
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 12 August 2021
Sec. Parasitology
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.698454
This article is part of the Research Topic Stowaways of a Stowaway: Parasites of Invasive Rodents View all 9 articles
The expansion and intensification of agriculture are driving profound changes in ecosystems worldwide, favoring the (re)emergence of many human infectious diseases. Muroid rodents are a key host group for zoonotic infectious pathogens and frequently invade farming environments, promoting disease transmission and spillover. Understanding the role that fluctuating populations of farm dwelling rodents play in the epidemiology of zoonotic diseases is paramount to improve prevention schemes. Here, we review a decade of research on the colonization of farming environments in NW Spain by common voles (Microtus arvalis) and its public health impacts, specifically periodic tularemia outbreaks in humans. The spread of this colonizing rodent was analogous to an invasion process and was putatively triggered by the transformation and irrigation of agricultural habitats that created a novel terrestrial-aquatic interface. This irruptive rodent host is an effective amplifier for the Francisella tularensis bacterium during population outbreaks, and human tularemia episodes are tightly linked in time and space to periodic (cyclic) variations in vole abundance. Beyond the information accumulated to date, several key knowledge gaps about this pathogen-rodent epidemiological link remain unaddressed, namely (i) did colonizing vole introduce or amplified pre-existing F. tularensis? (ii) which features of the “Francisella—Microtus” relationship are crucial for the epidemiology of tularemia? (iii) how virulent and persistent F. tularensis infection is for voles under natural conditions? and (iv) where does the bacterium persist during inter-epizootics? Future research should focus on more integrated, community-based approaches in order to understand the details and dynamics of disease circulation in ecosystems colonized by highly fluctuating hosts.
Biological invasions and natural range shifts leading to colonization are processes that involve dispersal movements of species outside their historical distribution limits or from a point of introduction (1). The main difference between invasion and natural range shift lies in whether or not the species' arrival of species to new locations is directly mediated by humans, either deliberately or accidentally, as is the case of invasions (1). Yet, human activities can facilitate and promote shifts in species distribution by creating new opportunities and conditions that allow species to overcome factors limiting natural colonization and settlement (2). Human-driven distributional shifts are therefore conceptually very similar to invasive processes because they would not have happened without human intervention. Globalization, anthropogenic climate change and land-use changes thus represent major factors that alter species distribution and movement (3, 4).
Agricultural expansion and intensification have driven profound environmental changes worldwide, triggered by increased global food demand for a fast-growing human population (5). Agricultural practices directly affect habitat conditions and modify biotic communities, including host-pathogen systems (hereafter, we use the term “pathogen” to refer to both micro- and macroparasites) (6–8). The alteration of host-pathogen assemblages and their relative abundances can significantly modify the dynamics of infectious diseases, including zoonoses of global public health concern (3, 4). Different aspects of agricultural intensification, including land conversion and irrigation, have been associated with more than half of all 150 zoonotic infectious diseases that have emerged in the last 80 years (9). They include SARS, ebola, HIV, malaria, yellow fever, filariasis or trypanosomiasis (5, 9). Global trends in emerging infectious diseases reveal several concerning hot spots in Europe and North America (10), although zoonotic diseases remain most often associated with developing countries and poverty (11). Zoonotic risk is therefore a pressing global issue that requires major scientific attention and quantification worldwide.
Zoonoses emerging from wildlife populations represent a significant and growing threat to the human population (12). The largest number of zoonotic hosts is found among rodents (13, 14), and co-infections are a frequent phenomenon (15). The high adaptability of rodents has favored their success under human-driven landscape changes (14, 16, 17). Some rodent species with irruptive population dynamics (i.e., multi-annual “boom-bust” fluctuations in abundance) can be involved in the disease amplification, spillover and transmission to humans (16, 18). The role of native rodent hosts with highly fluctuating population has been critical in the emergence of many zoonoses worldwide, such as hantaviruses (19, 20), Lassa virus (21, 22), Andes virus (23), Lyme disease and tick-borne encephalitis (24). The challenge goes beyond the identification of a new rodent host but lies in understanding when changes in rodent communities resulting from invasions or range shift can lead to the emergence of novel pathogens in ecosystems, as in the case of Yersinia pestis and invasive Rattus sp. in Madagascar (25). The invasion of a new rodent host could lead to the establishment and emergence of the pathogen if all the necessary conditions, other than the presence of an effective amplifying host, were met prior to the invasion. The invasive rodent has to be a competent host for the pathogen, and the transmission between hosts and the pathogen reservoir must be efficient; asymptomatic hosts further facilitate the spread of the infection. Low minimum infection dose, broad host range and high survival of the pathogen in the environment all favor potential transmission. Environmental changes that increase the number and incidence of reservoirs and that enhance the probability of human contact are usually associated with emerging diseases. Anthropogenic causes including demographics, behavior, land use and health measures have been traditionally considered in epidemiology. The confluence of a new host, pathogen, reservoir, environmental and human features also play an important role here (26). From the current paradigm of a “One-Health” perspective, it is critical to understand the role that rodents invasions and fluctuations in abundance play in the epidemiology and dynamics of zoonotic diseases.
The common vole (Microtus arvalis) is the most widespread rodent in European agro-ecosystems (27, 28) and is characterized by its fast generation time, precocial reproduction, and cyclic fluctuations in abundance (29). Overabundances during population peak years periodically lead to economic losses (30), food and veterinary risks (29, 31, 32), cyclic zoonotic outbreaks (33), and recurring social conflicts (34). Many zoonotic agents are known to use common voles as hosts, from helminths (35) to viruses (36), along with a wide range of bacteria, protozoans and even fungi (37–40). This vole species can act as a vector of hantavirus (41), a reservoir of Listeria, Babesia and Toxoplasma (41, 42), and an intermediate host of Echinococcus multilocularis (43). Identifying the possible pool of pathogens, co-occurrence patterns and role of the host within the pathogen cycle is essential in order to uncover interspecific relationships that can modify the effect of those diseases on the host and the rest of the parasite community (44).
