- 1College of Veterinary Medicine, Henan Agricultural University, Zhengzhou, China
- 2Institute for Animal Husbandry and Veterinary Research, Henan Academy of Agricultural Sciences, Zhengzhou, China
- 3Department of Veterinary Medicine, Centre for Infection Medicine, Institute of Microbiology and Epizootics, Freie Universität Berlin, Berlin, Germany
The aim of this study was to gain insight into the resistance determinants conferring resistance to tigecycline in Streptococcus (S.) suis and to investigate the genetic elements involved in their horizontal transfer. A total of 31 tetracycline-resistant S. suis isolates were screened for tigecycline resistance by broth microdilution. S. suis isolate SC128 was subjected to whole genome sequencing with particular reference to resistance determinants involved in tigecycline resistance. Transferability of genomic island (GI) GISsuSC128 was investigated by transformation. The roles of tet(L) or tet(M) in contributing to tigecycline resistance in S. suis were confirmed by transformation using different tet(L)- or tet(M)-carrying constructs. Only S. suis SC128 showed a tigecycline resistance phenotype. A tet(L)-tet(M) and catA8 co-carrying GISsuSC128 was identified in this isolate. After transfer of the novel GI into a susceptible recipient, this recipient showed the same tigecycline resistance phenotype. Further transfer experiments with specific tet(L)- or tet(M)-carrying constructs confirmed that only tet(M), but not tet(L), contributes to resistance to tigecycline. Protein sequence analysis identified a Tet(M) variant, which is responsible for tigecycline resistance in S. suis SC128. It displayed 94.8% amino acid identity with the reference Tet(M) of Enterococcus faecium DO plasmid 1. To the best of our knowledge, this is the first time that a tet(M) variant conferring resistance to tigecycline was identified in S. suis. Its location on a GI will accelerate its transmission among the S. suis population.
Introduction
Tigecycline, a semisynthetic antibiotic that binds to 16S rRNA and prevents translation of mRNA, is one of the last-resort antibiotics to treat complicated infections caused by multidrug-resistant Gram-negative and Gram-positive bacteria (1, 2). It belongs to the subgroup of tetracyclines called glycylcyclines (1). The presence of high-level tigecycline resistance genes, including several tet(X) variants such as tet(X3), tet(X4), tet(X6), and tet(X14), severely compromised the efficacy of tigecycline (3–8). In addition, several plasmid-mediated tetracycline resistance genes, such as variants of tet(A) and tet(L), have been reported to confer elevated tigecycline minimum inhibitory concentration (MICs) resulting in non-susceptibility (9, 10).
In production and breeding, the carrying rate of S. suis in pigs is as high as 80%. Despite a high carrier rate, morbidity rarely exceeds 5%, although it can reach more than 50% in cases of poor hygiene and concurrent disease (11). S. suis is also an important zoonotic pathogen. S. suis poses a threat to public health. In Vietnam, S. suis is the most important pathogen causing meningitis in adults (12). S. suis will not only cause huge economic losses to the breeding industry, but also have great harm to public health, especially the health of related practitioners.
In S. suis, the tetracycline resistance genes tet(O), tet(M), tet(W), tet(O/32/O), tet(S), tet(O/W/32/O), tet(40) and tet(B) have frequently been described (13–20). However, the tigecycline resistance mechanism in S. suis has not yet been explored.
In this study, the tetracycline resistance genes among S. suis isolates were investigated, with particular reference to the mechanism(s) involved in tigecycline resistance, and the genetic elements involved and their horizontal transfer.
Materials and Methods
Bacterial Strains and Plasmids
During 2011–2018, a total of 31 non-duplicate tetracycline-resistant S. suis strains were isolated and identified on Todd-Hewitt Agar (THA) plates containing 10 mg/L tetracycline from individual diseased pigs in four provinces (Henan, Shanxi, Shandong and Guangdong) in China. The S. suis-Escherichia coli shuttle plasmid vector pSET2s (SPE+) was used for cloning of the tet(L) gene. The serotype 2 strain S. suis P1/7 served as recipient strain in the transformation experiments. All S. suis strains were cultured in Todd-Hewitt Broth (THB) at 37°C.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing (AST) was performed by broth microdilution according to the recommendations given by EUCAST and the results were categorized according to the EUCAST breakpoint tables for interpretation of MICs and zone diameters, Version 11.0 (21). Streptococcus pneumoniae ATCC 49619 served as the quality control strain.
