
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci., 18 October 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.719834
This article is part of the Research TopicEmerging Zoonoses and Transboundary InfectionsView all 27 articles
Coronaviruses are single-stranded RNA viruses that affect humans and a wide variety of animal species, including livestock, wild animals, birds, and pets. These viruses have an affinity for different tissues, such as those of the respiratory and gastrointestinal tract of most mammals and birds and the hepatic and nervous tissues of rodents and porcine. As coronaviruses target different host cell receptors and show divergence in the sequences and motifs of their structural and accessory proteins, they are classified into groups, which may explain the evolutionary relationship between them. The interspecies transmission, zoonotic potential, and ability to mutate at a higher rate and emerge into variants of concern highlight their importance in the medical and veterinary fields. The contribution of various factors that result in their evolution will provide better insight and may help to understand the complexity of coronaviruses in the face of pandemics. In this review, important aspects of coronaviruses infecting livestock, birds, and pets, in particular, their structure and genome organization having a bearing on evolutionary and zoonotic outcomes, have been discussed.
Coronaviruses (CoVs) form the subfamily Orthocoronavirinae of family Coronaviridae under order Nidovirales and realm Riboviria. These are pleomorphic, enveloped, single molecule of linear, positive-sense, single-stranded RNA viruses containing genome sizes of around 30 kb among known RNA viruses (1). The club-shaped peplomers composed of large viral glycoprotein (spike or S protein responsible for attachment to cells) projecting from the envelope give a crown-like appearance of the virus under a transmission electron microscope, thus named corona meaning crown. CoV was considered a minor pathogen of the respiratory tract until 2002 in humans (2). The increased interest in its replication, transmission, pathogenesis, and distribution was pursued after an outbreak linked to the emergence of a new CoV [severe acute respiratory syndrome (SARS)-CoV] causing SARS after 2002 (2–5). Another virus called Middle East respiratory syndrome CoV (MERS-CoV) in 2014, distinct from SARS-CoV, was isolated from an outbreak of severe respiratory infection in the Middle East (6). On the other hand, an acute respiratory infection caused by an avian CoV, later named as infectious bronchitis virus with high mortality (40–90%), had shown up in the late 1920s and was the earliest report of a CoV infection in animals (7, 8). Currently, the SARS-CoV-2 and its mutated strains predominantly infecting humans with contentious animal origin have created a platform for researchers to study its genomics in-depth. Animal species play an important role as a host or reservoir in the transmission cycle of CoV, and specific receptors on their cell provide essential factors for replication and mutations within the genome of CoVs. CoVs infect various animal species ranging from livestock, poultry, cats, dogs, mice, bats, pangolins, wild felids, and other species of animals such as minks, rabbits, ducks, guinea fowls, gooses, beluga whales, etc., (9–12). These mammals are frequently studied for understanding their coevolution with human CoVs, interspecies transmission, and the emergence of new mutant strains. CoV infection in animals is mainly associated with respiratory and gastrointestinal systems resulting in mild to fatal diseases. The bovine, avian, and porcine animal groups form a major part of production industries, and in like manner, the canine and feline species have paramount importance as pets commercially due to their high demand in society. The incidence of diseases in these animal species represents a threat to the animal welfare, environment, public health, and economy, reflecting as losses in productivity, trade, market value, control costs, and food security. In this review, CoVs infecting important livestock, poultry, and pets have been discussed in relation to their structure and genome organization having a bearing on evolutionary and zoonotic outcomes.
Virions are roughly spherical and enveloped with marked spike (S) proteins that identify various specific host cell receptors and co-receptors for attachment, fusion, and entry of the virus into the cell. In addition to S proteins, other structural proteins are nucleocapsid (N) proteins, the most abundant membrane (M) proteins, envelope (E) proteins, and other non-glycosylated envelope proteins present in lower quantities, which help in the formation of an envelope. The flexible nucleocapsid within the envelope consists of genomic RNA linked to the nucleoprotein (Figure 1).
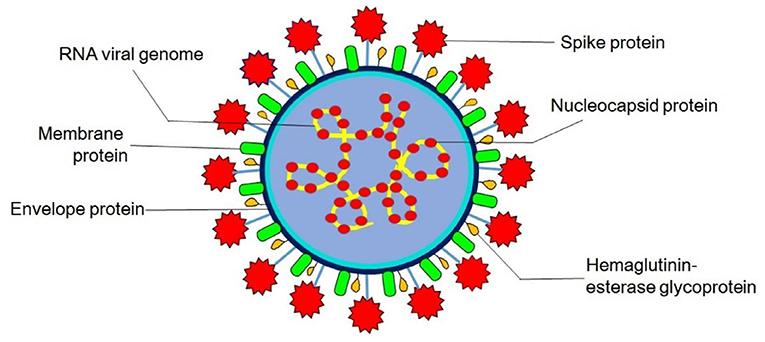
Figure 1. Structure of coronavirus. Hemagglutinin-esterase glycoprotein is exclusively present in members of Betacoronavirus (human and bovine coronavirus).
The functions of these major structural proteins of CoV are stated in Table 1.
Initially, the classification was based on their serological and antigenic properties—groups 1, 2, and 3 as opposed to newly revised taxonomy based on the level of viral genetic phylogeny. The phylogenetic analysis for classification of CoV is usually acquired by using short fragments of several conserved genes that are present in all CoV genomes and are of a significant length, such as Pol (RNA-dependent RNA polymerase), N (nucleoprotein), S (spike protein), and chymotrypsin-like protease and helicase. The envelope and membrane genes are not used in phylogenetic studies due to their short lengths (1, 22). Furthermore, complete genome sequence and proteomic approaches are also carried out to construct the phylogenetic tree of CoVs. Now, the subfamily Orthocoronavirinae is classified into four genera: alpha, beta, gamma, and delta CoVs infecting a wide variety of animal and avian species (23, 24). Betacoronavirus genus is further classified into lineages A, B, C, and D (1) and other subgenus Hibecovirus (25). The list of important CoV species classified under individual genera is given later (Table 2). Apart from this, several other animal species that harbor the CoVs are rodents, rabbits, bats, pangolin, ferrets, mink, snake, frogs, marmots, hedgehogs, and many other wild animals, as carriers or reservoirs that may need attention regarding zoonotic interventions (26–33).
The viral replication cycle of all the CoVs is confined to the cytoplasm (Figure 2); additionally, murine CoVs can also replicate in enucleated cells (34–36).
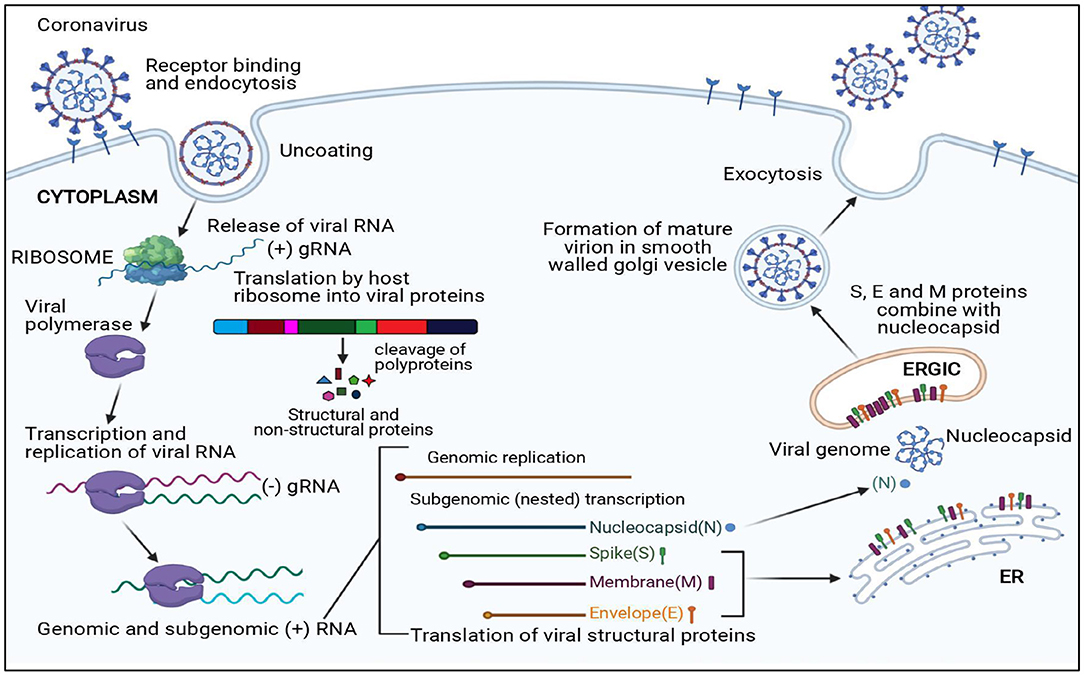
Figure 2. Replication cycle of coronavirus. N, Nucleocapsid; S, Spike protein; M, Membrane protein; E, Envelope protein; ER, Endoplasmic reticulum; ERGIC, Endoplasmic reticulum–Golgi intermediate compartment.
The first essential step of the viral replication cycle is host cell receptor recognition by S protein and its attachment to the cell. The membrane fusion event results in penetration of virion aided by “fusion peptide,” which is exposed after variable rearrangement of S protein initiated by proteolytic cleavage of spike protein and acidic pH. This is followed by synthetic events such as translation of replicase gene from viral genome and formation of polyproteins, transcription, and RNA synthesis. After replication and RNA synthesis, the S, E, and M viral structural proteins are translated and inserted into the endoplasmic reticulum. Both M and E proteins function together to form envelope and virus-like proteins. The N protein binds to viral RNA and is later accompanied by M protein, which keeps the N protein and RNA complex stable. This interaction facilitates the assembly of virus particles on the membrane of the endoplasmic reticulum–Golgi intermediate compartment and initiates the budding process. Mature virions formed within the membrane-bound vesicles are released by exocytosis. The viroporin of E protein with ion channel activity promotes virus release by altering cell secretory pathways.
The organization of large 20–32-kb size, capped and polyadenylated genome of CoV contains seven common genes in the following order, 5′-leader-untranslated region (UTR)-replicase-Spike (S)-Envelope (E)-Membrane (M)-Nucleocapsid (N)-3′ UTR-poly (A) tail.
The receptor-binding S1 subunit of spike proteins contains two distinct domains, the N-terminal domain (S1-NTD) and the C-terminal domain (S1-CTD). These domains recognize at least four protein receptors and three sugar receptors of the host cell (Figure 3) and, thus, can form the basis for classification according to the host cell recognition pattern (35, 37). The open reading frame (ORF) 1a/b encompasses a much larger section, i.e., the initial two-thirds of the genome encoding two viral replicase polyproteins—pp1a and pp1ab. These polyproteins are then further processed into 16 non-structural proteins (nsp1–16) by viral proteases and assemble to form a membrane-associated viral replicase–transcriptase complex (38–40). These are conserved among the subgroups of CoVs and thus share their relative position in the genome (41–44). Structural and some accessory proteins occupy only the last third of the coding capacity of the genome (45, 46) despite their range of complexity and function (40, 47).
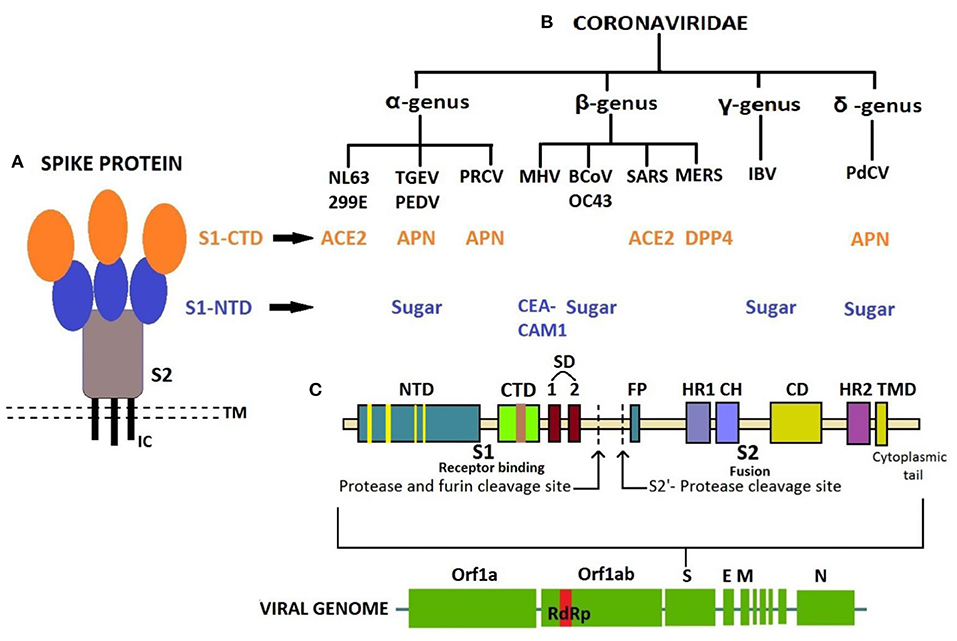
Figure 3. (A) Structure of Spike protein. (B) Classification of coronaviruses based on host cell recognition pattern by spike protein. (C) Genome organization of coronavirus and single S protein, from N- to C-terminus in left-to-right orientation. N-terminal domain in blue with receptor-binding motif (RBM) in yellow; C-terminal domain in green with RBM in brown. CTD, C-terminal domain; NTD, N-terminal domain; TMD, Transmembrane domain; IC, Intracellular tail; ACE2, Angiotensin-converting enzyme 2; APN, Aminopeptidase N; CEACAM1, Carcinoembryonic antigen-related cell adhesion molecule 1; DPP4, Dipeptidyl peptidase 4; SD, Subdomain; FP, Fusion peptide; HR1, Heptad repeat 1; HR2, Heptad repeat 2; CH, Central helix; CD, Connector domain.
Divergence in the sequence and motifs or residues of these proteins among CoVs may corroborate in classifying them in groups. Many researchers have demonstrated the phylogenetic relationships among their genomes based on the analysis of ORF1b replicase protein, 3C-like proteinase, polymerase, and structural proteins, which confirm the presence of different CoV group clusters (48–51). For instance, pairwise alignments of the corresponding ORFs and proteins of HCoV-OC43, bovine CoV (BCoV), PHEV, ECoV, and MHV suggest sequence similarity among them under the β-CoV group (50, 51). ORF8, a highly variable accessory gene and showing structural changes, plays a significant role in the evolution of SARS-related CoVs (52). The absence of a 29-nucleotide (nt) sequence in ORF8 and the presence of characteristic motif of single-nucleotide variations located in the S gene were observed in later phases of the SARS-CoV outbreak in 2002–2003 (53, 54).
The interaction between receptor-binding domain (RBD) and its host cell receptor helps in determining the CoV host range and cross-species infection (55, 56). This is dependent on the topology of RBD, its receptor-binding motif (RBM), and virus-binding motifs on specific proteins or sugars that complement each other in shape and chemical details. Both the distinctive domains, S1-NTD and S1-CTD of receptor-binding S1 subunit of CoV spike protein, can function as RBDs (57).
CoVs have been shown to mutate with high rate and recombination frequencies in their RNA genome (~10−4 nucleotide substitution/site/year). The mutations in the RBD of the spike gene are of significance, along with errors in the O-linked glycans and furin cleavage site, enzymes such as replicase and RNA-dependent RNA polymerase (58–60). The Indian SARS-CoV-2 Consortium on Genomics working on genome sequencing of new variants of SARS-CoV-2 has been reporting mutations and deletions in the amino acid sequence of spike protein since the SARS-CoV-2 pandemic. These mutational changes have led to the emergence of new double and triple mutants, alpha, beta, gamma, and delta variants with the capability of immune escape, increased virulence, transmissibility, and changes in clinical disease presentation (61).
The S1 subunits from the same genus share significant sequence similarity, whereas those from different genera have little sequence similarity (57). However, the speculation on members placed in different genera identifying the same receptor protein or those in the same genera identifying different receptor proteins still holds, despite evidence of a common evolutionary origin for the S1 subunit. The studies reveal that viral RBDs of CoVs of the same genus have a conserved CTD core structure but marked structural variations in their RBMs that enforce recognition of different receptors (62–65). Also, several other studies demonstrate that the viral RBDs of CoVs from different genus can bind to the same protein receptor due to the presence of a common virus-binding hot spot on the protein (62, 66). Thus, the data mentioned earlier provide an insight into an extensive divergent evolution of CoV S1-CTDs (67).
The crystal structure of β-genus MHV S1-NTD complexed with mouse CEACAM1 protein and BCoV S1-NTD with a sialic acid (SA) named Neu5, 9Ac2 (5-N-acetyl-9-O-acetylneuraminic acid) have the same structural fold in its core structure as for human galectins (galactose-binding lectins). Nevertheless, the BCoV S1-NTD is determined by its sugar-binding site instead of protein due to subtle changes in the conformations of their RBM loops and mutagenesis (68). This suggests that the ancestral CoVs inserted the host galectin gene into 5′ end of their spike gene, which resulted in CoV S1-NTD. After that, CoV S1-NTDs underwent divergent evolution in α, β, and γ genera, out of which S1-NTDs of β-genus BCoV, α-genus transmissible gastroenteritis virus (TGEV), and γ-genus infectious bronchitis virus (IBV) evolved their lectin activity and specificity for a different sugar receptor other than galactose. On the other hand, β-genus MHV S1-NTD subsequently lost its lectin activity and evolved specificity for a novel protein receptor, CEACAM1 (62). The S1-NTD of newly identified porcine delta coronavirus (PdCoV) (1, 69) shares a similar structure as α-CoV, β-CoV, and host galectins; thus, it recognizes sugar as its potential receptor and binds to sugar moiety of mucin to facilitate initial viral attachment, whereas the S1-CTD has the same structural fold as α-CoV S1-CTDs, but it differs from that of S1-CTDs of β-CoV (70). The PdCoV S1-CTD has a significant affinity for pig cells known to express aminopeptidase N (APN) as efficiently as TGEV-S1. Therefore, the porcine APN acts as a functional cross-genus receptor for both enteropathogenic PdCoV and TGEV for cellular entry (71). Such similarities suggest a close relationship between PdCoV, α-CoV, and β-CoV evolutionarily; however, PdCoV belongs to Deltacoronavirus owing to its genomic similarities with the avian species suggesting an ancestral avian origin (24).
The evolution of the spike protein of CoVs has also been proposed to help the virus in surviving against host immune response similar to the influenza virus (72, 73). The S1-NTDs of CoVs have also evolved the ceiling-like structure on top of its core to protect these sites and to evade the immune surveillance by the host immune system (74). As per the location of S1-NTDs and S1-CTDs on spike protein, the tips with S1-CTD are the protruding region and most exposed directly to the host immune system and therefore evolves at an increased pace to combat the host immune surveillance. Based on the immune pressure and different receptors (some of which are still unidentified) recognized by different animal CoV groups, it is clear that the S1-CTD exhibits a common evolutionary origin and has undergone divergent evolution. Moreover, the monoclonal antibodies directed against spike protein demonstrate common antigenic determinants for β-CoVs, especially the members of subgroup embecovirus, i.e., BCoV, PHEV, and HCoV-OC43, which corresponds to a close antigenic relationship (75, 76). This can put forward some hypotheses concerning the origin of β-CoV members, adaptation to a human host, and recombination events leading to novel CoVs with different species specificity responsible for emergence.
The crystal structure of the N-terminal domain of nucleocapsid protein of MHV shares a similar topology structure with that of SARS-CoV and IBV containing five short β-strands (arranged as β4-β2-β3-β1-β5) across a U-shaped β-platform (Figure 4) but differs in its potential surface, indicating a possible varied RNA-binding module (77). The three residues, Arg-125 and Tyr-127 on the β3 strand and Tyr-190 on the β5 strand, provide a key role in transcriptional regulatory sequence RNA binding and helix destabilization essential for replication. These residues are totally invariant in betacoronavirus N proteins and incisively occupy analogous positions on the fold of each NTD, therefore likely to define similar RNA binding grooves between them (78). On the other hand, the sequence comparison of the C-terminal domain of N protein, also referred to as the dimerization domain as its residues form homodimers and homo-oligomers (oligomerization) (79, 80), shows that the domain is conserved at least among the alpha, beta, and gamma groups of CoVs, suggesting a common role for this domain.
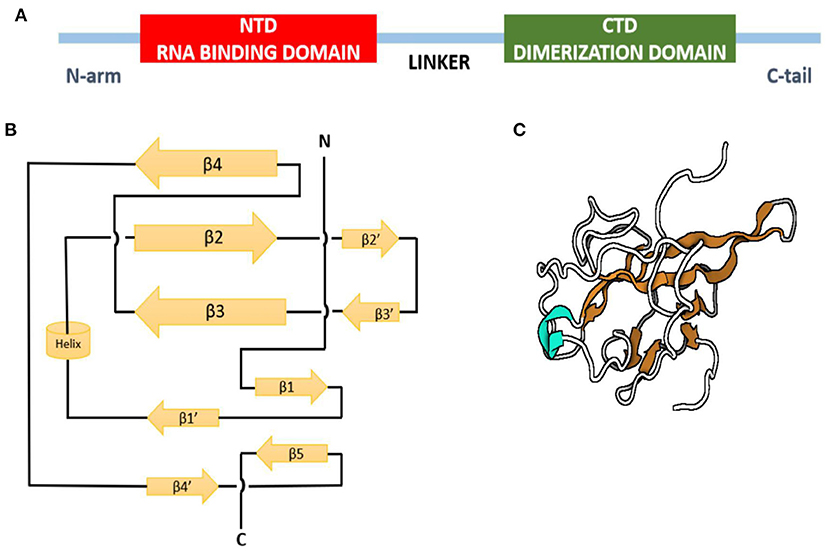
Figure 4. Nucleocapsid protein of infectious bronchitis virus. (A) Domain structure of N protein. (B) Topology diagram of N protein. (C) Three-dimensional structure of N protein. Beta strands in orange, and helix is shown in cyan.
The positively charged groove formed by the presence of the eight positively charged lysine and arginine residues of CTD is similar in SARS-related viruses and IBV-N CTD, except that the positively charged surface area in the SARS-CoV is larger than IBV (81) due to the absence of two lysine residues and the presence of additional negatively charged residues in the IBV N protein (82). Oligomerization and interaction of proteins with the viral genome is required for packaging of the genome by CoV N proteins to form ribonucleoprotein complexes for viral assembly (83). These functions of N proteins are performed similarly by SARS-CoV, IBV, and MHV. Thus, the overall similarity in the topology of the NTD and CTD domains of the N protein from SARS-CoV, IBV, and MHV fortifies a conserved mechanism of nucleocapsid formation for CoVs (74).