Here, we review a decade of research conducted on the biological interaction between the zoonotic bacterium Francisella tularensis and a colonizing rodent Microtus arvalis that massively invaded agricultural landscapes in NW Spain during the 1970–1990s (45). We use this case study to highlight how the irruptive population dynamics of a newly arrived rodent host are fundamental to our understanding of human tularemia outbreaks. The settlement of the fluctuating vole in NW Spain has triggered cascading effects via ecological interactions across several trophic levels, strongly influencing the dynamics of predators (46), ectoparasites (47, 48) and infectious pathogens (47–53). The biological consequences of this invasion and its repercussions on human public health have been the main focus of recent empirical work (33, 45–52, 54–59). In this review, we will first summarize the main research findings on the ecological and epidemiological interaction between F. tularensis and M. arvalis in intensively farmed landscapes from NW Spain and, subsequently, we will identify the main knowledge gaps of this pathogen-rodent dynamic system, which has so far officially affected more than 1,500 people in Spain since 1997 (Red Nacional de Vigilancia Epidemiológica [RENAVE]).
The common vole is a small herbivorous rodent with a strong preference for protein-rich plants and well-adapted to grasslands and steppe habitats (29). In NW Spain, common vole populations were historically restricted to mountainous habitats until the early 1970s (45) (Figure 1A). Populations in this historical range show density fluctuations with wide enough amplitude to influence rodent predator demography (60). The range expansion during the 1970–90s putatively emanated from surrounding wet mountainous habitat into drier lowland landscape (45), resulting in a geographical range expansion and colonization of ca. 5 million hectares of intensively cultivated agricultural areas (45, 59). Dramatic land-use changes had occurred prior to and during the colonization period: a network of irrigation infrastructure was built throughout the region, followed by a large increase in irrigated crop surface (Figure 1B) and the production of permanent fodder crops, especially alfalfa (55, 59). According to the possible dispersal pathways of Wilson et al. (2), the range expansion of the native common vole population followed an invasion pathway in which dispersal could be facilitated by the creation of corridors (habitats linked to the irrigation system network) interconnecting new suitable vole refuge habitats (permanent alfalfa crops). Aridity and summer droughts were likely to have been the main constraints preventing the common vole from colonizing xeric Mediterranean habitats (55). However, such natural limitation may have been lifted by increases in irrigation and protein-rich crop cultivation, providing suitable conditions for voles during the warmest and driest months of the year. Jareño et al. (55) determined that other concomitant changes, such as increased fall precipitation and higher winter temperatures, have also occurred but they may have played a less decisive role in the vole colonization. All available evidence supports that the massive range shift experienced by common vole populations in NW Spain was a human-driven event (55, 59).
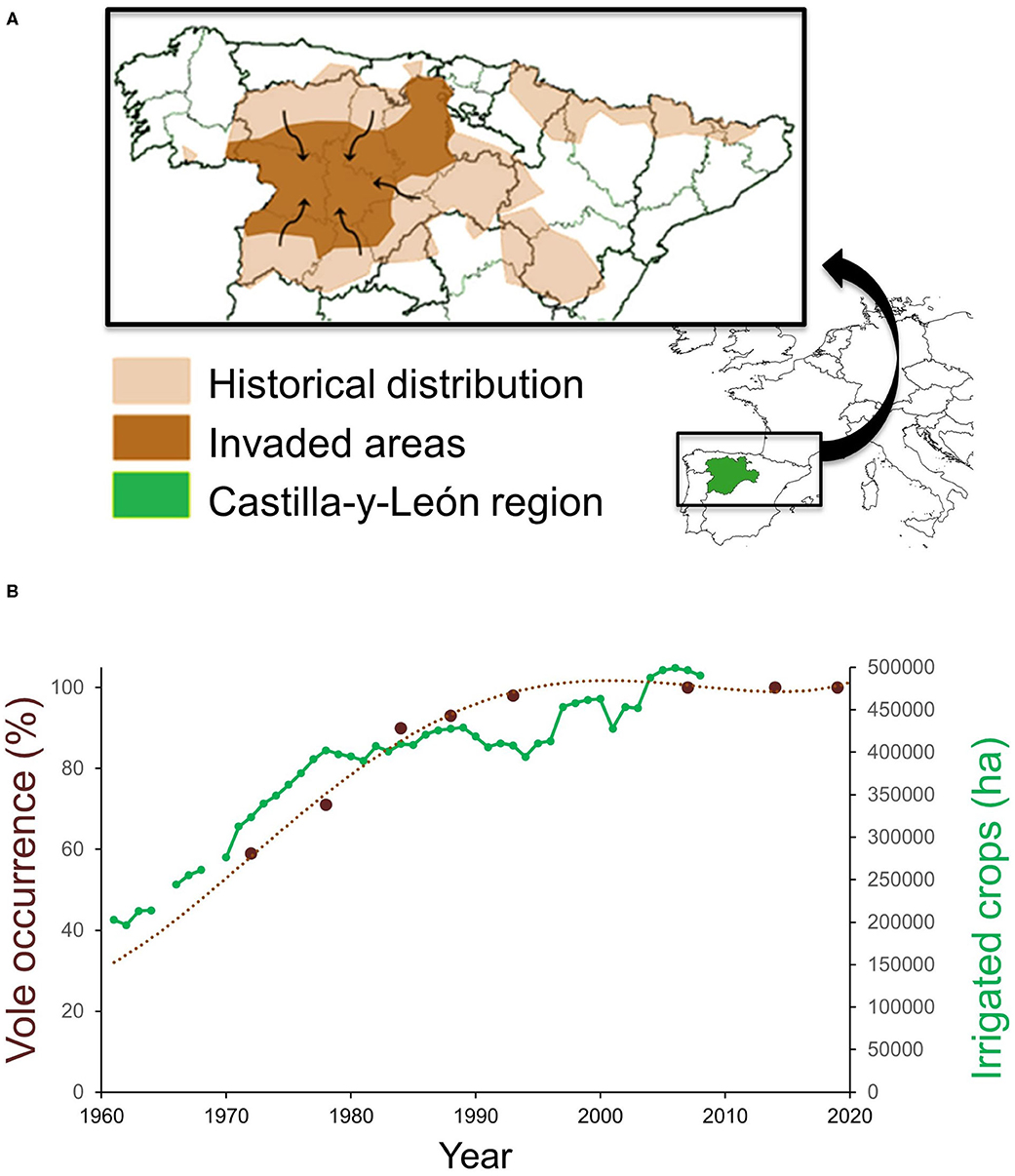
Figure 1. (A) Map of NW Spain showing the historical distribution and area invaded by common voles during the 1970–90s, and (B) temporal association between vole occurrence (percentage of agricultural counties with vole presence) and the extent of irrigated crops planted in the Castilla-y-León region, NW Spain. Adapted from Jareño et al. (55) and Luque-Larena et al. (59).