PCR Analyses
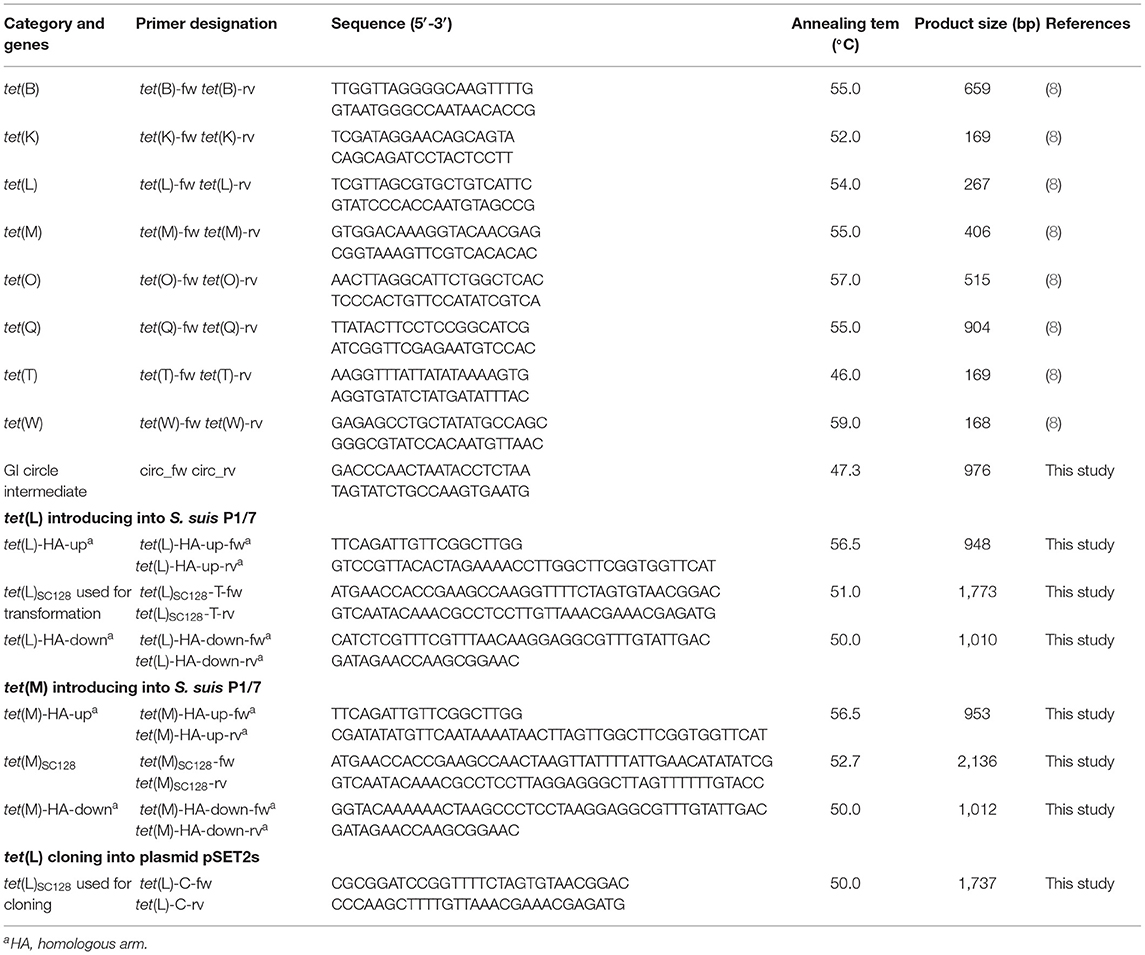
The S. suis isolates were investigated for the presence of the genes tet(K), tet(L), tet(M), tet(O), tet(Q), tet(T), tet(W), and tet(B) by PCR using the primers listed in Table 1 (22). The presence of a genomic island (GI) circular intermediate in S. suis SC128 and its transformant was detected by PCR using the primers circ_fw and circ_rv listed in Table 1.