The primary sequence of the E proteins shows large variations in sequence and size among the groups with <30% identity and conserved membrane amino acid residues (84). Multiple membrane topologies of E proteins have been determined between different CoVs depending on the level of protein expression and oligomerization (84). The experimental studies have shown that the IBV E protein exhibits topology of cytoplasmic C-terminus while N-terminus in the lumen of the Golgi complex (85). Conversely, the TGEV E protein has a luminal C-terminus and N-terminus located cytoplasmically (86, 87). The CoV E proteins of only IBV, SARS-CoV, and MHV function for palmitoylation, i.e., modulation of protein–protein interactions, subcellular trafficking of proteins across the membrane, and membrane anchoring (88–91). A protein-binding motif located at the end of the C-terminus is highly conserved in α- and β-CoVs and is not found in the γ-CoVs (92).
The primary M protein sequence varies, although the secondary structures and an amphipathic region of the transmembrane domain are also conserved in almost all the members of the family (93). The type of glycosylation in the M protein of α- and δ-CoVs is N-linked, whereas O-linked glycosylation is found in the β-CoVs, but it is not critical for the viral assembly (19, 94–96).
The hemagglutinin-esterase (HE) gene is exclusively present in members of β-CoVs. CoV HE proteins were firstly identified from the PHEV, BCoV, and HCoV-OC43 bearing SA-9-O-acetylesterases similar to a hemagglutinin-esterase fusion protein of influenza C virus (97). The HE gene of CoV shares 30% sequence identity with the subunit of a HE fusion protein and has been found to be acquired by independent, non-homologous recombination events or evolutionary trajectories between influenza virus, torovirus, and CoV (98–100). All the CoV HEs are O-acetyesterases, whereas BCoV and HCoV-OC43 have dual activity of both hemagglutination and acetylesterase (101). Both these CoVs can agglutinate chicken erythrocytes, whereas purified HE protein of BCoV only agglutinates Neu5, 9Ac2-enriched erythrocytes of rodents. On the contrary, purified S glycoprotein can agglutinate chicken erythrocytes (102). This indicates that the major hemagglutinin is the S protein that also acts as the major SA-binding protein. The function of the hemagglutinin-esterase enzyme relies on the distinctive carbohydrate-binding domain as lectin and receptor-destroying enzyme domain (Figure 5).
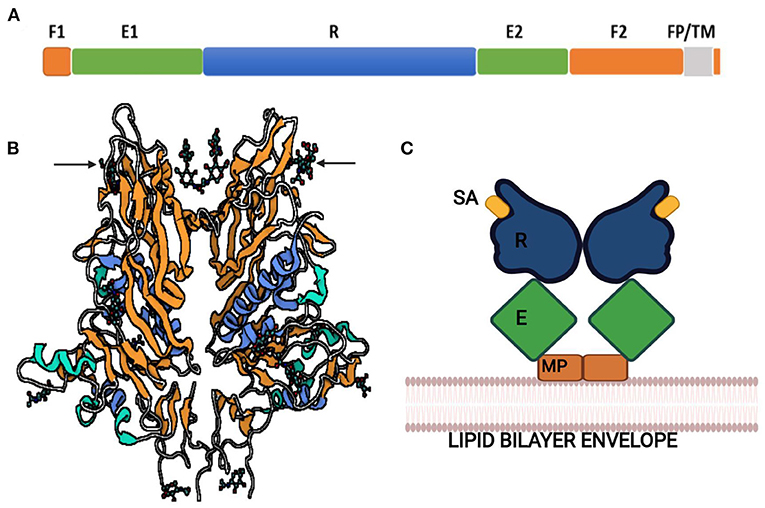
Figure 5. Hemagglutinin-esterase protein of bovine coronavirus. (A) Linear order of sequence segments HE protein. F1 and F2, Fusion domains; E1 and E2, Enzyme domains; R, Receptor domain; FP, Fusion peptide; TM, Transmembrane domain. (B) Ribbon representation of HE protein. Lectin domain in orange, esterase domain in blue, and structures shown by arrows are sialic acids (sialic acid-9-O-acetylesterase) bound to lectin domain. (C) Schematic illustration of HE dimer. SA, Sialic acid; R, Receptor domain; E, Enzyme domain; MP, Membrane-proximal domain.
HE protein with these domains and SA-O-acetylesterase activity mediates viral entry with S glycoprotein and attachment to the O-acetylated SA receptors on the host cell. The acetylesterase of murine CoVs prefers to esterize 4-O-acetyl-NeuAc and thus has different substrate-binding specificity than BCoV and HCoV-OC43, which targets 9-O-acetyl-SA (103). The combined activity of S glycoprotein and HE is specific for human CoV attachment to SA-associated receptors on the host cell (104), but the role of HE protein in HCoV other than HCoV-OC43 is not known much. However, the HE protein of SARS-CoV-2 also acts as the classical glycan-binding lectin and receptor-destroying enzyme and may show evolutionary adaption toward recognition of O-acetylated SA and virus entry for viral–host interaction (105).
BCoV belonging to genera Betacoronaviruses subgroup A along with swine HEV, canine respiratory CoV, feline enteric CoV, human CoV-OC43, and HKU1 is associated with three major clinical syndromes: neonatal calf diarrhea, hemorrhagic winter dysentery in adult cattle, and respiratory infections in cattle of different ages (106–118). It is an important livestock pathogen having an economic impact on the cattle industry worldwide (119). It is a leading cause of enteritis in combination with other enteric bacterial, viral, parasitic, and protozoal pathogens and is also found to be involved in the bovine respiratory disease complex in feedlot cattle since its discovery in 1993 (106). The host range includes all breeds of cattle and wildlife ruminants. The SA receptor for BCoV reflects wide tissue tropism due to the presence of sugars in abundance for interaction between viral spike glycoprotein and a specific carbohydrate receptor (102).
The BCoV variants, which are genetically and/or antigenically related, have also been isolated from other animal species along with humans representing a similar respiratory and enteric form of the disease (9, 120–123). Despite antigenic variations between different strains and interspecies transmission, only a single serotype is evident (124).
Infection is probably distributed worldwide—Africa, Asia, Europe, Oceania, and North and South America (125). The virus is shed in feces and nasal secretions predominantly. A study on naturally and experimentally infected animals revealed an excess of virus load isolated from nasal swabs and massive replication in airways, whereas the fecal shedding started later (126). It is readily transmitted by the feco-oral or respiratory route indirectly and directly by direct contact or aerosols on farms, maintained by a clinically normal cow and calves where adult animals can act as carriers. Calves of 1 week to 3 months of age are highly susceptible due to inadequate maternal antibodies. The adults are usually subclinically affected, and the virus may be excreted intermittently at low titer (127).
The virus replicates in the epithelial cells of the upper (nasal turbinate, trachea) and lower (terminal bronchioles, lungs) respiratory tract: intestinal tract mainly along the lining of villi and crypts of epithelial cells. These cells are capable of resisting viruses and have the ability to replace the damaged cells (106), and thus, calves may recover from infection. The replication results in the destruction of mature absorptive cells lining the villi and mucosal surface in the large intestine, necrosis of cells in mesenteric lymph nodes and payer's patches, and subsequently viremia (125). This diminishes the absorption in the gut, failure to secrete digestive enzymes impairing the glucose and lactose metabolism and causing malabsorptive diarrhea. Pathological lesions such as marked intestinal hemorrhages and extensive cell necrosis within crypts are observed, whereas lesions of the respiratory system include hemorrhages, atelectasis, intestinal pneumonia, and emphysema (128).
Infection in calves depends on age and their immune status. Coinfection with Campylobacter jejuni, enterotoxigenic or enteropathogenic Escherichia coli, and Rotavirus may expedite the severity of the disease. The morbidity rate is 20–100% in affected animals, but mortality is 1–2% depending on the level of maternally or actively derived antibodies and severity of dehydration (119). The incubation period in calves is 24–48 h, and the clinical signs include profuse diarrhea, which subsequently results in dehydration, acidosis, and death in uncontrolled cases (123). In adult animals, the incubation period is 2–7 days and is the cause of acute sporadic enteritis prevalent during winter months, thus named winter dysentery. The disease is characterized by explosive, often hemorrhagic diarrhea, anorexia, emaciation, and unthriftiness along with decreased milk production along and frequent respiratory signs including fever, rhinitis, dyspnea, rales, pneumonia in 2–6 months old calves, and serious respiratory distress followed by death (125). The pneumotropic strains of the virus in adults are the precipitating cause of the bovine respiratory disease complex that exacerbate the fatality when manifested by superimposed environmental or managemental stress.
Infectious bronchitis is an acute, highly contagious disease responsible for the economic impact on the poultry industry. The majority of CoV of avian species are classified into genera gamma- and delta-CoVs, within which IBV is of significance belonging to the gamma genus. Chickens and pheasants are the natural hosts but have also been detected in turkey, duck, guinea fowl, pigeon, peafowl, goose, teal, and partridge (129). IBV has a primary affinity for the respiratory system, accompanied by infection in the reproductive, renal, and alimentary systems. IBV occurs in various antigenic variants with a difference in virulence and tissue tropism as a result of mutations and recombination in its genome. The multiple serotypes of IBV based on S1 spike protein difference present a challenge in establishing an effective vaccination program, as cross-protection is found to be poor (130–132).
The distribution is worldwide, but some may have restricted geographical spread where different antigenic variants can co-circulate in a given region (133). It is of significant concern in poultry industries due to poor weight gains in broilers and suboptimal downgrading of egg production in layers (134). Birds of all ages are susceptible to the infection, but the severity and clinical signs may vary (135). In the acute phase of infection, IBV is copiously shed in respiratory secretions, tracheobronchial exudate, and feces and is spread by aerosols, ingestion of contaminated feed, drinking water with feces, and indirect transmission between birds at the farm over long distances through fomites (136). The vertical transmission is not clearly understood; however, the virus was isolated from day-old chicks and recovered from the semen of cockerels after inoculation (137, 138). The excretion and persistence of some strains of virus for a considerable time in target sites such as kidney or alimentary tract, particularly cecal tonsils followed by re-excretion, is suspected to be influenced by adverse environmental or changed physiological conditions, which suggests carrier state or latency (134). The strain of virus, route of exposure, age, diet, nutrition (level of calcium), and external factors such as cold stress in winter, poor ventilation, and coinfection with enteric bacteria provoke the disease; also, breed factor, such as some heavier birds become more susceptible, may be related to immune response (129). Although the morbidity rate is high as 100%, the mortality rate can vary from 20 to 30% and more depending on vast tissue tropism, secondary bacterial infection, and standards of management in an infected flock (139).
The initial replication occurs in ciliated epithelial cells of the respiratory tract, which cause histopathological lesions mainly in the trachea, such as ciliary loss, desquamation, epithelial hyperplasia, edema, marked lymphoplasmocytic inflammation, and mononuclear and heterophilic cell infiltration of the submucosa, which resumes in 14–21 days after infection (140, 141). A succinct viremia within 1–2 days eventually leads to extensive spread to the reproductive system, kidneys, and intestinal tract, but the damage is minimal, and bursa of fabricius may be the cause of immunosuppression. The main attachment factor for IBV is the receptor-binding domain in S1 spike glycoprotein and SA glycans (142, 143) widely distributed in host tissues; thus, variation in the glycoprotein and glycans partly determine the virulence, tissue binding, and tropism. The gross pathological findings include congested respiratory tract with serous or catarrhal exudate in nasal passages, trachea, extrapulmonary bronchi, and air sacs. The main bronchi get blocked with caseous casts in young chicks, the probable cause of death. The epithelial cells of the oviduct, mainly the goblet cells, become cuboidal, hypoglandular oviduct, ovarian regression, and congestion; sometimes, the ova may rupture, resulting in free yolk in the abdominal cavity (144, 145). Extensive tubular degeneration, interstitial inflammatory response characterized by the pale, enlarged, or marbled kidney, ureters distended with deposits of urates, and large uroliths are seen in the chronic stage of nephritis (141, 146, 147). Certain IBV strains also induce pathological lesions in deep and superficial pectoral muscles, i.e., bilateral myopathy in broilers and breeders (148).
The incubation period is 18–48 h; the course of the disease lasts for 5–7 days and, in outbreaks, up to 14 days (146, 149). Chicks with an age of 2–6 weeks are severely affected, although birds of all age groups are susceptible. The main three clinical manifestations are respiratory, reproductive disorder, and nephritis (134). The most conspicuous clinical findings are the initial respiratory signs—gasping, tracheal rales, dyspnea, swollen sinus, conjunctivitis, profuse lacrimation, cellulitis of periorbital tissues, and coughing with or without nasal discharge. This is followed by lethargy, ruffled feathers, anorexia, rapid weight loss, stooped stance, scouring, excessive water intake, and characteristic wet litter implying nephritis. The reproductive disorder shows signs of rales followed by a marked decline in egg production up to 50–70%, usually within 8–12 days, which differs depending on the stage of lay at infection, hampering the hatchability rate (150). The external and internal quality of the egg is highly affected, exhibiting misshapen eggs, thin, soft, or no shell, ridging and distortions, watery albumen, which may resume within 8 weeks or more. It may also lead to permanent damage to immature oviduct resulting in so-called false layer syndrome, as the layers or breeders never resume the loss of egg production. The concurrent secondary infection with E. coli, avian mycoplasma species, etc., or nephropathogenic strain of IBV may expedite the infection causing air-sacculitis and interstitial nephritis (151). Chicks may die suddenly by occlusion in bronchi as a probable cause of death.
Among few porcine CoVs known, the clinical disease is mainly associated with the TGEV. It is highly contagious among young pigs and is found to be a significant cause of economic loss more in breeding herds than the rearing and finishing herds, primarily due to piglet mortality (152). The porcine epidemic diarrhea virus (PEDV) is clinically similar but serologically unrelated to TGEV and comparatively spreads slowly in the herd (11). The porcine respiratory coronavirus (PRCV) is a non-pathogenic respiratory variant of TGEV (153, 154). It can cause subclinical mild respiratory disease and serologically cross-reacts with TGEV, but tests are available to distinguish them. These porcine CoVs are grouped in genus Alphacoronavirus; however, a new porcine delta-CoV genetically distinct from TGEV and PEDV associated with enteric disease in pigs has recently been found (155).
TGEV is reported all over the world, affecting the global pork industry (156). It spreads between and within farms by shedding infected feces for up to 2–3 weeks. The virus may also spread through fomites, aerosols at least for short distances, or mechanical spread by animals, insects, or birds, particularly starlings and in milk or feces to the piglets (157). The infection occurs throughout the year but mainly follows a seasonal pattern with a higher incidence in colder months (158). Infection results in two different clinical presentations: epidemic and endemic (159). In epidemics, when a virus enters a naive herd, pigs of all ages are affected, particularly the newborn piglets, whereas the infection is self-limiting in farrowing and finishing herds. The endemic disease is observed in farms after the epidemic phase due to incomplete all-in-all-out management or continuous movement of naive gilts in breeding farms. TGEV can end up showing mild disease, thus presenting high morbidity up to 100% in neonatal piglets but low mortality (11).
The virus enters through the oro-nasal route and replicates in enterocytes of the small and large intestines. The replication causes shortening and blunting of villi, mainly in jejunum and ileum, due to the segmental nature of lesions followed by malabsorption, disruption of cellular transport of nutrients and electrolytes, and increased osmolarity, thinning of the gut wall, and diarrhea (160, 161). The crypts of epithelial cells usually remain uninfected; thus, recovery of function of villi is rather rapid. In neonates and piglets, a combination of these factors coupled with the slow regeneration time of epithelial cells results in death. TGEV has also been found to replicate in extra-intestinal tissues, including lungs and mammary gland, causing imprecise pneumonia and agalactia, respectively (162, 163).
The incubation period is short 12 to 72 h, i.e., up to 3 days (164). TGEV presents a mild disease except in piglets ≤3 weeks of age that may succumb to death and in sows infected at or near farrowing. Vomition is the initial sign in a non-immune herd followed by profuse watery diarrhea, rapid dehydration, weight loss, marked thirst, and agalactia with recovery within 5–10 days (11, 165). As the disease progress in unweaned piglets, feces often contain curds of undigested milk and may approach 100% mortality due to the slow replacement rate of villous cells. The course of disease in porcine CoV infections does not exceed 3–4 weeks normally due to rapid herd immunity; thus, the mortality is low, but morbidity is high (159). In some herds, the TGEV remains subclinical, although there may be short episodes of clinical reemerging infection particularly due to the purchase and replacement of breeding pigs and their litters.
Feline CoVs are classified into two biotypes based on the pathogenicity referred to as feline enteric coronavirus (FECV) and feline infectious peritonitis virus (FIPV) belonging to genus betacoronavirus and alphacoronavirus, respectively. Higher sequence similarity in both the biotypes indicates a close relationship but with distinct virulence properties (166–169). The FIPV is primarily observed in a cat population that is tenaciously infected with FECV (170, 171). These recurrent observations and animal experiments have led to the widely accepted theory of “internal mutation” that suggests an evolution of FIPV from non-pathogenic FECV by specific mutation(s) occurring in the viral genomes (172–179). The sequence differences in spike and membrane protein, mutations in theS1/S2 locus, furin recognition site, and disrupted NSP3c genes may further contribute to the risk of FIP in the individual (180). However, very less is known about the stage at which mutation(s) occur during the development of FIP. The FCoVs are further separated into two serotypes based on the serological properties of the virus (181). Some independent studies provide constant evidence that the emergence of serotype II viruses is via double homologous recombination between serotype I FCoV and canine enteric coronavirus (CCoV) (170, 182–186). Both serotypes I and II can cause FIP and clinically inapparent FECV infections.
FCoV infection is widely disseminated in the domestic and wild feline population. The seropositivity varies from 20 to 60% and approaching up to 90% in domiciled cats, multi-cat households, catteries, and animal shelters, according to the global data (171, 187–189). The serotype I FCoVs are mostly responsible for natural infections (187, 190–192). Serotype I FCoV strains are vastly isolated from the United States and Europe (80–95%), whereas serotype II predominates in Asia in up to 25% (192–196) analysis. The seroprevalence studies had demonstrated high incidences and seropositivity in cats from 3 months to 3 years old and in adult individuals (190, 197, 198). There is no significant difference in seropositivity related to sex and breed of cats; however, genetic predisposition can affect the reproductive condition, hereditary factors and systemically manifest the disease (171, 199). FIPVs in animals are less likely to be transmitted horizontally, and infection due to contact with feces from diseased cats (172, 178, 200–202) is thought to be limited. However, immunosuppression favored by stress or coinfections with feline immunodeficiency virus and feline leukemia virus may trigger the progression of FIP in some cases (171). In contrast, FECV is highly contagious and transmitted horizontally through the fecal–oral route (167, 171, 188). The infected cats can continually shed FECVs in their feces for a longer period and even in postinfection, which may last for several months but with low virus load (167, 203).
The main site of FECV replication is the apical epithelium of the villi from the lower portion of the small intestines extending to the cecum (167, 203). In addition, the viral RNA can be recovered from blood and different tissues as well, suggesting the capability of FECV to infect peripheral monocytes, albeit less efficiently (179, 203–207). FIPV presents an altered cell tropism and infects both monocytes and macrophages (170, 206, 208, 209). The distribution of macrophages in the body results in viral dissemination from the intestine to the spleen, liver, and central nervous system. Thus, it is considered as an immune complex disease involving activation of these cells (210) and expression of tumor necrosis factor-α, interleukin-1β, adhesion molecules, matrix metalloproteinase-9, vascular endothelial growth factor, vasoactive amines, and inflammatory mediators (210–217). These factors, along with less susceptible leukocytes activated by an unknown mechanism during FIPV infection, induce capillary endothelial cell retraction, increased vascular permeability, and hence protein-rich effusion in body cavities (218, 219). Therefore, the FIPV infection is characterized by fibrinous and granulomatous serositis, protein-rich serous exudates in body cavities, and/or pyogranulomas (213, 220–223).
The infection caused by FECV remains persistent and asymptomatic and/or induces mild and transient diarrhea and occasionally causes severe enteritis (224). Feline infectious peritonitis is an immune-mediated, systemic, and fatal disease (190). The infection can be clinically distinguished into three forms based on the presence or absence of protein-rich effusions in the pleural and abdominal cavities—wet (effusive), dry (non-effusive), and a combination referred to as mixed form (171, 188, 213, 225, 226). The clinical progression of the disease is believed to be dependent on the host cellular and humoral immune responses. The wet form is associated with weak cellular but robust B cell responses, whereas the dry form is caused by strong T cell immune responses (171, 188). It has been observed that wet form is more prevalent in natural infections than other forms and frequently develops in the terminal stage of dry form resulting in subsidence of the immune system (171, 188). The wet form is characterized by abdominal, thoracic, or pericardial effusions leading to fluidic waves in the abdomen, visceral and omental adhesions or enlarged mesenteric lymph nodes, dyspnea, or tachypnea, cyanotic mucous membranes, and muffled sounds in the lungs and heart. The dry form is characterized by granulomatous changes in several organs, including the central nervous system and the eye. The ocular lesions include white sheathing of retinal vasculature, mild uveitis, keratin deposition in the cornea, and hemorrhages in the anterior chamber and retina. In cats with FIP, the neurological signs are variable and can induce multifocal lesions. Ataxia with subsequent seizures, tremors, nystagmus, incoordination, and hyperesthesia is the most common clinical sign. When FIP lesions are associated with cranial nerve, visual impairment and loss of menace response are observed, whereas lameness or paresis can be seen in peripheral nerve involvement.
