- 1Laboratory of Biomedical Embryology, Department of Health, Animal Science and Food Safety and Centre for Stem Cell Research, UniStem, Università Degli Studi di Milano, Milan, Italy
- 2Laboratory of Biomedical Embryology, Department of Agricultural and Environmental Sciences-Production, Landscape, Agroenergy and Centre for Stem Cell Research, UniStem, Università Degli Studi di Milano, Milan, Italy
The first differentiation event in mammalian embryos is the formation of the trophectoderm, which is the progenitor of the outer epithelial components of the placenta, and which supports the fetus during the intrauterine life. However, the epigenetic and paracrine controls at work in trophectoderm differentiation are still to be fully elucidated and the creation of dedicated in vitro models is desirable to increase our understanding. Here we propose a novel approach based on the epigenetic conversion of adult dermal fibroblasts into trophoblast-like cells. The method combines the use of epigenetic erasing with an ad hoc differentiation protocol. Dermal fibroblasts are erased with 5-azacytidine (5-aza-CR) that confers cells a transient high plasticity state. They are then readdressed toward the trophoblast (TR) phenotype, using MEF conditioned medium, supplemented with bone morphogenetic protein 4 (BMP4) and inhibitors of the Activin/Nodal and FGF2 signaling pathways in low O2 conditions. The method here described allows the generation of TR-like cells from easily accessible material, such as dermal fibroblasts, that are very simply propagated in vitro. Furthermore, the strategy proposed is free of genetic modifications that make cells prone to instability and transformation. The TR model obtained may also find useful application in order to better characterize embryo implantation mechanisms and developmental disorders based on TR defects.
Introduction
Reproduction in eutherian mammals strictly depends on the placenta that is required for both maintenance of pregnancy and fetal development. The placenta is a transient extraembryonic organ that nourishes and supports the fetus during intrauterine life. It anchors the fetus to the uterine wall, provides immune protection and ensures transport and exchange of nutrients, gases, waste and hormones between the mother and the fetus (1). Trophoblast (TR) cells represent the major placental cell type and are highly specialized in each of the above mentioned functions (2, 3). These cells originate from a simple epithelial sheet that surrounds the blastocoel and the inner cell mass (ICM) at the blastocyst stage, namely the embryonic trophectoderm (TE). Comparative studies on placental morphology revealed that TR tissue can range from a single layer of cells to a complex multilayered organization, based on the different types of placentation (4). Specifically, in species exhibiting hemochorial placentation, such as the murine and the human, during embryo implantation, the outer trophectoderm layer attaches to the uterine endometrial epithelial cells and differentiates into the two main subpopulations, namely cytotrophoblast (CT) and syncytiotrophoblast (ST) cells. The former directly derives from trophoblast stem cells and constitutes an undifferentiated non-polarized proliferative cell population. CT cells are considered the major invasive component of the placenta, since they aggregate into multilayered columns at the tips of villi, forming their basement membrane, and migrate from this site into the maternal endometrium, through both the uterine stroma and the lumen of maternal arteries (5). In addition, in hemochorial placentae, villous CT cells fuse together and differentiate into multinucleated ST cells (6, 7), which cover the villi and make direct contact with maternal blood, becoming the major site of maternal–fetal gas and nutrient exchange (8). Conversely, animals with non-invasive epitheliochorial placentation, namely horses, camels, pigs, hippopotami, and cetaceans, do not form an ST layer (9) and CT cells directly interact with maternal uterine luminal epithelium, forming the trophoblast-endometrial epithelial bilayer (also known as “interareolar region”) and serving analogous functions of nutrition and gas exchange (10, 11).
To date, TR cells have been extensively described in the mouse (12, 13), where they can be easily derived from blastocysts or from extra-embryonic ectoderm (14). In contrast, a more limited amount of information is available in large mammals where they can be isolated with poor efficiency (15, 16). Furthermore, although the transcriptional regulation driving TR cell differentiation processes are beginning to be understood, experimental limitations exist to investigate the early events taking place during the first stages of pregnancy (2, 17). The creation of a large mammal in vitro model that mimics the initial steps of placenta development is therefore needed in order to increase our understanding and better elucidate both the epigenetic mechanisms and the paracrine controls at work during TR specification (18). It is interesting to note that porcine induced TR cells spontaneously generated during standard reprogramming processes (19) and formed floating spheres consisting of a single epithelial sheet, whose cells expressed transcription factors associated with TR lineage (20).
Here we describe an alternative approach based on the epigenetic conversion of adult porcine dermal fibroblasts into TR-like cells. The method combines the use of the epigenetic eraser 5-azacytidine (5-aza-CR) that pushes fibroblasts toward a less committed high plasticity state. Erased cells are then readdressed toward the TR lineage, using MEF conditioned medium supplemented with bone morphogenetic protein 4 (BMP4) and inhibitors of the Activin/Nodal and FGF2 signaling pathways.
Materials and Methods
All reagents were purchased from Thermo Fisher Scientific unless otherwise indicated.
Ethical Statement
This study was authorized by the Ethical Committee of the University of Milan, Italy. All methods were carried out in accordance with the approved guidelines.
Isolation and Culture of Adult Porcine Dermal Fibroblasts
Porcine dermal fibroblasts were isolated from fresh abdominal skin biopsies of 3 female and 2 male adult individuals. Tissues were cut in small fragments of ~2 mm3, transferred onto 0.1% gelatin (Sigma) pre-coated Petri dishes (Sarstedt) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% Fetal Bovine Serum (FBS), 2 mM glutamine (Sigma) and antibiotics. After 6 days of culture, fibroblasts started to grow out of the explant fragments and the latter were carefully removed. One primary cell line was established from each skin biopsy. Fibroblasts were maintained in the medium described above, grown in 5% CO2 at 37°C and passaged twice a week in a 1:3 ratio. Primary cell lines obtained from each individual (n = 5) were used at least in triplicate in 3 independent experiments.
Treatment of Adult Porcine Dermal Fibroblasts With 5-Aza-CR
Fibroblasts at passages between 6 and 8 after isolation were plated in 4-well multidishes (Nunc) previously coated with 0.1% gelatin (Sigma) at a concentration of 7.8 × 104 cells/cm2. Twenty-four hours after seeding, cells were exposed to the epigenetic eraser 5-aza-CR (Sigma) at 1 μM concentration for 18 h. Concentration and time of exposure were selected based on our previous studies (21–27).