Since farming landscapes were colonized by common vole populations, these have strongly fluctuated in abundance and region-wide outbreaks have occurred approximately every 5 years (45), with associated crop damages and socio-economical conflicts (34, 45). Vole outbreaks are recurrent in the study area (46) where local vole fluctuations have a 3-years periodicity since monitoring began in 2009 (46) (dashed line in Figure 2). Particularly relevant are the human public health concerns due to periodic tularemia outbreaks among the human population of the region that shares this agricultural area with common voles (34, 45).
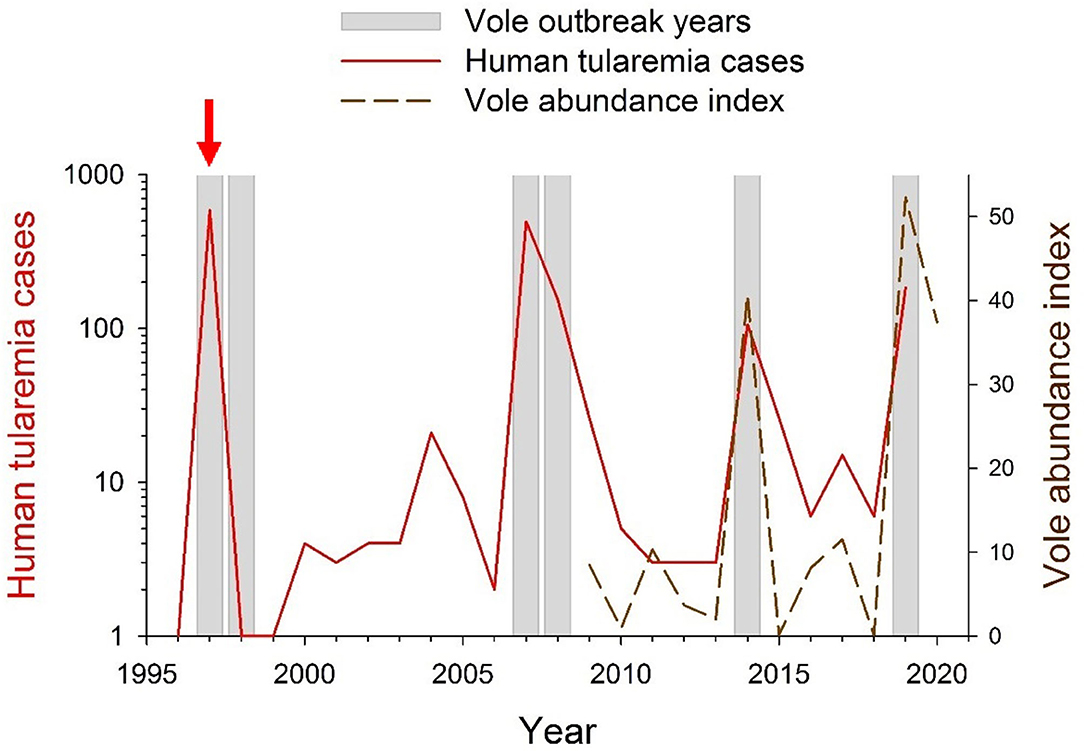
Figure 2. Yearly occurrence of vole and tularemia outbreaks in NW Spain between 1996 and 2020. During 1996–2008, vole outbreak years were identified by Luque-Larena et al. (45). During 2008–2020 vole outbreak years were identified based on common vole abundance indices obtained from live-trapping monitoring [(45, 48, 49), and unpublished data]. Data on yearly numbers of human tularemia cases (in red; log-transformed) were obtained from the Red Nacional de Vigilancia Epidemiológica (RENAVE; Instituto de Salud Carlos III, Madrid, Spain). The red vertical arrow indicates the year 1997, when tularemia became a notifiable disease in Spain.