CoVs of Canidae family fall in two groups— CCoV in group 1 alphacoronavirus and canine respiratory coronavirus (CRCoV) in group 2 betacoronavirus. CCoV has been described since 1971 (227) and exists in two closely related serotypes—CCoV-I and CCoV-II based on random point mutations and recombination associated with the spike protein and distinct serological properties (182). Type I and II CCoVs and FCoVs have been proposed to be closely related based on their evolution through recombination events from a common but unknown genetic source (186, 228). CCoV-II can be further classified into CCoV-IIa and CCoV-IIb as a result of the recombinant origin of CCoV with NTD of spike protein homologous to TGEV (229, 230). A novel CCoV with CCoV-I- or FCoV-I-like NTD was discovered in 2014 and was referred to as CCoV-IIc but has not been classified into any clade yet (231–233). CCoV generally causes mild and self-limiting diarrhea with low mortality and high morbidity (234, 235). Virulent and pantropic strains of CCoV causing severe enteric and fatal systemic diseases in the absence of coinfection with canine adenovirus type I and canine parvovirus type 2 have also been reported (236–244). Therefore, CCoV is now considered a significant pathogen in the dog population due to its ability to evolve into variants with altered tissue tropism and pathogenicity.
CRCoV was newly recognized in 2003 from the tracheal and lung samples of dogs facing enzootic respiratory disease (9). It is one of the members of multiple etiologies causing canine infectious respiratory disease along with canine parainfluenza virus, canine adenovirus (CAV) type 2, canine herpesvirus and canine influenza virus, Bordetella bronchiseptica, Streptococcus equi subsp. zooepidemicus, and Mycoplasma spp (245, 246). CRCoV was found to carry an additional gene encoding for HE protein; thus, it shows high sequence identity up to 98% with BCoV and human CoV OC43 along with similarities in spike protein and polymerase gene sequences (9, 247).
Both CCoV and CRCoV are common infections of the canine population with worldwide distribution (121, 248–258). Single, as well as multiple infections, have been reported with more than one genotype of CCoV that indicates co-circulating of CCoV–I, CCoV-IIa, and CCoV-IIb strains in prevalent regions (259). Canine CoVs are highly prevalent in dogs living in dense populations such as shelters, kennels, or grouped environments and thus exhibit rapid transmission through feco-oral and naso-oral routes (260). Dogs are likely to act as clinically normal carriers maintaining the infection in the canine population due to long time shedding of CCoV after postinfection and clinical resolution (261, 262). Apart from the domestic dogs, canine CoV infection has also been reported in foxes, wolves, and raccoon dogs (263, 264). The infection occurs throughout the year in dogs, whereas CRCoV is frequently detected during the fall to winter months (265). Canine CoV infections can occur in all age groups, significantly more in young puppies for CCoV in contrast to CRCoV, which is most prevalent in dogs more than 1 year age (254, 255).
The pathogenesis of CCoV is similar to that of other enteric pathogens. It replicates in the apical and lateral mature epithelial cells of intestinal villi resulting in villous atrophy and consequently malabsorption and diarrhea (233). The severe form of enteritis represents gross pathology as moderate, diffuse, segmental hemorrhagic and necrotic enteritis, ileo-cecal intussusception, along with infiltration of lymphocytes and plasmacytes (244). Systemic infection caused by pantropic CCoV produces lesions in several organs, including infarction in the renal cortex, fibrinopurulent bronchopneumonia, fatty change in the centrilobular zone of the liver, multifocal hemorrhages in the spleen, and depletion of gut-associated lymphoid tissue (266).
The pathogenesis of CRCoV is mainly associated with the trachea, nasal cavity, and nasal tonsil with less severity in the lower respiratory tract. It causes distortion of the ciliated respiratory epithelium and infiltration of inflammatory cells resulting in failure to clear the particulate matter in the lungs, bronchi, etc., (267). The virus has also been isolated from the colon, mesenteric lymph nodes, and spleen (121, 257). This suggests dual tissue tropism similar to BCoV, although the ability to replicate in tissues other than the respiratory tract needs further investigation. Furthermore, the possible interaction of multiple pathogens during the canine infectious respiratory disease complex needs to be considered as contributing factor in the pathogenesis of CRCoV.
CCoV generally causes mild and self-limiting diarrhea in dogs. The severity of enteric disease increases when infected with multiple pathogens or pantropic strains (CCoV-IIa biotype) of CCoV. The clinical signs thus include gastrointestinal distress, hemorrhagic diarrhea, along with neurological signs (268, 269). The infection with CRCoV exhibits common and mild clinical signs associated with the upper respiratory tract, including sneezing, coughing, and nasal and ocular discharge (267), which may progress to bronchopneumonia and multisystemic illness depending on the involvement of other organs.
Enteric and respiratory infections of CoVs are mainly associated with the shedding of the virus through feces and nasal secretions, respectively. The clinical samples for diagnosis include feces, intestinal contents, nasal secretions, tracheobronchial lavage fluids, and postmortem specimens comprising of nasal, pharyngeal, tracheal, lungs, and tissues from different regions of the gut focusing primarily on the distal small intestine. In addition, trachea, kidney, proventriculus, tonsil, and oviduct specimens for IBV and aqueous humor, whole blood and fine-needle aspiration, a biopsy of the liver, spleen, and mesenteric lymph nodes for FIP are obtained for diagnosis depending on the clinical presentation of the animal. Direct detection of virus in clinical samples by transmission electron microscopy, immunofluorescence, immunoperoxidase, or immunohistochemical staining of tissues using hyperimmune antiserum or monoclonal antibodies provides definitive diagnosis (171, 213, 221, 270–284). Immunohistochemistry using an antibody directed against BCoV can also help in detecting CRCoV (285). The laboratory tests using effusions have more diagnostic value than blood tests for FIP (286, 287). Cytological and macroscopic examination, along with cell count and biochemical properties of effusions, can be carried out for differential diagnosis (283, 286, 288–290). A simple, quick, and inexpensive “Rivalta's test” with more than 90% sensitivity and 66–81% specificity for differentiating transudate from an exudate can be useful to exclude FIP and rule out other causes for the effusions (287, 291).
Tracheal organ culture, McClurkin swine testicle (ST) cell line, human rectal tumor HRT-18 cells, Vero, and other cell lines derived from specific host species can be used for primary and secondary virus isolation and propagation favoring syncytia, plaque, or cytolysis induction (111, 112, 116, 118, 231, 291–297). To improve the detection of the typical cytopathic effect, it may require the addition of pancreatin or trypsin to the cell culture along with additional blind passages. The allantoic cavity of 9–10-day-old chicken embryo inoculated with IBV-infected material exhibits curling and dwarfism as characteristic IBV lesions observed in the embryo (298, 299). On the other hand, virus isolation of CCoV-I and CRCoV is often unsuccessful and, even if achieved, does not produce cytopathic effects (121, 183, 184).
Serological assays such as enzyme-linked immunosorbent assay (ELISA), virus neutralization test, immunofluorescence antibody test, rapid immunochromatographic tests, and blocking ELISA using monoclonal antibodies to differentiate between strains and serotypes of particular CoV are used (300–309). Serological tests are of limited value in IBV, CCoV, and FCoV, as they fail to discriminate between several serotypes. The BCoV antigens can also be used against canine sera instead of CRCoV in ELISA, serum neutralization, or hemagglutination inhibition test (9, 238, 256). The supernatant from CRCoV-infected cell culture was able to agglutinate chicken erythrocytes at 4°C, which means that hemagglutination assays can be optimized to detect CRCoV (310).
The highly sensitive molecular assays including serotype-specific reverse transcription-polymerase chain reaction (RT-PCR), nested PCR, real-time quantitative RT-PCR using conserved gene regions—UTR, N-gene, S1 gene, or HE gene (257), reverse transcription loop-mediated isothermal amplification assay, reverse transcription recombinase polymerase amplification assay, pan CoV RT-PCR are used providing high detection rates than other assays (176, 261, 286, 302, 311–339). The next-generation sequencing to decipher the whole genome within a short period is currently being used in advanced laboratories (340, 341).
With an increasing rate at which serious infections with new variant strains of human CoVs, especially SARS-CoV-2 spread, the need was felt to deploy rapid, accurate, and precise diagnostic tools for laboratory and point-of-care (POC)-based settings. This includes nucleic acid amplification testing such as the centralized laboratory-based real-time RT-PCR (the current gold standard for etiological diagnosis), rapid POC based tests such as lateral flow assays, rapid serological (antibody or antigen) tests, LAMP test, and serological assays such as ELISA and automated EIA (342–345). Some novel strategies for SARS-CoV-2 detection include CRISPR/Cas (reliable on-site diagnostic method)-based paper strip test, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry combined with artificial intelligence, surface-enhanced Raman scattering spectroscopy, metabolomic approaches, aptamer (third-generation molecular probe)-based diagnostic tests, proteome microarray, optical biosensor, antigen-Au/Ag nanoparticle-based electrochemical biosensor, and surface plasmon resonance (346–350). Despite rapid refinement in existing tools and deployment of novel strategies, the advancement in diagnostics of animal CoVs comparatively fall behind and mostly rely on clinical diagnosis, detection, and titration of CoV particles (plaque assay, electron microscopy, and transmission electron microscopy), detection of CoV antibodies (ELISA and EIA), detection of CoV antigen (monoclonal- and polyclonal-based ELISA, rapid Ag detection tests, immunofluorescence, and immunochromatographic assays), and nucleic acid-based assays (RT-PCR, real-time PCR, and loop-mediated isothermal amplification-PCR). Although these molecular and serology-based methods provide accurate results, they require well-trained technicians, specific types of equipment, and ample time and effort and are not convenient for use on farms or by breeders. For this purpose, various studies have been undertaken to develop efficient and appropriate diagnostic tools for animal CoVs as has been seen with SARS-CoV-2. The diagnosis for a newly identified pathogen, porcine delta CoV, was made possible by developing a cost-effective fluorescent microsphere immunoassay as used for PEDV (351) that could detect antibodies against multiple target antigens of porcine delta CoV for better efficiency and sero-surveillance on a herd level (352). Recently, a europium (III) chelate microparticle-based lateral flow test strip was developed for identification and epidemiological surveillance of PEDV with high reliability and sensitivity (353). A novel multiplex PCR-electronic microarray assay for rapid and comprehensive detection of bovine respiratory and enteric pathogens, including BCoV, was also developed (354). A highly specific plaque reduction neutralization test was combined with two sensitive molecular methods: real-time RT-PCR and Sanger sequencing, to investigate SARS-CoV-2 infection in cats and dogs in Brazil (355). Similarly, a Luciferase Immunoprecipitation System assay using the fragments of spike protein and nucleoprotein as antigens was used to detect antibodies against SARS-CoV-2 in dogs and cats and MERS-CoV in dromedary camels and monkeys (356–358). For Luciferase Immunoprecipitation System assay, there is no need for a BSL-3 laboratory set up and species-specific labeled secondary antibodies for detection as required for other tests such as microneutralization, immunofluorescence assay, and the plaque reduction neutralization test. Moreover, many researchers have envisaged the use of lateral flow assays for diagnosis and pseudovirus-based neutralization assay for evaluation of antiviral mechanism for SARS-CoV-2 in animals as well (359, 360). As far as the specific and sensitive diagnosis of animal CoVs is concerned, currently available methods might not resolve the unprecedented challenges associated with CoVs due to rapid mutations, interspecies jumps, and the emergence of new variants. This warrants continual efforts to develop robust, sensitive, and rapid tests, POC devices, and multiple diagnostic techniques to achieve a cost-effective and multidimensional diagnostic efficiency for clinical and epidemiological investigations.
The CoV infection in animals is routinely managed by the whole cell-based inactivated or modified live vaccines (361–370). The inactivated vaccines give rise to a high titer of circulating antibodies, whereas modified or attenuated live vaccines provide stimulation of cell-mediated immunity and accelerate IgA response against local mucosal infection and thus are of more value to commercial farms. The protection against animal CoVs is mainly dependent on IgA production; thus, parental vaccination is not favored generally in many of these infections (362, 371–373). The intranasal vaccination in adult cattle entering the feedlots using live attenuated enteric CoV against diverse field strains causing BCoV infections has been suggested as an ideal strategy to develop a protective immune response at the site of entry (oro-nasal) of the virus (123). Currently, the attenuated live vaccines used in broilers, layers, and brooders provide relatively inferior protection due to the presence of several serotypes of IBV and poor cross-protection. Therefore, a protectotype vaccine strategy based on shared antigens among variants has been proposed based on knowledge about serotypes, immunity, and the prevalence of variant strains of IBV (374). These protectotypes are quite effective in inducing cross-protection against heterologous serotypes (375). The feline CoV vaccination may enhance the risk of immune-mediated FIP; therefore, activation of IgA response is more relevant than IgG production. The administration of a modified live intranasal vaccine (temperature-sensitive), which activates IgA in the oropharynx as a consequence of replication of a temperature-sensitive mutant of the FCoV strain, is found to be more effective in FCoV (362). Canine CoV infection is mild and self-limiting and thus discourages the wide application of vaccines even if inactivated and live attenuated vaccines have been successfully developed for CCoV (369). In an experimental trial, a beta-propiolactone-inactivated MF59-adjuvanted vaccine was developed against the CCoV/TGEV recombinant, which failed to prevent the shedding of the virus totally but was considered to be safe (376). Novel experimental vaccines for TGEV and PEDV such as plasmid-vectored DNA vaccines encoding S, N, or M (377, 378) and recombinant vaccines or vectored vaccines using engineered swinepox virus or porcine adenoviruses to deliver the TGEV spike protein (379) were also developed. These vaccines were efficient in inducing both a systemic and a local humoral immune response with sufficient neutralizing antibodies. An RNA vaccine derived from Venezuelan equine encephalitis replicon expressing the PEDV spike gene was also developed against PEDV infections (379). Subunit vaccines have also been developed in an effort to express the S1 domain of spike protein in Baculovirus, yeast, or plant-based delivery system and have been found to work well in pigs (380–382). The oil adjuvanted vaccine comprising of a solubilized cell extract of BCoV-infected cells overexpressing viral hemagglutinin of BCoV was developed by Takamura et al. (383). This induced high hemagglutinating antibody titers without any adverse effects and was suggested to prevent winter dysentery in dairy cows (383). It is possible to combine two or more immunogenic strains of protectotype candidate against animal CoVs in a single vaccine with combined benefits such as the stimulation of cell-mediated immunity and higher IgA production for local mucosal immunity. Animal CoVs such as PEDV and IBV undergo frequent genetic shifts, and as a result, the protection level remains low despite using innovative vaccine strategies. Also, the vaccination in animals provides relatively a short duration of protective immunity with low efficacy due to focus mainly on whole-cell preparations rather than specific proteins or antigens for developing vaccines. Hence, effective and safe vaccines providing long immunity in animals still remain elusive. The scientists working on developing veterinary vaccines must take a cue from innovative and successful emerging technologies being used in developing SARS-CoV-2 vaccines so that suitable prophylaxis also becomes available for animals.
CoVs are capable of producing a broad spectrum of disease outcomes in mammals and avian species. These viruses are well-recognized to alter their tissue tropism and cross interspecies barriers to adapt to various ecological niches for abundance and persistence. The RBD of CoVs can recognize the same receptor present in multiple animal species. These virus–receptor interactions, evolutionary selection, and viral genetic diversity by frequent homologous recombination, inherent point mutations enable the virus to jump the species barrier and rapidly adapt to a new host species. It has been observed that CoVs, apart from infecting their primary host, do cause occasional infections in other animal species, per se dog and cat CoV, pig CoV, and TGEV can cross-infect, resulting in different disease outcomes suggesting host range mutants of an ancestral CoV. The BCoV is an excellent example that crosses interspecies barriers. Variants having genetic and antigenic relatedness with BCoV have been identified in the respiratory secretions of dogs infected subclinically (121–123), humans (120), and diverse groups of other domestic (124) and wild ruminant species such as camelids, waterbuck, sambar deer, white-tailed deer, and giraffe. Moreover, it can experimentally infect and cause enteric diseases in avian hosts, including turkey poults (384) that were found to transmit viruses to the control birds. These variant strains with established cross-species transmission are termed as Bovine-like CoVs (385) and classified as host range variants rather than a distinct virus. Furthermore, the BCoV has also been shown to procure new genes via recombination, i.e., acquisition of an influenza C-like hemagglutinin that may have a possible role in binding to different cell types (386).
An amino acid composition of APN receptor of human, feline, and porcine CoVs shows strong 78% identity. However, the APN receptor used by α-CoVs is species-specific, and the feline APN is a functional receptor for other members of α-CoVs, including FIPV and FECV, TGEV, canine CoV, and HCoV-229E. Due to this property of feline APN, cats can get infected by TGEV, CCoV, or HCoV-229E with or without developing symptoms. Similarly, the sequence comparisons reveal that TGEV has resulted from the host species jump of CCoV-II from dogs to pigs (40). The generation of less virulent porcine respiratory CoV from TGEV by spike gene deletion (153, 154) accounted for altered tissue tropism from the enteric to the respiratory system (387). The FIPV type II and CCoV-IIb strains from double recombination events between FIPV type I-CCoV and CCoV-II-TGEV, respectively, are also the results of genomic characteristics within CoVs for rapid adaption to novel CoV species suggesting coinfection in at least one host species.
A chimeric virus with spike protein of PEDV and backbone of TGEV was identified as a variant genotype of TGEV strain with unique deletions and distinct amino acid changes similar to PRCV, suggesting a recombination event between the variant TGEV, PEDV, and PRCV (388–391).
In humans, CoVs can cause infections ranging from the common cold to highly pathogenic diseases such as SARS and MERS. The seven human CoVs known to date are HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1 causing mild symptoms, and the highly pathogenic MERS-CoV, SARS-CoV, and SARS-CoV-2 causing adverse lower respiratory tract infection. The phylogenetic analysis has shown that bats, mice, or domestic animals serve as gene sources for all the seven HCoV (392). The HCoVs triggering common cold circulate in the human population without any need of an animal reservoir, whereas SARS-CoV and MERS-CoV need to maintain and propagate in their zoonotic intermediate host for possible transmission to human targets. Also, the frequent crossing of species by CoV has led to the emergence of important human pathogens. The key determinants for CoV–host specificity, such as variability in spike glycoprotein, its RBD in different species, and identical nucleotide sequence, provide a framework to understand the switch of hosts and positive selection during inter- and intraspecies transmission events.
The complete genome sequence analysis suggests that BCoV is possibly related to HCoV OC43 (50). It is believed that an interspecies jump was responsible for this genetic similarity, albeit there are no case reports suggesting infection resulting in disease by BCoV in humans. The findings of various studies suggest cross-protection among HCoV and BCoV and evidence of viral RNA in the human nasal mucosa for a short duration after exposure to BCoV (393). A human enteric CoV strain HECV-4408 isolated from a child suffering from acute diarrhea was passaged four times in HRT-18 cells and was inoculated orally in four gnotobiotic calves followed by challenging with BCoV-DB2 strain. The calves inoculated with HECV-4408 developed diarrhea and mild clinical signs along with detection of virus in the feces and nasal shedding by RT-PCR, whereas after BCoV inoculation, no diarrhea or virus shedding was observed in the calves. This experiment fairly suggests that (i) the less severe clinical signs in calves inoculated with HECV-4408 may be due to naturally lower virulence of HECV in calves or that the virulence was altered by a passage in cell culture, as it affects the efficiency of agglutination of RBCs (120, 127), its intestinal replication (128), antigenic composition (394), and associated mutations in the genome (395). (ii) Fecal and serum IgG titers were detected, which either remained the same or increased 2-fold after the challenge. This indicated that the HECV-4408 inoculation developed a protective immune response such that no replication of the virus was observed after challenging with the BCoV-DB2 strain.
There is no risk suspected yet to the public health from IBV. Humans are not considered as a reservoir for replication of IBV. There is no evidence of transmission of virus between humans and from humans to animals. Various mechanical means leading to infections in chicks have been reported. The persons working in commercial poultry farms may be at risk of getting an infection, but the significance is not known (396). IBV has also been detected in wild birds, which may serve as a vector for transmission between free-living and domestic birds (397). Therefore, the wild birds could feature as essential for recombination events that may further lead to the emergence of variants of concerns for humans. The isolation of avian IBV such as viruses from a man with a morphological resemblance with 229E strain of human CoV was reported, which was later confirmed to be an isolate of HCoV and not otherwise (398). This illustrates a likely possibility of the origin of avian and human CoVs from a common gene source resulting in divergent evolution.
The PEDV is genetically closely related to HCoV-229E than to other α-CoVs, and it can also be cultured in Vero cells such as SARS-CoV (399). It is relevant to mention that the swine had always been a predominant species for the evolution of outbreak-causing viruses such as new strains of influenza A virus and are also found to be infected by bat CoVs. The role of pigs and the possibility of the emergence of new strains of viruses, including human CoVs, should not be overlooked and need explorative studies (32, 400, 401). However, Shi et al. (402) had reported that there is no significant susceptibility of pigs to SARS-CoV-2.
The wild mammals and birds harbor CoV resulting in undetected transmission. Genetically similar human CoVs were identified in civets, raccoons, and horseshoe bats (403), and an experimental infection caused disease in macaques, ferrets, and subclinical infection in cats (404, 405). The CoVs in these animals show nucleotide sequence homology (88–90%) with other HCoVs and use ACE2 receptors to bind to spike glycoprotein for entry as seen with SARS-CoV and SARS-CoV-2 (406, 407). These findings suggest their role as intermediate or reservoir hosts. Bats are found to be reservoir hosts for pathogenic HCoV but not as immediate hosts due to few sequence divergence (408). Similarly, the highest genome sequence homology has been found in RBDs of SARS-CoV-2, pangolins, and bats, but still, there is no direct supportive evidence for their origin from a common ancestor, and thus, the evolutionary pathway remains obscure (392). However, a study on analysis of the evolutionary origin of SARS using phylogenetic analysis suggested an avian-like origin for nucleocapsid and matrix protein, mammalian-like origin for replicase proteins, and a mosaic of mammalian–avian-like origin for spike protein. The spike gene was also subjected to a bootscan recombination analysis that revealed high nucleotide sequence similarity between the SARS virus and FIPV (409).
MERS also serves as another example where interspecies transmission is important. The dromedary camels of Middle East countries in Asia and Africa, such as Egypt and Qatar, were detected with MERS-CoV isolated from their nasal swabs and were found seropositive with neutralizing antibodies for MERS-CoV species (410–416). Also, the study on experimental MERS infection in camels revealed massive shedding of a large amount of virus through not only respiratory route but also feco-oral route and milk, suggesting the risk of foodborne transmission to humans and occupational exposure (417–419). This indicates that the camels serve as the bona fide reservoir host of MERS-CoV. In a case report, the full genome sequences of two isolates obtained from a dromedary camel with rhinorrhea and a human in close contact with the camel were identical and positive for MERS-CoV RNA (420). The rate of secondary transmission within humans is also observed to be low, only up to 5% (421). A case of a 39-year-old male who developed fever and cough with a history of close contact with dromedary camels at his farm tested positive for MERS-CoV by RT-PCR (422). However, other study reports suggest there is no contact history with camels before the onset of symptoms in many confirmed cases of MERS in humans (423), which attributed to either human-to-human transmission or other transmission routes involving unknown animal species paving the way for further investigations.