Trophoblast Induction
Post 5-aza-CR treatment, cells were cultured in embryonic stem cell (ESC) medium consisting of DMEM-low glucose:HAM'S F10 (1:1) supplemented with 5% FBS, 10% K.O. serum, 2 mM glutamine, 0.1 mM β-mercaptoethanol, nucleoside mix, 1% non-essential amino acids, 1,000 IU/ml ES-growth (LIF, Chemicon), 5 ng/ml b-FGF (R&D System) (22, 28, 29) for 3 h at 37°C in 5% CO2. Culture dish were then incubated at 37°C in low O2 condition (5% O2, 5% CO2, and 90% N2 atmosphere) for 21 h. TR differentiation was induced using mouse embryonic fibroblast (MEF) conditioned medium obtained by culturing inactivated MEF in ESC culture medium without b-FGF for 24 h (see below) supplemented with 10 ng/ml bone morphogenetic protein 4 (BMP4, Sigma), 1 μM activin/nodal signaling inhibitor (A83-01, Sigma) and 0.1 μM FGF2 signaling inhibitor (PD173074, Sigma) (30). Cells were maintained in low O2 conditions at 37°C and culture medium was refreshed every other day, until day 11, when culture was arrested. Phenotype induction was daily scored using a Nikon Eclipse TE200 microscope and specific analysis were carried out at day 2, 5, and 11, in agreement with previously published results (2, 30).
Preparation of MEF Conditioned Medium
MEFs were routinely cultured in complete MEF medium consisting of DMEM supplemented with 10% FBS, 2 mM glutamine and antibiotics. Once confluence was reached, cells were plated at a seeding density of 20.000 cells/cm2. Twenty-four hours after plating, cells were inactivated with 10 μg/ml mitomycin-C for 3 h and subsequently cultured in ESC culture medium without b-FGF for 24 h. Conditioned medium was harvested, filtered, and stored at −20°C till further use.
Global DNA Methylation Analysis
Genomic DNA was extracted using the PureLink® Genomic DNA Kit following the manufacturer's protocol. Double-stranded DNA was denatured by incubation at 95°C for 5 min, followed by rapid chilling on ice. It was digested to nucleosides using nuclease P1 in 20 mM sodium acetate (pH 5.2) for 2 h at 37°C. Alkaline phosphatase in 100 mM Tris (pH 7.5) was added and an 1 h incubation at 37°C was performed. Samples were centrifuged and the supernatant was used for ELISA assay using Global DNA Methylation ELISA Kit (5′-methyl-2′-deoxycytidine Quantitation; CELL BIOLABS) according to the manufacturer's instructions.
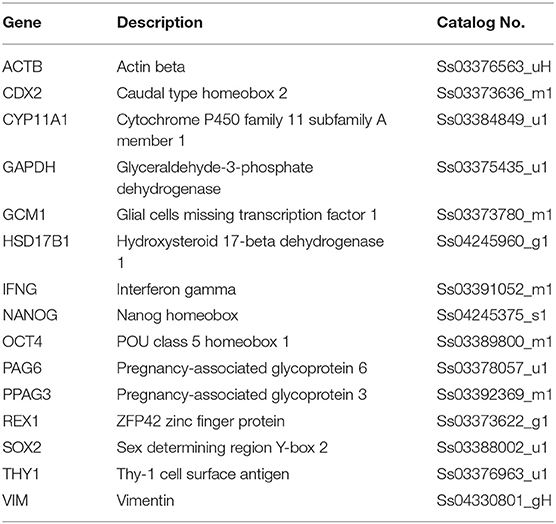
Gene Expression Analysis
RNA was extracted using the TaqManGene Expression Cells to Ct kit (Applied Biosystems) and DNase I was added in lysis solution at 1:100 concentration, as indicated by the manufacturer's instructions. Quantitative PCR was performed using the CFX96 Real-Time PCR (Bio-Rad Laboratories) and pre-designed gene-specific primers and probe sets from TaqManGene Expression Assays (Table 1). GAPDH and ACTB were used as internal reference genes. Target gene quantification was carried out with CFX Manager software (Bio-Rad Laboratories).
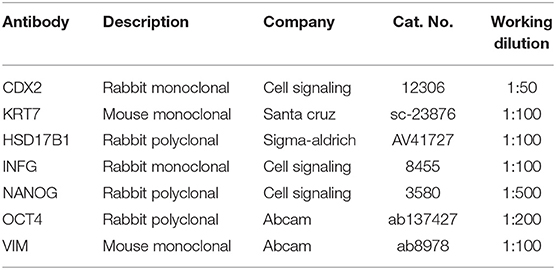
Immunocytochemical Analysis
Cells were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature, washed three times in PBS and permeabilized with 0.1% Triton X-100 (Sigma) in PBS for 30 min. Samples were then treated with a blocking solution containing 10% goat serum (Sigma) in PBS for 30 min. Primary antibodies were incubated overnight at +4°C (for working dilutions see Table 2) followed by staining with suitable secondary antibodies (1:250, Alexa Fluor) for 1 h at room temperature. Nuclei were counterstained with 4′,6-diamidino-2- phenylindole (DAPI, Sigma). Samples were observed under a Nikon Eclipse TE200 microscope.
Cell Counting
The number of CDX2, KRT7, HSD17B1, and INFG immuno-positive cells was counted in 10 randomly selected fields at 200X total magnification. A minimum of 500 cells were scored in three independent replicates. The number of positively stained cells was expressed as a percentage of the total cell counted.
Statistical Analysis
Statistical analysis was performed using the Student t-test (SPSS 19.1; IBM). Data were presented as mean ± standard deviation (SD). Differences of p ≤ 0.05 were considered significant and were indicated with different superscripts. The data were presented as mean of 3 separate experiments performed using triplicates of each of the 5 primary cultures isolated.
Results
Characterization of Adult Porcine Dermal Fibroblasts
Fibroblasts obtained from skin biopsies grew out of the original explants within 6 days of culture (Figure 1A, left panel) and formed a monolayer (Figure 1A, right panel). Cells displayed the standard morphology, elongated in shape, and a uniform immunopositivity for the fibroblast specific marker vimentin (VIM, Figure 1B). No signal for the pluripotency-related (OCT4 and NANOG) and the early (CDX2) and mature TR (KRT7, HSD17B1 and INFG) markers was detected (Figure 1B). In agreement with this, molecular analysis of untreated fibroblasts (T0) demonstrated a strong expression of the THY and VIM genes (Figure 2A), while pluripotency-related transcripts (OCT4, NANOG, REX1, and SOX2) were undetectable (Figure 2A). No expression was found for TR specific genes, namely CDX2, GCM1, PPAG3, PAG6, HSD17B1, CYP11A1, and IFNG (Figure 2A).