Transformation Experiments
The transformation experiments were performed as described in previous studies (23, 24). The peptide (GNWGTWVEE) was used as a pheromone for the transformation. The detailed protocols for the transformation were as follows. The tetracycline-susceptible recipient strain S. suis P1/7 was grown to exponential phase at 37°C under 5% CO2. Then, the logarithmic P1/7 strains were diluted 1:50 into THY medium and grown at 37°C without shaking. When they reached an OD600 between 0.035 and 0.058, the chromosomal DNA (1.0 μg) of the donor S. suis SC128 and the synthetic peptide (250 μM) were added to 100 μL aliquots of the recipient strain. After 2 h, the samples were diluted, plated on THA plates containing 10 mg/L tetracycline, and incubated overnight at 37°C. Colonies were further confirmed by AST and multilocus sequence typing (MLST) following previous research (25).
Whole Genome Sequencing and Protein Sequence Analysis
Whole genome DNA of S. suis SC128 was sequenced using the PacBio RS and Illumina MiSeq platforms (Shanghai Personal Biotechnology Co., Ltd., China). The PacBio sequence reads were assembled with HGAP4 and CANU (Version 1.6), and corrected by Illumina MiSeq with pilon (Version 1.22). The prediction of ORFs and their annotations were performed using Glimmer 3.0. Three methods were used to identify genomic islands as previously described, including GC content analysis (26, 27), whether it contains integrase (28), and PCR amplification of circular intermediates. Protein sequence analysis of the tet(M) gene was performed using the software DNAMAN 8.0.
Introduction of the tet(L) and tet(M) Genes Into the Recipient by Homologous Recombination
In order to demonstrate the contribution of tet(L) or tet(M) located on GISsuSC128 to tigecycline resistance, tet(L)-carrying or tet(M)-carrying transformants were obtained by introduction of the tet(L) or the tet(M) genes into the tetracycline-susceptible S. suis P1/7, based on the protocol described previously (29). Briefly, as shown in Table 1, the two pairs of primers tet(L)-HA-up-fw/-rv and tet(L)-HA-down-fw/-rv were designed to amplify the upstream and downstream homologous fragments, which targets the glutamate dehydrogenase (GDH) gene and the dihydroorotate dehydrogenase (DHODH) gene from the recipient strain S. suis P1/7. The primers tet(L)SC128-T-fw/-rv were designed to amplify the intact copy of tet(L) including their promoter and translational attenuator from the donor strain S. suis SC128. The three amplification products were ligated by fusion PCR with primers tet(L)-HA-up-fw and tet(L)-HA-down-rv (30), and the recombination fragments were then introduced into S. suis P1/7 by transformation according to the method described in section “Transformation Experiments.” A concentration of 10 mg/L tetracycline was used for selecting the tet(L)-carrying transformants. In addition, the tet(M)-carrying transformants were generated in a similar way using the primers described in Table 1 and the same tetracycline concentration for selecting transformants.
Functional Cloning of the tet(L) Gene
A 1,732 base pair (bp) fragment containing BamHI and HindIII restriction sites in the non-coding regions at the 5' and 3' termini of the tet(L) gene was amplified by PCR using a pair of primers tet(L)-C-fw/rv listed in Table 1. Then, the amplified fragment was ligated into the BamHI- and HindIII-digested S. suis-E. coli shuttle vector pSET2s. The recombinant plasmid pSET2s_tet(L) was then transformed into E. coli DH5α. After identification, the recombinant plasmid pSET2s_tet(L) was extracted and transformed into S. suis P1/7 by natural transformation as described above and the transformants were selected on the medium containing 100 mg/L of spectinomycin and grown at 37°C under 5% CO2 (23, 24).
Results
A Novel tet(L)-tet(M) Co-carrying GI Was Identified in S. suis
MIC results revealed that all 31 S. suis strains were tetracycline-resistant, which were further screened for the presence of tetracycline resistance genes using the primers listed in Table 1. Of them, tet(O) was detected in 30/31 of the total strains, with tet(M) in 1/30, and tet(L) in 1/30. Among them, the strain SC128 was positive for the combination of tet(L) and tet(M) and proved to be tigecycline resistant (Table 2). According to the interpretation criteria of MICs in the version 9.0 issued by EUCAST in 2021, S. suis is susceptible to tigecycline at a breakpoint of ≤0.125 mg/L, which means that a MIC > 0.125 mg/L is considered resistant (21). This strain was then selected for WGS. The GC percentage of SC128 was 41.03%, while that of the GISsuSC128 was 36.10%. GISsuSC128 contains an integrase. A circular intermediate was successfully amplified by PCR. These results indicated that GISsuSC128 was a genomic island. As shown in Figure 1, a novel tet(L)-tet(M) and catA8 co-carrying GI was identified, designated ICESsuSC128, which has a size of 36.097 kbp. It was inserted at the rplL locus, which is one of the common insertion hotspots of mobile genetic elements (MGEs) in S. suis, forming 15 bp imperfect target site duplications at the integration site (5′-AGACCTGGTTTTTTA-3′ and 5′- AGCCCTGGTTTCTTA-3′) (31, 32). The DNA sequence of integrase in GISsuSC128 was compared to those deposited the NCBI GenBank, and the BLASTn result was that it had 100% identity and 100% coverage rate with the integrase in the S. suis NC28-6 genomic island. However, the 29.661 kbp GISsuNC286 in S. suis strain NC28-6 carried the resistance genes spw_like, aadE, lnu(B) and lsa(E), but not tet(L) or tet(M), which is present in GISsuSC128 (33).