The phylogenetic analyses on the molecular level reveal that SARS-CoV-2 is close to SARS-CoV with bat origin and shares 50–51.8% and 79% identity with MERS-CoV and SARS-CoV, respectively (32, 424–426). The recent reports on the detection of SARS-CoV-2 with mild respiratory illness in animals such as cats, dogs, and tigers in contact with humans suggest true infection caused by human-to-animal transmission (427). The two dogs reported from Hong Kong and a dog in New York living in close contact with their SARS-CoV-2 positive owners tested positive through RT-PCR in both nasal and oral samples (428–430). Seroconversion in dogs was observed when the blood samples were tested in the later stages, with weak positive results indicating low viral infection that resulted in the production of antibodies against SARS-CoV-2. Similarly, cats can be found susceptible to SARS-CoV-2 through experimental inoculation and can spread infection through droplets (402, 431). A serological study on the immune response of cats against SARS-CoV-2 revealed high titers of neutralizing antibodies (432). The big feline such as tigers in proximity to asymptomatic positive zookeepers of Bronx Zoo in New York City tested positive for SARS-CoV-2 after showing mild respiratory symptoms (433). Recently, two cases of human-to-cat transmission of SARS-CoV-2 were identified during a screening program of a feline population of households in the United Kingdom (434). The first case of transmission of SARS-CoV-2 from an asymptomatic human carrier, probably caretakers, to eight Asiatic lions at Hyderabad's Nehru Zoological Park was reported in India (435).
The high percent identity between feline and human ACE2 protein sequences, particularly within the receptor-binding interface region, may explain the incidence of cross-species transmission between them. The ferrets were also found to harbor SARS-CoV-2 infection with isolation of virus from the upper respiratory tract and development of mild clinical signs during experimental investigations (402). Therefore, SARS-CoV-2 infection in these companion animals demonstrates their susceptibility under natural and experimental settings. Other farmed animals such as minks, when infected with SARS-CoV-2, may show respiratory and gastrointestinal signs with increased mortality (436, 437). There is evidence that SARS-CoV-2 has evolved at the genetic level into a variant strain in minks in Denmark, and mink-to-mink and mink-to-human transmissions have been observed on farms of Denmark and the Netherlands, whereas ferrets have been reported to transmit the virus to other ferrets in an experimental study (432, 437). Natural SARS-CoV-2 infection in gorillas of San Diego Zoo in California (438) and Asian small-clawed otters (439) have also been reported with high susceptibility to the infection and clinical signs. Experimental investigations on SARS-CoV-2 infection in raccoon dogs, rabbits, pigs, cattle, poultry, hamsters, fruit bats, white-tailed deer, macaques, voles, and mice suggested low to high susceptibility, none to mild clinical signs, and pieces of evidence of low transmissibility between respective animals (440–447). These findings, however, do not hold any substantial evidence for zoonoses, as the transmissibility is low due to insufficient viral load and its replication (427, 431). The viruses in these animals do not satisfy Koch's postulates, and thus, it is inconclusive to declare these species as reservoir hosts for SARS-CoV-2 (448).
The novel animal and human CoVs are the outcomes of recombination, divergence, and subsequent evolution. An efficient genomic, phylogenetic, evolutionary rate, and divergence time analyses are now possible with improved bioinformatics tools and the availability of a large pool of CoVs discovered over time. This is done using different approaches incorporating molecular clock analysis or similar techniques that evaluate mutation rates of biomolecules such as DNA, RNA, or amino acid sequences for proteins. The ecological approaches focus on the spatial distribution of pathogen and host populations and their interactions with each other and their environment. Molecular approaches rely on genetic and cellular aspects of the host–pathogen relationship at the individual and population levels. A combination of these approaches, epidemiological data, and whole-genome sequencing along with some anticipative strategies are essential to clarify the mechanisms by which the virus jumps from one species to another. Interspecies transmission and emergence of novel CoVs are studied through molecular epidemiology, which describes the distribution of genes or their variants by considering parameters such as place, time, and population. Moreover, phylogenomic analysis intersects genomics, origin, and evolution of CoVs carried out exclusively after outbreaks of SARS in humans. This analysis provided evidence for interspecies transmission events such as the emergence of HCoV-OC43 from bovines to humans, TGEV from CCoV-II, FCoV-II, and CCoV-II from recombination between early CCoV-I and FCoV-I and porcine CoV HKU15 likely from a sparrow. Many researchers use the Rob Lanfear's method (449) and the maximum likelihood method with FastTree software (MicrobesOnline, Berkeley, CA, USA) to construct a phylogenetic tree and sequence alignment with the best setting determined by Global Initiative for Sharing All Influenza Data database (https://www.gisaid.org/) and Nextstrain (https://nextstrain.org) (450). Although Bayesian Evolutionary Analysis Sampling Trees is the most widely used approach, it uses different genes (RdRp, ORF1ab, S, N, and helicase genes) and molecular datasets for the purpose to estimate the considerable difference in the life history and evolution of CoVs (1, 22, 451). Klompus et al. demonstrated interspecies cross-reactivity mediated by reactive monoclonal antibodies that bind to antigens from human CoVs (hCoV-OC43 and hCoV-HKU1) and several animal CoVs with shared motifs to SARS-CoV-2. This serological strategy based on cross-reactive antibody responses along with DNA sequencing and RT-qPCR testing has been a dominant methodology and potential diagnostic strategy for infections of novel CoV spillovers (452).
CoV has a wide host range affecting domestic livestock, wild animals, and birds, often resulting in serious disease outcomes and economic losses. Furthermore, pets and various livestock species share a common environment with humans and continue to pose a threat to the public at risk. The presence of antibodies against animal CoVs in humans could merely be a normal immune response to an occasional or occupational exposure without any apparent disease. However, frequent and prolonged exposure to animal CoVs and the need for the virus to mutate to bypass the protective host-immune responses would likely result in newer CoV strains with a better ability to infect immunocompromised human hosts. Interestingly, it has been observed that companion animals are susceptible hosts for human CoVs such as SARS-CoV-2. The similarities in the CoV key proteins having a role in initiating infections, together with genetic and evolutionary relatedness and habitat sharing by diverse hosts, will drive future disease outcomes and virus evolution. At present, there is no strong evidence suggesting the role of livestock, poultry, and pets' CoVs to cause serious disease in humans; however, ample scope to undergo mutations and cross-jump species barrier to cause life-threatening illnesses exists. The enigmatic nature of CoVs to cause alteration in tissue tropism, jump species barriers, and form variants is remarkable and needs elucidation. CoVs tend to rapidly adapt to changing ecological niches and enhance the possibility of the emergence of novel CoVs due to high mutational errors particularly in the RBD of spike protein, along with inconstancy of replication enzymes and cleavage sites. These errors in S protein, important for host receptor usage, are essential for the emergence of mutants and the establishment of an effective and productive human infection. Consequently, the occurrence of double and triple mutants has made researchers focus on establishing epidemiological linkage to correlate these variants of concerns with the existing public health scenario. Furthermore, the proven propensity of CoVs for interspecies transmission, to emerge potentially from unknown reservoirs and to genetically relate with CoVs from different hosts, indicates the continued introduction of animal CoVs into the human population. The outbreaks of SARS-CoV in the year 2002 and MERS-CoV in 2012 in the form of severe acute respiratory distress had less impact than the current pandemic caused by SARS-CoV-2 that accounted for nearly 216.30 million confirmed cases with a death toll nearing four million by August 30, 2021, globally (453). Although these numbers are way higher, the case fatality rate of SARS-CoV-2 is around 4%, which is still lower than SARS-CoV and MERS-CoV outbreaks, which ranged between 0 and 50% (454–456). The genome of novel SARS-COV-2 is chimeric in the sense that the majority of it shares homology with the bat CoV genome, whereas the RBD portion bears a sequence similar to pangolin CoV. It is this specific RBD sequence that enables SARS-COV-2 to bind with high affinity to hACE2, resulting in productive infection and deaths. This is an unambiguous example of how recombination and mutation events result in highly virulent strains claiming lives and damaging economies.
The definitive understanding of the origin of SARS-CoV-2 will have marked repercussions on how humans interact with the ecosystem and on laboratory practice policies and biosafety regulations. Therefore, the emergence and evolution of novel CoVs because of their widespread association and frequent infections in animal hosts, both livestock and wild, combined with a high mutation rate, need to be contemplated to meet future challenges. There is an impending need to implement an effective strategy and monitor animal coronavirome as a potential reservoir of CoV reintroduction to humans. Large-scale sampling, metagenomic sequencing, and comparative genome analysis from animals of different geographical locations, especially those in frequent contact with human and wildlife habitats, should be considered. Baseline samples of unexposed individuals, preferably from a pre-pandemic population, need to be collected to compare with the individuals infected with new spillovers. The establishment of phage antibody libraries for profiling antibody responses against novel CoVs and enhancing the immediate availability of animal CoV serological assays will assist in the screening of numerous antigens in a critically early phase of future outbreaks (452). Despite the availability of established and novel diagnostic technologies, the detection of animal CoVs is still based on conventional, time-consuming, and less-sensitive molecular techniques. Therefore, there is a need to drive efforts for the development of rapid, cost-effective, and sensitive laboratory or POC diagnostic tools with a multi-prong approach to potentially increase the efficiency and specificity of diagnosis. These upgraded testing efforts will help in limiting the spread of animal CoV infections in farms, multi-pet households, and wildlife niches. The commonly used whole cell-based vaccine strategies to combat animal CoV infections on the field deliver poor protection in farm and pet animals. This calls for the exploitation of viral-vectored and nucleic acid-based vaccines as promising candidates. The knowledge about factors responsible for the success or failure of animal CoV vaccines can also provide better insight into the design of vaccines for humans against pandemic-causing CoVs. The standard measures used for the prevention of infectious diseases at farms and elsewhere should be followed besides maintaining personal hygiene and avoiding contact with wildlife to restrict exposure to animal CoVs. Needless to mention that the development of novel diagnostics, vaccines, detailed epidemiological studies, and sequencing of variants of interest and concern will, of course, need budgetary provisions.
PP: reviewed the literature and wrote the manuscript. SV: finalized the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. (2009) 234:1117–27. doi: 10.3181/0903-MR-94
2. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. (2003) 348:1953–66. doi: 10.1056/NEJMoa030781
3. Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. (2003) 348:1967–76. doi: 10.1056/NEJMoa030747
4. Zhong NS, Zheng BJ, Li YM, Poon LL, Xie ZH, Chan KH, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. (2003) 362:1353–8. doi: 10.1016/S0140-6736(03)14630-2
5. Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, et al. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. (2003) 423:240. doi: 10.1038/423240a
6. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. (2012) 367:1814–20. doi: 10.1056/NEJMoa1211721
7. Estola T. Coronaviruses, a new group of animal RNA viruses. Avian Dis. (1970) 14:330–6. doi: 10.2307/1588476
8. Fabricant J. The early history of infectious bronchitis. Avian Dis. (1998) 42:648–50. doi: 10.2307/1592697
9. Erles K, Toomey C, Brooks H, Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. (2003) 310:216–23. doi: 10.1016/S0042-6822(03)00160-0
10. Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, et al. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA. (2018) 115:E5135–43. doi: 10.1073/pnas.1802879115
11. Wang Q, Vlasova AN, Kenney SP, Saif LJ. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. (2019) 34:39–49. doi: 10.1016/j.coviro.2018.12.001
12. Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. (2020) 7:1012–23. doi: 10.1093/nsr/nwaa036
13. McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. (2014) 6:2991–3018. doi: 10.3390/v6082991
14. Liang FY, Lin LC, Ying TH, Yao CW, Tang TK, Chen YW, et al. Immunoreactivity characterisation of the three structural regions of the human coronavirus OC43 nucleocapsid protein by Western blot: implications for the diagnosis of coronavirus infection. J Virol Methods. (2013) 187:413–20. doi: 10.1016/j.jviromet.2012.11.009
15. Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. (2003) 77:8801–11. doi: 10.1128/JVI.77.16.8801-8811.2003
16. Woo PCY, Huang Y, Lau SKP, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. (2010) 2:1804–20. doi: 10.3390/v2081803
17. Venkatagopalan P, Daskalova SM, Lopez LA, Dolezal KA, Hogue BG. Coronavirus envelope (E) protein remains at the site of assembly. Virology. (2015) 478:75–85. doi: 10.1016/j.virol.2015.02.005
18. Kuo L, Hurst-Hess KR, Koetzner CA, Masters PS. Analyses of coronavirus assembly interactions with interspecies membrane and nucleocapsid protein chimeras. J Virol. (2016) 90:4357–68. doi: 10.1128/JVI.03212-15
19. Arndt AL, Larson BJ, Hogue BG. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J Virol. (2010) 84:11418–28. doi: 10.1128/JVI.01131-10
20. Zeng Q, Langereis MA, van Vliet ALW, Huizinga EG, de Groot RJ. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci USA. (2008) 105:9065–9. doi: 10.1073/pnas.0800502105
21. Lang Y, Li W, Li Z, Koerhuis D, van den Burg ACS, Rozemuller E, et al. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc Natl Acad Sci USA. (2020) 117:25759–70. doi: 10.1073/pnas.2006299117
22. Nieto-Torres JL, DeDiego ML, Álvarez E, Jiménez-Guardeño JM, Regla-Nava JA, Llorente M, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. (2011) 415:69–82. doi: 10.1016/j.virol.2011.03.029
23. ICTV Virus Taxonomy. (2009). Available online at: http://ictvonline.org/virusTaxonomy.asp?version=2009 (accessed August 1, 2010).
24. Woo PCY. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. (2012) 86:3995–4008. doi: 10.1128/JVI.06540-11
25. Wong ACP, Li X, Lau SKP, Woo PCY. Global epidemiology of bat coronaviruses. Viruses. (2019) 11:174. doi: 10.3390/v11020174
26. World Health Organization. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS). (2003). Available online at: https://apps.who.int/iris/handle/10665/70863
27. Dhama K, Pawaiya RVS, Chakraborty S, Tiwari R, Saminathan M, Verma AK. Coronavirus infection in equines: a review. Asian J Anim Vet Adv. (2014) 9:164–76. doi: 10.3923/ajava.2014.164.176
28. Dhama K, Singh SD, Rajamani B, Desingu P, Sandip C, Ruchi T, et al. Emergence of avian infectious bronchitis virus and its variants need better diagnosis, prevention and control strategies: a global perspective. Pak J Biol Sci. (2014) 17:751–67. doi: 10.3923/pjbs.2014.751.767
29. Monchatre-Leroy E, Boué F, Boucher J-M, Renault C, Moutou F, Ar Gouilh M, et al. Identification of alpha and beta coronavirus in wildlife species in France: bats, rodents, rabbits, and hedgehogs. Viruses. (2017) 9:364. doi: 10.3390/v9120364
30. Dhama K, Khan S, Tiwari R, Dadar M, Malik Y, Singh K, et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics and therapeutics—a mini-review. Hum Vaccin Immunother. (2020) 16:1232–8. doi: 10.1080/21645515.2020.1735227
31. Ji W, Wang W, Zhao X, Zai J, Li X. Cross species transmission of the newly identified coronavirus 2019 nCoV. J Med Virol. (2020) 92:433–40. doi: 10.1002/jmv.25682
32. Malik YS, Sircar S, Bhat S, Sharun K, Dhama K, Dadar M, et al. Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. (2020) 40:68–76. doi: 10.1080/01652176.2020.1727993
33. Xu Y. Genetic diversity and potential recombination between Ferret coronaviruses from European and American lineages. J Infect. (2020) 80:350–71. doi: 10.1016/j.jinf.2020.01.016
34. Wilhelmsen KC, Leibowitz JL, Bond CW, Robb JA. The replication of murine coronaviruses in enucleated cells. Virology. (1981) 110:225–30. doi: 10.1016/0042-6822(81)90027-1
35. Fehr A, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. (2015) 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1
36. Xian Y, Zhang J, Bian Z, Zhou H, Zhang Z, Lin Z, et al. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharm Sin B. (2020) 10:1163–74. doi: 10.1016/j.apsb.2020.06.002
37. Pyrc K, Berkhout B, van der Hoek L. The novel human coronaviruses NL63 and HKU1. J Virol. (2007) 81:3051–7. doi: 10.1128/JVI.01466-06
38. Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. (2006) 80:5927–40. doi: 10.1128/JVI.02501-05
39. Fang SG, Shen H, Wang J, Tay FP, Liu DX. Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells. Virology. (2008) 379:175–80. doi: 10.1016/j.virol.2008.06.038
40. Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. (2009) 7:439–50. doi: 10.1038/nrmicro2147
41. Brown TDK, Brierley I. The coronavirus nonstructural proteins. In Siddell SG, editor The Coronaviridae. The Viruses. Boston, MA: Springer (1995). p. 191–217. doi: 10.1007/978-1-4899-1531-3_10
42. de Vries AF, Horzinek MC, Rottier PJM, de Groot RJ. The genome organization of the nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. (1997) 8:33–47. doi: 10.1006/smvy.1997.0104
43. Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. (1997) 48:1–100. doi: 10.1016/S0065-3527(08)60286-9
44. Lai MMC, Holmes KV. Coronaviridae their replication. In: Knipe D, et al., editors. Fields' Virology. Philadelphia, PA: Lippincott Williams & Wilkins (2001). p. 1163–85.
45. Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. (2003) 331:991–1004. doi: 10.1016/S0022-2836(03)00865-9
46. Thiel V, Ivanov KA, Putics Á, Hertzig T, Schelle B, Bayer S, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. (2003) 84:2305–15. doi: 10.1099/vir.0.19424-0
47. Reguera J, Mudgal G, Santiago C, Casasnovas JM. A structural view of coronavirus-receptor interactions. Virus Res. (2014) 194:3–15. doi: 10.1016/j.virusres.2014.10.005
48. Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, et al. The Genome sequence of the SARS-associated coronavirus. Science. (2003) 300:1399–404. doi: 10.1126/science.1085953
49. Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. (2003) 300:1394–9. doi: 10.1126/science.1085952
50. Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. (2005) 79:1595–604. doi: 10.1128/JVI.79.3.1595-1604.2005
51. Vijgen L, Keyaerts E, Lemey P, Moës E, Li S. Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology. (2005) 337:85–92. doi: 10.1016/j.virol.2005.04.010
52. Alkhansa A, Lakkis G, El Zein L. Mutational analysis of SARS-CoV-2 ORF8 during six months of COVID-19 pandemic. Gene Rep. (2021) 23:101024. doi: 10.1016/j.genrep.2021.101024
53. Lan YC, Liu TT, Yang JY, Lee CM, Chen YJ, Chan YJ, et al. Molecular epidemiology of severe acute respiratory syndrome-associated coronavirus infections in Taiwan. J Infect Dis. (2005) 191:1478–89. doi: 10.1086/428591
54. Lan YC, Liu TT, Yang JY, Lee CM, Chen YJ, Chan YJ. Characterizing 56 complete SARS-CoV S-gene sequences from Hong Kong. J Clin Virol. (2007) 38:19–26. doi: 10.1016/j.jcv.2006.10.001
55. Li WH, Wong SK, Li F, Kuhn JH, Huang IC, Choe H, et al. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol. (2006) 80:4211–9. doi: 10.1128/JVI.80.9.4211-4219.2006
56. Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. (2013) 100:246–54. doi: 10.1016/j.antiviral.2013.08.014
57. Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J Virol. (2012) 86:2856–8. doi: 10.1128/JVI.06882-11
58. Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. (2016) 24:490–502. doi: 10.1016/j.tim.2016.03.003
59. Chen J. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microbes Infect. (2020) 22:69–71. doi: 10.1016/j.micinf.2020.01.004
60. Patel A, Jernigan DB, 2019-nCoV CDC Response Team. Initial public health response and interim clinical guidance for the 2019 novel coronavirus outbreak - United States, December 31, 2019-February 4, 2020. MMWR Morb Mortal Wkly Rep. (2020) 69:140–6. doi: 10.15585/mmwr.mm6905e1
61. INSACOG Power Point Presentation. INSACOG: Indian SARS-CoV-2 Genomics Consortium (2021). Available online at: https://dbtindia.gov.in/sites/default/files/INSACOG%20ppt%2023-06-2021.pdf (accessed June 23, 2021).
62. Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. (2015) 89:1954–64; doi: 10.1128/JVI.02615-14
63. Tusell SM, Schittone SA, Holmes KV. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. J Virol. (2007) 81:1261–73. doi: 10.1128/JVI.01510-06
64. Wu K, Li W, Peng G, Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci USA. (2009) 106:19970–4. doi: 10.1073/pnas.0908837106
65. Reguera J, Santiago C, Mudgal G, Ordoño D, Enjuanes L, Casasnovas JM. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. (2012) 8:e1002859. doi: 10.1371/journal.ppat.1002859
66. Wu K, Chen L, Peng G, Zhou W, Pennell CA, Mansky LM, et al. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J Virol. (2011) 85:5331–7. doi: 10.1128/JVI.02274-10
67. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. (2016) 3:237–61. doi: 10.1146/annurev-virology-110615-042301
68. Peng G, Xu L, Lin YL, Chen L, Pasquarella JR, Holmes KV, et al. Crystal structure of bovine coronavirus spike protein lectin domain. J Biol Chem. (2012) 287:41931–8. doi: 10.1074/jbc.M112.418210
69. Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z, Wang Q, et al. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol. (2015) 53:1537–48. doi: 10.1128/JCM.00031-15
70. Shang J, Zheng Y, Yang Y, Liu C, Geng Q, Tai W, et al. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J Virol. (2018) 92:e01556–17. doi: 10.1128/JVI.01556-17
71. Wang B, Liu Y, Ji C, Yang Y, Liang Q, Zhao P, et al. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J Virol. (2018) 92:e00318–18. doi: 10.1128/JVI.00318-18
72. Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. (1981) 289:366–73. doi: 10.1038/289366a0
73. Wang J. Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends Biochem Sci. (2002) 27:122–6. doi: 10.1016/S0968-0004(01)02038-2
74. Rossmann MG. The canyon hypothesis. Hiding the host-cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem. (1989) 264:14587–90. doi: 10.1016/S0021-9258(18)63732-9
75. Vautherot JF, Laporte J. Utilization of monoclonal antibodies for antigenic characterization of coronaviruses. Ann Rech Vet. (1983) 14:437–44.