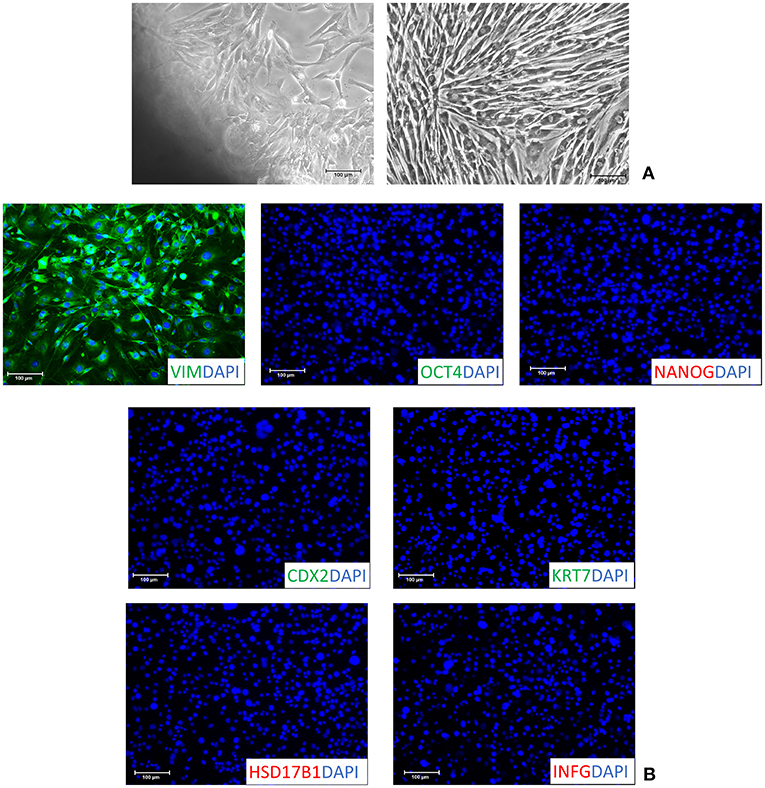
Figure 1. Characterization of adult porcine dermal fibroblasts. (A) Fibroblasts grew out of dermal tissue fragments within 6 days (left panel) and formed a monolayer (right panel). Cells displayed a standard morphology elongated in shape (right panel). Scale bars: 100 μm. (B) Isolated fibroblasts showed a uniform immuno-positivity for vimentin (VIM) and the complete absence of the pluripotent- (OCT4 and NANOG) and TR- (CDX2, KRT7, HSD17B1 and INFG) related markers. Nuclei were stained with DAPI (Scale bars: 100 μm).
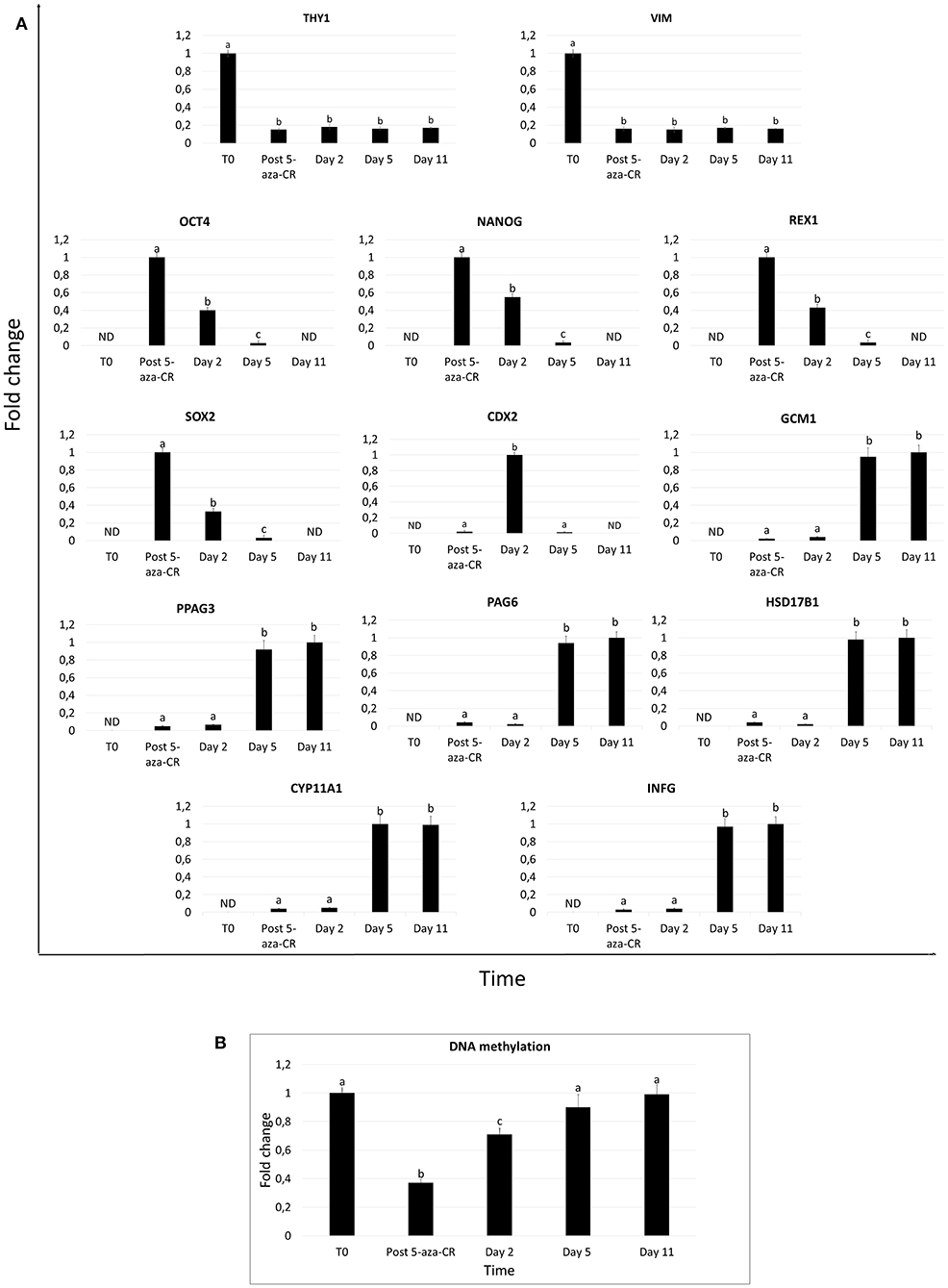
Figure 2. Gene expression profile and DNA methylation changes in adult porcine dermal fibroblasts exposed to 5-aza-CR and subjected to TR induction. (A) Expression pattern of fibroblast- specific (THY1, VIM), pluripotency-related (OCT4, NANOG, REX2, and SOX2), and early (CDX2) and mature TR (GCM1, PPAG3, PAG6, HSD17B1, CYP11A1, and IFNG) markers in untreated fibroblasts (T0), in fibroblast exposed to 5-aza-CR (Post 5-aza-CR), and at different days of TR induction (day 2, 5, and 11). Values are reported with highest expression set to 1 and all other times relative to this. Different superscripts denote significant differences (P ≤ 0.05). (B) Global DNA methylation levels in untreated fibroblasts (T0), in cells exposed to 5-aza-CR (Post 5-aza-CR) and during TR induction period (day 2, 5, and 11). Highest level set to 1 and all other relative to this. Bars represent the mean ± SD of three independent replicates. Different superscripts (a,b,c) denote significant differences (P ≤ 0.05).
