ORF3, and ORF4. ORF1 encodes a viral replication-related protein with a length of 945nt, which is located on the sense strand of the genome, and its primary transcript is alternatively spliced to produce two variants of rep (35.7 kDa) and rep' (20 kDa) (5). ORF2 is located on the antisense strand of the genome and encodes the Cap protein with 233–236 amino acids. It is the only Cap protein of the virus with a size of about 27.8 kDa. The Cap protein is also an important immunogenic protein of the virus and can induce the body to generate neutralizing antibodies against PCV2 (6). ORF3 is reverse-encoded in ORF1 and encodes a non-structural protein of the virus, which encodes 104 amino acids (11.9 kDa). Studies have shown that the ORF3-encoded protein induces cell apoptosis in vitro by activating the caspase 3 and 8 pathways (7). ORF3 is related to the pathogenicity of the virus in animals, and the pathogenicity of PCV2 mutants lacking ORF3 is lower than that of wild-type PCV2 in mice and pigs (7). The protein encoded by ORF4 is not necessary for virus replication. It can inhibit caspase activity and regulate the role of CD4+ and CD8+ lymphocytes during PCV2 infection (8).
Based on the PCV2 whole genome or PCV2-ORF2 sequence system evolution analysis, PCV2 mainly has five genotypes; namely, PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e, among which PCV2a/2b/2d subtypes have attracted wide attention from researchers (9). Before 2003, the virus was dominated by the PCV2a subtype. From 2004 to 2013, it was dominated by the PCV2b subtype. After 2013, it was dominated by the PCV2d subtype, and other subtypes still existed in pig herds (10). The PCV2c subtype is only found in Denmark and Brazil and the number of cases is small. Retrospective studies have shown that PCV2d was first discovered in Switzerland in 1998. Chinese scholars named the newly discovered PCV2 subtype PCV2d for the first time in 2010, and it is now the main popular subtype of PCV2 (11). PCV2e was first discovered in Mexico, and subsequent retrospective studies have shown that this virus subtype also exists in the United States. The earliest gene sequence information was released in 2006 (12). PCV2 can cause multiple disease syndromes in infected pigs, such as PMWS, PDNS, porcine respiratory disease complex, sow reproduction disorders, nervous system damage, and proliferative and necrotizing pneumonia, these disease syndromes are collectively called PCV diseases (PCVD) in Europe and PCV-associated diseases, PCVAD) in North America (13). Early research suggests that the epidemic strain has changed from PCV2a to PCV2b and gradually formed an epidemic worldwide since 2003 (14). A previous study found that 55% (22/40 samples) of their PCV2-positive samples were PCV2d (15), which implied that PCV2d is an ongoing and dominant PCV2 subtype in China. Subsequently, another study also found that PCV2d made up most of the PCV-positive isolates (68.2%; 45/66 samples) in southern China from 2011 to 2012 (16). These data provide evidence that a dynamic genotype shift occurs from PCV2b to PCV2d and that PCV2d dominates worldwide, including in America, Europe, Korea, and Thailand (10, 17, 18).
Although vaccination can effectively reduce the prevalence of PCV2 and alleviate the clinical signs of PCVAD, its continuous mutation through point mutations and genetic recombination causes concern that it may escape vaccination. The PCV2 vaccine needs to be updated regularly, including switching to a new strain. Therefore, obtaining genetic variation information related to PCV2 in pig herds can be crucial for vaccine development and the prevention of PCVAD. The main purpose of this study was to design and develop excellent candidate protein vaccines on the basis of the genetic variation and phylogenetic characteristics of PCV2 in China from 2018 to 2020.
This study was conducted in accordance with the Chinese Laboratory Animal Administration Act of 1988. All animal experimental procedures were performed in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China). Animal experiments in this study were subject to approval by the Hubei Province Science and Technology Department, concerning experimental animal ethics. The experiments were carried out under the supervision and inspection of the Scientific Ethical Committee for Experimental Animals of Huazhong Agricultural University, Wuhan, China. The field studies did not involve endangered protected species.
The 6,827 clinical samples, including those of the lungs, kidneys, lymph nodes, and spleen, were collected from commercial farms in China (including different provinces) from 2018 to 2020 and stored at −80 °C. Next, the DNA was extracted according to the manufacturer's instructions of the viral DNA extraction kit (TransGen, China) and stored at −80 °C.
Primers were designed based on the PCV2 sequence registered in GenBank (GenBank accession number: AY424401.1). Primers F1 and R1 were used to detect the PCV and primers F2 and R2 were used to amplify the ORF2 gene of the PCV. The primers are shown in Table 1.
To detect PCV-positive samples, the PCR reactions were prepared in a total volume of 20 μL:10 μL of 2 × Easy Taq PCR SuperMix (TransGen Biotech, Beijing, China), 1 μL of primer pairs (F1 and R1), 6 μL of ddH2O, and 2 μL of DNA template. PCR amplification was initiated at a predenaturation stage of 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s extension at 72 °C for 30 s, and 10 min extension at 72 °C. Amplified PCR products were subjected to agarose gel electrophoresis.
Next, 62 positive PCV samples were randomly selected for amplification of the PCV ORF2 gene. The PCR reactions were prepared in a total volume of 20 μL:10 μL of 2 × Easy Taq PCR SuperMix, 1 μL of primer pairs (F2 and R2), 5 μL of ddH2O, and 3 μL of DNA template. PCR amplification was initiated at a predenaturation stage of 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 53 °C for 40 s, extension at 72 °C for 30 s, followed by an extension at 72 °C for 10 min. The PCR product was then sent to Sangon Biotechnology (Beijing, China) for sequencing.