76. Vieler E, Schlapp T, Anders C, Herbst W. Genomic relationship of porcine hemagglutinating encephalomyelitis virus to bovine coronavirus and human coronavirus OC43 as studied by the use of bovine coronavirus S gene-specific probes. Arch Virol. (1995) 140:1215–23. doi: 10.1007/BF01322747
77. Ma Y, Tong X, Xu X, Li X, Lou Z, Rao Z. Structures of the N- and C-terminal domains of MHV-A59 nucleocapsid protein corroborate a conserved RNA-protein binding mechanism in coronavirus. Protein Cell. (2010) 1:688–97. doi: 10.1007/s13238-010-0079-x
78. Keane SC, Liu P, Leibowitz JL, Giedroc DP. Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. J Biol Chem. (2012) 287:7063–73. doi: 10.1074/jbc.M111.287763
79. Chang CK, Sue SC Yu TH, Hsieh CM, Tsai CK, Chiang YC, et al. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. (2006) 13:59–72. doi: 10.1007/s11373-005-9035-9
80. Lo YS, Lin SY, Wang SM, Wang CT, Chiu YL, Huang TH, et al. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. (2013) 587:120–7. doi: 10.1016/j.febslet.2012.11.016
81. Jayaram H, Fan H, Bowman BR, Ooi A, Jayaram J, Collisson EW, et al. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J Virol. (2006) 80:6612–20. doi: 10.1128/JVI.00157-06
82. Chen CY, Chang CK, Chang YW, Sue SC, Bai HI, Riang L, et al. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. (2007) 368:1075–86. doi: 10.1016/j.jmb.2007.02.069
83. Zlotnick A. Theoretical aspects of virus capsid assembly. J Mol Recogn. (2005) 18:479–90. doi: 10.1002/jmr.754
84. Ruch TR, Machamer CE. The coronavirus E protein: assembly and beyond. Viruses. (2012) 4:363–82. doi: 10.3390/v4030363
85. Kuo L, Hurst KR, Masters PS. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J Virol. (2007) 81:2249–62. doi: 10.1128/JVI.01577-06
86. Corse E, Machamer CE. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J Virol. (2000) 74:4319–26. doi: 10.1128/JVI.74.9.4319-4326.2000
87. Godet M, L'Haridon R, Vautherot JF, Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. (1992) 188:666–75. doi: 10.1016/0042-6822(92)90521-P
88. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. (2019) 16:69. doi: 10.1186/s12985-019-1182-0
89. Corse E, Machamer CE. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J Virol. (2002) 76:1273–84. doi: 10.1128/JVI.76.3.1273-1284.2002
90. Liao Y, Yuan Q, Torres J, Tam JP, Liu DX. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. (2006) 349:264–75. doi: 10.1016/j.virol.2006.01.028
91. Lopez LA, Riffle AJ, Pike SL, Gardner D, Hogue BG. Importance of conserved cysteine residues in the coronavirus envelope protein. J Virol. (2008) 82:3000–10. doi: 10.1128/JVI.01914-07
92. Salaun C, Greaves J, Chamberlain LH. The intracellular dynamic of protein palmitoylation. J Cell Biol. (2010) 191:1229–38. doi: 10.1083/jcb.201008160
93. Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML, et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. (2014) 10:e1004320. doi: 10.1371/journal.ppat.1004320
94. de Haan CA, Kuo L, Masters PS, Vennema H, Rottier PJ. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. (1998) 72:6838–50. doi: 10.1128/JVI.72.8.6838-6850.1998
95. Oostra M, de Haan CA, de Groot RJ, Rottier PJ. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J Virol. (2006) 80:2326–36. doi: 10.1128/JVI.80.5.2326-2336.2006
96. Rohrbach BW, Legendre AM, Baldwin CA, Lein DH, Reed WM, Wilson RB. Epidemiology of feline infectious peritonitis among cats examined at veterinary medical teaching hospitals. J Am Vet Med Assoc. (2001) 218:1111–5. doi: 10.2460/javma.2001.218.1111
97. Vlasak R, Luytjes W, Spaan W, Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA. (1988) 85:4526–9. doi: 10.1073/pnas.85.12.4526
98. Luytjes W, Bredenbeek PJ, Noten AFH, Horzinek MC, Spaan WJM. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. (1988) 166:415–22. doi: 10.1016/0042-6822(88)90512-0
99. Snijder EJ, den Boon JA, Horzinek MC, Spaan WJ. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology. (1991) 180:448–52. doi: 10.1016/0042-6822(91)90056-H
100. de Groot RJ. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj J. (2006) 23:59–72. doi: 10.1007/s10719-006-5438-8
101. Schultze B, Wahn K, Klenk HD, Herrler G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology. (1991) 180:221–8. doi: 10.1016/0042-6822(91)90026-8
102. Schultze B, Gross HJ, Brossmer R, Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J Virol. (1991) 65:6232–7. doi: 10.1128/jvi.65.11.6232-6237.1991
103. Smits SL, Gerwig GJ, van Vliet AL, Lissenberg A, Briza P, Kamerling JP, et al. Nidovirus sialate-O-acetylesterases: evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J Biol Chem. (2005) 280:6933–41. doi: 10.1074/jbc.M409683200
104. Tortorici MA, Walls AC, Lang Y, Wang C, Li Z, Koerhuis D, et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. (2019) 26:481–9. doi: 10.1038/s41594-019-0233-y
105. Kim CH. SARS-CoV-2 Evolutionary adaptation toward host entry and recognition of receptor o-acetyl sialylation in virus-host interaction. Int J Mol Sci. (2020) 21:4549. doi: 10.3390/ijms21124549
107. Van Kruiningen HJ, Khairallah LH, Sasseville VG, Wyand MS, Post JE. Calfhood coronavirus enterocolitis: a clue to the etiology of winter dysentery. Vet Pathol. (1987) 24:564–7. doi: 10.1177/030098588702400616
108. Heckert RA, Saif LJ, Hoblet KH, Agnes AG. A longitudinal study of bovine coronavirus enteric and respiratory infections in dairy calves in two herds in Ohio. Vet Microbiol. (1990) 22:187–201. doi: 10.1016/0378-1135(90)90106-6
109. Saif LJ. A review of evidence implicating bovine coronavirus in the etiology of winter dysentery in cows: an enigma resolved? Cornell Vet. (1990) 80:303–11.
110. Tsunemitsu H, Saif LJ. Antigenic and biological comparisons of bovine coronaviruses derived from neonatal calf diarrhea and winter dysentery of adult cattle. Arch Virol. (1995) 140:1303–11. doi: 10.1007/BF01322757
111. Storz J, Stine L, Liem A, Anderson GA. Coronavirus isolation from nasal swab samples in cattle with signs of respiratory tract disease after shipping. J Am Vet Med Assoc. (1996) 208:1452–5.
112. Hasoksuz M, Lathrop SL, Gadfield KL, Saif LJ. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am J Vet Res. (1999) 60:1227–33.
113. Tsunemitsu H, Smith DR, Saif LJ. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch Virol. (1999) 144:167–75. doi: 10.1007/s007050050493
114. Cho KO, Halbur PG, Bruna JD, Sorden SD, Yoon KJ, Janke BH, et al. Detection and isolation of coronavirus from feces of three herds of feedlot cattle during outbreaks of winter dysentery-like disease. J Am Vet Med Assoc. (2000) 217:1191–4. doi: 10.2460/javma.2000.217.1191
115. Lathrop SL, Wittum TE, Loerch SC, Perino LJ, Saif LJ. Antibody titers against bovine coronavirus and shedding of the virus via the respiratory tract in feedlot cattle. Am J Vet Res. (2000) 61:1057–61. doi: 10.2460/ajvr.2000.61.1057
116. Storz J, Purdy CW, Lin X, Burrell M, Truax RE, Briggs RE, et al. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp from cattle involved in two natural outbreaks of shipping fever. J Am Vet Med Assoc. (2000) 216:1599–604. doi: 10.2460/javma.2000.216.1599
117. Tråvén M, Näslund K, Linde N, Linde B, Silván A, Fossum C, et al. Experimental reproduction of winter dysentery in lactating cows using BCV – comparison with BCV infection in milk-fed calves. Vet Microbiol. (2001) 81:127–51. doi: 10.1016/S0378-1135(01)00337-6
118. Hasoksuz M, Hoet AE, Loerch SC, Wittum TE, Nielsen PR, Saif LJ. Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in an Ohio feedlot. J Vet Diagn Invest. (2002) 14:308–13. doi: 10.1177/104063870201400406
119. Boileau MJ, Kapil S. Bovine coronavirus associated syndromes. Vet Clin North Am Food Anim Pract. (2010) 26:123–46. doi: 10.1016/j.cvfa.2009.10.003
120. Zhang XM, Herbst W, Kousoulas KG, Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J Med Virol. (1994) 44:152–61. doi: 10.1002/jmv.1890440207
121. Decaro N, Desario C, Elia G, Mari V, Lucente MS, Cordioli P, et al. Serological and molecular evidence that canine respiratory coronavirus is circulating in Italy. Vet Microbiol. (2007) 121:225–30. doi: 10.1016/j.vetmic.2006.12.001
122. Lorusso A, Desario C, Mari V, Campolo M, Lorusso E, Elia G, et al. Molecular characterization of a canine respiratory coronavirus strain detected in Italy. Virus Res. (2009) 141:96–100. doi: 10.1016/j.virusres.2008.12.011
123. Saif LJ. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract. (2010) 26:349–64. doi: 10.1016/j.cvfa.2010.04.005
124. Hasoksuz M, Lathrop S, Al-dubaib MA, Lewis P, Saif LJ. Antigenic variation among bovine enteric coronaviruses (BECV) and bovine respiratory coronaviruses (BRCV) detected using monoclonal antibodies. Arch Virol. (1999) 144:2441–7. doi: 10.1007/s007050050656
125. Bovine Coronavirus Infection. Invasive Species Compendium. Centre for Agriculture and Biosciences International (2019). Available online at: https://www.cabi.org/isc/datasheet/91704 (accessed November 21, 2019).
126. Saif LJ, Redman DR, Moorhead PD, Theil KW. Experimentally induced coronavirus infections in calves: viral replication in the respiratory and intestinal tracts. Am J Vet Res. (1986) 47:1426–32.
127. Kapil S, Richardson KL, Maag TR, Goyal SM. Characterization of bovine coronavirus isolates/from eight different states in the USA. Vet Microbiol. (1999) 67:221–30. doi: 10.1016/S0378-1135(99)00042-5
128. Kapil S, Trent AM, Goyal SM. Excretion and persistence of bovine coronavirus in neonatal calves. Arch Virol. (1990) 115:127–32. doi: 10.1007/BF01310629
129. Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. (2007) 38:281–97. doi: 10.1051/vetres:2006055
130. Cavanagh D, Davis PJ, Mockett AP. Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. (1988) 11:141–50. doi: 10.1016/0168-1702(88)90039-1
131. Cavanagh D, Davis PJ, Darbyshire JH, Peters RW. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J Gen Virol. (1986) 67:1435–42. doi: 10.1099/0022-1317-67-7-1435
132. Casais R, Dove B, Cavanagh D, Britton P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J Virol. (2003) 77:9084–9. doi: 10.1128/JVI.77.16.9084-9089.2003
133. Ramakrishnan S, Kappala D. Avian infectious bronchitis virus. Recent Adv Anim. Virol. (2019) 301–19. doi: 10.1007/978-981-13-9073-9_16
134. Ignjatović J, Sapats S. Avian infectious bronchitis virus. Rev Sci Tech. (2000) 19:493–508. doi: 10.20506/rst.19.2.1228
135. Bande B, Arshad SS, Omar AR, Bejo MH, Abubakar MS, Abba Y. Pathogenesis and diagnostic approaches of avian infectious bronchitis. Adv Virol. (2016) 11:4621659. doi: 10.1155/2016/4621659
136. Jackwood MW. Review of infectious bronchitis virus around the world. Avian Dis. (2012) 56:634–41. doi: 10.1637/10227-043012-Review.1
137. McFerran JB, Clarke JK, Curran WL. The application of negative contrast electron microscopy to routine veterinary virus diagnosis. Res Vet Sci. (1971) 12:253–7. doi: 10.1016/S0034-5288(18)34187-0
138. Gallardo RA, Hoerr FJ, Berry WD, van Santen VL, Toro H. Infectious bronchitis virus in testicles and venereal transmission. Avian Dis. (2011) 55:255–8. doi: 10.1637/9592-102910-Reg.1
139. Bisgaard M. Infektiøs bronchitis virus indflydelse på aeglaegning, befrugtnings og klaekningsprocent og afgang af høner [The influence of infectious bronchitis virus on egg production, fertility, hatchability and mortality rate in chickens (author's transl)]. Nord Vet Med. (1976) 28:368–76.
140. Purcell DA, McFerran JB. The histopathology of infectious bronchitis in the domestic fowl. Res Vet Sci. (1972) 13:116–22. doi: 10.1016/S0034-5288(18)34055-4
141. Boroomand Z, Asasi K, Mohammadi A. Pathogenesis and tissue distribution of avian infectious bronchitis virus isolate IRFIBV32 (793/B serotype) in experimentally infected broiler chickens. Sci World J. (2012) 2012:402537. doi: 10.1100/2012/402537
142. Babcock GJ, Esshaki DJ, Thomas WD, Ambrosino DM. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. (2004) 78:4552–60. doi: 10.1128/JVI.78.9.4552-4560.2004
143. Promkuntod N, van Eijndhoven RE, de Vrieze G, Gröne A, Verheije MH. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. (2014) 448:26–32. doi: 10.1016/j.virol.2013.09.018
144. Chousalkar KK, Roberts JR. Ultrastructural study of infectious bronchitis virus infection in infundibulum and magnum of commercial laying hens. Vet Microbiol. (2007) 22:223–6. doi: 10.1016/j.vetmic.2007.01.021
145. Chousalkar KK, Roberts JR, Reece R. Comparative histopathology of two serotypes of infectious bronchitis virus (T and N1/88) in laying hens and cockerels. Poult Sci. (2007) 86:50–8. doi: 10.1093/ps/86.1.50
146. Cavanagh D, Gelb J. Infectious bronchitis. In: Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE, editors. Diseases of Poultry. Ames, IA: Blackwell (2008). p. 117–35.
147. Cong F, Liu X, Han Z, Shao Y, Kong X, Liu S, et al. Transcriptome analysis of chicken kidney tissues following coronavirus avian infectious bronchitis virus infection. BMC Genomics. (2013) 14:743. doi: 10.1186/1471-2164-14-743
148. Gough RE, Randall CJ, Dagless M, Alexander DJ, Cox WJ, Pearson D, et al. ‘New' strain of infectious bronchitis virus infecting domestic fowl in Great Britain. Vet Rec. (1992) 130:493–4. doi: 10.1136/vr.130.22.493
149. Alexander DJ, Gough RE. Isolation of avian infectious bronchitis virus from experimentally infected chickens. Res Vet Sci. (1977) 23:344–7. doi: 10.1016/S0034-5288(18)33129-1
150. Boltz DA, Nakai M, Bahra JM. Avian infectious bronchitis virus: a possible cause of reduced fertility in the rooster. Avian Dis. (2004) 48:909–15. doi: 10.1637/7192-040808R1
151. Liu H, Yang X, Zhang Z, Li J, Zou W, Zeng F, et al. Comparative transcriptome analysis reveals induction of apoptosis in chicken kidney cells associated with the virulence of nephropathogenic infectious bronchitis virus. Microb Pathog. (2017) 113:451–9. doi: 10.1016/j.micpath.2017.11.031
152. Zuñiga S, Pascual-Iglesias A, Sanchez CM, Sola I, Enjuanes L. Virulence factors in porcine coronaviruses and vaccine design. Virus Res. (2016) 226:142–51. doi: 10.1016/j.virusres.2016.07.003
153. Laude H, Van Reeth K, Pensaert M. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet Res. (1993) 24:125–50.
154. Schultze B, Krempl C, Ballesteros ML, Shaw L, Schauer R, Enjuanes L, et al. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J Virol. (1996) 70:5634–7. doi: 10.1128/jvi.70.8.5634-5637.1996
155. Chen Q, Gauger P, Stafne M, Thomas J, Arruda P, Burrough E, et al. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. (2015) 482:51–9. doi: 10.1016/j.virol.2015.03.024
156. Chen F, Knutson TP, Rossow S, Saif LJ, Marthaler DG. Decline of transmissible gastroenteritis virus and its complex evolutionary relationship with porcine respiratory coronavirus in the United States. Sci Rep. (2019) 9:3953. doi: 10.1038/s41598-019-40564-z
157. Niederwerder MC, Hesse RA. Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transb Emerg Dis. (2018) 65:660–75. doi: 10.1111/tbed.12823
158. Piñeyro PE, Lozada MI, Alarcón LV, Sanguinetti R, Cappuccio JA, Pérez EM, et al. First retrospective studies with etiological confirmation of porcine transmissible gastroenteritis virus infection in Argentina. BMC Vet Res. (2018) 14:292. doi: 10.1186/s12917-018-1615-9
159. Laber KE, Whary MT, Bingel SA, Goodrich JA, Smith AC, Swindle MM. Biology and diseases of swine. Lab Anim Med. (2002) 615–73. doi: 10.1016/B978-012263951-7/50018-1
160. Moon HW. Mechanisms in the pathogenesis of diarrhea: a review. J Am Vet Med Assoc. (1978) 172:443–8.
161. Bohl EH, Kohler EM, Saif LJ, Cross RF, Agnes AG, Theil KW. Rotavirus as a cause of diarrhea in pigs. J Am Vet Med Assoc. (1978) 172:458–63.
162. Kemeny LJ, Wiltsey VL, Riley JL. Upper respiratory infection of lactating sows with transmissible gastroenteritis virus following contact exposure to infected piglets. Cornell Vet. (1975) 65:352–62.
163. Vlasova AN, Wang Q, Jung K, Langel SN, Malik YS, Saif LJ. Porcine coronaviruses. Emerg Transbound Anim Viruses. (2020) 79–110. doi: 10.1007/978-981-15-0402-0_4
164. MacLachlan JN, Dubovi EJ. Coronaviridae. In MacLachlan JN, Dubovi EJ, editor The Fenner's Veterinary Virology. 5th ed. Academic Press (2017). p. 435–61.
165. Underdahl NR, Mebus CA, Torres-Medina A. Recovery of transmissible gastroenteritis virus from chronically infected experimental pigs. Am J Vet Res. (1975) 36:1473–6.
166. Herrewegh AA, Vennema H, Horzinek MC, Rottier PJ, de Groot RJ. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology. (1995) 212:622–31. doi: 10.1006/viro.1995.1520
167. Pedersen NC, Boyle JF, Floyd K, Fudge A, Barker J. An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am J Vet Res. (1981) 42:368–77.
168. Poland AM, Vennema H, Foley JE, Pedersen NC. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J ClinMicrobiol. (1996) 34:3180–4. doi: 10.1128/jcm.34.12.3180-3184.1996
169. Vennema H, Poland A, Foley J, Pedersen NC. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. (1998) 243:150–7. doi: 10.1006/viro.1998.9045
170. Haijema BJ, Rottier PJM, de Groot RJ. Feline Coronaviruses: a tale of two-faced types. In: Thiel V, editor. Coronaviruses: Molecular and Cellular Biology. Caister: Academic Press (2007). p. 182–203.