Effect of 5-Aza-CR Exposure on Cell Plasticity
After 5-aza-CR exposure, cell phenotype changed, and fibroblast elongated morphology was replaced by an oval or round shape. Treated cells became smaller with larger nuclei and granular and vacuolated cytoplasm (Post 5-aza-CR; Figure 3A). These changes were functionally accompanied by a significant decrease in global DNA methylation levels compared to untreated fibroblasts (T0, Figure 2B). In parallel, exposure to 5-aza-CR (Post 5-aza-CR) resulted in the onset of the pluripotency-related genes OCT4, NANOG, REX1, and SOX2, originally undetectable in untreated fibroblasts (T0; Figures 1B, 2A). In agreement with these observations, erased cells significantly downregulated the typical fibroblast markers THY and VIM (Figure 2A).
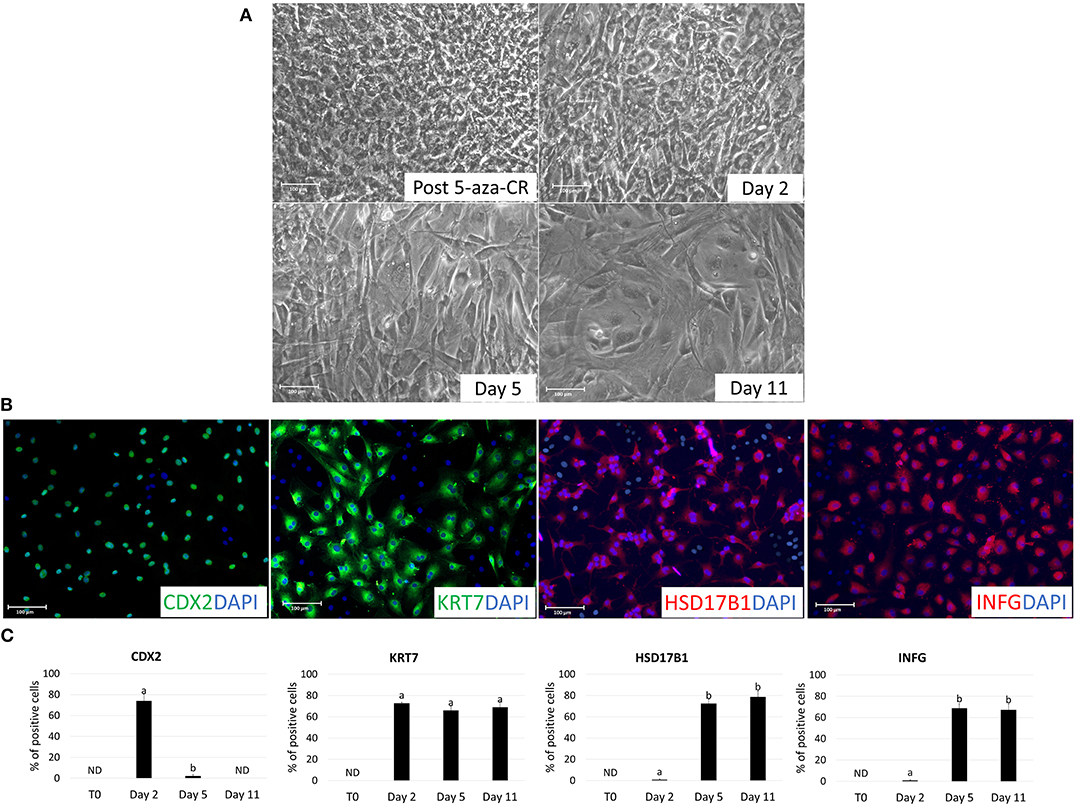
Figure 3. Morphological changes in adult porcine dermal fibroblasts after 5-aza-CR exposure and during TR induction. (A) Fibroblasts lost their typical elongated shape and become smaller with larger nuclei and granular cytoplasm in response to 18 h exposure to 5-aza-CR (Post 5-aza-CR). At day 2 of TR induction, cells acquired a tight adherent epithelial morphology (Day 2). By day 5 of differentiation, TR-like cells showed mature phenotype exhibiting round or ellipsoid shape with round nuclei and well-defined borders (Day 5 and 11). Scale bars: 100 μm (B) TR-like cells displayed immuno-positivity for early (CDX2) and mature TR (KRT7, HSD17B1, and IFNG) markers. Nuclei were stained with DAPI. Scale bars: 100 μm. (C) CDX2, KRT7, HSD17B1, and IFNG immunopositive cell rates in untreated fibroblasts (T0) and at different days of TR induction (day 2, 5, and 11). Bars represent the mean ± SD of three independent replicates. Different superscripts (a,b) denote significant differences (P ≤ 0.05).
Trophoblast Induction
After 2 days of culture with TR induction medium, cells acquired the tight adherent epithelial morphology typical of TR stem cells (Figure 3A). This change was accompanied by downregulation of the pluripotency-related genes (OCT4, NANOG, REX1, and SOX2; Figure 2A) and by an increase in global DNA methylation levels (Figure 2B). Consistent with this, active transcription of the early TR gene CDX2 was detected (Figure 2A). Immunocytochemical analysis confirmed this result, showing 74.05 ± 6.42% CDX2 and 72.8 ± 0.65% KRT7 immuno-positive cells (Figures 3B,C).