Tularemia is an endemic zoonosis of the northern hemisphere caused by the highly infectious Francisella tularensis, classified as a Class A biothreat agent by the Center for Disease Control and Prevention (CDC) (61). There are four subspecies, but only two have human health implications: F. tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B) (62). Type B is rarely fatal, but Type A is much more virulent for humans and, without treatment, it has an average mortality of 8% (up to 50% in some clinical forms) (63). There are five clinical manifestations (ulceroglandular, oculoglandular, oropharyngeal, pneumonic and typhoidal form) depending on the route of infection (skin inoculation, eye inoculation, ingestion, inhalation and undefined route, respectively) (62). This pathogen is a facultative intracellular Gram-negative bacterium known to be the zoonotic agent that infects the widest range of animal hosts (62). Lagomorphs and rodents have been long considered as main reservoirs and spreaders, while ticks, mosquitoes and flies could act as vectors (62, 64); the bacterium can also persist in watercourses hosted by the protozoan Acanthamoeba castellanii (65). Rodents have been associated with tularemia in different parts of the world since the disease was first reported (66, 67). In fact, the disease was initially named “plague-like disease of rodents” (68). Humans become infected by arthropods bites, by direct contact with infected animals, or by ingestion/inhalation of infective material (62, 64). Two different cycles have been described in Europe for tularemia: a terrestrial cycle, which includes lagomorphs, rodents and ectoparasite vectors such as ticks; and an aquatic cycle, which includes water, mosquitos, crayfish and semi-aquatic rodents such as beavers or muskrats (64, 66, 69). However, evidence from NW Spain suggests that both cycles are probably intertwined, as key terrestrial and aquatic agents effectively coexist in time and space (33).
Between 1992 and 2019, more than 20,000 human cases of type B tularemia were reported across Europe (64, 70, 71), mostly as discrete outbreak episodes spaced by interepizootic periods (54). In Spain, tularemia has been a notifiable disease since 1997, when the first large tularemia epidemic broke out in the Castilla-y-León region (Figure 1). Since then, three other large human outbreaks of tularemia have occurred in the region (Figure 2). The largest epidemics (with >500 confirmed human cases per outbreak) took place in 1997–1998 and 2007–2008 and were followed by relatively milder outbreaks during 2014 and 2019 (ca. 100 cases per outbreak). All human epidemic outbreaks recorded in NW Spain, without exception, closely coincide in time and space with large abundances of common voles in the environment (i.e., peak phase of their “boom-bust” cycles) [(55) and Figure 2]. When vole amplitudes did not reach a certain density threshold (as during the populations peaks of 2011 or 2017) there seems to be no spillover and no human epidemics were declared (Figure 2). However, further analyses should be done to confirm this hypothesis. Since the first record of the epidemic incidence of tularemia in 1997, approximately 1,500 clinical cases of human tularemia have been officially reported by the National Network of Epidemiologic Surveillance of Spain [Red Nacional de Vigilancia Epidemiológica (RENAVE), Instituto de Salud Carlos III, Madrid, Spain]. Most of these cases (97%) occurred in the intensive agricultural landscapes of Castilla-y-León, accumulating 92–100% of cases during epidemics (RENAVE). A serological survey carried out just before the first outbreak in 1997 revealed that the human exposure to F. tularensis in the region was very limited, with an average seroprevalence of 0.19% (72). This indicates that prior to common vole expansion the incidence of the disease in the region was negligible. Nevertheless, how seroprevalence has evolved among the human population since regular exposure to tularemia in 1997 and the length of seropositivity remains unaddressed.
Recent studies have empirically demonstrated that fluctuating common vole populations in NW Spain contribute to amplifying F. tularensis in the environment, subsequently increasing the potential transmission routes and spillover to humans. For instance, during a large vole outbreak recorded between 2013 and 2015 (vole abundance peaked during summer 2014), the proportion of voles infected with F. tularensis directly increased with their abundance (58). This means that the more voles in the environment, the largest the population of zoonotic bacteria. Once vole numbers have collapsed, the pathogen was not detected among rodents in the study area (58). However, the sample size of tested voles during low-density periods (n = 35 from four trapping sessions) was small and provided limited evidence to establish whether F. tularensis infection is enzootic in voles or not. In NW Spain, tularemia epidemics in humans always occurred immediately after the number of infected voles has reached outbreak density, with no temporal delays (33, 54, 58).
In this case, tularemia epidemics are fuelled by vole numbers, which increase the pathogen pressure, the pathogen exposure to humans and the probability of infection in the ecosystem. The mean prevalence of F. tularensis among voles during a complete fluctuation can be as high as 33% at the maximum high-density phase (58). Thus, a direct density-dependent relationship between vole density and tularemia prevalence in voles has been thus established, supporting the hypothesis that common voles may play an important role in the amplification of the bacterium (33, 54, 58). This would increase pathogen pressure on voles and other competent hosts or vectors with which they share the agricultural landscape (i.e., ticks, fleas, hares). The high abundance of infected rodents during vole outbreaks also leads to a higher pathogen exposure to humans via direct contact or environmental contamination: voles drowning in irrigation canals contaminate water; voles dying in fields contaminate soils; and voles contributing to the cross-species transmission that eventually come in contact with humans (pets, livestock, crayfish, game species or invertebrate vectors) (50, 73). The use of anticoagulant rodenticides has been a common practice to attempt to control vole numbers during population outbreaks (45). It has been suggested that this practice could have worsened the epidemic situation in the past, concentrating the number of infected vole corpses in fields and thereby facilitating environmental contamination with infected material (50). Noteworthy, tularemia has been experimentally reported as both highly infective and lethal among voles (74), which underlies a significantly high probability of infection among voles.
Arthropod vectors are usually pointed as relevant agents involved in tularemia transmission elsewhere, including mosquitoes from the genera Aedes, Culex and Anopheles; ticks from of the genera Ixodes and Dermacentor; and flies from the genera Chrysops, Tabanus and Chrysozona (62). Francisella tularensis has been detected in other arthropods, but their vectoring role is still unclear (62). In NW Spain, common voles harbor mainly fleas (68% prevalence), and more rarely ticks (2% prevalence) (48). Human epidemics of several vector-borne zoonoses involving rodents typically show a one-year delayed pattern in other geographical areas (24, 75), including tularemia (76). However, in Spain, there is no evidence of any time delay between vole outbreaks and tularemia epidemics in humans (33) which, along with a very low mean prevalence of F. tularensis among fleas (collected from infected voles during the maximum density period), suggests a minor role of fleas in terms of tularemia circulation in our studied system (48).