171. Pedersen NC. A review of feline infectious peritonitis virus infection: 1963–2008. J Feline Med Surg. (2009) 11:225–58. doi: 10.1016/j.jfms.2008.09.008
172. Bank-Wolf BR, Stallkamp I, Wiese S, Moritz A, Tekes G, Thiel HJ. Mutations of 3c and spike protein genes correlate with the occurrence of feline infectious peritonitis. Vet Microbiol. (2014) 173:177–88. doi: 10.1016/j.vetmic.2014.07.020
173. Barker EN, Tasker S, Gruffydd-Jones TJ, Tuplin CK, Burton K, Porter E, et al. Phylogenetic analysis of feline coronavirus strains in an epizootic outbreak of feline infectious peritonitis. J Vet Intern Med. (2013) 27:445–50. doi: 10.1111/jvim.12058
174. Chang HW, de Groot RJ, Egberink HF, Rottier PJ. Feline infectious peritonitis: insights into feline coronavirus pathobiogenesis and epidemiology based on genetic analysis of the viral 3c gene. J Gen Virol. (2010) 91:415–20. doi: 10.1099/vir.0.016485-0
175. Chang HW, Egberink HF, Halpin R, Spiro DJ, Rottier PJ. Spike protein fusion peptide and feline coronavirus virulence. Emerg Infect Dis. (2012) 18:1089–95. doi: 10.3201/eid1807.120143
176. Lewis CS, Porter E, Matthews D, Kipar A, Tasker S, Helps CR, et al. Genotyping coronaviruses associated with feline infectious peritonitis. J Gen Virol. (2015) 96:1358–68. doi: 10.1099/vir.0.000084
177. Licitra BN, Millet JK, Regan AD, Hamilton BS, Rinaldi VD, Duhamel GE, et al. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg Infect Dis. (2013) 19:1066–73. doi: 10.3201/eid1907.121094
178. Pedersen NC, Liu H, Scarlett J, Leutenegger CM, Golovko L, Kennedy H, et al. Feline infectious peritonitis: role of the feline coronavirus 3c gene in intestinal tropism and pathogenicity based upon isolates from resident and adopted shelter cats. Virus Res. (2012) 165:17–28. doi: 10.1016/j.virusres.2011.12.020
179. Porter E, Tasker S, Day MJ, Harley R, Kipar A, Siddell SG, et al. Amino acid changes in the spike protein of feline coronavirus correlate with systemic spread of virus from the intestine and not with feline infectious peritonitis. Vet Res. (2014) 45:49. doi: 10.1186/1297-9716-45-49
180. Tekes G, Thiel HJ. Feline coronaviruses: pathogenesis of feline infectious peritonitis. Adv Virus Res. (2016) 96:193–218. doi: 10.1016/bs.aivir.2016.08.002
181. Fiscus SA, Teramoto YA. Antigenic comparison of feline coronavirus isolates: evidence for markedly different peplomer glycoproteins. J Virol. (1987) 61:2607–13. doi: 10.1128/jvi.61.8.2607-2613.1987
182. Decaro N, Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology. Vet Microbiol. (2008) 132:221–34. doi: 10.1016/j.vetmic.2008.06.007
183. Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ, de Groot RJ. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. (1998) 72:4508–14. doi: 10.1128/JVI.72.5.4508-4514.1998
184. Lin CN, Chang RY, Su BL, Chueh LL. Full genome analysis of a novel type II feline coronavirus NTU156. Virus Genes. (2013) 46:316–22. doi: 10.1007/s11262-012-0864-0
185. Terada Y, Matsui N, Noguchi K, Kuwata R, Shimoda H, Soma T, et al. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS ONE. (2014) 9:e106534. doi: 10.1371/journal.pone.0106534
186. Lorusso A, Decaro N, Schellen P, Rottier PJ, Buonavoglia C, Haijema BJ, et al. Gain, preservation, and loss of a group 1a coronavirus accessory glycoprotein. J Virol. (2008) 82:10312–7. doi: 10.1128/JVI.01031-08
187. Hohdatsu T, Okada S, Ishizuka Y, Yamada H, Koyama H. The prevalence of types I and II feline coronavirus infections in cats. J Vet Med Sci. (1992) 54:557–62. doi: 10.1292/jvms.54.557
188. Pedersen NC. An update on feline infectious peritonitis: virology and immunopathogenesis. Vet J. (2014) 201:123–32. doi: 10.1016/j.tvjl.2014.04.017
189. Brown MA, Troyer JL, Pecon-Slattery J, Roelke ME, O'Brien SJ. Genetics and pathogenesis of feline infectious peritonitis virus. Emerg Infect Dis. (2009) 15:1445–52. doi: 10.3201/eid1509.081573
190. Addie DD, Schaap IAT, Nicolson L, Jarrett O. Persistence and transmission of natural type I feline coronavirus infection. J Gen Virol. (2003) 84:2735–44. doi: 10.1099/vir.0.19129-0
191. Kennedy M, Citino S, McNabb AH, Moffatt AS, Gertz K, Kania S. Detection of feline coronavirus in captive Felidae in the USA. J Vet Diagn Invest. (2002) 14:520–2. doi: 10.1177/104063870201400615
192. Kummrow M, Meli ML, Haessig M, Goenczi E, Poland A, Pedersen NC, et al. Feline coronavirus serotypes 1 and 2: seroprevalence and association with disease in Switzerland. Clin Diagn Lab Immunol. (2005) 12:1209–15. doi: 10.1128/CDLI.12.10.1209-1215.2005
193. Benetka V, Kübber-Heiss A, Kolodziejek J, Nowotny N, Hofmann-Parisot M, Möstl K. Prevalence of feline coronavirus types I and II in cats with histopathologically verified feline infectious peritonitis. Vet Microbiol. (2004) 99:31–42. doi: 10.1016/j.vetmic.2003.07.010
194. Amer A, Siti Suri A, Abdul Rahman O, Mohd HB, Faruku B, Saeed S, et al. Isolation and molecular characterization of type I and type II feline coronavirus in Malaysia. Virol J. (2012) 9:278. doi: 10.1186/1743-422X-9-278
195. An DJ, Jeoung HY, Jeong W, Park JY, Lee MH, Park BK. Prevalence of Korean cats with natural feline coronavirus infections. Virol J. (2011) 8:455. doi: 10.1186/1743-422X-8-455
196. Sharif S, Arshad SS, Hair-Bejo M, Omar AR, Zeenathul NA, Fong LS, et al. Descriptive distribution and phylogenetic analysis of feline infectious peritonitis virus isolates of Malaysia. Acta Vet Scand. (2010) 52:1. doi: 10.1186/1751-0147-52-1
197. Akkan HA, Karaca M. Studies on the seroprevalence, age, and gender on the distribution of feline coronavirus in van cat kept in a multiple-cat environment. Bull Vet Inst Pulawy. (2009) 53:183–6.
198. Addie DD, McLachlan SA, Golder M, Ramsey I, Jarrett O. Evaluation of an in-practice test for feline coronavirus antibodies. J Feline Med Surg. (2004) 6:63–7. doi: 10.1016/j.jfms.2004.01.001
199. Almeida A, Galdino MV, Araújo JP. Seroepidemiological study of feline coronavirus (FCoV) infection in domiciled cats from small animal diseases. Pesq Vet Bras. (2019) 39:129–33. doi: 10.1590/1678-5150-pvb-5706
200. Bell ET, Malik R, Norris JM. The relationship between the feline coronavirus antibody titre and the age, breed, gender and health status of Australian cats. Aust Vet J. (2006) 84:2–7. doi: 10.1111/j.1751-0813.2006.tb13114.x
201. Tekes G, Spies D, Bank-Wolf B, Thiel V, Thiel HJ. A reverse genetics approach to study feline infectious peritonitis. J Virol. (2012) 86:6994–8. doi: 10.1128/JVI.00023-12
202. Thiel V, Thiel HJ, Tekes G. Tackling feline infectious peritonitis via reverse genetics. Bioengineered. (2014) 5:396–400. doi: 10.4161/bioe.32133
203. Vogel L, Van der Lubben M, Te Lintelo EG, Bekker CP, Geerts T, Schuijff LS, et al. Pathogenic characteristics of persistent feline enteric coronavirus infection in cats. Vet Res. (2010) 41:71. doi: 10.1051/vetres/2010043
204. Herrewegh AA, Mähler M, Hedrich HJ, Haagmans BL, Egberink HF, Horzinek MC, et al. Persistence and evolution of feline coronavirus in a closed cat-breeding colony. Virology. (1997) 234:349–63. doi: 10.1006/viro.1997.8663
205. Kipar A, Meli ML, Baptiste KE, Bowker LJ, Lutz H. Sites of feline coronavirus persistence in healthy cats. J Gen Virol. (2010) 91:1698–707. doi: 10.1099/vir.0.020214-0
206. Dewerchin HL, Cornelissen E, Nauwynck HJ. Replication of feline coronaviruses in peripheral blood monocytes. Arch Virol. (2005) 150:2483–500. doi: 10.1007/s00705-005-0598-6
207. Meli M, Kipar A, Müller C, Jenal K, Gönczi E, Borel N, et al. High viral loads despite absence of clinical and pathological findings in cats experimentally infected with feline coronavirus (FCoV) type I and in naturally FCoV-infected cats. J Feline Med Surg. (2004) 6:69–81. doi: 10.1016/j.jfms.2003.08.007
208. Rottier PJ, Nakamura K, Schellen P, Volders H, Haijema BJ. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J Virol. (2005) 79:14122–30. doi: 10.1128/JVI.79.22.14122-14130.2005
209. Stoddart CA, Scott FW. Intrinsic resistance of feline peritoneal macrophages to coronavirus infection correlates with in vivo virulence. J Virol. (1989) 63:436–40. doi: 10.1128/jvi.63.1.436-440.1989
210. Regan AD, Cohen RD, Whittaker GR. Activation of p38 MAPK by feline infectious peritonitis virus regulates pro-inflammatory cytokine production in primary blood-derived feline mononuclear cells. Virology. (2009) 384:135–43. doi: 10.1016/j.virol.2008.11.006
211. Kipar A, Baptiste K, Barth A, Reinacher M. Natural FCoV infection: cats with FIP exhibit significantly higher viral loads than healthy infected cats. J Feline Med Surg. (2006) 8:69–72. doi: 10.1016/j.jfms.2005.07.002
212. Kipar A, Meli ML, Failing K, Euler T, Gomes-Keller MA, Schwartz D, et al. Natural feline coronavirus infection: differences in cytokine patterns in association with the outcome of infection. Vet Immunol Immunopathol. (2006) 112:141–55. doi: 10.1016/j.vetimm.2006.02.004
213. Kipar A, Meli ML. Feline infectious peritonitis: still an enigma? Vet Pathol. (2014) 51:505–26. doi: 10.1177/0300985814522077
214. Kiss I, Poland AM, Pedersen NC. Disease outcome and cytokine responses in cats immunized with an avirulent feline infectious peritonitis virus (FIPV)-UCD1 and challenge-exposed with virulent FIPV-UCD8. J Feline Med Surg. (2004) 6:89–97. doi: 10.1016/j.jfms.2003.08.009
215. Takano T, Azuma N, Satoh M, Toda A, Hashida Y, Satoh R, et al. Neutrophil survival factors (TNF-alpha, GM-CSF, and G-CSF) produced by macrophages in cats infected with feline infectious peritonitis virus contribute to the pathogenesis of granulomatous lesions. Arch Virol. (2009) 154:775–81. doi: 10.1007/s00705-009-0371-3
216. Takano T, Hohdatsu T, Hashida Y, Kaneko Y, Tanabe M, Koyama H, et al. “Possible” involvement of TNF-alpha in apoptosis induction in peripheral blood lymphocytes of cats with feline infectious peritonitis. Vet Microbiol. (2007) 119:121–31. doi: 10.1016/j.vetmic.2006.08.033
217. Takano T, Hohdatsu T, Toda A, Tanabe M, Koyama H. TNF-alpha, produced by feline infectious peritonitis virus (FIPV)-infected macrophages, upregulates expression of type II FIPV receptor feline aminopeptidase N in feline macrophages. Virology. (2007) 364:64–72. doi: 10.1016/j.virol.2007.02.006
218. Takano T, Ohyama T, Kokumoto A, Satoh R, Hohdatsu T. Vascular endothelial growth factor (VEGF), produced by feline infectious peritonitis (FIP) virus-infected monocytes and macrophages, induces vascular permeability and effusion in cats with FIP. Virus Res. (2011) 158:161–8. doi: 10.1016/j.virusres.2011.03.027
219. Olyslaegers DA, Dedeurwaerder A, Desmarets LM, Vermeulen BL, Dewerchin HL, Nauwynck HJ. Altered expression of adhesion molecules on peripheral blood leukocytes in feline infectious peritonitis. Vet Microbiol. (2013) 166:438–49. doi: 10.1016/j.vetmic.2013.06.027
220. Hayashi T, Goto N, Takahashi R, Fujiwara K. Systemic vascular lesions in feline infectious peritonitis. Nihon JuigakuZasshi. (1977) 39:365–77. doi: 10.1292/jvms1939.39.365
221. Kipar A, Bellmann S, Kremendahl J, Köhler K, Reinacher M. Cellular composition, coronavirus antigen expression and production of specific antibodies in lesions in feline infectious peritonitis. Vet Immunol Immunopathol. (1998) 65:243–57. doi: 10.1016/S0165-2427(98)00158-5
222. Kipar A, May H, Menger S, Weber M, Leukert W, Reinacher M. Morphologic features and development of granulomatous vasculitis in feline infectious peritonitis. Vet Pathol. (2005) 42:321–30. doi: 10.1354/vp.42-3-321
223. Weiss RC, Scott FW. Pathogenesis of feline infetious peritonitis: pathologic changes and immunofluorescence. Am J Vet Res. (1981) 42:2036–48.
224. Kipar A, Kremendahl J, Addie DD, Leukert W, Grant CK, Reinacher M. Fatal enteritis associated with coronavirus infection in cats. J Comp Pathol. (1998) 119:1–14. doi: 10.1016/S0021-9975(98)80067-4
225. Drechsler Y, Alcaraz A, Bossong FJ, Collisson EW, Diniz PP. Feline coronavirus in multicat environments. Vet Clin North Am Small Anim Pract. (2011) 41:1133–69. doi: 10.1016/j.cvsm.2011.08.004
226. Hartmann K. Feline infectious peritonitis. Vet Clin North Am Small Anim Pract. (2005) 35:39–79, vi. doi: 10.1016/j.cvsm.2004.10.011
227. Binn LN, Lazar EC, Keenan KP, Huxsoll DL, Strano AJ. Recovery and characterization of a coronavirus from military dogs with diarrhea. Proc Annu Meet US Anim Health Assoc. (1974) 78:359–66.
228. Pratelli A, Martella V, Decaro N, Tinelli A, Camero M, Cirone F, et al. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J Virol Methods. (2003) 110:9–17. doi: 10.1016/S0166-0934(03)00081-8
229. Decaro N, Mari V, Campolo M, Lorusso A, Camero M, Elia G, et al. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of Swine are circulating in dogs. J Virol. (2009) 83:1532–7. doi: 10.1128/JVI.01937-08
230. Decaro N, Mari V, Elia G, Addie DD, Camero M, Lucente MS, et al. Recombinant canine coronaviruses in dogs, Europe. Emerg Infect Dis. (2010) 16:41–7. doi: 10.3201/eid1601.090726
231. Regan AD, Millet JK, Tse LP, Chillag Z, Rinaldi VD, Licitra BN, et al. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology. (2012) 430:90–9. doi: 10.1016/j.virol.2012.04.013
232. Escutenaire S, Isaksson M, Renström LH, Klingeborn B, Buonavoglia C, Berg M, et al. Characterization of divergent and atypical canine coronaviruses from Sweden. Arch Virol. (2007) 152:1507–14. doi: 10.1007/s00705-007-0986-1
233. Licitra BN, Duhamel GE, Whittaker GR. Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins. Viruses. (2014) 6:3363–76. doi: 10.3390/v6083363
234. Keenan KP, Jervis HR, Marchwicki RH, Binn LN. Intestinal infection of neonatal dogs with canine coronavirus 1-71: studies by virologic, histologic, histochemical, and immunofluorescent techniques. Am J Vet Res. (1976) 37:247–56.
235. Tennant BJ, Gaskell RM, Kelly DF, Carter SD, Gaskell CJ. Canine coronavirus infection in the dog following oronasal inoculation. Res Vet Sci. (1991) 51:11–8. doi: 10.1016/0034-5288(91)90023-H
236. Pratelli A, Tempesta M, Roperto FP, Sagazio P, Carmichael L, Buonavoglia C. Fatal coronavirus infection in puppies following canine parvovirus 2b infection. J Vet Diagn Invest. (1999) 11:550–3. doi: 10.1177/104063879901100615
237. Pratelli A, Martella V, Elia G, Tempesta M, Guarda F, Capucchio MT, et al. Severe enteric disease in an animal shelter associated with dual infections by canine adenovirus type 1 and canine coronavirus. J Vet Med B Infect Dis Vet Public Health. (2001) 48:385–92. doi: 10.1046/j.1439-0450.2001.00466.x
238. Decaro N, Martella V, Elia G, Campolo M, Desario C, Cirone F, et al. Molecular characterisation of the virulent canine coronavirus CB/05 strain. Virus Res. (2007) 125:54–60. doi: 10.1016/j.virusres.2006.12.006
239. Ntafis V, Xylouri E, Mari V, Papanastassopoulou M, Papaioannou N, Thomas A, et al. Molecular characterization of a canine coronavirus NA/09 strain detected in a dog's organs. Arch Virol. (2012) 157:171–5. doi: 10.1007/s00705-011-1141-6
240. Zicola A, Jolly S, Mathijs E, Ziant D, Decaro N, Mari V, et al. Fatal outbreaks in dogs associated with pantropic canine coronavirus in France and Belgium. J Small Anim Pract. (2012) 53:297–300. doi: 10.1111/j.1748-5827.2011.01178.x
241. Buonavoglia C, Decaro N, Martella V, Elia G, Campolo M, Desario C, et al. Canine coronavirus highly pathogenic for dogs. Emerg Infect Dis. (2006) 12:492–4. doi: 10.3201/eid1203.050839
242. Alfano F, Fusco G, Mari V, Occhiogrosso L, Miletti G, Brunetti R, et al. Circulation of pantropic canine coronavirus in autochthonous and imported dogs, Italy. Transbound Emerg Dis. (2020) 67:1991–9. doi: 10.1111/tbed.13542
243. Alfano F, Dowgier G, Valentino MP, Galiero G, Tinelli A, Nicola D, et al. Identification of pantropic canine coronavirus in a wolf (Canis lupus italicus) in Italy. J Wildl Dis. (2019) 55:504–8. doi: 10.7589/2018-07-182
244. Evermann JF, Abbott JR, Han S. Canine coronavirus-associated puppy mortality without evidence of concurrent canine parvovirus infection. J Vet Diagn Invest. (2005) 17:610–4. doi: 10.1177/104063870501700618
245. Decaro N, Mari V, Larocca V, Losurdo M, Lanave G, Lucente MS, et al. Molecular surveillance of traditional and emerging pathogens associated with canine infectious respiratory disease. Vet Microbiol. (2016) 192:21–5. doi: 10.1016/j.vetmic.2016.06.009
246. Lavan R, Knesl O. Prevalence of canine infectious respiratory pathogens in asymptomatic dogs presented at US animal shelters. J Small Anim Pract. (2015) 56:572–6. doi: 10.1111/jsap.12389
247. Erles K, Brownlie J. Canine respiratory coronavirus: an emerging pathogen in the canine infectious respiratory disease complex. Vet Clin North Am Small Anim Pract. (2008) 38:815–25. doi: 10.1016/j.cvsm.2008.02.008
248. Soma T, Ohinata T, Ishii H, Takahashi T, Taharaguchi S, Hara M. Detection and genotyping of canine coronavirus RNA in diarrheic dogs in Japan. Res Vet Sci. (2011) 90:205–7. doi: 10.1016/j.rvsc.2010.05.027
249. Costa EM, de Castro TX, Bottino Fde O, Garcia Rde C. Molecular characterization of canine coronavirus strains circulating in Brazil. Vet Microbiol. (2014) 168:8–15. doi: 10.1016/j.vetmic.2013.10.002
250. Ntafis V, Mari V, Decaro N, Papanastassopoulou M, Pardali D, Rallis TS, et al. Canine coronavirus, Greece. Molecular analysis and genetic diversity characterization infect. Genet Evol. (2013) 16:129–36. doi: 10.1016/j.meegid.2013.01.014
251. Wang X, Li C, Guo D, Wang X, Wei S, Geng Y, et al. Co-circulation of canine coronavirus I and IIa/b with high prevalence and genetic diversity in Heilongjiang Province, Northeast China. PLoS ONE. (2016) 11:e0146975. doi: 10.1371/journal.pone.0146975
252. van Nguyen D, Terada Y, Minami S, Yonemitsu K, Nagata N, Le TD, et al. Characterization of canine coronavirus spread among domestic dogs in Vietnam. J Vet Med Sci. (2017) 79:343–9. doi: 10.1292/jvms.16-0538
253. Naylor MJ, Walia CS, McOrist S, Lehrbach PR, Deane EM, Harrison GA. Molecular characterization confirms the presence of a divergent strain of canine coronavirus (UWSMN-1) in Australia. J Clin Microbiol. (2002) 40:3518–22. doi: 10.1128/JCM.40.9.3518-3522.2002
254. Priestnall SL, Brownlie J, Dubovi EJ, Erles K. Serological prevalence of canine respiratory coronavirus. Vet Microbiol. (2006) 115:43–53. doi: 10.1016/j.vetmic.2006.02.008
255. Priestnall SL, Pratelli A, Brownlie J, Erles K. Serological prevalence of canine respiratory coronavirus in southern Italy and epidemiological relationship with canine enteric coronavirus. J Vet Diagn Invest. (2007) 19:176–80. doi: 10.1177/104063870701900206
256. Kaneshima T, Hohdatsu T, Satoh K, Takano T, Motokawa K, Koyama H. The prevalence of a group 2 coronavirus in dogs in Japan. J Vet Med Sci. (2006) 68:21–5. doi: 10.1292/jvms.68.21
257. Yachi A, Mochizuki M. Survey of dogs in Japan for group 2 canine coronavirus infection. J Clin Microbiol. (2006) 44:2615–8. doi: 10.1128/JCM.02397-05
258. Knesl O, Allan FJ, Shields S. The seroprevalence of canine respiratory coronavirus and canine influenza virus in dogs in New Zealand. N Z Vet J. (2009) 57:295–8. doi: 10.1080/00480169.2009.58624
259. He HJ, Zhang W, Liang J, Lu M, Wang R, Li G, et al. Etiology and genetic evolution of canine coronavirus circulating in five provinces of China, during 2018-2019. Microb Pathog. (2020) 145:104209. doi: 10.1016/j.micpath.2020.104209
260. Stavisky J, Pinchbeck GL, German AJ, Dawson S, Gaskell RM, Ryvar R, et al. Prevalence of canine enteric coronavirus in a cross-sectional survey of dogs presenting at veterinary practices. Vet Microbiol. (2010) 140:18–24. doi: 10.1016/j.vetmic.2009.07.012
261. Decaro N, Martella V, Ricci D, Elia G, Desario C, Campolo M, et al. Genotype-specific fluorogenic RT-PCR assays for the detection and quantitation of canine coronavirus type I and type II RNA in faecal samples of dogs. J Virol Methods. (2005) 130:72–8. doi: 10.1016/j.jviromet.2005.06.005
262. Pratelli A, Elia G, Martella V, Tinelli A, Decaro N, Marsilio F, et al. M Gene evolution of canine coronavirus in naturally infected dogs. Vet Rec. (2002) 151:758–61. doi: 10.1136/vr.151.25.758
263. Ma GG, Lu CP. [Two genotypes of Canine coronavirus simultaneously detected in the fecal samples of healthy foxes and raccoon dogs]. Wei Sheng Wu Xue Bao. (2005) 45:305–8.