The general analysis, including the trimming and assembling of raw sequencing data, conversion into reverse complement sequences, and conceptual translation into protein sequences, was performed using CLC Main Workbench 7 software (Qiagen). The final length of the analysis sequence was 482 bp, and 20 PCV2 reference strain whole genomes and PCV2 ORF2 sequences were downloaded from GenBank. The information of 20 PCV2 strain reference sequences is shown in Table 2. Multiple alignments of nucleotides and deduced amino acid sequences diversity analysis were performed by MEGAlign software. The phylogenetic tree was constructed by the neighbor-joining method using the maximum composite likelihood model with MEGA v6 software. The reliability of the cluster separated in the tree was evaluated by performing 500 bootstrap replicates.
Spodoptera frugiperda (Sf9) cells (ExpiSf9; Gibco) were cultured in Sf-900™ II SFM medium (Gibco) at 28 °C. PCV2b (GenBank accession number: FJ598044) was used for amplification of the nucleotide sequence of the recombinant Cap protein by PCR. The PCV2d-based Cap protein nucleotide sequence came from the PCV2d (HB16 strain). PK-15 cells without PCV infection were cultured in a modified serum-free medium (Celkey®CDPK15, China) and cultured at 37 °C under 5% CO2, which was used to propagate PCV2b and PCV2d.
The recombinant baculovirus of PCV2b and PCV2d Cap proteins were constructed by the Bac-to-Bac Baculovirus Expression System (Invitrogen, USA) following the manufacturer's instructions. In short, the Cap protein genes of PCV2b and PCV2d were amplified with PrimeSTAR HS DNA polymerase (Takara, Japan). Primers are shown in Table 1. The first PCV2 Cap gene amplified by the F3/R3 primers was digested with BamHI and SacI (Takara, Japan) and cloned into the pFastBacDual vector (Invitrogen) containing the baculovirus polyhedrin gene promoter. The second PCV2 Cap gene amplified by the F4/R4 primers was digested with KpnI and NcoI and cloned into the downstream position controlled by the P10 promoter. Recombinant pFBD-2b Cap and pFBD-2d Cap were transformed into chemically competent Escherichia coli DH10Bac cells (Invitrogen). Recombinant bacmids were screened by blue and white spots and further verified by sequencing (Sangon, Wuhan, China). The recombinant bacmid DNA was transfected into Sf9 cells using CellfectinII reagent (Invitrogen), and rBac-2bCap and rBac-2dCap were generated. The recombinant baculovirus titers were computed by immunofluorescence assay.
Sf9 cells were infected with rBac-2bCap and rBac-2dCap in the MOI of 0.5 and then harvested at 7 days post-infection (dpi). The expressed recombinant Cap proteins were purified by anion exchange chromatography columns. In short, at 7 dpi, the harvested culture mixture was centrifuged at 10,000 × g for 30 min under 4 °C. Then, the supernatants were passed through a 0.45 μm polyethersulfone syringe filter. The harvested supernatants were purified by the Q Sepharose Fast Flow Ion Exchange Chromatography Columns (GE Healthcare, USA). The purified sample was assembled into virus-like particles (VLPs), which were observed under a transmission electron microscope (TEM; Hitachi, Japan) at the Huazhong Agricultural University.
The purified PCV2b and PCV2d Cap proteins were subjected to 12% SDS-PAGE and then transferred to nitrocellulose membranes (Merck, Germany) using semidry transfer apparatus (Bio-Rad). A porcine anti-PCV2 antibody (diluted 1:500) was used to prove the purified protein by western blot, and horseradish peroxidase (HRP)-conjugated goat anti-porcine IgG antibody (diluted 1:5,000) was the secondary antibody.
Seventy 8-week-old female BALB/c mice were randomly divided into the following seven groups with 10 mice per group: phosphate-buffered saline (PBS), UnVac-2b/Ch, Vac2b-2b/Ch, Vac2d-2b/Ch, UnVac-2d/Ch, Vac2b-2d/Ch, and Vac2d-2d/Ch groups. At 28 days post-challenge (dpc, 8 weeks of age), 18 μg of purified PCV2b Cap protein was mixed with MontanideTM ISA 201 VG (Seppic, France) adjuvant at a ratio of 1:1 (w/w) and then injected into the leg muscles of the BALB/c mice. Similarly, the immunization dose of the PCV2d Cap protein was the same as that of PCV2b. at −14 dpc, the second immunization was performed, the dosage and method of protein immunization were the same as those of the first immunization. At 0 dpc, 500 μL of PCV2 virus solution was injected intraperitoneally, and the BALB/c mice in the negative control group were injected with 500 μL of PBS. At 0 and 14 dpc, blood samples were collected and stored at −80 °C for the following experiments.
Thirty 8-week-old female BALB/c mice were randomly divided into three groups (PBS, Vac PCV2b Cap, and Vac PCV2b Cap groups) and the 10× protein immunization dose (180 μg of purified protein) was injected into the mice to evaluate the safety of the vaccine. The clinical symptoms of the mice were monitored daily in the following 14 days after immunization. In short, the score was defined as follows: 0 (normal), 1 (rough haircoat), 2 (rough haircoat and dyspnea), 4 (severe dyspnea and abdominal breathing), and 6 (death). The observer was unaware of the different vaccination statuses of the mice.
An indirect ELISA was used to test the PCV2-specific antibodies of 0 dpc in the serum samples. In short, a 96-well microtiter plate was coated with 50 ng/well of purified Bac-2b Cap or Bac-2d Cap protein at 4 °C overnight. Each well was washed with PBS 0.05% Tween 20 (PBS-T), then 200 μ L of PBS solution containing 2% bovine serum albumin was added to each well and incubated at 37 °C for 2 h. Subsequently, the plate was washed with PBS-T and 100 μ L aliquots of diluted serum samples (1:100) were added to each well and then incubated at 37 °C for 2 h. After washing with PBS-T, the plate was incubated with HRP-conjugated goat anti-mouse IgG (1:10,000; SouthernBiotech, USA) at 37 °C for 1 h. Subsequently, the plate was washed with PBS-T and TMB one-component HRP microwell substrate (SouthernBiotech, USA) was added, followed by incubation of the wells in the dark for 5 min. Finally, the reaction was stopped with 2N H2SO4, and then the OD450 nm value was determined by a microplate reader (Thermo Fisher Scientific, USA).