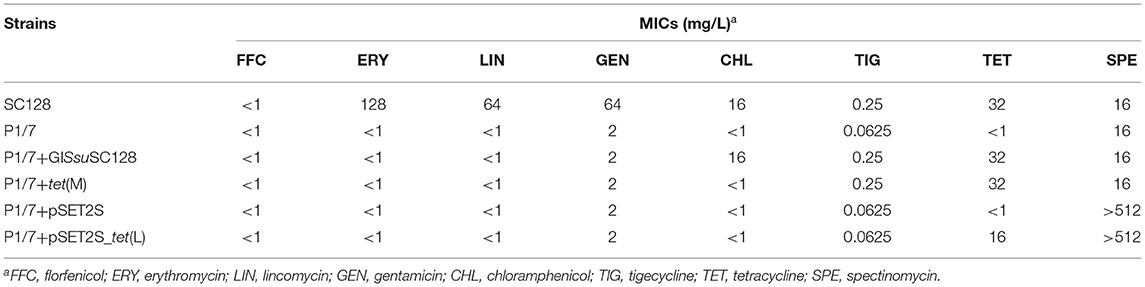
Table 2. MICs of the tet(L)-tet(M) co-carrying S. suis strain, the recipient S. suis strain P1/7 and their transformants.
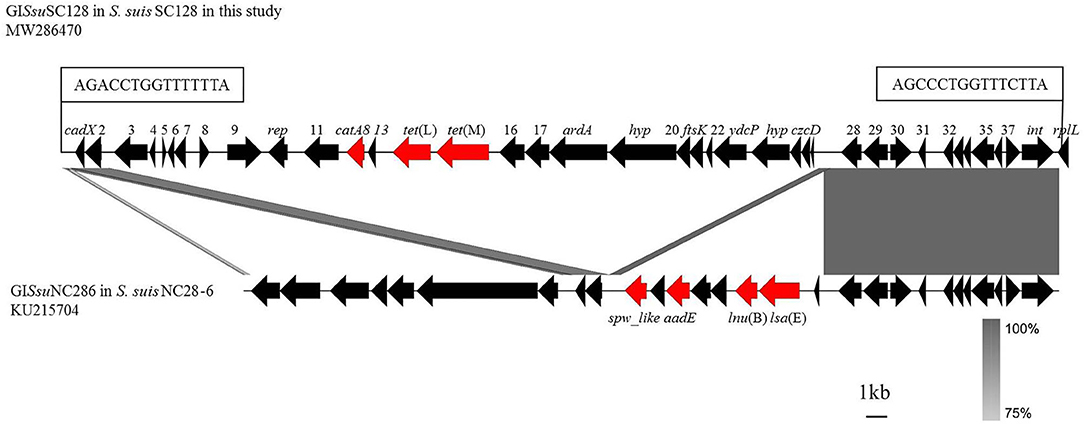
Figure 1. Comparison of GISsuSC128 in this study with GISsuNC286 in S. suis NC28-6 described previously. Analysis and creation of the image were performed by the software Easyfig2.2.3, the resistance genes are shown in red and other genes are shown in black. The locations of the PCR primers for the detection of circularizable forms of GI are indicated by arrows. The perfect 15 bp target site duplications at both termini are shown in boxes. Regions with more than 75% nucleotide sequence identity are shaded gray.