264. Zarnke RL, Evermann J, Ver Hoef JM, McNay ME, Boertje RD, Gardner CL, et al. Serologic survey for canine coronavirus in wolves from Alaska. J Wildl Dis. (2001) 37:740–5. doi: 10.7589/0090-3558-37.4.740
265. Arsevska E, Priestnall SL, Singleton DA, Jones PH, Smyth S, Brant B, et al. Small animal disease surveillance: respiratory disease 2017. Vet Rec. (2018) 182:369–73. doi: 10.1136/vr.k1426
266. Zappulli V, Caliari D, Cavicchioli L, Tinelli A, Castagnaro M. Systemic fatal type II coronavirus infection in a dog: pathological findings and immunohistochemistry. Res Vet Sci. (2008) 84:278–82. doi: 10.1016/j.rvsc.2007.05.004
267. Mitchell JA, Brooks HW, Szladovits B, Erles K, Gibbons R, Shields S, et al. Tropism and pathological findings associated with canine respiratory coronavirus (CRCoV). Vet Microbiol. (2013) 162:582–94. doi: 10.1016/j.vetmic.2012.11.025
268. Decaro N, Campolo M, Lorusso A, Desario C, Mari V, Colaianni ML, et al. Experimental infection of dogs with a novel strain of canine coronavirus causing systemic disease and lymphopenia. Vet Microbiol. (2008) 128:253–60. doi: 10.1016/j.vetmic.2007.10.008
269. Decaro N, Cordonnier N, Demeter Z, Egberink H, Elia G, Grellet A, et al. European surveillance for pantropic canine coronavirus. J Clin Microbiol. (2013) 51:83–8. doi: 10.1128/JCM.02466-12
270. Saif LJ, Brock KV, Redman DR, Kohler EM. Winter dysentery in dairy herds: electron microscopic and serological evidence for an association with coronavirus infection. Vet Rec. (1991) 128:447–9. doi: 10.1136/vr.128.19.447
271. Patterson S, Bingham RW. Electron microscope observations on the entry of avian infectious bronchitis virus into susceptible cells. Arch Virol. (1976) 52:191–200. doi: 10.1007/BF01348016
272. Bezuidenhout A, Mondal SP, Buckles EL. Histopathological and immunohistochemical study of air sac lesions induced by two strains of infectious bronchitis virus. J Comp Pathol. (2011) 145:319–26. doi: 10.1016/j.jcpa.2011.01.011
273. Arshad SS, Al-Salihi KA. Immunohistochemical detection of infectious bronchitis virus antigen in chicken respiratory and kidney tissues. In: Proceedings of the 12th Federation of Asian Veterinary Associations Congress/14th Veterinary Association Malaysia Scientific Congress (2002).
274. Abdel-Moneim AS, Zlotowski P, Veits J, Keil GM, Teifke JP. Immunohistochemistry for detection of avian infectious bronchitis virus strain M41 in the proventriculus and nervous system of experimentally infected chicken embryos. Virol J. (2009) 6:15. doi: 10.1186/1743-422X-6-15
275. Yagyu K, Ohta S. Detection of infectious bronchitis virus antigen from experimentally infected chickens by indirect immunofluorescent assay with monoclonal antibody. Avian Dis. (1990) 34:246–52. doi: 10.2307/1591405
276. Saif LJ, Bohl EH, Kohler EM, Hughes JH. Immune electron microscopy of transmissible gastroenteritis virus and rotavirus (reovirus-like agent) of swine. Am J Vet Res. (1977) 38:13–20.
277. Pensaert M, Haelterman EO, Burnstein T. Transmissible gastroenteritis of swine: virus-intestinal cell interactions. I Immunofluorescence, histopathology and virus production in the small intestine through the course of infection. Arch Gesamte Virusforsch. (1970) 31:321–34. doi: 10.1007/BF01253767
278. Shoup DI, Swayne DE, Jackwood DJ, Saif LJ. Immunohistochemistry of transmissible gastroenteritis virus antigens in fixed paraffin-embedded tissues. J Vet Diagn Invest. (1996) 8:161–7. doi: 10.1177/104063879600800204
279. Lanza I, Shoup DI, Saif LJ. Lactogenic immunity and milk antibody isotypes to transmissible gastroenteritis virus in sows exposed to porcine respiratory coronavirus during pregnancy. Am J Vet Res. (1995) 56:739–48.
280. Sestak K, Lanza I, Park SK, Weilnau PA, Saif LJ. Contribution of passive immunity to porcine respiratory coronavirus to protection against transmissible gastroenteritis virus challenge exposure in suckling pigs. Am J Vet Res. (1996) 57:664–71.
281. Sestak K, Lanza I, Park SK, Weilnau PA, Saif LJ. Contribution of passive immunity to porcine respiratory coronavirus to protection against transmissible gastroenteritis virus challenge exposure in suckling pigs. Am J Vet Res. (1996) 57:664–71.
282. van Nieuwstadt AP, Cornelissen JB, Zetstra T. Comparison of two methods for detection of transmissible gastroenteritis virus in feces of pigs with experimentally induced infection. Am J Vet Res. (1988) 49:1836–43.
283. Giori L, Giordano A, Giudice C, Grieco V, Paltrinieri S. Performances of different diagnostic tests for feline infectious peritonitis in challenging clinical cases. J Small Anim Pract. (2011) 52:152–7. doi: 10.1111/j.1748-5827.2011.01042.x
284. Addie DD, Paltrinieri S, Pedersen NC. Recommendations from workshops of the second international feline coronavirus/feline infectious peritonitis symposium. J Feline Med Surg. (2004) 6:125–30. doi: 10.1016/j.jfms.2003.12.009
285. Ellis JA, McLean N, Hupaelo R, Haines DM. Detection of coronavirus in cases of tracheobronchitis in dogs: a retrospective study from 1971 to 2003. Can Vet J. (2005) 46:447–8.
286. Stranieri A, Giordano A, Paltrinieri S, Giudice C, Cannito V, Lauzi S. Comparison of the performance of laboratory tests in the diagnosis of feline infectious peritonitis. J Vet Diagn Investig. (2018) 30:459–63. doi: 10.1177/1040638718756460
287. Hartmann K, Binder C, Hirschberger J, Cole D, Reinacher M, Schroo S, et al. Comparison of different tests to diagnose feline infectious peritonitis. J Vet Intern Med. (2003) 17:781–90. doi: 10.1111/j.1939-1676.2003.tb02515.x
288. Hirschberger J. Zytologievon Körperhöhlenergüssen [Cytology of body cavity effusions]. TierarztlPrax. (1995) 23:192–9.
289. Hirschberger J, Koch S. Validation of the determination of the activity of adenosine deaminase in the body effusions of cats. Res Vet Sci. (1995) 59:226–9. doi: 10.1016/0034-5288(95)90007-1
290. Hirschberger J, Sauer G. Clinical and chemical investigations of body cavity effusions. TieraerztlichePrax. (1991) 19:431–4.
291. Bohl EH, Kumagai T. The use of cell cultures for the study of swine transmissible gastroenteritis virus. In: United States Livestock Sanitary Association Meeting. (1965). p. 343–50.
292. Laude H, Gelfi J, Aynaud JM. In vitro properties of low- and high-passaged strains of transmissible gastroenteritis coronavirus of swine. Am J Vet Res. (1981) 42:447–9.
293. Witte KH. Isolation of the virus of transmissible gastroenteritis (TGE) from naturally infected piglets in cell culture. Zentralbl Veterinarmed B. (1971) 18:770–8. doi: 10.1111/j.1439-0450.1971.tb01654.x
294. McClurkin AW, Norman JO. Studies on transmissible gastroenteritis of swine. II Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can J Comp Med Vet Sci. (1966) 30:190–8.
295. Otsuki K, Yamamoto H, Tsubokura M. Studies on avian infectious bronchitis virus (IBV). I. Resistance of IBV to chemical and physical treatments. Arch Virol. (1979) 60:25–32. doi: 10.1007/BF01318094
296. Jones BV, Hennion RM. The preparation of chicken tracheal organ cultures for virus isolation, propagation, and titration. Methods Mol Biol. (2008) 454:103–7. doi: 10.1007/978-1-59745-181-9_9
297. Bhattacharjee PS, Naylor CJ, Jones RC. A simple method for immunofluorescence staining of tracheal organ cultures for the rapid identification of infectious bronchitis virus. Avian Pathol. (1994) 23:471–80. doi: 10.1080/03079459408419017
298. Beaudette F, Hudson C. Cultivation of the virus of infectious bronchitis. J Am Vet Med Assoc. (1937) 90:51–60.
299. Loomis LN, Cunningham CH, Gray ML, Thorp F. Pathology of the chicken embryo infected with infectious bronchitis virus. Am J Vet Res. (1950) 11:245–51.
300. Bernard S, Bottreau E, Aynaud JM, Have P, Szymansky J. Natural infection with the porcine respiratory coronavirus induces protective lactogenic immunity against transmissible gastroenteritis. Vet Microbiol. (1989) 21:1–8. doi: 10.1016/0378-1135(89)90013-8
301. Garwes DJ, Stewart F, Cartwright SF, Brown I. Differentiation of porcine coronavirus from transmissible gastroenteritis virus. Vet Rec. (1988) 122:86–7. doi: 10.1136/vr.122.4.86
302. Callebaut P, Pensaert MB, Hooyberghs J. A competitive inhibition ELISA for the differentiation of serum antibodies from pigs infected with transmissible gastroenteritis virus (TGEV) or with the TGEV-related porcine respiratory coronavirus. Vet Microbiol. (1989) 20:9–19. doi: 10.1016/0378-1135(89)90003-5
303. Pedersen NC. Serologic studies of naturally occurring feline infectious peritonitis. Am J Vet Res. (1976) 37:1449–53.
304. Addie DD, Jarrett O. Feline coronavirus antibodies in cats. Vet Rec. (1992) 131:202–3. doi: 10.1136/vr.131.9.202-a
305. Addie DD, le Poder S, Burr P, Decaro N, Graham E, Hofmann-Lehmann R, et al. Utility of feline coronavirus antibody tests. J Feline Med Surg. (2015) 17:152–62. doi: 10.1177/1098612X14538873
306. Osterhaus AD, Horzinek MC, Reynolds DJ. Seroepidemiology of feline infectious peritonitis virus infections using transmissible gastroenteritis virus as antigen. Zentralbla Veterinärmed B. (1977) 24:835–41. doi: 10.1111/j.1439-0450.1977.tb00976.x
307. Horzinek MC, Osterhaus AD. Feline infectious peritonitis: a worldwide serosurvey. Am J. Vet. Res. (1979) 40:1487–92.
308. Pratelli A. Comparison of serologic techniques for the detection of antibodies against feline coronaviruses. J Vet Diagn Investig. (2008) 20:45–50. doi: 10.1177/104063870802000108
309. Chen L, Li F. Structural analysis of the evolutionary origins of influenza virus hemagglutinin and other viral lectins. J Virol. (2013) 87:4118–20. doi: 10.1128/JVI.03476-12
310. Erles K, Shiu KB, Brownlie J. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. (2007) 124:78–87. doi: 10.1016/j.virusres.2006.10.004
311. Qiao J, Meng Q, Cai X, Chen C, Zhang Z, Tian Z. Rapid detection of Betacoronavirus 1 from clinical fecal specimens by a novel reverse transcription loop-mediated isothermal amplification assay. J Vet Diagn Invest. (2012) 24:174–7. doi: 10.1177/1040638711425937
312. Amer HM, Abd El Wahed A, Shalaby MA, Almajhdi FN, Hufert FT, Weidmann M, et al. new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J Virol Methods. (2013) 193:337–40. doi: 10.1016/j.jviromet.2013.06.027
313. Stephensen CB, Casebolt DB, Gangopadhyay NN. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. (1999) 60:181–9. doi: 10.1016/S0168-1702(99)00017-9
314. Lin Z, Kato A, Kudou Y, Umeda K, Ueda S. Typing of recent infectious bronchitis virus isolates causing nephritis in chicken. Arch Virol. (1991) 120:145–9. doi: 10.1007/BF01310957
315. Zwaagstra KA, van der Zeijst BA, Kusters JG. Rapid detection and identification of avian infectious bronchitis virus. J Clin Microbiol. (1992) 30:79–84. doi: 10.1128/jcm.30.1.79-84.1992
316. Kwon HM, Jackwood MW, Gelb J. Differentiation of infectious bronchitis virus serotypes using polymerase chain reaction and restriction fragment length polymorphism analysis. Avian Dis. (1993) 37:194–202. doi: 10.2307/1591474
317. Keeler CL, Reed KL, Nix WA, Gelb J. Serotype identification of avian infectious bronchitis virus by RT-PCR of the peplomer (S-1) gene. Avian Dis. (1998) 42:275–84. doi: 10.2307/1592477
318. Lin Z, Kato A, Kudou Y, Ueda S. A new typing method for the avian infectious bronchitis virus using polymerase chain reaction and restriction enzyme fragment length polymorphism. Arch Virol. (1991) 116:19–31. doi: 10.1007/BF01319228
319. Mardani K, Noormohammadi AH, Ignatovic J, Browning GF. Typing infectious bronchitis virus strains using reverse transcription-polymerase chain reaction and restriction fragment length polymorphism analysis to compare the 3' 75 kb of their genomes. Avian Pathol. (2006) 35:63–9. doi: 10.1080/03079450500465817
320. Montassier MDFS, Brentano L, Montassier HJ, Richtzenhain LG. Genetic grouping of avian infectious bronchitis virus isolated in Brazil based on RT-PCR/RFLP analysis of the S1 gene. Pesqui Vet Bras. (2008) 28:190–4 doi: 10.1590/S0100-736X2008000300011
321. Chousalkar KK, Cheetham BF, Roberts JR. LNA probe-based real-time RT-PCR for the detection of infectious bronchitis virus from the oviduct of unvaccinated and vaccinated laying hens. J Virol Methods. (2009) 155:67–71. doi: 10.1016/j.jviromet.2008.09.028
322. Acevedo AM, Perera CL, Vega A, Ríos L, Coronado L, Relova D, et al. A duplex SYBR green I-based real-time RT-PCR assay for the simultaneous detection and differentiation of massachusetts and non-massachusetts serotypes of infectious bronchitis virus. Mol Cell Probes. (2013) 27:184–92. doi: 10.1016/j.mcp.2013.06.001
323. Meir R, Maharat O, Farnushi Y, Simanov L. Development of a real-time TaqMan RT-PCR assay for the detection of infectious bronchitis virus in chickens, and comparison of RT-PCR and virus isolation. J Virol Methods. (2010) 163:190–4. doi: 10.1016/j.jviromet.2009.09.014
324. Chen HW, Wang CH. A multiplex reverse transcriptase-PCR assay for the genotyping of avian infectious bronchitis viruses. Avian Dis. (2010) 54:104–8. doi: 10.1637/8954-060609-Reg.1
325. Chen HT, Zhang J, Ma YP, Ma LN, Ding YZ, Liu XT, et al. Reverse transcription loop-mediated isothermal amplification for the rapid detection of infectious bronchitis virus in infected chicken tissues. Mol Cell Probes. (2010) 24:104–6. doi: 10.1016/j.mcp.2009.10.001
326. Kim SY, Song DS, Park BK. Differential detection of transmissible gastroenteritis virus and porcine epidemic diarrhea virus by duplex RT-PCR. J Vet Diagn Invest. (2001) 13:516–20. doi: 10.1177/104063870101300611
327. Masuda T, Tsuchiaka S, Ashiba T, Yamasato H, Fukunari K, Omatsu T, et al. Development of one-step real-time reverse transcriptase-PCR-based assays for the rapid and simultaneous detection of four viruses causing porcine diarrhea. Jpn J Vet Res. (2016) 64:5–14.
328. Ogawa H, Taira O, Hirai T, Takeuchi H, Nagao A, Ishikawa Y, et al. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J Virol Methods. (2009) 160:210–4. doi: 10.1016/j.jviromet.2009.05.010
329. Longstaff L, Porter E, Crossley VJ, Hayhow SE, Helps CR, Tasker S. Feline coronavirus quantitative reverse transcriptase polymerase chain reaction on effusion samples in cats with and without feline infectious peritonitis. J Feline Med Surg. (2017) 19:240–5. doi: 10.1177/1098612X15606957
330. Felten S, Leutenegger CM, Balzer HJ, Pantchev N, Matiasek K, Sangl L, et al. Evaluation of a discriminative realtimert-PCR in cerebrospinal fluid for the diagnosis of feline infectious peritonitis. In: Proceedings of the 27th ECVIM-CA Congress. St. Julian's (2017). p. 14–6.
331. Felten S, Emmler L, Matiasek K, Balzer H-J, Pantchev N, Leutenegger CM, et al. Detection of mutated and non-mutated feline coronavirus in tissues and body fluids of cats with feline infectious peritonitis. In: Proceedings of the 2018 ISCAID Symposium. Portland, OR (2018).
332. Felten S, Sangl L, Matiasek K, Leutenegger CM, Pantchev N, Balzer H-J. Comparison of different diagnostic methods in aqueous humor to diagnose feline infectious peritonitis. In: Proceedings of the 2016 ISCAID Symposium. Bristol (2016).
333. Li X, Scott FW. Detection of feline coronaviruses in cell cultures and in fresh and fixed feline tissues using polymerase chain reaction. Vet Microbiol. (1994) 42:65–77. doi: 10.1016/0378-1135(94)90078-7
334. Barker EN, Stranieri A, Helps CR, Porter EL, Davidson AD, Day MJ, et al. Limitations of using feline coronavirus spike protein gene mutations to diagnose feline infectious peritonitis. Vet Res. (2017) 48:60. doi: 10.1186/s13567-017-0467-9
335. Freiche GM, Guidez CL, Duarte M, Le Poder YB. Sequencing of 3c and spike genes in feline infectious peritonitis: which samples are the most relevant for analysis? A retrospective study of 33 cases from 2008 to 2014. J Vet Intern Med. (2016) 30:436.
336. Felten S, Weider K, Doenges S, Gruendl S, Matiasek K, Hermanns W, et al. Detection of feline coronavirus spike gene mutations as a tool to diagnose feline infectious peritonitis. J Feline Med Surg. (2017) 19:321–35. doi: 10.1177/1098612X15623824
337. Felten S, Leutenegger CM, Balzer HJ, Pantchev N, Matiasek K, Wess G, et al. Sensitivity and specificity of a real-time reverse transcriptase polymerase chain reaction detecting feline coronavirus mutations in effusion and serum/plasma of cats to diagnose feline infectious peritonitis. BMC Vet Res. (2017) 13:228. doi: 10.1186/s12917-017-1147-8
338. Decaro N, Pratelli A, Campolo M, Elia G, Martella V, Tempesta M, et al. Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT-PCR. J Virol Methods. (2004) 119:145–50. doi: 10.1016/j.jviromet.2004.03.012
339. Gizzi AB, Oliveira ST, Leutenegger CM, Estrada M, Kozemjakin DA, Stedile R, et al. Presence of infectious agents and co-infections in diarrheic dogs determined with a real-time polymerase chain reaction-based panel. BMC Vet Res. (2014) 10:23. doi: 10.1186/1746-6148-10-23
340. Zulperi ZM, Omar AR, Arshad SS. Sequence and phylogenetic analysis of S1, S2, M, and N genes of infectious bronchitis virus isolates from Malaysia. Virus Genes. (2009) 38:383–91. doi: 10.1007/s11262-009-0337-2
341. Abro SH, Renström LH, Ullman K, Isaksson M, Zohari S, Jansson DS, et al. Emergence of novel strains of avian infectious bronchitis virus in Sweden. Vet Microbiol. (2012) 155:237–46. doi: 10.1016/j.vetmic.2011.09.022
342. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. (2020) 323:1843–4. doi: 10.1001/jama.2020.3786
343. Yu L, Wu S, Hao X, Dong X, Mao L, Pelechano V, et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem. (2020) 66:975–7. doi: 10.1093/clinchem/hvaa102
344. Vashist SK. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics. (2020) 10:202. doi: 10.3390/diagnostics10040202
345. Younes N, Al-Sadeq DW, Al-Jighefee H, et al. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. (2020) 12:582. doi: 10.3390/v12060582
346. Rao VK. Point of care diagnostic devices for rapid detection of novel coronavirus (SARS-nCoV19) pandemic: a review. Front. Nanotechnol. (2021) 2:2673–3013 doi: 10.3389/fnano.2020.593619
347. Samson R, Navale GR, Dharne MS. Biosensors: frontiers in rapid detection of COVID-19. Biotech. (2020) 10:385. doi: 10.1007/s13205-020-02369-0
348. Wang X, Zhong M, Liu Y, Ma P, Dang L, Meng Q, et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci Bull. (2020) 65:1436–9. doi: 10.1016/j.scib.2020.04.041
349. Chen Z, Wu Q, Chen J, Ni X, Dai J, A DNA. Aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol Sin. (2020) 35:351–4. doi: 10.1007/s12250-020-00236-z
350. Fraser DD, Slessarev M, Martin CM, Daley M, Patel MA, Miller MR, et al. Metabolomics profiling of critically ill coronavirus disease 2019 patients: identification of diagnostic and prognostic biomarkers. Crit Care Explor. (2020) 2:e0272. doi: 10.1097/CCE.0000000000000272
351. Okda F, Liu X, Singrey A, Clement T, Nelson J, Christopher-Hennings J, et al. Development of an indirect ELISA, blocking ELISA, fluorescent microsphere immunoassay and fluorescent focus neutralization assay for serologic evaluation of exposure to North American strains of porcine epidemic diarrhea virus. BMC Vet Res. (2015) 11:180. doi: 10.1186/s12917-015-0500-z
352. Okda F, Lawson S, Liu X, Singrey A, Clement T, Hain K, et al. Development of monoclonal antibodies and serological assays including indirect ELISA and fluorescent microsphere immunoassays for diagnosis of porcine deltacoronavirus. BMC Vet Res. (2016) 12:95. doi: 10.1186/s12917-016-0716-6
353. Liu J, Shi H, Cong G, Chen J, Zhang X, Shi D, et al. Development of a rapid and sensitive europium (III) chelate microparticle-based lateral flow test strip for the detection and epidemiological surveillance of porcine epidemic diarrhea virus. Arch Virol. (2020) 165:1049–56. doi: 10.1007/s00705-020-04566-x
354. Thanthrige-Don N, Lung O, Furukawa-Stoffer T, Buchanan C, Joseph T, Godson DL, et al. A novel multiplex PCR-electronic microarray assay for rapid and simultaneous detection of bovine respiratory and enteric pathogens. J Virol Methods. (2018) 261:51–62. doi: 10.1016/j.jviromet.2018.08.010
355. Calvet GA, Pereira SA, Ogrzewalska M, Pauvolid-Corrêa A, Resende PC, Tassinari W, et al. Investigation of SARS-CoV-2 infection in dogs and cats of humans diagnosed with COVID-19 in Rio de Janeiro, Brazil. PLoS ONE. (2021) 16:e0250853. doi: 10.1371/journal.pone.0250853
356. Haljasmägi L, Remm A, Rumm AP, Krassohhina E, Sein H, Tamm A, et al. LIPS method for the detection of SARS-CoV-2 antibodies to spike and nucleocapsid proteins. Eur J Immunol. (2020) 50:1234–6. doi: 10.1002/eji.202048715
357. Temmam S, Barbarino A, Maso D, Behillil S, Enouf V, Huon C, et al. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Health. (2020) 10:100164. doi: 10.1016/j.onehlt.2020.100164
358. Wang W, Wang T, Deng Y, Niu P, Ruhan A, Zhao J, et al. A novel luciferase immunosorbent assay performs better than a commercial enzyme-linked immunosorbent assay to detect MERS-CoV specific IgG in humans and animals. Biosaf Health. (2019) 1:134–43. doi: 10.1016/j.bsheal.2019.12.006
359. Csiszar A, Jakab F, Valencak TG, Lanszki Z, Tóth GE, Kemenesi G, et al. Companion animals likely do not spread COVID-19 but may get infected themselves. Geroscience. (2020) 42:1229–36. doi: 10.1007/s11357-020-00248-3
360. Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. (2020) 9:680–6. doi: 10.1080/22221751.2020.1743767
361. Decaro N, Campolo M, Mari V, Desario C, Colaianni ML, Di Trani L, et al. A candidate modified-live bovine coronavirus vaccine: safety and immunogenicity evaluation. New Microbiol. (2009) 32:109–13.