The F1 and R1 primers were used to detect PCV2 and to amplify the PCV2 DNA in serum at 14 dpc. The experimental method was the same as the identification PCR experiment method. Primers are shown in Table 1.
The 14 dpc blood samples were collected and used to extract viral DNA by using a viral DNA/RNA extraction kit (TransGen, China) according to the manufacturer's instructions. Then, the concentration of viral nucleic acid was determined by a Nanodrop spectrophotometer. Then, qPCR was used for amplification using PCV2 ORF2-specific primers (PCV2_ORF2_F and R) and probing with PerfectStart™ II Probe qPCR SuperMix (TransGen, China) as shown in Table 1. The number of PCV2 DNA copies was calculated.
The 14 dpc sera were analyzed using mouse interferon-gamma (IFN-γ) ELISA kits (Multisciences, China) according to the manufacturer's instructions. The mean values of the cytokines were expressed as p g/m L.
Data were analyzed using one-way analysis of variance with Dunnett's multiple comparisons test using GraphPad Prism 7 software (GraphPad Software, USA). p values < 0.05 were considered to indicate significance.
A PCR assay was used to detect the PCV2 in the samples. As shown in Figure 1, the positive samples were able to amplify a band of about 496 bp. As shown in Table 3, among the 6,827 clinical pig samples collected from different pig farms in China and tested by PCR from 2018 to 2020, PCV2 was identified in 53% of them. At the sample level, the prevalence rate of PCV2 decreased continuously from 2018 to 2020; the positive rate in 2018 was 65.8% (2,379/4,164), in 2019 was 41.9% (450/1,071), and in 2020 was 26.2% (430/1,637). However, the unbalanced number of samples submitted each year may affect the positive rate.
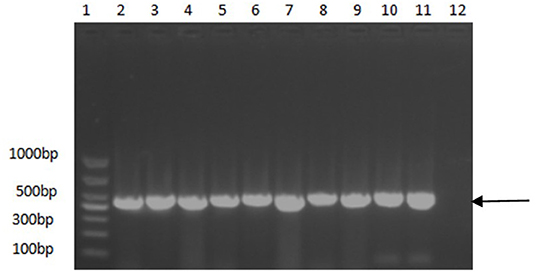
Figure 1. PCR identification results of some positive samples. 1, DNA molecular size marker; 2–10, Partial positive samples; 11, Positive control; 12, Negative control. Arrow indicates the position of the predicted DNA size.
Among the collected samples, 62 with brighter PCR bands were selected for the cloning of the PCV2 ORF2 gene sequence and further sequence analysis. Through identity analysis of the nucleotide sequences by MegAlign software, we found that the identity of the ORF2 nucleotide sequences of 62 strains was 84.4–100%, and that of 62 strains with 20 reference sequences was 72.9–99.8%. The identity comparison results of some strains are shown in Figure 2. Multisequence alignment of the 62 amino acid sequences encoded by ORF2 genes was conducted using HQ395032 as the reference sequence. Although the 62 ORF2-encoded Cap protein sequences were relatively conservative (Figure 3), there were some scattered variable amino acid positions, such as 8, 30, 169, 210, and 230 aa. The Cap protein sequence encoded by HuN-XSY, HeN-SY, and HuB-TS strains were different at the epitope. This may affect the infectivity of the virus and the activity of neutralizing antibodies. Overall, 62 PCV2 isolate strain sequences were compared with 20 PCV2 reference strains, and the phylogenetic tree was constructed by MEGA V6.0 software. As shown in Figure 4, it was concluded that PCV2d accounted for 79% (49/62 samples), PCV2a for 12.9% (8/62 samples), PCV2b for 8% (5/62 samples), and PCV2c and PCV2e genotypes were not found, which proves that the PCV2d genotype was the epidemic strain of PCV2 in China from 2018 to 2020.

Figure 3. Multiple sequence alignment of ORF2-encoded Cap protein amino acids of some strains. Epitopes are boxed.