The GISsuSC128 Is Transmissible
Using the primers and PCR conditions listed in Table 1, a 976 bp amplicon was detected, which suggested that GISsuSC128 has the ability to excise from the S. suis chromosomal DNA and to form a circular translocatable unit. Transferability of GISsuSC128 was investigated by natural transformation using S. suis P1/7 as the recipient. The transformant, designated P1/7+GISsuSC128, was successfully obtained. MICs for the S. suis strain SC128, the recipient strain S. suis P1/7, and the transformant P1/7+GISsuSC128 were shown in Table 2. The transformant P1/7-GISsuSC128 displayed a 4-fold increase in the MICs of tigecycline compared with the recipient strain S. suis P1/7.
tet(M) but Not tet(L) in GISsuSC128 Confers Resistance to Tigecycline
To identify the individual role of either tet(L) or tet(M) alone in conferring resistance to tigecycline in S. suis, homologous recombination fragments were constructed, which consisted of the upstream and downstream homologous arm targeting the glutamate dehydrogenase (GDH) gene and the dihydroorotate dehydrogenase (DHODH) gene, respectively, from the recipient strain S. suis P1/7 and the intact copy of tet(L) or tet(M) including their putative regulatory regions from the donor strain S. suis SC128. These constructs were then transformed into S. suis P1/7 by homologous recombination. The tet(M)-carrying transformant was successfully obtained, and designated S. suis P1/7+tet(M). It displayed a 4-fold increase in the MICs of tigecycline compared with the parent strain S. suis P1/7 (Table 2), revealing that tet(M) in GISsuSC128 alone can confer resistance to tigecycline in S. suis. However, the introduction of tet(L) into S. suis P1/7 failed as no tet(L)-carrying transformants were obtained in three independent attempts.
To further eliminate the influence of experimental factors and to confirm the contributing role of tet(L) in GISsuSC128 to tigecycline resistance in S. suis, the tet(L) gene from GISsuSC128 was cloned into the plasmid vector pSET2s and then transformed into S. suis P1/7. The positive clone was designated S. suis P1/7+pSET2s_tet(L). MICs for S. suis P1/7, S. suis P1/7+pSET2s and S. suis P1/7+pSET2s_tet(L) were shown in Table 2. The transformant P1/7-GISsuSC128 displayed a 4-fold increase in the MICs of tigecycline, compared with the recipient strain S. suis P1/7, S. suis P1/7+pSET2s, and S. suis P1/7+pSET2s_tet(L), which which suggested tet(L) in GISsuSC128 has no influence on tigecycline resistance in S. suis. These data confirmed that it is tet(M), not tet(L), in GISsuSC128, which confers resistance to tigecycline in S. suis SC128.
tet(M) in GISsuSC128 Is a Variant
Previous studies showed that mutations in tet(M), which go along with deletions or amino acid substitutions, are associated with elevated tigecycline MICs, ranging from 0.0625 to 0.25 mg/L (34). In addition, a single deletion of L505 in the tet(M) caused the highest increase of tigecycline MIC (0.25 mg/L) (34). Protein sequence analysis with the software DNAMAN8.0 indicated that there are 33 amino acid substitutions occurring in the deduced amino acid sequence of the Tet(M) protein from S. suis SC128 (Identity, 94.8%), compared with the reference Tet(M) from E. faecium DO plasmid 1 (accession number YP_006377310.1) (Figure 2) (35). Thus, this GI-borne tet(M) gene is a naturally occurring variant.

Figure 2. Amino acid substitutions occurring in the deduced amino acid sequence of the Tet(M) protein from S. suis SC128, compared with the reference Tet(M) from E. faecium DO plasmid 1 (accession number YP_006377310.1).
Discussion
Tigecycline has strong antibacterial activity against most Gram-positive and Gram-negative bacteria except Proteus and Pseudomonas, including Enterobacteriaceae, Acinetobacter baumannii, Staphylococcus aureus, Enterococcus, Streptococcus pneumoniae, etc. (36). Tigecycline has gradually become the most effective treatment for various bacterial infections among medical clinical antibacterial drugs due to its wide antibacterial spectrum, strong antibacterial activity, and low drug resistance rate, and it is also a last line of defense drug (37). However, with the continuous use of tigecycline in clinical anti-infective therapy, some researchers have isolated bacteria that are clinically resistant to tigecycline (38). The mechanism of bacterial resistance to tigecycline is very complex, mainly including the efflux pump mechanism, cell membrane pore channel protein variation, drug binding site changes and drug enzymatic degradation. Tet(M) is a ribosome protective protein, which prevents tetracycline from binding to 23S rRNA. Tet(L) is a member of the MFS efflux pump family, which can export tetracycline but is not effective against tigecycline (32).