363. Zhang Y, Huang S, Zeng Y, Xue C, Cao Y. Rapid development and evaluation of a live-attenuated QX-like infectious bronchitis virus vaccine. Vaccine. (2018) 36:4245–54. doi: 10.1016/j.vaccine.2018.05.123
364. Bohl EH, Gupta RP, Olquin MF, Saif LJ. Antibody responses in serum, colostrum, and milk of swine after infection or vaccination with transmissible gastroenteritis virus. Infect Immun. (1972) 6:289–301. doi: 10.1128/iai.6.3.289-301.1972
365. Saif LJ, Bohl E. Passive immunity to transmissible gastroenteritis virus: intramammary viral inoculation of sows. Ann N Y Acad Sci. (1983) 409:708. doi: 10.1111/j.1749-6632.1983.tb26910.x
366. Baek PS, Choi HW, Lee S, Yoon IJ, Lee YJ, Lee du S, et al. Efficacy of an inactivated genotype 2b porcine epidemic diarrhea virus vaccine in neonatal piglets. Vet Immunol Immunopathol. (2016) 174:45–9. doi: 10.1016/j.vetimm.2016.04.009
367. Sato T, Takeyama N, Katsumata A, Tuchiya K, Kodama T, Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes. (2011) 43:72–8. doi: 10.1007/s11262-011-0617-5
368. Gerdts V, Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet Microbiol. (2017) 206:45–51. doi: 10.1016/j.vetmic.2016.11.029
369. Pratelli A, Tinelli A, Decaro N, Martella V, Camero M, Tempesta M, et al. Safety and efficacy of a modified-live canine coronavirus vaccine in dogs. Vet Microbiol. (2004) 99:43–9. doi: 10.1016/j.vetmic.2003.07.009
370. Pratelli A. Genetic evolution of canine coronavirus and recent advances in prophylaxis. Vet Res. (2006) 37:191–200. doi: 10.1051/vetres:2005053
371. Zhang D, Huang X, Zhang X, Cao S, Wen X, Wen Y, et al. Construction of an oral vaccine for transmissible gastroenteritis virus based on the TGEV N gene expressed in an attenuated Salmonella typhimurium vector. J Virol Methods. (2016) 227:6–13. doi: 10.1016/j.jviromet.2015.08.011
372. Yuan X, Lin H, Fan H. Efficacy and immunogenicity of recombinant swinepox virus expressing the A epitope of the TGEV S protein. Vaccine. (2015) 33:3900–6. doi: 10.1016/j.vaccine.2015.06.057
373. Orr-Burks N, Gulley SL, Toro H, van Ginkel FW. Immunoglobulin A as an early humoral responder after mucosal avian coronavirus vaccination. Avian Dis. (2014) 58:279–86. doi: 10.1637/10740-120313-Reg.1
374. Jordan B. Vaccination against infectious bronchitis virus: a continuous challenge. Vet Microbiol. (2017) 206:137–43. doi: 10.1016/j.vetmic.2017.01.002
375. Smialek M, Tykalowski B, Dziewulska D, Stenzel T, Koncicki A. Immunological aspects of the efficiency of protectotype vaccination strategy against chicken infectious bronchitis. BMC Vet Res. (2017) 13:44. doi: 10.1186/s12917-017-0963-1
376. Decaro N, Mari V, Sciarretta R, Colao V, Losurdo M, Catella C, et al. Immunogenicity and protective efficacy in dogs of an MF59™-adjuvanted vaccine against recombinant canine/porcine coronavirus. Vaccine. (2011) 29:2018–23. doi: 10.1016/j.vaccine.2011.01.028
377. Zhang Y, Zhang X, Liao X, Huang X, Cao S, Wen X, et al. Construction of a bivalent DNA vaccine co-expressing S genes of transmissible gastroenteritis virus and porcine epidemic diarrhea virus delivered by attenuated Salmonella typhimurium. Virus Genes. (2016) 52:354–64. doi: 10.1007/s11262-016-1316-z
378. Meng F, Ren Y, Suo S, Sun X, Li X, Li P, et al. Evaluation on the efficacy and immunogenicity of recombinant DNA plasmids expressing spike genes from porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. PLoS ONE. (2013) 8:e57468. doi: 10.1371/journal.pone.0057468
379. Tuboly T, Nagy É. Construction and characterization of recombinant porcine adenovirus serotype 5 expressing the transmissible gastroenteritis virus spike gene. J Gen Virol. (2001) 82:183–90. doi: 10.1099/0022-1317-82-1-183
380. Oh J, Lee KW, Choi HW, Lee C. Immunogenicity and protective efficacy of recombinant S1 domain of the porcine epidemic diarrhea virus spike protein. Arch Virol. (2014) 159:2977–87. doi: 10.1007/s00705-014-2163-7
381. Makadiya N, Brownlie R, van den Hurk J, Berube N, Allan B, Gerdts V, et al. S1 domain of the porcine epidemic diarrhea virus spike protein as a vaccine antigen. Virol J. (2016) 13:57. doi: 10.1186/s12985-016-0512-8
382. Lamphear BJ, Jilka JM, Kesl L, Welter M, Howard JA, Streatfield SJ, et al. corn-based delivery system for animal vaccines: an oral transmissible gastroenteritis virus vaccine boosts lactogenic immunity in swine. Vaccine. (2004) 22:2420–4. doi: 10.1016/j.vaccine.2003.11.066
383. Takamura K, Matsumoto Y, Shimizu Y. Field study of bovine coronavirus vaccine enriched with hemagglutinating antigen for winter dysentery in dairy cows. Can J Vet Res. (2002) 66:278–81.
384. Ismail MM, Cho KO, Ward LA, Saif LJ, Saif YM. Experimental bovine coronavirus in turkey poults and young chickens. Avian Dis. (2001) 45:157–63. doi: 10.2307/1593023
385. Tsunemitsu H, el-Kanawati ZR, Smith DR, Reed HH, Saif LJ. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J Clin Microbiol. (1995) 33:3264–9. doi: 10.1128/jcm.33.12.3264-3269.1995
386. Brian DA, Hogue BG, Kienzle TE. The coronavirus hemagglutinin esterase glycoprotein. In: Siddell S, editor. The Coronaviridae. New York, NY: Plenum Press (1995). p. 165–79.
387. Ballesteros ML, Sánchez CM, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. (1997) 227:378–88. doi: 10.1006/viro.1996.8344
388. Saif L, Wesley R. Transmissible gastroenteritis virus. In: Straw BE, D'Allaire S, Mengeling WL, Taylor D, editors. Diseases of Swine. Ames, IA: Iowa State University Press (1998).
389. Akimkin V, Beer M, Blome S, Hanke D, Höper D, Jenckel M, et al. New chimeric porcine coronavirus in swine feces, Germany, 2012. Emerg Infect Dis. (2016) 22:1314–5. doi: 10.3201/eid2207.160179
390. Belsham GJ, Rasmussen TB, Normann P, Vaclavek P, Strandbygaard B, Bøtner A. Characterization of a novel chimeric swine enteric coronavirus from diseased pigs in Central Eastern Europe in 2016. Transbound Emerg Dis. (2016) 63:595–601. doi: 10.1111/tbed.12579
391. Boniotti MB, Papetti A, Lavazza A, Alborali G, Sozzi E, Chiapponi C, et al. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. (2016) 22:83–7. doi: 10.3201/eid2201.150544
392. Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. (2020) 16:1686–97. doi: 10.7150/ijbs.45472
393. Oma VS, Klem T, Tråvén M, Alenius S, Gjerset B, Myrmel M, et al. Temporary carriage of bovine coronavirus and bovine respiratory syncytial virus by fomites and human nasal mucosa after exposure to infected calves. BMC Vet Res. (2018) 14:22. doi: 10.1186/s12917-018-1335-1
394. Hussain KA, Storz J, Kousoulas KG. Comparison of bovine coronavirus (BCV) antigens: monoclonal antibodies to the spike glycoprotein distinguish between vaccine and wild-type strains. Virology. (1991) 183:442–5. doi: 10.1016/0042-6822(91)90163-6
395. Poon LLM, Chu DKW, Chan KH, Wong OK, Ellis TM, Leung YHC, et al. Identification of a novel coronavirus in bats. J Virol. (2005) 79:2001–9. doi: 10.1128/JVI.79.4.2001-2009.2005
396. Promkuntod N. Dynamics of avian coronavirus circulation in commercial and non-commercial birds in Asia—a review. Vet Q. (2016) 36:30–−44, doi: 10.1080/01652176.2015.1126868
397. Miłek J, Blicharz-Domańska K. Coronaviruses in avian species—review with focus on epidemiology and diagnosis in wild birds. J Vet Res. (2018) 62:249–55. doi: 10.2478/jvetres-2018-0035
398. McIntosh K, Kapikian AZ, Hardison KA, Hartley JW, Chanock RM. Antigenic relationships among the coronaviruses of man and between human and animal coronaviruses. J Immunol. (1969) 102:1109–18.
399. Hofmann M, Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol. (1988) 26:2235–9. doi: 10.1128/jcm.26.11.2235-2239.1988
400. Brown IH. The pig as an intermediate host for influenza a viruses between birds and humans. Int Congr Ser. (2001) 1219:173–8. doi: 10.1016/S0531-5131(01)00666-5
401. Dhama K, Verma AK, Rajagunalan S, Deb R, Karthik K, Kapoor S, et al. Swine flu is back again: a review. Pak J Biol Sci. (2012) 15:1001–9. doi: 10.3923/pjbs.2012.1001.1009
402. Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. (2020) 368:1016–20. doi: 10.1126/science.abb7015
403. Decaro N, Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet Microbiol. (2020) 244. doi: 10.1016/j.vetmic.2020.108693
404. Martina BE, Haagmans BL, Kuiken T, Fouchier RA, Rimmelzwaan GF, Van Amerongen G, et al. Virology: SARS virus infection of cats and ferrets. Nature. (2003) 425:915. doi: 10.1038/425915a
405. Bosco-Lauth AM, Hartwig AE, Porter SM, Gordy PW, Nehring M, Byas AD, et al. Experimental infection of domestic dogs and cats with SARS-CoV-2: pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci USA. (2020) 117:26382–8. doi: 10.1073/pnas.2013102117
406. Ge XY Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. (2013) 503:535–8. doi: 10.1038/nature12711
407. Benton DJ, Wrobel AG, Xu P, et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. (2020) 588:327–30. doi: 10.1038/s41586-020-2772-0
408. Zhou H, Chen X, Hu T, Song H, Liu Y, Wang P, et al. A novel bat coronavirus closely related to SARS- CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike proteins. Curr Biol. (2020) 30:2196–203.e3 doi: 10.1016/j.cub.2020.05.023
409. Stavrinides J, Guttman D. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J Virol. (2004) 78:76–82. doi: 10.1128/JVI.78.1.76-82.2004
410. Hemida MG, Perera RA, Wang P, Alhammadi MA, Siu LY Li M, et al. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill. (2013) 18:20659. doi: 10.2807/1560-7917.ES2013.18.50.20659
411. Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke GJ, Meyer B, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. (2013) 13:859–66. doi: 10.1016/S1473-3099(13)70164-6
412. Briese T, Mishra N, Jain K, Zalmout IS, Jabado OM, Karesh WB. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio. (2014) 5:e01146-14. doi: 10.1128/mBio.01146-14
413. Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. (2014) 14:140–5. doi: 10.1016/S1473-3099(13)70690-X
414. Meyer B, Müller MA, Corman VM, Reusken CB, Ritz D, Godeke GJ, et al. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis. (2014) 20:552–9. doi: 10.3201/eid2004.131746
415. Raj VS, Farag EA, Reusken CB, Lamers MM, Pas SD, Voermans J, et al. Isolation of MERS coronavirus from a dromedary camel, Qatar, 2014. Emerg Infect Dis. (2014) 20:1339–42. doi: 10.3201/eid2008.140663
416. Reusken CB, Messadi L, Feyisa A, Ularamu H, Godeke GJ, Danmarwa A, et al. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg Infect Dis. (2014) 20:1370–4. doi: 10.3201/eid2008.140590
417. Reusken CB, Farag EA, Jonges M, Godeke GJ, El-Sayed AM, Pas SD, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels, Qatar, April 2014. Euro Surveill. (2014) 19:20829. doi: 10.2807/1560-7917.ES2014.19.23.20829
418. Farag EA, Reusken CB, Haagmans BL, Mohran KA, Raj VS, Pas SD, et al. High proportion of MERS-CoV shedding dromedaries at slaughterhouse with a potential epidemiological link to human cases, Qatar 2014. Infect Ecol Epidemiol. (2015) 5:28305. doi: 10.3402/iee.v5.28305
419. Reusken CB, Farag EA, Haagmans BL, Mohran KA, Godeke GJ, Raj VS, et al. Occupational Exposure to Dromedaries and Risk for MERS-CoV Infection, Qatar, 2013-2014. Emerg Infect Dis. (2015) 21:1422–5. doi: 10.3201/eid2108.150481
420. Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. (2014) 370:2499–505. doi: 10.1056/NEJMoa1401505
421. Drosten C, Meyer B, Müller MA, Corman VM, Al-Masri M, Hossain R, et al. Transmission of MERS-coronavirus in household contacts. Engl J Med. (2014) 371:828–35. doi: 10.1056/NEJMoa1405858
422. World Health Organization. Middle East Respiratory Syndrome Coronavirus (MERS-CoV)—United Arab Emirates. (2021). Available online at: https://www.who.int/emergencies/disease-outbreak-news/item/2021-DON314 (accessed March 17, 2021).
423. Samara EM, Abdoun KA. Concerns about misinterpretation of recent scientific data implicating dromedary camels in epidemiology of Middle East respiratory syndrome (MERS). MBio. (2014) 5:e01430-14. doi: 10.1128/mBio.01430-14
424. Mohd HA, Al-Tawfiq JA, Memish ZA. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir. Virol J. (2016) 13:87. doi: 10.1186/s12985-016-0544-0
425. Ramadan N, Shaib H. Middle East respiratory syndrome coronavirus (MERS-CoV): a review. Germs. (2019) 9:35–42. doi: 10.18683/germs.2019.1155
426. Ren LL, Wang YM, Wu ZQ, Xiang ZC, Guo L, Xu T, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. (2020) 133:1015–24. doi: 10.1097/CM9.0000000000000722
427. Almendros A, Gascoigne E. Can companion animals become infected with Covid-19? Vet Rec. (2020) 186:419–20. doi: 10.1136/vr.m1322
428. Almendros A. Can companion animals become infected with Covid-19? Vet Rec. (2020) 186:388–9. doi: 10.1136/vr.m1194
429. American Veterinary Medical Association. SARS-CoV-2 in Animals, Including Pets. (2020). Available online at: https://www.avma.org/resources-tools/animal-health-and-welfare/covid-19/sars-cov-2-animals-including-pets (accessed April 11, 2020).
430. USDA APHIS. Confirmation of COVID-19 in Pet Dog in New York. (2020). Available online at: https://www.aphis.usda.gov/aphis/newsroom/stakeholder-info/sa_by_date/sa-2020/sa-06/sars-cov-2-dog (accessed Jun 2, 2020).
431. Mallapaty S. Coronavirus can infect cats - dogs, not so much. Nature. (2020) doi: 10.1038/d41586-020-00984-8
432. Zhang Q, Zhang H, Gao J, Huang K, Yang Y, Hui X, et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg Microbes Infect. (2020) 9:2013–9. doi: 10.1080/22221751.2020.1817796
433. Daly N. Seven More Big Cats Test Positive for Coronavirus at Bronx Zoo Animals, Coronavirus Coverage. (2020). Available online at: https://www.nationalgeographic.com/animals/2020/04/tiger-coronavirus-covid19-positive-test-bronx-zoo (accessed April 23, 2020).
434. UK scientists find evidence of human-to-cat Covid transmission. The Guardian. (2021). Available online at: https://www.theguardian.com/lifeandstyle/2021/apr/23/uk-scientists-find-evidence-of-human-to-cat-covid-transmission
436. Middlemiss C, Voas S, Glossop C, Huey R. SARS-CoV-2 in ferrets. Vet Rec. (2021) 188:133. doi: 10.1002/vetr.104
437. World Health Organization. SARS-CoV-2 Mink-Associated Variant Strain – Denmark. (2020). Available online at: https://www.who.int/csr/sars/en/WHOconsensus.pdf (accessed November 6, 2020).
438. USDA APHIS. USDA Animal and Plant Health Inspection Service. Confirmation of COVID-19 in Gorillas at a California Zoo. (2021). Available online at: https://www.aphis.usda.gov/aphis/newsroom/stakeholder-info/sa_by_date/sa-2021/sa-01/ca-gorillas-sars-cov-2 (accessed January 11, 2021).
439. Georgia Aquarium. Asian Small-Clawed Otters at Georgia Aquarium Test Positive for COVID-19. (2021). Available online at: https://news.georgiaaquarium.org/stories/releases-20210418#::~text=April%2018%2C%202021-,Asian%20Small%2DClawed%20Otters%20at%20Georgia,Test%20Positive%20for%20COVID%2D19&text=The%20Asian%20small%2Dclawed%20otters,%2C%20mild%20lethargy%2C%20and%20coughing
440. Cohen J. From mice to monkeys, animals studied for coronavirus answers. Science. (2020) 368:221–2. doi: 10.1126/science.368.6488.221
441. CDC. Coronavirus Disease (COVID-19)—Pets & Other Animals. (2019). Available online at: https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/positive-pet.html (accessed May 29, 2020).
442. Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. (2020) 1:e218–25. doi: 10.1016/S2666-5247(20)30089-6
443. Pickering BS, Smith G, Pinette MM, Embury-Hyatt C, Moffat E, et al. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. (2021) 27:104–12. doi: 10.3201/eid2701.203399
444. Mykytyn AZ, Lamers MM, Okba NMA, Breugem TI, Schipper D, van den Doel PB, et al. Susceptibility of rabbits to SARS-CoV-2. Emerg Microbes Infect. (2021) 10:1–7. doi: 10.1080/22221751.2020.1868951
445. Ulrich L, Wernike K, Hoffmann D, Mettenleiter TC, Beer M. Experimental infection of cattle with SARS-CoV-2. Emerg Infect Dis. (2020) 26:2979–81. doi: 10.3201/eid2612.203799
446. Freuling CM, Breithaupt A, Müller T, Sehl J, Balkema-Buschmann A, Rissmann M, et al. Susceptibility of raccoon dogs for experimental SARS-CoV-2 Infection. Emerg Infect Dis. (2020) 26:2982–5. doi: 10.3201/eid2612.203733
447. Michelitsch A, Wernike K, Ulrich L, Mettenleiter TC, Beer M. SARS-CoV-2 in animals: from potential hosts to animal models. Adv Virus Res. (2021) 110:59–102. doi: 10.1016/bs.aivir.2021.03.004
448. Brownlie J. Conclusive proof needed for animal virus reservoirs. Vet Rec. (2020) 186:354. doi: 10.1136/vr.m1076
449. Lanfear R. A global phylogeny of SARS-CoV-2 sequences from GISAID. Zenodo. (2020). doi: 10.5281/zenodo.3958883
450. Swaminathan S. The WHO's chief scientist on a year of loss and learning. Nature. (2020) 588:583–5. doi: 10.1038/d41586-020-03556-y
451. Chan JF, To KK, Tse H, Jin DY, Yuen KY. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. (2013) 21:544–55. doi: 10.1016/j.tim.2013.05.005
452. Klompus S, Leviatan S, Vogl T, Mazor RD, Kalka IN, Stoler-Barak L, et al. Cross-reactive antibodies against human coronaviruses and the animal coronavirome suggest diagnostics for future zoonotic spillovers. Sci Immunol. (2021) 6:eabe9950. doi: 10.1126/sciimmunol.abe9950
453. World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. (2021). Available online at: https://covid19.who.int
454. Sorci G, Faivre B, Morand S. Explaining among-country variation in COVID-19 case fatality rate. Sci Rep. (2020) 10:18909. doi: 10.1038/s41598-020-75848-2
455. World Health Organization. Update 49 - SARS Case-Fatality Ratio, Incubation Period. Available online at: https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON301#::~textŌn%205%20November%2C%20the%20Danish,from%20August%20to%20September%202020 (accessed May 7, 2003).
456. WHO EMRO. Situation Region Update. Middle East Respiratory Syndrome. Available online at: http://www.emro.who.int/fr/health-topics/mers-cov/situation-update.html
Keywords: coronavirus, evolution, interspecies transmission, host cell receptor, protein binding motif, zoonotic animal origin, classification, animal coronaviruses
Citation: Parkhe P and Verma S (2021) Evolution, Interspecies Transmission, and Zoonotic Significance of Animal Coronaviruses. Front. Vet. Sci. 8:719834. doi: 10.3389/fvets.2021.719834
Received: 03 June 2021; Accepted: 07 September 2021;
Published: 18 October 2021.
Edited by:
Yashpal S. Malik, Indian Veterinary Research Institute (IVRI), IndiaReviewed by:
Hicham Sid, Technical University of Munich, GermanyCopyright © 2021 Parkhe and Verma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subhash Verma, c3Zlcm1hOEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.