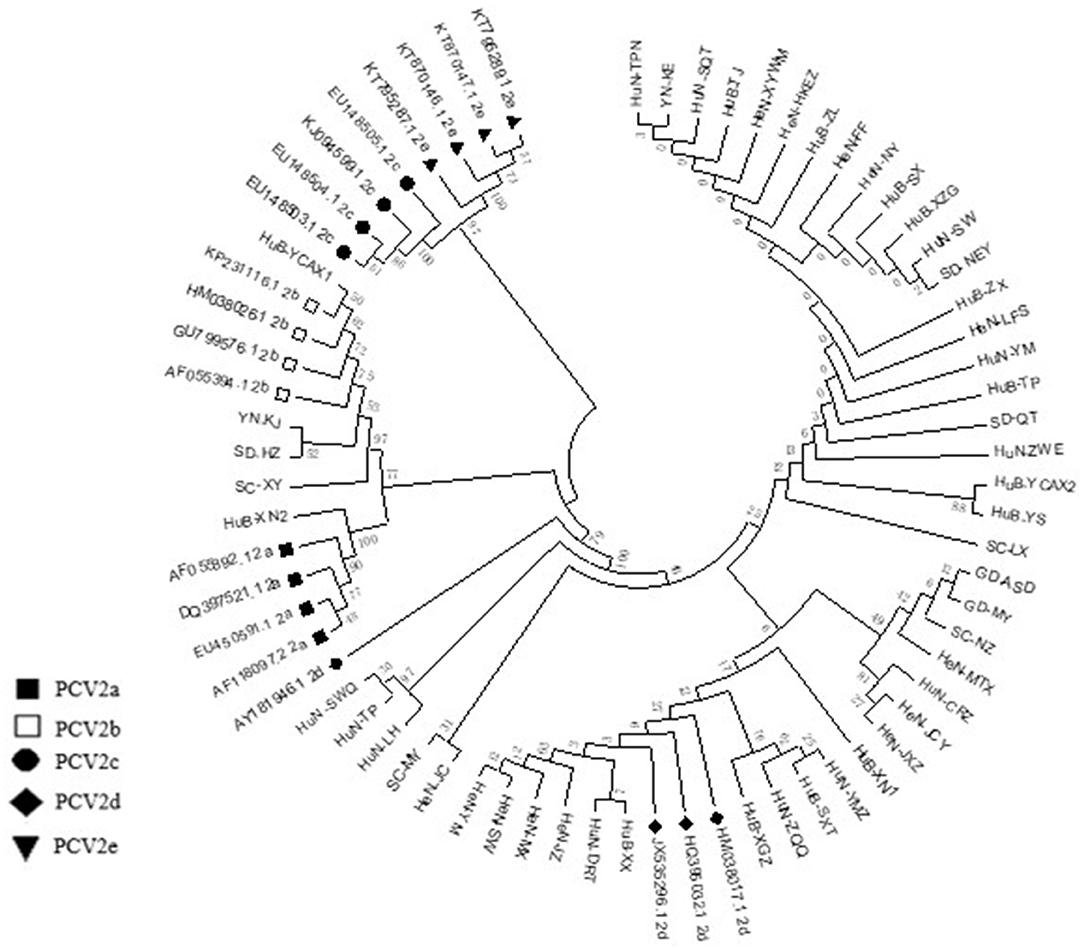
Figure 4. Phylogenetic tree based on 82 PCV2 nucleotide sequences, including the 62 samples obtained in this study (not marked) and 20 strains of the different genotypes available from GenBank (PCV2a to PCV2e) (marked). The phylogenetic tree was constructed by the neighbor-joining method using the maximum composite likelihood model with MEGA v6 software.
The recombinant PCV-2b and PCV-2d Cap proteins were expressed in Sf9 cells and purified by anion exchange chromatography (Figure 5A). Purified Bac-2b and Bac-2d Cap proteins were successfully self-assembled into VLPs and were observed by TEM. As shown in Figures 5B,C, the assembled VLPs showed the expected particle size and shape with a diameter of 17 nm.
The safety of the PCV2b and PCV2d Cap proteins was evaluated in mice. Clinical monitoring records and scoring results are shown in Table 4. No notable clinical signs and no local and systemic adverse reactions were observed in any of the groups throughout the study. This shows that these two proteins are safe for mice. The PCV2b Cap protein induces protective immunity in mice against challenge with PCV2b and PCV2d strains.
To evaluate whether the PCV2b Cap protein can protect pigs against infection with both PCV2b and PCV2d viruses, first, at 0 dpc, the PCV2b and PCV2d specific antibody levels in serum were determined by indirect ELISA using the Bac-2b and Bac-2d Cap antigens as a coating buffer. As shown in Figure 6A, VacPCV2b Cap maintained high levels of the anti-PCV2b- and the anti-PCV2d-specific IgG antibody. The anti-PCV2b-specific IgG antibody showed a mean OD value of 1.063, which was higher than that for the anti-PCV2d-specific IgG antibody at 0.844. The OD values of these were 17.69 (p < 0.001) and 16.88 (p < 0.001) times higher than that of the PBS group, respectively. Secondly, the 10 serum samples at 14 dpc were used to amplify PCV2 DNA by PCR (Figure 6B). The PCV2b and PCV2d challenge groups were all able to amplify positive bands, while Vac2b-2b/Ch showed a 2/10 and Vac2b-2d/Ch showed a 3/10 positive rate, showing that the PCV2b Cap protein can effectively resist the viremia of PCV2b and PCV2d. Thirdly, qPCR was performed using PCV2-specific primers for calculating the number of genomic DNA copies in the serum after the challenge (Figure 6C). Compared with the UnVac-2b/Ch group, the PCV2b viral load of the Vac2b-2b/Ch group decreased by 5,318 times, and the difference was significant (p < 0.001). Compared with the UnVac-2d/Ch group, the PCV2d viral load of the Vac2b-2d/Ch group decreased by 6,023 times, and the difference was significant (p < 0.001), which further verifies that PCV2b Cap protein is able to resist viremia caused by PCV2b and PCV2d. To evaluate whether the Bac-2bCap protein would elicit cellular immune responses, we analyzed the production of IFN-γ in serum (Figure 6D). At 14 dpc, high levels of IFN-γ were detected in all groups compared with the PBS group (p < 0.001). The UnVac-2b/Ch and UnVac-2d/Ch groups had slightly higher cytokine levels than the Vac2b-2b/Ch and Vac2b-2d/Ch groups, and the difference was not obvious, which indicated that PCV2b Cap protein stimulates TH1 cells to secrete IFN-γ cytokines. These results indicate that the PCV2b Cap protein can effectively activate the host's humoral and cellular immunity, thereby reducing the body's viremia caused by PCV2b and PCV2d.