People could be infected with the bacterium during high-risk activities such as crop harvesting or manipulation of infected animals (i.e., harvested crayfish, hunted lagomorphs). Transmission may occur through skin lesions, contact with infective material or inhalation (50, 62). The route of infection typically determines the most common clinical manifestations in humans (62), which varies between outbreaks in the region (54). In the first epidemic (1997–1998), 93% of human cases were likely infected through direct contact with contaminated hares handled for consumption, causing a glandular form of tularemia. However, during subsequent epidemics, the underlying cause was probably the inhalation of contaminated dust or water (45–59% of infected people), causing a respiratory form of tularemia (77). The numerous vole corpses lying in the fields during the harvest and decomposing in watercourses during the irrigation period (Figure 3) could be the origin of the contaminated aerosols and therefore the proximate cause of disease spillover.

Figure 3. Common vole corpses decomposing in watercourses during a peak year (summer 2019). Photos by J.J Luque-Larena.
The common voles that invaded the farmed landscapes of NW Spain are good candidates to be zoonotic “hyper-reservoirs” (14) and may harbor and modulate other public health risks beyond tularemia. When screening common voles for zoonotic bacteria other than F. tularensis (i.e., Anaplasma spp., Bartonella spp., Borrelia spp., Coxiella spp., and Rickettsia spp.), only several species of the Bartonella genus were detected in the study area (49). Bartonella prevalence reached maximum values during vole outbreaks, when nearly 70% of the animals were infected by this zoonotic bacterium. Co-infection with different Bartonella spp. occured in more than half of the voles tested, and vole abundance, flea prevalence and coexisting mouse density influenced pathogen prevalence. Additionally, co-infection with Bartonella spp. and F. tularensis occurred in 13% of voles, reaching 24% of co-infection rate during the phase of maximum vole density (49). Bartonella is a highly interactive bacterium involved in positive and negative associations with other co-infectious pathogens in similar rodent systems with multiannual dynamics (44). The implications of this co-occurrence in the studied vole populations are, however, still unknown. Noticeably, some recent screening of the same vole populations has detected Coxiella burnetii (prevalence 12.2 %) and Leishmania spp. (1.2%) (53, 78). Sampling to evaluate disease dynamics in complex multi-species systems should always consider quantitative ecological perspectives. Regarding fleas parasitizing common voles in NW Spain, five species have been identified (79): Nosopsyllus fasciatus, Leptopsylla taschenbergi, Ctenophthalmus apertus (subspecies gilcolladoi and apertus) C. baeticus and Rhadinopsylla beillardae. The first three species were shared with other sympatric small mammals (79). In these fleas collected from voles, Francisella and Bartonella have been detected (48) but, conversely to what occurs in voles, this co-infection has not been found in their fleas (48). Recent research showed that N. fasciatus could hold a specific vectoring role of Bartonella spp. among rodent populations, in accordance with previous studies (80). Implications of flea species shared between the small mammal guild should be further investigated.
Human-induced ecosystem modifications can lead to unexpected relationships between species that can result in the spread and spillover of zoonoses, as in common voles and tularemia in NW Spain (45, 54). This situation is in line with findings from a recent global meta-analysis showing that profound alterations due to agricultural intensification modify ecosystems and worsen zoonotic risks (81). The “Francisella–Microtus” case study has taught us the crucial role that a new rodent can play in disease dynamics when appropriate conditions are met, eventually triggering epidemics in humans. We have illustrated how the range shift of a native species at a regional scale is directly associated with the recent emergence of tularemia outbreaks among people. Our research provides convincing evidence that tularemia outbreaks in humans are closely linked with vole population outbreaks. However, there are a number of important knowledge gaps about the nature of the “Francisella–Microtus” epidemiological relationship under a dynamic host-density scenario, including the following key questions: (i) did colonizing voles introduce or amplified pre-existing F. tularensis?; (ii) what features of the “Francisella–Microtus” relationship are crucial for the epidemiology of tularemia?; (iii) what is the impact of F. tularensis infection on voles under natural conditions?; and (iv) where do bacterium populations remain when vole densities are very low (inter-epizootic period)?
A first key issue is to determine if F. tularensis was co-introduced with the invading voles, or was it native but latent and amplified by M. arvalis in the region. In other words, we need to understand whether or not conditions for tularemia transmission were met prior to the introduction of the new rodent. Unfortunately, we lack data about tularemia prevalence before and during the range shift, hindering the evaluation of the real effect that vole colonization had on the system. The unspecific symptomatology of tularemia (62) means that it could have been diagnosed as “fever of unknown origin” (FUO) until the first severe epidemic broke out in 1997 (and tularemia became notifiable). Many zoonoses cause FUO in humans (82) and are underreported because they are misdiagnosed (83) or masked by another disease (82). Sampling the rodent population or conducting serological surveys of humans in the historical mountainous range of Microtus arvalis would be informative to address this issue.
Lagomorphs are considered competent hosts for the bacterium (73) and the Iberian hare Lepus granatensis is a traditional game species in the colonized area. Although hares have been directly linked to tularemia epidemics throughout Europe (73), no official epidemic event had been reported before the common vole colonization in NW Spain. If the condition for disease establishment were not met before the fluctuating vole became widespread, then it could have been introduced secondarily to its main competent host. Several pathways of introduction of this bacterium are plausible. One hypothesis is that tularemia was introduced by translocation of infected hares from other European countries (73) since F. tularensis genotype is the same as can be found in central and Western Europe (84) and hare translocations for hunting purposes have been frequent. Alternatively, migratory birds, in particular waterbirds, could have hosted subadult tick species involved in the tularemia cycle or transported the bacterium itself (73, 85). In the absence of evidence of tularemia outbreaks until the colonization of common voles, we favor the secondary introduction coinciding or following vole invasion rather than the amplification of a pre-existing pathogen.