This study confirmed that the tet(M) variant is located on the genomic island of a S. suis strain. Three methods were used to identify genomic islands (26–28), GC content analysis, integrase detection, and PCR amplification of circular intermediates. The results indicated that GISsuSC128 was a genomic island. Integrases are critical to genomic islands, because the genomic islands of S. suis are often classified according to the integrase sequence. The DNA sequence of integrase in the genomic island in this study was compared those deposited in the NCBI GenBank, and the BLASTn result was that it had 100% identity and 100% coverage rate with the integrase in the S. suis NC28-6 genomic island (33). However, the 29.661 kbp GISsuNC286 in S. suis strain NC28-6 carried the resistance genes spw_like, aadE, lnu(B) and lsa(E), but not tet(L) or tet(M), which are present in GISsuSC128. This genomic island can undergo horizontal transfer. After the transfer, the MIC value of tigecycline to the recipient bacteria increased 4-fold, which made the recipient bacteria resistant to tigecycline. One limitation in this study is that the EUCAST breakpoints of Streptococcus groups A, B, C, and G were used to determine tigecycline resistance in S. suis. Although these breakpoints are currently the most accurate reference for resistance determination in S. suis according to EUCAST guidelines, S. suis does not belong to these groups. Species-specific breakpoints according to EUCAST or similar guidelines must be determined before tigecycline resistance of S. suis isolate SC128 can be finally confirmed.
By comparing and analyzing the amino acid sequence of Tet(M) in this study with the reference Tet(M) sequence from E. faecium DO plasmid 1 (35), it is found that Tet(M) in this study has multiple mutations. The results can be used for future research and provide a basis for the key amino acid sites of Tet(M) conferring resistance to tigecycline. The amino acid sequence of Tet(M) was compared with those deposited in the NCBI GenBank, and the BLASTp result indicated that it had 100% identity and 100% coverage rate with that in an unpublished Lactobacillales strain (WP_024406177). Whole-genome sequencing revealed the genetic environment of tet(M) and the mechanism of horizontal transfer, and provided a theoretical basis for controlling the spread of drug-resistance genes.
Conclusions
In this study, both the transformants P1/7+GISsuSC128 and P1/7+tet(M) displayed a tigecycline resistance phenotype. This observation indicated that tet(M) variants, whether they occurred naturally or were artificially induced, could confer resistance to tigecycline. The location of a tet(M) variant responsible for the tigecycline resistance on a GI will facilitate its transmission within the S. suis population.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW286470.
Author Contributions
Y-HS and X-DD designed the research and supervised the study. RY, YZ, YX, and X-SL performed the experiments and analyzed the data. RY, YZ, SS, and X-DD wrote the manuscript. All authors revised the manuscript and approved the final version for submission.
Funding
This work was supported by grants from the Program for Innovative Research Team (in Science and Technology) in University of Henan Province (No. 19IRTSTHN007), Zhongyuan high level talents special plan (No. 204200510015) and the German Federal Ministry of Education and Research (BMBF) under project number 01KI1727D as part of the Research Network Zoonotic Infectious Diseases.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Wanjiang Zhang (Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, P. R. China) for providing recipient strain S. suis P1/7.