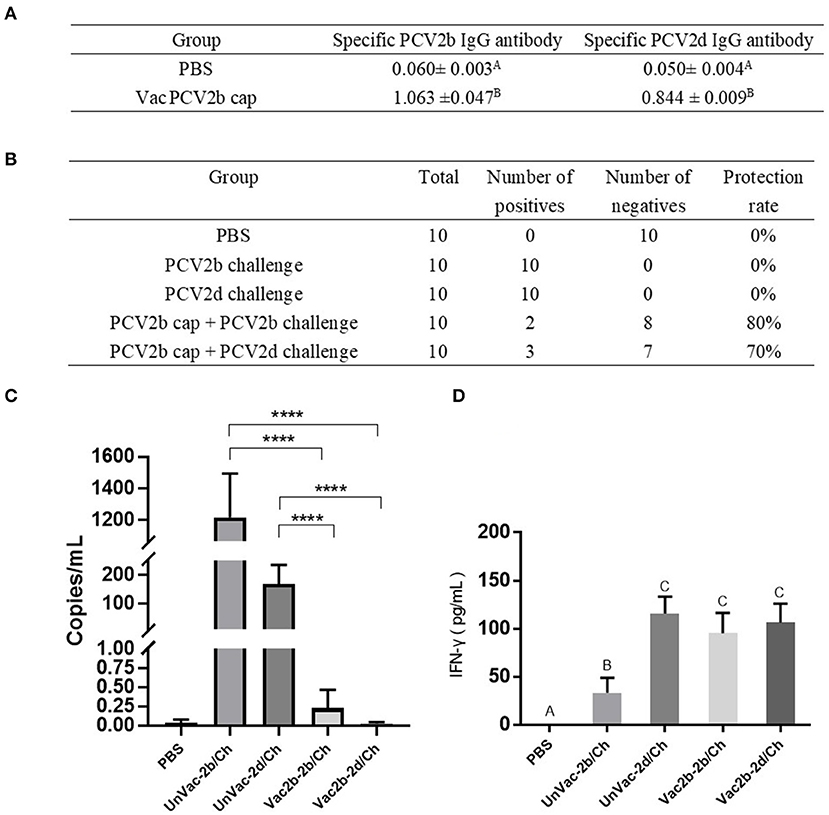
Figure 6. Homologous and heterologous immune response of PCV2d Cap protein against the PCV2b and PCV2dagainst the PCV2b and PCV2d. (A) Specific antibody levels for PCV2b and PCV2d Cap proteins at 0 dpc. (B) Detection of PCV2 DNA in serum by PCR at 14 dpc. (C) Detection of the PCV2 DNA load in serum by RT-PCR at 14 dpc. (D) Levels of IFN-γ in blood at 14 dpc. The data are presented as group mean optical density (MOD ± standard error). Significant differences are indicated by different superscripts (A–C) (p < 0.05). Data were analyzed using one-way analysis of variance with Dunnett's multiple comparisons test using GraphPad Prism 7 software (GraphPad Software, USA), ****p < 0.001.
To evaluate whether the PCV2d Cap protein can protect pigs against infection with both PCV2b and PCV2d viruses, first, at 0 dpc, the PCV2b- and PCV2d-specific antibody levels in serum were determined by indirect ELISA using the Bac-2b and Bac-2d Cap antigens as a coating buffer. As shown in Figure 7A, the VacPCV2b Cap group preserved high levels of the anti-PCV2b-2d-specific IgG antibody and the anti-PCV2d-specific IgG antibody with a mean OD value of 1.456, which was 30.33 (p < 0.001) times higher than that of the control group, and the anti-PCV2b-specific IgG antibody with a mean OD value of 0.885, which was 16.69 (p < 0.001) times higher than that in the control group. Secondly, at 14 dpc, serum samples from mice were used to detect PCV2 DNA by PCR. The unvaccinated group with the challenge by PCV2b and PCV2d showed a 10/10 positive rate for both, while Vac2d-2b/Ch showed a 2/10 and Vac2d-2d/Ch a 1/10 positive rate, which proved that the PCV2d Cap protein can effectively reduce the viremia of both PCV2b and PCV2d (Figure 7B). Thirdly, we assessed the levels of the virus in the sera of infected mice by qPCR (Figure 7C). Compared with the UnVac-2b/Ch group, the PCV2b viral load of the Vac2d-2b/Ch group decreased by 71 times, and the difference was significant (p < 0.001). Compared with the UnVac-2d/Ch group, the PCV2d viral load of the Vac2d-2d/Ch group decreased by 42 times, and the difference was significant (p < 0.001). To further evaluate the T cell immune response stimulated by our PCV2d Cap protein vaccine candidate, we analyzed the production of IFN-γ in serum (Figure 7D). At 14 dpc, compared with the control group, mice from the experimental groups had high levels of IFN-γ (p < 0.001). UnVac-2b/Ch had significantly less IFN-γ cytokine levels than UnVac-2d/Ch (p < 0.05), which may be caused by the difference in the challenge dose. These results indicate that PCV2d Cap protein can effectively activate the host's humoral and cellular immunity, thereby resisting the body's viremia caused by PCV2b and PCV2d.