By day 5 of TR differentiation, cells acquired a mature TR morphology, with cells exhibiting round or ellipsoid shape, round nuclei, and well-defined borders (Figure 3A). These changes were paralleled by a further increase in DNA methylation levels, that returned comparable to those detected in untreated fibroblasts (T0) (Figure 2B). Onset of mature TR related genes, namely GCM1, PPAG3, PAG6, HSD17B1, CYP11A1, and IFNG, was observed (Figure 2A). The transcription of these markers was detectable at day 5 and stably maintained until day 11, when culture was arrested (Figure 2A). This was further support by immunocytochemical results, showing cell positivity for the mature TR markers KRT7, HSD17B1 and INFG, both at day 5 (65.99 ± 3.98%, 72.56 ± 4.1%, and 68.71 ± 4.25%, respectively) and at day 11 (69.1 ± 4.22%, 78.61 ± 5.56%, and 67.14 ± 5.47%, respectively) (Figures 3B,C). Interestingly, expression of the early TR marker CDX2 (Figure 2A) and detection of the encoded protein (Figure 3C) was downregulated by day 5.
Discussion
In the present study we demonstrate the possibility to directly convert porcine adult dermal fibroblasts into TR-like cells, using the epigenetic eraser 5-aza-CR and an ad hoc lineage specific induction protocol. The possibility to generate in vitro cell colonies that display distinctive features of the TR lineage in the pig has been demonstrated before by Ezashi et al. (20). Here we further extend the feasibility to obtain a TR model in large mammalian species, starting from terminally differentiated cells.
5-aza-CR ability to reactivate silent genes and, as a consequence, modify the differentiation state of an eukaryotic cell, has been previously reported (31–33). Based on this, the epigenetic eraser has been extensively used to induce a high plasticity state in adult somatic cells by several authors (21–27, 34–41). In particular, the compound has been demonstrated to induce a global DNA hypomethylation, by actively modulating ten-eleven translocation (TET) gene transcription (27) and by indirectly inhibiting DNA methyltransferase (DNMT) activities (42). Consistent with these observations, in the present work, porcine adult dermal fibroblasts exposed to 5-aza-CR for 18 h, showed a significant decrease in global DNA methylation levels, compared to untreated fibroblasts (T0), suggesting the acquisition of a less committed state. Cell hypomethylation was paralleled by the onset of the main pluripotency-related gene transcription. In particular, OCT4, NANOG, REX1, and SOX2, which were originally undetectable in untreated fibroblasts (T0), were actively expressed after exposure to 5-aza-CR. Interestingly, the molecular changes described were supported by the morphological observations that showed the transition of the erased cell from the fibroblast typical elongated morphology to a round or oval shape, with larger nuclei and granular, vacuolated cytoplasm. These features closely resemble those previously identified in embryonic stem cells (ESCs) (43, 44) and induced pluripotent cells (iPS) (45), indicating the acquisition of morphological characteristics distinctive of a high plasticity phenotype.
Taking advantage of the acquired high permissivity window, cells were readdressed toward the TR lineage, using a well-established induction protocol that has been previously shown to successfully allow for the differentiation of human ESCs toward TR cells (30, 46–54). In particular, epigenetically erased fibroblasts were induced with MEF conditioned medium supplemented with BMP4 and inhibitors of the activin/nodal and FGF2 signaling pathways. The BMP is known to be involved in several differentiation processes, favoring the acquisition of multiple phenotypes belonging to both embryonic (55–57) and extraembryonic lineages (46, 58, 59). In addition, activin/nodal and FGF signaling have the ability to influence a broad range of BMP-mediated cellular events, including self-renewal and differentiation (46). In particular, the presence of FGF2, ACTIVIN-A, and BMP4 is able to drive lineage directionality in ESCs, selectively toward TR, mesoderm, or endoderm, depending on the relative concentration of each molecule in the culture medium (52). When FGF2 and activin-a/nodal signaling suppressors are added to the differentiation cocktail containing BMP4, the induction is reported to be completely and unidirectionally toward TR (2, 30, 52, 60). Although these observations were derived from experiments carried out with human cells, they are fully confirmed in the present work, where the use of appropriate concentrations of FGF2, ACTIVIN-A, and BMP4 allows for the generation of TR-like cells in the porcine specie. It must be however noted that the pig cell conversion efficiency obtained in our experiment resulted to be ~70%, which is lower than that scored in the human (higher than 90%) (30, 52, 61, 62). We have no clear explanation for this discrepancy. We hypothesize that species-specific differences may account for dissimilar lineage amenability. In addition, in the present work adult differentiated cells were used as a starting material, while the results described for the human were obtained from ESCs.
Interestingly, a further key aspect of the protocol adopted was represented by the use of low O2 conditions that mimic the physiological hypoxic environment of uterine milieu, favoring the correct lineage specification. Previous studies demonstrated that O2 tensions profoundly affect how quickly cells differentiate in response to BMP4 (2, 63) and tune the expression of several key TR-related genes (30, 60). The results presented in the present manuscript are consistent with these observations and demonstrate that the combination of the appropriate medium formulation and O2 tension was indeed able to induce the acquisition of a TR-like phenotype. In particular, after 2 days of induction, cells acquired a tight adherent epithelial morphology, comparable to that described for murine and human TR stem cells (64, 65). These changes were paralleled by the onset of the early TR gene CDX2 transcription and by downregulation of the pluripotency-related genes (OCT4, NANOG, REX1, and SOX2). Notably, these results are in agreement with the transcription pattern described during in vitro differentiation of ESCs toward the TR lineage that also report CDX2 gene expression peaking at day 2 of induction (30, 66–68). Consistent with this, by day 5 of induction, epigenetically erased cells showed a downregulated CDX2 expression and active transcription for the TR mature markers KRT7, GCM1, PPAG3, PAG6, HSD17B1, CYP11A1, and IFNG, that were stably maintained until day 11, when culture was arrested. Progression toward the new TR phenotype was accompanied by an increase in DNA methylation levels, that returned comparable to those of untreated fibroblasts (T0), suggesting cell transition from an hypomethylated state toward a higher methylation status, distinctive of a differentiated cell population (69, 70). The acquisition of a mature TR-like phenotype was further confirmed by the morphological observations, demonstrating that, by day 5 of induction, cells exhibited a round epithelial shape, round nuclei and well-defined borders. These features closely resemble those reported in Assheton in vivo study on porcine placental tissues (71). In addition, the cell phenotype obtained at the end of di induction period were highly reminiscent of TR cells isolated from in vitro-produced porcine blastocysts and parthenotes (16).