An interesting issue is whether the parasite-host interaction or transmission cycles are qualitatively different in the colonized range compared to those in the native range. Serologically positive dogs and sheep have been detected within the historical vole distribution area (77), suggesting that the bacterium was present, although no human epidemic event have been notified there. Common vole populations have a continuous distribution in NW Spain (86). Since they are competent hosts for the bacterium, F. tularensis could be present to a greater or lesser extent over the whole species' range, although it has not been assessed yet. A relevant difference between invaded and native vole populations might be related to the amplitude of the abundance fluctuations. Vole populations fluctuate in the native range dominated by pastures (60), but with a much reduced amplitude (45, 46). Extremely high common vole densities (>1,000 voles /ha) are frequently reported in intensive agricultural landscape, and in alfalfa crops in particular (56). Thus, the boom-bust dynamic and high amplitude cycle may be a key feature to understand the tularemia epidemiology, especially when conditions allow for extremely high vole densities enhancing disease spillover potential. Another as yet unexplored possibility is that hosts may be more susceptible in the invaded than in the native range. This is the case of plague and rodents from different areas: disease resistance in Asian rodents is not present in North American ones, where microevolutionary resistance has not yet developed (87). Invasive and native rodents can show different response to the same disease. For example, European red squirrels are highly susceptible to squirrelpox virus introduced with the non-native gray squirrel, causing the decline of the native squirrel populations (88). Another possible explanation for the occurrence of epidemics in colonized areas but not in the traditional vole range could be differences in the transmission routes to humans, likely linked to differences in the agricultural work (crop harvesting, irrigation water) or wildlife consumption (crayfish, hare hunting) (77). The native and invaded common vole populations inhabit different agricultural systems (pastures and extensive cattle production vs. intensive irrigated cereal and alfalfa production), where tularemia transmission pathways or contact rates between people and infected hosts or vectors will likely be different. Furthermore, the level of human exposure to the bacterium in the historical range could be lower owing to the human population characteristics (scattered settlements and low population density). Finally, the lack or scarcity of a crucial vector, spreader or reservoir needed to complete the bacterium cycle could also result in a lower probability of infection of voles and the overall reduction of tularemia prevalence.
Given the virulence of the F. tularensis, infected animals are expected to quickly die (66). Voles experimentally infected with tularemia in the laboratory are known to shed large quantities of bacteria through feces and urine and to die rapidly (74), but very little is known about the effects of tularemia in voles inhabiting natural environments. Laboratory research has found differences in pathogenicity and disease duration between different F. tularensis genotypes (89). Furthermore, some classical research suggests the possibility of chronic tularemia nephritis in rodents (90). Vole species can recover from several viral and bacterial diseases within a few weeks while other diseases, such as vole tuberculosis and babesiosis, become chronic (91). Under a theoretical scenario where the main hosts (i.e., hares and voles) die fast after infection, tularemia persistence in the focal area could be compromised when low-density periods in host populations and unfavorable environmental conditions for the bacterium coincide. During peak phases and population crashes, most dead voles are probably infected by F. tularensis (50), yet some live-trapped F. tularensis-infected voles were asymptomatic and healthy in external appearance (58). Maybe these individuals will irretrievably die of tularemia at some point, but survive longer than under experimental conditions; or maybe their immune system could allow them to overcome the infection; or maybe they remain chronically infected and excrete the bacterium at low rates. In the absence of further serological, pathological and molecular evidence, the outcome of voles when facing the disease is yet unknown. Unveiling the duration of the disease infection and the mortality rate among reservoir populations would help characterize the course of tularemia epidemics. Intermediate levels of pathogen virulence may maximize transmission by balancing replication and host survival at increasing densities, and natural selection may favor the bacterium becoming deadlier as replication is not compromised (peak densities).
A high density of voles amplifies F. tularensis but voles are not always so abundant, even in the post-colonization era (46). Thus, a key knowledge gap is where the bacterium persists during inter-epizootics (i.e., between human tularemia outbreaks), and whether those refugia existed and may have been functional before vole colonization. The emergence of tularemia outbreaks is linked to voles, and bacterium survival may have been favored by the agricultural intensification. New aquatic habitats were created in the previously arid areas, thereby removing putatively limiting conditions for the persistence of F. tularensis or voles, and adding new potential reservoirs and hosts. In these farming landscapes, both terrestrial and aquatic components coexist within the same habitats (Figure 4), suggesting that the terrestrial and aquatic tularemia cycles act as a single unified cycle (33). A new terrestrial-aquatic interface arose with the consequent development of the typical fauna of this ecosystem that offers new possible hosts, reservoirs and vectors for the bacterium (i.e., waterbirds, aquatic protozoa, semiaquatic mammals and arthropods linked to aquatic habitats). Little is known about the role played by animals other than voles and lagomorphs in tularemia circulation in Spain. However, preliminary results have detected the bacterium in water, sediment and ticks from hares from the same habitats (51). Water and sediment are typical elements involved in the aquatic cycle, while ticks, hares and voles are typical hosts of terrestrial cycles. These findings support the hypothesis of a unified tularemia cycle in the region (33). In Europe, F. tularensis has been frequently detected in other small mammals (68), predators (92, 93) and arthropod vectors (94). It has also been isolated in samples from domestic cats (95) and migratory birds (96). These could be “dead-end” hosts, but we do not know to what extent they could be competent hosts amplifying the infection and potentially spreading it. Therefore, the circulation cycle may be more complex than initially expected. The determination of all biotic and abiotic elements and their role in tularemia circulation still remains partially unknown, especially during inter-epizootics periods. Many possible actors could be involved (Figure 4). And there are several non-exclusive scenarios, which can be tested by doing a longitudinal survey of tularemia prevalence in all the candidate reservoirs: (i) the bacterium remains in the environment (sediment, water); and/or (ii) the bacterium survives in other hosts (vole predators) that outlive the voles; and/or (iii) the bacterium persists in vectors (e.g., ticks, fleas) that may also survive from a vole outbreak to the next.