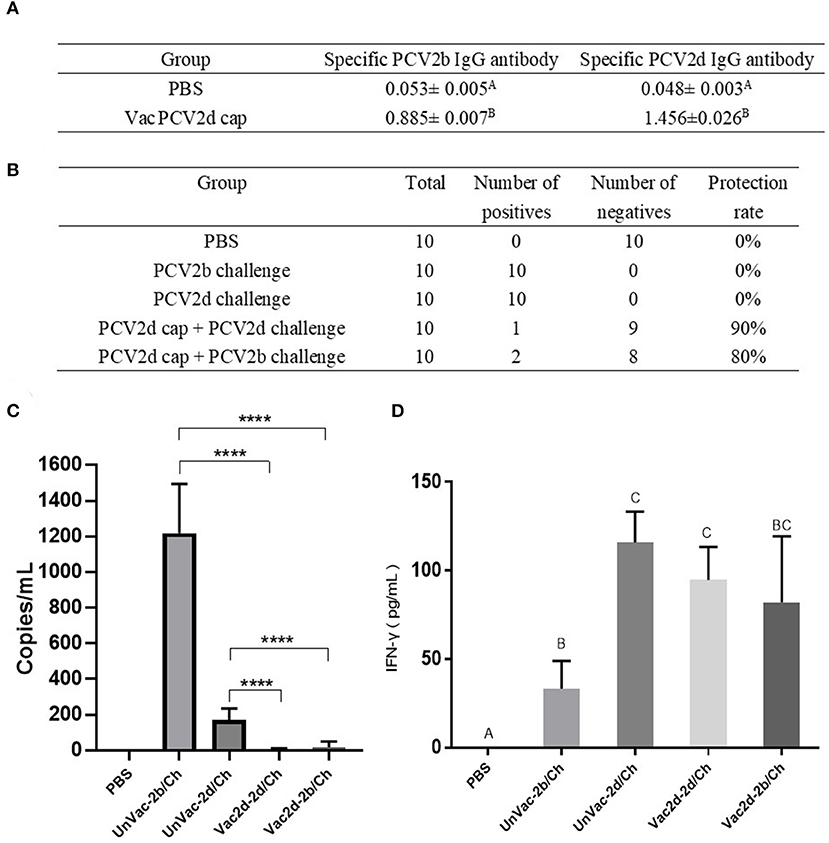
Figure 7. Homologous and heterologous immune response of PCV2d Cap protein against the PCV2b and PCV2dagainst the PCV2b and PCV2d. (A) Specific antibody levels for PCV2b and PCV2d Cap proteins at 0 dpc. (B) Detection of PCV2 DNA in serum by PCR at 14 dpc. (C) Detection of the PCV2 DNA load in serum by RT-PCR at 14 dpc. (D) Levels of IFN-γ in blood at 14 dpc. The data are presented as group mean optical density (MOD ± standard error). Significant differences are indicated by different superscripts (A–C) (p < 0.05). Data were analyzed using one-way analysis of variance with Dunnett's multiple comparisons test using GraphPad Prism 7 software (GraphPad Software, USA), ****p < 0.001.
Since the discovery of porcine circovirus type 2 (PCV2) in 1998 (19, 20), PCV2 has gradually spread worldwide and caused huge economic losses to the global swine industry. In recent years, epidemiological surveillance results have shown that the PCV2 infection rate is on the rise in China (21). In addition, many studies have reported that the co-infection of PCV with other porcine pathogens, such as porcine reproductive and respiratory syndrome virus, porcine parvovirus, swine influenza virus, Mycoplasma hyopneumoniae, and salmonella may aggravate PCV2 infection, resulting in serious cases of PCVD.
This study showed that the positive rate of PCV2 was 53% (3,619/6,827 samples) in China from 2018 to 2020, which is close to the 50.3% of Northeast China from 2015 to 2018 (46). Moreover, we found that the identity of the ORF2 gene between 62 PCV2 isolates in this study was 84.4–100%, indicating that there are differences among PCV2 molecules prevailing in China. Cap protein encoded by the ORF2 gene is the main structural protein and key epitope cluster of PCV2. Compared with the reference PCV2d Cap protein amino acid sequence (GenBank accession number: HQ395032.1), the 62 isolates had higher variations in several regions, and there were some scattered variable amino acid positions, such as 8, 30, 169, 210, and 230 aa. Changes in the amino acid sequence of different strains may reveal differences in virulence, which will bring some difficulties to the prevention and control of PCV2 disease. The immunodominant epitope on the amino acid Cap protein of PCV2 is recognized by the antibodies of PCV2-infected pigs. It is characterized by different A, B, C, and D regions, which roughly correspond to the amino acid fragments 65–87, 113–139, 169–183, and a few C-terminal amino acids (22, 23), respectively. the Cap protein sequence encoded by HuN-XSY, HeN-SY, and HuB-TS strains were variable at the epitopes A, B, and C, which may affect the effectiveness of antibodies produced by the vaccine.
Studies have reported that the PCV2d genotype has become the predominant genotype in some regions of China, including Yunnan province, Hunan province, Shandong province, and northwest China (24–28). The positive rate of the PCV2d genotype was as high as 79.0% in this study, indicating that PCV2d has been the prevalent strain in China. With the discovery of PCV2d and subsequent genotype shift from the previously predominant PCV2b to PCV2d, concerns over PCV2 vaccine efficacy have been raised. Although all current vaccines have been proven to be effective at preventing clinical signs and global economic loss due to PCVAD (29, 30), the emergence of the PCV2d subtype cannot be ignored (31). A study pointed out that the PCV2b-based Cap protein expressed by baculoviruses can induce the body to produce greater humoral and cellular immunity, and this subunit vaccine can effectively reduce the viremia of pigs naturally infected with PCV2d (32). Logically, PCV2d Cap protein can provide better protection against PCV2d than PCV2b Cap protein. Therefore, the main purpose of this study was to explore the protection ability of the PCV2b and PCV2d Cap proteins against the PCV2b and PCV2d virus challenge.
In this study, after immunizing twice, the PCV2b and PCV2d Cap protein immune groups both contained higher levels of PCV2b and PCV2d IgG antibodies, and the titer of antibodies against the PCV2d Cap protein was slightly higher than that against the PCV2b Cap protein, but this difference was not significant.
PCV2 infection can cause severe viremia (31). PCR detection of PCV2 DNA in serum samples at 14 dpi found that the positive rate of UnVac-2b/Ch and UnVac-2d/Ch viral DNA reached 100%. However, the positive rates of viral DNA in the Vac2b-2b/Ch and Vac2b-2d/Ch groups were 20 and 30%, respectively, and those of viral DNA in the Vac2d-2b /Ch and Vac2d-2d/Ch groups were 20 and 10%, respectively.