Conclusions
Altogether, our findings demonstrate that it is possible to convert terminally differentiated adult dermal fibroblasts into porcine TR-like cells. The use of a well-establish differentiation cocktail containing BMP4 in combination with activin/nodal and FGF2 signaling inhibitors in low O2 conditions was shown to allow TR differentiation, ensuring high efficiency, and supporting and extending to the porcine species data previously reported for the human.
The TR in vitro model generated in the present experiments may find useful applications in order to increase our knowledge on embryo implantation mechanisms and to better elucidate developmental disorders based on TR defects. It may also represent a valuable tool for drug discovery and veterinary regenerative medicine.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
SA performed the experiments. GP performed the experiments and drafted the manuscript. FG and TB designed and coordinated the study and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Carraresi Foundation and is part of the project MIND FoodS HUB (Milano Innovation District Food System Hub): Innovative concept for the eco-intensification of agricultural production and for the promotion of dietary patterns for human health and longevity through the creation in MIND of a digital Food System Hub cofunded by POR FESR 2014–2020_BANDO Call HUB Ricerca e Innovazione, Regione Lombardia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are members of the COST Action CA16119 in vitro 3-D total cell guidance and fitness (CellFit) and of the Trans-COST Actions Task-Force on Covid-19.
References
1. Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. (2001) 2:538–48. doi: 10.1038/35080570
2. Schulz L, Ezashi T, Das P, Westfall S, Livingston K, Roberts R. Human embryonic stem cells as models for trophoblast differentiation introduction: the trophoblast lineage and its emergence. Placenta. (2009) 29:1–12. doi: 10.1016/j.placenta.2007.10.009
3. Sheridan MA, Fernando RC, Gardner L, Hollinshead MS, Burton GJ, Moffett A, et al. Establishment and differentiation of long-term trophoblast organoid cultures from the human placenta. Nat Protoc. (2020) 15:3441–63. doi: 10.1038/s41596-020-0381-x
4. Ramsoondar J, Christopherson RJ, Guilbert LJ, Wegmann TG. A porcine trophoblast cell line that secretes growth factors which stimulate porcine macrophages. Biol Reprod. (1993) 49:681–94. doi: 10.1095/biolreprod49.4.681
5. Pijnenborg R. Implantation and immunology: maternal inflammatory and immune cellular responses to implantation and trophoblast invasion. Reprod Biomed Online. (2002) 4(Suppl. 3):14–7. doi: 10.1016/S1472-6483(12)60110-2
6. Georgiades P, Fergyson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. (2002) 23:3–19. doi: 10.1053/plac.2001.0738
7. Malassiné A, Cronier L. Hormones and human trophoblast differentiation: a review. Endocrine. (2002) 19:3–11. doi: 10.1385/ENDO:19:1:3
8. Benirschke K. Anatomical relationship between fetus and mother. Ann N Y Acad Sci. (1994) 731:9–20. doi: 10.1111/j.1749-6632.1994.tb55744.x
9. Imakawa K, Nakagawa S. The Phylogeny of Placental Evolution Through Dynamic Integrations of Retrotransposons. Prog Mol Biol Transl Sci. (2017) 145:89–109. doi: 10.1016/bs.pmbts.2016.12.004
10. Renegar RH, Bazer FW, Roberts RM. Placental transport and distribution of uteroferrin in the fetal pig. Biol Reprod. (1982) 27:1247–60. doi: 10.1095/biolreprod27.5.1247
11. Hong L, Xu X, Huang J, Lei M, Xu D, Zhao S, Yu M. Difference in expression patterns of placental cholesterol transporters, ABCA1 and SR-BI, in Meishan and Yorkshire pigs with different placental efficiency. Sci Rep. (2016) 6:20503. doi: 10.1038/srep20503
12. Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion to trophoblast stem cell proliferation by FGF4. Science. (1998) 282:2072–5. doi: 10.1126/science.282.5396.2072
13. Roberts RM, Fisher SJ. Trophoblast stem cells. Biol Reprod. (2011) 84:412–21. doi: 10.1095/biolreprod.110.088724
14. Tanaka S. Derivation and culture of mouse trophoblast stem cells in vitro. Methods Mol Biol. (2006) 329:35–44. doi: 10.1385/1-59745-037-5:35
15. Enders AC. Trophoblasts-uterine interactions in the first days of implantation: models for the study of implantation events in the human. Semin Reprod Med. (2000) 18:255–63. doi: 10.1055/s-2000-12563
16. Hou D, Su M, Li X, Li Z, Yun T, Zhao Y, et al. The efficient derivation of trophoblast cells from porcine in vitro fertilized and parthenogenetic blastocysts and culture with ROCK inhibitor Y-27632. PLoS ONE. (2015) 10:e0142442. doi: 10.1371/journal.pone.0142442
17. Ross JW, Ashworth MD, Stein DR, Couture OP, Tuggle CK, Geisert RD. Identification of differential gene expression during porcine conceptus rapid trophoblastic elongation and attachment to uterine luminal epithelium. Physiol Genomics. (2009) 36:140–8. doi: 10.1152/physiolgenomics.00022.2008
18. Arcuri S, Gandolfi F, Somigliana E, Brevini TAL. A two-step protocol to erase human skin fibroblasts and convert them into trophoblast-like cells. In: Methods in Molecular Biology. Totowa, NJ: Humana Press Inc. (2021). p. 151–8.
19. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. (2006) 126:663–76. doi: 10.1016/j.cell.2006.07.024
20. Ezashi T, Matsuyama H, Telugu BPVLL, Roberts RM. Generation of colonies of induced trophoblast cells during standard reprogramming of porcine fibroblasts to induced pluripotent stem cells. Biol Reprod. (2011) 85:779–87. doi: 10.1095/biolreprod.111.092809
21. Pennarossa G, Maffei S, Campagnol M, Tarantini L, Gandolfi F, Brevini TAL. Brief demethylation step allows the conversion of adult human skin fibroblasts into insulin-secreting cells. Proc Natl Acad Sci USA. (2013) 110:8948–53. doi: 10.1073/pnas.1220637110
22. Pennarossa G, Maffei S, Campagnol M, Rahman MM, Brevini TAL, Gandolfi F. Reprogramming of pig dermal fibroblast into insulin secreting cells by a brief exposure to 5-aza-cytidine. Stem Cell Rev Rep. (2014) 10:31–43. doi: 10.1007/s12015-013-9477-9
23. Pennarossa G, Santoro R, Manzoni EFM, Pesce M, Gandolfi F, Brevini TAL. Epigenetic erasing and pancreatic differentiation of dermal fibroblasts into insulin-producing cells are boosted by the use of low-stiffness substrate. Stem Cell Rev Rep. (2018) 14:398–411. doi: 10.1007/s12015-017-9799-0
24. Brevini TA, Pennarossa G, Rahman MM, Paffoni A, Antonini S, Ragni G, et al. Morphological and molecular changes of human granulosa cells exposed to 5-azacytidine and addressed toward muscular differentiation. Stem Cell Rev Rep. (2014) 10:633–42. doi: 10.1007/s12015-014-9521-4
25. Brevini TAL, Pennarossa G, Acocella F, Brizzola S, Zenobi A, Gandolfi F. Epigenetic conversion of adult dog skin fibroblasts into insulin-secreting cells. Vet J. (2016) 211:52–6. doi: 10.1016/j.tvjl.2016.02.014
26. Brevini TAL, Pennarossa G, Maffei S, Zenobi A, Gandolfi F. Epigenetic conversion as a safe and simple method to obtain insulinsecreting cells from adult skin fibroblasts. J Vis Exp. (2016) 18:53880. doi: 10.3791/53880
27. Manzoni EFM, Pennarossa G, DeEguileor M, Tettamanti G, Gandolfi F, Brevini TAL. 5-azacytidine affects TET2 and histone transcription and reshapes morphology of human skin fibroblasts. Sci Rep. (2016) 6:37017. doi: 10.1038/srep37017
28. Brevini TAL, Pennarossa G, Attanasio L, Vanelli A, Gasparrini B, Gandolfi F. Culture conditions and signalling networks promoting the establishment of cell lines from parthenogenetic and biparental pig embryos. Stem Cell Rev Rep. (2010) 6:484–95. doi: 10.1007/s12015-010-9153-2
29. Brevini TAL, Pennarossa G, Maffei S, Gandolfi F. Pluripotency network in porcine embryos and derived cell lines. Reprod Domest Anim. (2012) 47:86–91. doi: 10.1111/j.1439-0531.2012.02060.x
30. Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA. (2013) 110:E1212–21. doi: 10.1073/pnas.1303094110
31. Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. (1979) 17:771–9. doi: 10.1016/0092-8674(79)90317-9
32. Jones PA. Effects of 5-azacytidine and its 2'-deoxyderivative on cell differentiation and DNA methylation. Pharmacol Ther. (1985) 28:17–27. doi: 10.1016/0163-7258(85)90080-4
33. Glover TW, Coyle-Morris J, Pearce-Birge L, Berger C, Gemmill RM. DNA demethylation induced by 5-azacytidine does not affect fragile X expression. Am J Hum Genet. (1986) 38:309–18.
34. Brevini TA, Pennarossa G, Maffei S, Gandolfi F. Phenotype switching through epigenetic conversion. Reprod Fertil Dev. (2015) 27:776–83. doi: 10.1071/RD14246
35. Pennarossa G, Manzoni EFM, Ledda S, DeEguileor M, Gandolfi F, Brevini TAL. Use of a PTFE micro-bioreactor to promote 3D cell rearrangement and maintain high plasticity in epigenetically erased fibroblasts. Stem Cell Rev Rep. (2019) 15:82–92. doi: 10.1007/s12015-018-9862-5
36. Pennarossa G, Ledda S, Arcuri S, Gandolfi F, Brevini TAL. A two-step strategy that combines epigenetic modification and biomechanical cues to generate mammalian pluripotent cells. J Vis Exp. (2020) 29:162. doi: 10.3791/61655
37. Brevini TAL, Pennarossa G, Manzoni EFM, Gandolfi F. Safety and Efficacy of Epigenetically Converted Human Fibroblasts Into Insulin-Secreting Cells: A Preclinical Study. Cham: Springer. (2018). p. 151–62.
38. Tan SJ, Fang JY, Wu Y, Yang Z, Liang G, Han B. Muscle tissue engineering and regeneration through epigenetic reprogramming and scaffold manipulation. Sci Rep. (2015) 5:16333. doi: 10.1038/srep16333
39. Mirakhori F, Zeynali B, Kiani S, Baharvand H. Brief azacytidine step allows the conversion of suspension human fibroblasts into neural progenitor-like cells. Cell J. (2015) 17:153–8. doi: 10.22074/cellj.2015.522
40. Chandrakanthan V, Yeola A, Kwan JC, Oliver RA, Qiao Q, Kang YC, et al. PDGF-AB and 5-Azacytidine induce conversion of somatic cells into tissue-regenerative multipotent stem cells. Proc Natl Acad Sci USA. (2016) 113:E2306–15. doi: 10.1073/pnas.1518244113
41. Diomede F, Zini N, Pizzicannella J, Merciaro I, Pizzicannella G, D'Orazio M, et al. 5-Aza exposure improves reprogramming process through embryoid body formation in human gingival stem cells. Front Genet. (2018) 9:e00419. doi: 10.3389/fgene.2018.00419
42. Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2'-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. (2008) 28:752–71. doi: 10.1128/MCB.01799-07
43. Niwa H. How is pluripotency determined and maintained? Development. (2007) 134:635–46. doi: 10.1242/dev.02787
44. Lai D, Wang Y, Sun J, Chen Y, Li T, Wu Y, et al. Derivation and characterization of human embryonic stem cells on human amnion epithelial cells. Sci Rep. (2015) 5:10014. doi: 10.1038/srep10014
45. Courtot AM, Magniez A, Oudrhiri N, Feraud O, Bacci J, Gobbo E, et al. Morphological analysis of human induced pluripotent stem cells during induced differentiation and reverse programming. Biores Open Access. (2014) 3:206–16. doi: 10.1089/biores.2014.0028
46. Sudheer S, Bhushan R, Fauler B, Lehrach H, Adjaye J. FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast. Stem Cells Dev. (2012) 21:2987–3000. doi: 10.1089/scd.2012.0099