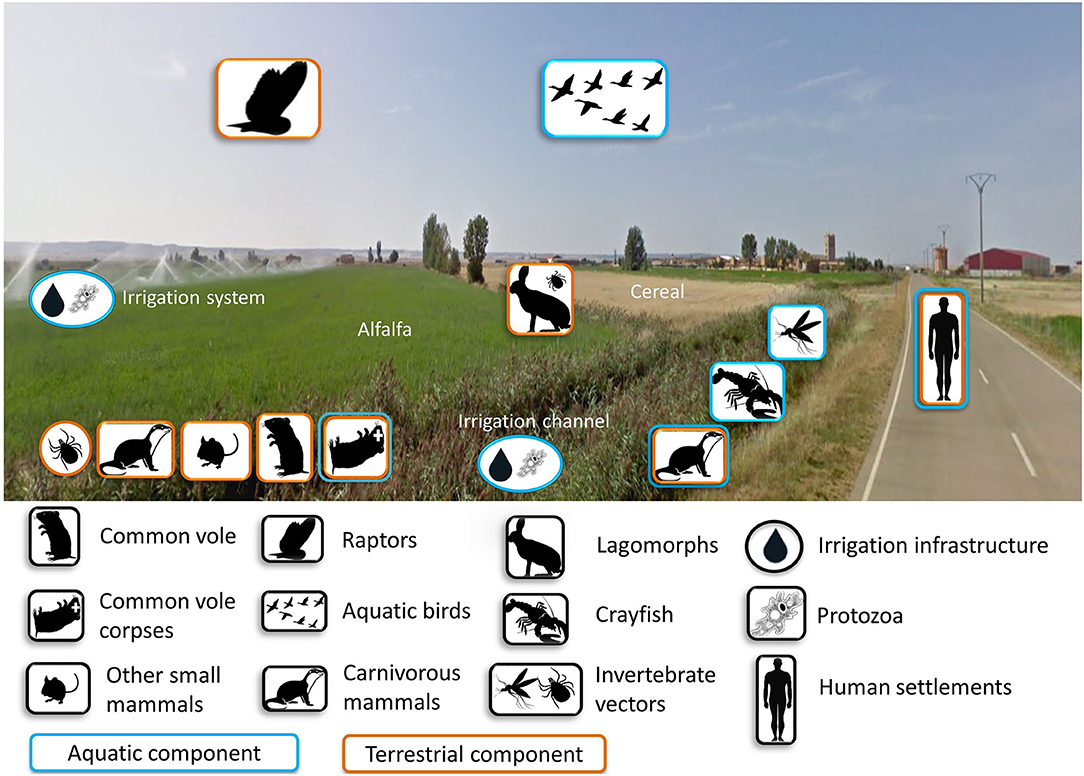
Figure 4. Components potentially involved in the cycle of tularemia either as amplifying, maintenance, spillover or dead-end hosts in intensive farming landscapes of NW Spain. Photo by GoogleMaps.
Determining the key reservoirs of F. tularensis is crucial to uncover how voles become infected during low-density phases. More research is needed to understand how voles get infected in the first place, before disease spillover, and for how long infected voles survive and spread the bacteria. Three main, non-exclusive, scenarios could occur. One possibility is that tularemia persists in surviving voles during inter-epizootics within small spatially limited refuges, which are hard to detect. Another alternative is that voles become recurrently reinfected by F. tularensis spill-over from alternative hosts, especially those species with larger lifespan, such as carnivores. Enzootic cycles involving lagomorphs and ticks are common in some tularemia endemic areas of the USA (97). In NW Spain, mean seroprevalence in lagomorphs during inter-epizootic periods (n = 515) varied between 0 and 14.6% (77), but their role remains understudied in depth. A third option would be that voles become reinfected from environmental reservoirs (Figure 4) as they widely spread through space. Contaminated water is a usual tularemia source in endemic areas of Northern and Eastern Europe (98). Once this first infection of voles has happened, high contact rates and transmission without lag in outbreaking vole populations may facilitate disease spread across the landscape. A second infection step from voles to the ecosystem would trigger the disease spillover during the common vole high-density phases, opening new transmission routes to humans. Future work should elucidate the mechanistic pathway explaining cyclical tularemia epidemics in NW Spain including how voles get infected before disease spillover, and how infected voles spread the bacteria to the system.
Human-induced ecosystem modifications can favor unexpected relationships between species that result in the spread and spillover of zoonoses, as in common voles and tularemia in NW Spain (45, 54). The “Francisella–Microtus” case study shows us that the range shift of a native species at a regional scale can lead to the emergence of zoonotic epidemics when suitable conditions are met. Convincing evidence points that highly fluctuating dynamics of common voles are closely linked with tularemia outbreaks in humans. However, there are still some knowledge gaps that need to be investigated to further understand the epidemiology of tularemia in the focused area, such as the susceptibility and impact of F. tularensis infection on voles under natural conditions; uncovering all key vectors and reservoir hosts involved in the bacterium cycle; determining where the bacterium persists during inter-epizootics, and where it was before the first epidemic occurred. Future research should focus on more integral, community-based disease knowledge considering sympatric species, vole predators, ectoparasitic vectors, and alternative potential hosts and reservoirs. This will help to better comprehend the circulation of zoonoses infecting voles, forecasting new zoonotic disease emergencies, and elucidating possible effects of pathogens on vole populations.