References
1. Pournaras S, Koumaki V, Spanakis N, Gennimata V, Tsakris A. Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. J Antimicrob Agents. (2016) 48:11–8. doi: 10.1016/j.ijantimicag.2016.04.017
2. Dryden M. Tigecycline: an antibiotic for the twenty-first century. J Antimicrob Chemother. (2013) 68 (Suppl. 2):ii3–4. doi: 10.1093/jac/dkt139
3. Sun J, Chen C, Cui CY, Zhang Y, Liu X, Cui ZH, et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol. (2019) 4:1457–64. doi: 10.1038/s41564-019-0496-4
4. He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbio. (2019) 4:1450–6. doi: 10.1038/s41564-019-0445-2
5. Zhang R, Dong N, Zeng Y, Shen Z, Lu J, Liu C, et al. Chromosomal and plasmid-borne tigecycline resistance genes tet(X3) and tet(X4) in dairy cows on a Chinese farm. Antimicrob Agents Chemother. (2020) 64:e00674–20. doi: 10.1128/AAC.00674-20
6. Liu D, Zhai W, Song H, Fu Y, Schwarz S, He T, et al. Identification of the novel tigecycline resistance gene tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin. J Antimicrob Chemother. (2020) 75:1428–31. doi: 10.1093/jac/dkaa037
7. Li R, Lu X, Peng K, Liu Z, Li Y, Liu Y, et al. Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems. (2020) 5:e00134–20. doi: 10.1128/mSystems.00134-20
8. Cheng Y, Chen Y, Liu Y, Guo Y, Zhou Y, Xiao T, et al. Identification of novel tetracycline resistance gene tet(X14) and its co-occurrence with tet(X2) in a tigecycline-resistant and colistin-resistant Empedobacter stercoris. Emerg Microbes Infect. (2020) 9:1843–52. doi: 10.1080/22221751.2020.1803769
9. Yao H, Cheng J, Li A, Yu R, Zhao W, Qin S, et al. Molecular characterization of an IncFIIk plasmid co-harboring blaIMP−26 and tet(A) variant in a clinical Klebsiella pneumoniae isolate. Front Microbiol. (2020) 11:1610. doi: 10.3389/fmicb.2020.01610
10. Yao H, Qin S, Chen S, Shen J, Du X. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. (2018) 18:25. doi: 10.1016/S1473-3099(17)30628-X
11. Dutkiewicz J, Sroka J, Zajac V, Wasiński B, Cisak E, Sawczyn A, et al. Streptococcus suis: a re-emerging pathogen associated with occupational exposure to pigs or pork products. Part I - Epidemiology. Ann Agric Environ Med. (2017) 24:683–95. doi: 10.26444/aaem/79813
12. Mai NT, Hoa NT, Nga TV, Linh le D, Chau TT, Sin DX, et al. Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis. (2008) 46:659–67. doi: 10.1086/527385
13. Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Micro Mol Biol Rev. (2001) 65:232–60. doi: 10.1128/MMBR.65.2.232-260.2001
14. Morse SA, Johnson SR, Biddle JW, Roberts MC. High-level tetracycline resistance in Neisseria gonorrhoeae is result of acquisition of streptococcal tetM determinant. Antimicrob Agents Chemother. (1986) 30:664–70. doi: 10.1128/AAC.30.5.664
15. Hillen W, Schollmeier K. Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. (1983) 11:525–39. doi: 10.1093/nar/11.2.525
16. Barbosa TM, Scott KP, Flint HJ. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ Microbiol. (1999) 1:53–64. doi: 10.1046/j.1462-2920.1999.00004.x
17. Chander Y, Oliveira SR, Goyal SM. Identification of the tet(B) resistance gene in Streptococcus suis. Vet J. (2011) 189:359–60. doi: 10.1016/j.tvjl.2010.07.004
18. Chen L, Song Y, Wei Z, He H, Zhang A, Jin M. Antimicrobial susceptibility, tetracycline and erythromycin resistance genes, and multilocus sequence typing of Streptococcus suis isolates from diseased pigs in China. J Vet Med Sci. (2013) 75:583–7. doi: 10.1292/jvms.12-0279
19. Marini E, Palmieri C, Magi G, Facinelli B. Recombination between Streptococcus suis ICESsu32457 and Streptococcus agalactiae ICESa2603 yields a hybrid ICE transferable to Streptococcus pyogenes. Vet Microbiol. (2015) 178:99–104. doi: 10.1016/j.vetmic.2015.04.013
20. Haenni M, Lupo A, Madec JY. Antimicrobial resistance in Streptococcus spp. Microb Spectrum. (2018) 6:1–25. doi: 10.1128/microbiolspec.ARBA-0008-2017
21. EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 11.0 (2021). Available online at: https://eucast.org/clinical_breakpoints/