Compared with the 2b/Ch and 2d/Ch groups, the viral load of PCV2b and PCV2d in the Vac2b group decreased by 5,318 and 6,023 times, respectively, while the viral load of PCV2b and PCV2d in the Vac2d group decreased by 71 and 42 times, respectively. This shows that both proteins can reduce the virus infection of the body and, under the same protein immune dose, the effect of the PCV2b Cap protein in reducing viremia is better than that of the PCV2d protein. This is inconsistent with the results of conventional PCR. Conventional PCR can only be qualitative but not quantitative, and the criteria for judging negative and positive have certain subjective characteristics, while RT-PCR can perform absolute quantification of viral DNA, and its results will be more reliable than the former.
Increased secretion of IFN-γ is responsible for the reduction of PCV2 viremia (33, 34), where the level is considered the measurement of protective immunity (35, 36). The UnVac-PCV2b/Ch group had more PCV2 genome copy numbers than the UnVac-PCV2d/Ch group, while the levels of IFN-γ of the UnVac-PCV2b/Ch group were lower than the latter, which is consistent with the previous conclusion. Compared with the blank control group, the levels of IFN-γ were higher in all experimental groups, indicating the improvement of the IFN-γ secretion level induced by the TH1 cells after immunization with protein.
In summary, PCV2d was found to be the current circulating strain in pigs in China. PCV2b and PCV2d Cap proteins expressed by the baculovirus system can both stimulate the level of humoral immunity and cellular immune response in mice. Interestingly, these two proteins had certain homologous and heterologous immune response against the PCV2b and PCV2d strains, and the effect of the PCV2b Cap protein in reducing viremia was better than that of the PCV2d Cap protein. Therefore, this study has significance for the subsequent prevention and control of PCV and the preparation of a suitable vaccine.
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by Animal Experimental Ethical Inspection of Laboratory Animal Centre, Huazhong Agriculture University.
YH designed and performed the experiments, analyzed the data, and wrote the manuscript. XC, YL, LY, WS, JL, QL, GL, and DY performed the experiments. CH and XT conceived the project, analyzed the data, and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study received funding from Wuhan Keqian Biology Co., Ltd. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
All authors are employed by Wuhan Keqian Biology Co., Ltd.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. (1982) 295:64–6. doi: 10.1038/295064a0
2. Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. (2016) 91:e01879–16. doi: 10.1128/JVI.01879-16
3. Albina E, Truong C, Hutet E, Blanchard P, Cariolet R, L'Hospitalier R, et al. An experimental model for post-weaning multisystemic wasting syndrome (PMWS) in growing piglets. J Comp Pathol. (2001) 125:292–303. doi: 10.1053/jcpa.2001.0508
4. Hamel AL, Lin LL, Nayar GP. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. (1998) 72:5262–7. doi: 10.1128/JVI.72.6.5262-5267.1998
5. Mankertz A, Mankertz J, Wolf K, Buhk HJ. Identification of a protein essential for replication of porcine circovirus. J Gen Virol. (1998) 79:381–4. doi: 10.1099/0022-1317-79-2-381
6. Nawagitgul P, Harms PA, Morozov I, Thacker BJ, Sorden SD, Lekcharoensuk C, et al. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin Diagn Lab Immunol. (2002) 9:33–40. doi: 10.1128/CDLI.9.1.33-40.2002
7. Liu J, Chen I, Kwang J. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol. (2005) 79:8262–74. doi: 10.1128/JVI.79.13.8262-8274.2005
8. He J, Cao J, Zhou N, Jin Y, Wu J, Zhou J. Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J Virol. (2013) 87:1420–9. doi: 10.1128/JVI.01443-12
9. He W, Zhao J, Xing G, Li G, Wang R, Wang Z, et al. Genetic analysis and evolutionary changes of Porcine circovirus 2. Mol Phylogenet Evol. (2019) 139:106520. doi: 10.1016/j.ympev.2019.106520
10. Xiao CT, Harmon KM, Halbur PG, Opriessnig T. PCV2d-2 is the predominant type of PCV2 DNA in pig samples collected in the U.S. during 2014–2016. Vet Microbiol. (2016) 197:72–7. doi: 10.1016/j.vetmic.2016.11.009
11. Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J. (2010) 7:273. doi: 10.1186/1743-422X-7-273
12. Harmon KM, Gauger PC, Zhang J, Piñeyro PE, Dunn DD, Chriswell AJ. Whole-genome sequences of novel porcine circovirus type 2 viruses detected in swine from Mexico and the United States. Genome Announc. (2015) 3:e01315–15. doi: 10.1128/genomeA.01315-15
13. Segalés J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. (2012) 164:10–9. doi: 10.1016/j.virusres.2011.10.007
14. Xiao CT, Halbur PG, Opriessnig T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol. (2015) 96:1830–41. doi: 10.1099/vir.0.000100
15. Cai L, Ni J, Xia Y, Zi Z, Ning K, Qiu P, et al. Identification of an emerging recombinant cluster in porcine circovirus type 2. Virus Res. (2012) 165:95–102. doi: 10.1016/j.virusres.2012.01.008
16. Wei C, Zhang M, Chen Y, Xie J, Huang Z, Zhu W, et al. Genetic evolution and phylogenetic analysis of porcine circovirus type 2 infections in southern China from 2011 to 2012. Infect Genet Evol. (2013) 17:87–92. doi: 10.1016/j.meegid.2013.03.041