47. Lee YL, Fong SW, Chen ACH, Li T, Yue C, Lee CL, et al. Establishment of a novel human embryonic stem cell-derived trophoblastic spheroid implantation model. Hum Reprod. (2015) 30:2614–26. doi: 10.1093/humrep/dev223
48. Yang Y, Adachi K, Sheridan MA, Alexenko AP, Schust DJ, Schulz LC, et al. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc Natl Acad Sci USA. (2015) 112:E2337–46. doi: 10.1073/pnas.1504778112
49. Yabe S, Alexenko AP, Amita M, Yang Y, Schust DJ, Sadovsky Y, et al. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci USA. (2016) 113:E2598–607. doi: 10.1073/pnas.1601630113
50. Jain A, Ezashi T, Roberts RM, Tuteja G. Deciphering transcriptional regulation in human embryonic stem cells specified towards a trophoblast fate. Sci Rep. (2017) 7:17257. doi: 10.1038/s41598-017-17614-5
51. Koel M, Võsa U, Krjutškov K, Einarsdottir E, Kere J, Tapanainen J, et al. Optimizing bone morphogenic protein 4-mediated human embryonic stem cell differentiation into trophoblast-like cells using fibroblast growth factor 2 and transforming growth factor-β/activin/nodal signalling inhibition. Reprod Biomed Online. (2017) 35:253–63. doi: 10.1016/j.rbmo.2017.06.003
52. Roberts RM, Ezashi T, Sheridan MA, Yang Y. Specification of trophoblast from embryonic stem cells exposed to BMP4. Biol Reprod. (2018) 99:212–24. doi: 10.1093/biolre/ioy070
53. Sheridan MA, Yang Y, Jain A, Lyons AS, Yang P, Brahmasani SR, et al. Early onset preeclampsia in a model for human placental trophoblast. Proc Natl Acad Sci USA. (2019) 116:4336–45. doi: 10.1073/pnas.1816150116
54. Karvas RM, McInturf S, Zhou J, Ezashi T, Schust DJ, Roberts RM, et al. Use of a human embryonic stem cell model to discover GABRP, WFDC2, VTCN1 and ACTC1 as markers of early first trimester human trophoblast. Mol Hum Reprod. (2020) 26:425–40. doi: 10.1093/molehr/gaaa029
55. Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. (2003) 102:906–15. doi: 10.1182/blood-2003-03-0832
56. Boyd NL, Dhara SK, Rekaya R, Godbey EA, Hasneen K, Rao RR, et al. BMP4 promotes formation of primitive vascular networks in human embryonic stem cell-derived embryoid bodies. Exp Biol Med. (2007) 232:833–43. doi: 10.3181/00379727-232-2320833
57. Zhang P, Li J, Tan Z, Wang C, Liu T, Chen L, et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. (2008) 111:1933–41. doi: 10.1182/blood-2007-02-074120
58. Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. (2002) 20:1261–4. doi: 10.1038/nbt761
59. Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, et al. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J Cell Sci. (2004) 117:1269–80. doi: 10.1242/jcs.00970
60. Ezashi T, Telugu BPVL, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res. (2012) 349:809–24. doi: 10.1007/s00441-012-1371-2
61. Li Y, Moretto-Zita M, Soncin F, Wakeland A, Wolfe L, Leon-Garcia S, et al. BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a ΔNp63+ cytotrophoblast stem cell state. Dev. (2013) 140:3965–76. doi: 10.1242/dev.092155
62. Horii M, Li Y, Wakeland AK, Pizzo DP, Nelson KK, Sabatini K, et al. Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc Natl Acad Sci USA. (2016) 113:E3882–91. doi: 10.1073/pnas.1604747113
63. Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA, Roberts RM. Effects of FGF2 and oxygen in the BMP4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res. (2007) 1:61–74. doi: 10.1016/j.scr.2007.09.004
64. Ohinata Y, Tsukiyama T. Establishment of trophoblast stem cells under defined culture conditions in mice. PLoS ONE. (2014) 9:107308. doi: 10.1371/journal.pone.0107308
65. Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, et al. Derivation of human trophoblast stem cells. Cell Stem Cell. (2018) 22:50–63.e6. doi: 10.1016/j.stem.2017.11.004
66. Roberts RM, Ezashi T, Das P. Trophoblast gene expression: transcription factors in the specification of early trophoblast. Reprod Biol Endocrinol. (2004) 2:47. doi: 10.1186/1477-7827-2-47
67. Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N, et al. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem. (2008) 283:24991–5002. doi: 10.1074/jbc.M803893200
68. Erb TM, Schneider C, Mucko SE, Sanfilippo JS, Lowry NC, Desai MN, et al. Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: the role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev. (2011) 20:1601–14. doi: 10.1089/scd.2010.0281
69. Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. (2013) 20:311–6. doi: 10.1038/nsmb.2510
70. Suelves M, Carrió E, Núñez-Álvarez Y, Peinado MA. DNA methylation dynamics in cellular commitment and differentiation. Brief Funct Genomics. (2016) 15:443–53. doi: 10.1093/bfgp/elw017
71. Assheton R. The morphology of the ungulate placenta, particularly the development of that organ in the sheep, and notes upon the placenta of the elephant and hyrax on JSTOR. In: Philosophical Transactions of the Royal Society of London. Series B, Containing Papers of a Biological Character. London: JSTOR. (1906). p. 143–220.
Keywords: 5-azacytidine, epigenetic conversion, fibroblasts, porcine, trophoblast-like cells
Citation: Arcuri S, Pennarossa G, Gandolfi F and Brevini TAL (2021) Generation of Trophoblast-Like Cells From Hypomethylated Porcine Adult Dermal Fibroblasts. Front. Vet. Sci. 8:706106. doi: 10.3389/fvets.2021.706106
Received: 06 May 2021; Accepted: 25 June 2021;
Published: 19 July 2021.
Edited by:
Fabiana Fernandes Bressan, University of São Paulo, BrazilReviewed by:
Lleretny Rodriguez-Alvarez, University of Concepcion, ChileMario Baratta, University of Turin, Italy
Rafael Sampaio, Université de Montréal, Canada
Copyright © 2021 Arcuri, Pennarossa, Gandolfi and Brevini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georgia Pennarossa, Z2VvcmdpYS5wZW5uYXJvc3NhQHVuaW1pLml0
†These authors have contributed equally to this work and share first authorship
 Sharon Arcuri
Sharon Arcuri Georgia Pennarossa
Georgia Pennarossa Fulvio Gandolfi
Fulvio Gandolfi Tiziana A. L. Brevini1
Tiziana A. L. Brevini1