22. Villedieu A, Diaz-Torres ML, Hunt N, McNab R, Spratt DA, Wilson M, et al. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob Agents Chemother. (2003) 47:878–82. doi: 10.1128/AAC.47.3.878-882.2003
23. Zaccaria E, van Baarlen P, de Greeff A, Morrison DA, Smith H, Wells JM. Control of competence for DNA transformation in Streptococcus suis by genetically transferable pherotypes. PLoS ONE. (2014) 9:e99394. doi: 10.1371/journal.pone.0099394
24. Zhu Y, Dong W, Ma J, Zhang Y, Pan Z, Yao H. Utilization of the ComRS system for the rapid markerless deletion of chromosomal genes in Streptococcus suis. Future Microbiol. (2019) 14:207–22. doi: 10.2217/fmb-2018-0279
25. King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, et al. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol. (2002) 40:3671–80. doi: 10.1128/JCM.40.10.3671-3680.2002
26. Zhang R, Zhang CT. A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinformatics. (2004) 20:612–22. doi: 10.1093/bioinformatics/btg453
27. Gao F, Zhang CT. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. (2006) 34:686–91. doi: 10.1093/nar/gkl040
28. Mantri Y, Williams KP. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. (2004) 32:55–8. doi: 10.1093/nar/gkh059
29. Xiao G, Wu Z, Zhang S, Tang H, Wang F, Lu C. Mac protein is not an essential virulence factor for the virulent reference strain Streptococcus suis P1/7. Curr Microbiol. (2017) 74:90–6. doi: 10.1007/s00284-016-1160-3
30. Hilgarth RS, Lanigan TM. Optimization of overlap extension PCR for efficient transgene construction. MethodsX. (2019) 7:100759. doi: 10.1016/j.mex.2019.12.001
31. Huang J, Liang Y, Guo D, Shang K, Ge L, Kashif J, et al. Comparative genomic analysis of the ICESa2603 family ICEs and spread of erm(B)- and tet(O)-carrying transferable 89K-subtype ICEs in swine and bovine isolates in China. Front Microbiol. (2016) 7:55. doi: 10.3389/fmicb.2016.00055
32. Huang J, Ma J, Shang K, Hu X, Liang Y, Li D, et al. Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: a probable mobile genetic elements reservoir for other streptococci. Front Cell Infect Microbiol. (2016) 6:118. doi: 10.3389/fcimb.2016.00118
33. Huang K, Zhang Q, Song Y, Zhang Z, Zhang A, Xiao J, et al. Characterization of spectinomycin resistance in Streptococcus suis leads to two novel insights into drug resistance formation and dissemination mechanism. Antimicrob Agents Chemother. (2016) 60:6390–2. doi: 10.1128/AAC.01157-16
34. Linkevicius M, Sandegren L, Andersson DI. Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob Agents Chemother. (2015) 60:789–96. doi: 10.1128/AAC.02465-15
35. Fiedler S, Bender JK, Klare I, Halbedel S, Grohmann E, Szewzyk U, et al. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J Antimicrob Chemother. (2016) 71:871–81. doi: 10.1093/jac/dkv420
36. Garrison MW, Neumiller JJ, Setter SM. Tigecycline: an investigational glycylcycline antimicrobial with activity against resistant gram-positive organisms. Clin Ther. (2005) 27:12–22. doi: 10.1016/j.clinthera.2005.01.007
37. Fritzenwanker M, Imirzalioglu C, Herold S, Wagenlehner FM, Zimmer KP, Chakraborty T. Treatment options for carbapenem-resistant gram-negative infections. Dtsch Arztebl Int. (2018) 21:345–52. doi: 10.3238/arztebl.2018.0345
Keywords: Streptococcus suis, genomic island, tigecycline, resistance, tet(M) variant
Citation: Yu R, Zhang Y, Xu Y, Schwarz S, Li X-S, Shang Y-H and Du X-D (2021) Emergence of a tet(M) Variant Conferring Resistance to Tigecycline in Streptococcus suis. Front. Vet. Sci. 8:709327. doi: 10.3389/fvets.2021.709327
Received: 29 May 2021; Accepted: 27 July 2021;
Published: 19 August 2021.
Edited by:
Dirk Werling, Royal Veterinary College (RVC), United KingdomReviewed by:
Hanna Marti, University of Zurich, SwitzerlandMariela Srednik, University of Buenos Aires, Argentina
Copyright © 2021 Yu, Zhang, Xu, Schwarz, Li, Shang and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang-Dang Du, eGRkdUBoZW5hdS5lZHUuY24=; Yan-Hong Shang, c2hhbmd5YW5ob25nQGhlbmF1LmVkdS5jbg==
 Rui Yu
Rui Yu Yue Zhang1
Yue Zhang1 Stefan Schwarz
Stefan Schwarz Xiang-Dang Du
Xiang-Dang Du