17. Franzo G, Cortey M, Segalés J, Hughes J, Drigo M. Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol Phylogenet Evol. (2016) 100:269–80. doi: 10.1016/j.ympev.2016.04.028
18. Saporiti V, Huerta E, Correa-Fiz F, Grosse Liesner B, Duran O, Segalés J, et al. Detection and genotyping of Porcine circovirus 2 (PCV-2) and detection of Porcine circovirus 3 (PCV-3) in sera from fattening pigs of different European countries. Transbound Emerg Dis. (2020) 67:2521–31. doi: 10.1111/tbed.13596
19. Morozov I, Sirinarumitr T, Sorden SD, Halbur PG, Morgan MK, Yoon KJ, et al. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol. (1998) 36:2535–41. doi: 10.1128/JCM.36.9.2535-2541.1998
20. Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, et al. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. (1998) 10:3–10. doi: 10.1177/104063879801000102
21. Shuai J, Wei W, Li X, Chen N, Zhang Z, Chen X, et al. Genetic characterization of porcine circovirus type 2 (PCV2) from pigs in high-seroprevalence areas in southeastern China. Virus Genes. (2007) 35:619–27. doi: 10.1007/s11262-007-0121-0
22. Mahé D, Blanchard P, Truong C, Arnauld C, Le Cann P, Cariolet R, et al. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J Gen Virol. (2000) 81:1815–24. doi: 10.1099/0022-1317-81-7-1815
23. Lekcharoensuk P, Morozov I, Paul PS, Thangthumniyom N, Wajjawalku W, Meng XJ. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J Virol. (2004) 8:8135–45. doi: 10.1128/JVI.78.15.8135-8145.2004
24. Xia D, Huang L, Xie Y, Zhang X, Wei Y, Liu D, et al. The prevalence and genetic diversity of porcine circovirus types 2 and 3 in Northeast China from 2015 to 2018. Arch Virol. (2019) 164:2435–49. doi: 10.1007/s00705-019-04336-4
25. Lv N, Zhu L, Li W, Li Z, Qian Q, Zhang T, et al. Molecular epidemiology and genetic variation analyses of porcine circovirus type 2 isolated from Yunnan Province in China from 2016-2019. BMC Vet Res. (2020) 16:96. doi: 10.1186/s12917-020-02304-8
26. Wang S, Xin C, Wu X, Shi J, Peng Z, Sun P, et al. Genetic characterization of Porcine circovirus type 2 from 2013 to 2018 in Shandong Province, China. Vet Med Sci. (2020) 6:76–81. doi: 10.1002/vms3.196
27. Qu T, Li R, Yan M, Luo B, Yang T, Yu X. High prevalence of PCV2d in Hunan province, China: a retrospective analysis of samples collected from 2006 to 2016. Arch Virol. (2018) 163:1897–906. doi: 10.1007/s00705-018-3823-9
28. Zheng G, Lu Q, Wang F, Xing G, Feng H, Jin Q, et al. Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China. BMC Vet Res. (2020) 16:6. doi: 10.1186/s12917-019-2193-1
29. Karuppannan AK, Opriessnig T. Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology. Viruses. (2017) 9:99. doi: 10.3390/v9050099
30. Kwon T, Lee DU, Yoo SJ, Je SH, Shin JY, Lyoo YS. Genotypic diversity of porcine circovirus type 2 (PCV2) and genotype shift to PCV2d in Korean pig population. Virus Res. (2017) 228:24–9. doi: 10.1016/j.virusres.2016.11.015
31. Shen HG, Halbur PG, Opriessnig T. Prevalence and phylogenetic analysis of the current porcine circovirus 2 genotypes after implementation of widespread vaccination programmes in the USA. J Gen Virol. (2012) 93:1345–55. doi: 10.1099/vir.0.039552-0
32. Opriessnig T, Karuppannan AK, Halbur PG, Calvert JG, Nitzel GP, Matzinger SR, et al. Porcine circovirus type 2a or 2b based experimental vaccines provide protection against PCV2d/porcine parvovirus 2 co-challenge. Vaccine. (2020) 38:1975–81. doi: 10.1016/j.vaccine.2020.01.013
33. Kim K, Hahn TW. Evaluation of novel recombinant porcine circovirus type 2d (PCV2d) vaccine in pigs naturally infected with PCV2d. Vaccine. (2021) 39:529–35. doi: 10.1016/j.vaccine.2020.12.013
34. Opriessnig T, Castro AMMG, Karuppanan AK, Gauger PC, Halbur PG, Matzinger SR, et al. A Porcine circovirus type 2b (PCV2b)-based experimental vaccine is effective in the PCV2b-Mycoplasma hyopneumoniae coinfection pig model. Vaccine. (2019) 37:6688–95. doi: 10.1016/j.vaccine.2019.09.029
35. Meerts P, Misinzo G, Lefebvre D, Nielsen J, Bøtner A, Kristensen CS. Correlation between the presence of neutralizing antibodies against porcine circovirus 2 (PCV2) and protection against replication of the virus and development of PCV2-associated disease. BMC Vet Res. (2006) 2:6. doi: 10.1186/1746-6148-2-6
Keywords: epidemiological analysis, PCV2b Cap, PCV2d Cap, homologous protection, heterologous protection
Citation: Huang Y, Chen X, Long Y, Yang L, Song W, Liu J, Li Q, Liang G, Yu D, Huang C and Tang X (2021) Epidemiological Analysis From 2018 to 2020 in China and Prevention Strategy of Porcine Circovirus Type 2. Front. Vet. Sci. 8:753297. doi: 10.3389/fvets.2021.753297
Received: 04 August 2021; Accepted: 19 October 2021;
Published: 16 November 2021.
Edited by:
Enric M. Mateu, Universitat Autònoma de Barcelona, SpainReviewed by:
Himani Dhanze, Indian Veterinary Research Institute (IVRI), IndiaCopyright © 2021 Huang, Chen, Long, Yang, Song, Liu, Li, Liang, Yu, Huang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Huang, Mzk0NTExMzM1QHFxLmNvbQ==; Xibiao Tang, dGFuZ3Jlbjc3QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.