SH-C drafted the manuscript. FM, XL, and JL-L critically revised the paper. All authors read and approved the final manuscript.
This work contributes to the projects ECOTULA (CGL2015-66962-C2-1-R) and BOOMRAT (PID2019-109327RB-I00) funded by the Government of Spain, and regional project GESINTTOP (co-funded by Instituto Tecnológico Agrario de Castilla y León—Junta de Castilla y León (ITACYL-JCYL), Diputación Provincial de Palencia and Diputación Provincial de Valladolid). SH-C was supported by a Ph.D. studentship from Junta de Castilla-y-León (co-funded by European Social Fund, Orden 10/11/2016).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to all the people that contributed to the long-term vole monitoring over the last 10 years (Leticia Arroyo, Daniel Jareño, Jose-Luis Guzmán, Ruth Rodríguez-Pastor, Juan Romairone, Deon Roos, and Cristina Marín) or contributed to the study of the Francisella—Microtus interactions (Dolors Vidal, Raquel Escudero, Pedro Anda, and Beatriz Arroyo).
1. Wilson JRU, García-Díaz P, Cassey P, Richardson DM, Pyšek P, Blackburn TM. Biological invasions and natural colonisations are different—the need for invasion science. NeoBiota. (2016) 31:87–98. doi: 10.3897/neobiota.31.9185
2. Wilson JRU, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM. Something in the way you move: dispersal pathways affect invasion success. Trends Ecol Evol. (2009) 24:136–44. doi: 10.1016/j.tree.2008.10.007
3. Dunn AM, Hatcher MJ. Parasites and biological invasions: parallels, interactions, and control. Trends Parasitol. (2015) 31:189–99. doi: 10.1016/j.pt.2014.12.003
4. Kilpatrick AM, Randolph SE. Drivers, dynamics, and control of emerging vector-borne zoonotic diseases. Lancet. (2012) 380:1946–55. doi: 10.1016/S0140-6736(12)61151-9
5. Jones BA, Grace D, Kock R, Alonso S, Rushton J, Said MY, et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc Natl Acad Sci. (2013) 110:8399–404. doi: 10.1073/pnas.1208059110
6. Patz JA, Daszak P, Tabor GM, Aguirre AA, Pearl M, Epstein J, et al. Unhealthy landscapes: policy recommendations on land use change and infectious disease emergence. Environ Health Perspect. (2004) 112:1092–8. doi: 10.1289/ehp.6877
7. Matson PA, Parton WJ, Power AG, Swift MJ. Agricultural intensification and ecosystem properties. Science. (1997) 277:504–9. doi: 10.1126/science.277.5325.504
8. McMahon BJ, Morand S, Gray JS. Ecosystem change and zoonoses in the Anthropocene. Zoonoses Public Health. (2018) 65:755–65. doi: 10.1111/zph.12489
9. Rohr JR, Barrett CB, Civitello DJ, Craft ME, Delius B, DeLeo GA, et al. Emerging human infectious diseases and the links to global food production. Nat Sustain. (2019) 2:445–56. doi: 10.1038/s41893-019-0293-3
10. Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. (2008) 451:990–3. doi: 10.1038/nature06536
11. Grace D. Zoonoses of poverty: measuring and managing the multiple burdens of zoonoses and poverty. In: Sing A, editor. Zoonoses—Infections Affecting Humans and Animals. Dordrecht: Springer, Netherlands (2015). p. 1127–37.
12. Gibb R, Redding DW, Chin KQ, Donnelly CA, Blackburn TM, Newbold T, et al. Zoonotic host diversity increases in human-dominated ecosystems. Nature. (2020) 584:398–402. doi: 10.1038/s41586-020-2562-8
13. Han BA, Kramer AM, Drake JM. Global patterns of zoonotic disease in mammals. Trends Parasitol. (2016) 32:565–77. doi: 10.1016/j.pt.2016.04.007
14. Han BA, Schmidt JP, Bowden SE, Drake JM. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci. (2015) 112:7039–44. doi: 10.1073/pnas.1501598112
15. Vaumourin E, Vourc'h G, Gasqui P, Vayssier-Taussat M. The importance of multiparasitism: examining the consequences of co-infections for human and animal health. Parasit Vectors. (2015) 8:545. doi: 10.1186/s13071-015-1167-9
16. Bordes F, Blasdell K, Morand S. Transmission ecology of rodent-borne diseases: new frontiers. Integr Zool. (2015) 10:424–35. doi: 10.1111/1749-4877.12149
17. Auffray J-C, Renaud S, Claude J. Rodent biodiversity in changing environments. Kasetsart J Natural Sci. (2009) 43:83–93.
18. Karesh WB, Dobson A, Lloyd-Smith JO, Lubroth J, Dixon MA, Bennett M, et al. Ecology of zoonoses: natural and unnatural histories. Lancet. (2012) 380:1936–45. doi: 10.1016/S0140-6736(12)61678-X
19. Sauvage F, Langlais M, Pontier D. Predicting the emergence of human hantavirus disease using a combination of viral dynamics and rodent demographic patterns. Epidemiol Infect. (2007) 135:46–56. doi: 10.1017/S0950268806006595
20. Kallio ER, Begon M, Henttonen H, Koskela E, Mappes T, Vaheri A, et al. Cyclic hantavirus epidemics in humans—Predicted by rodent host dynamics. Epidemics. (2009) 1:101–7. doi: 10.1016/j.epidem.2009.03.002
21. Leirs H, Verhagen R, Verheyen W, Mwanjabe P, Mbise T. Forecasting rodent outbreaks in Africa: an ecological basis for Mastomys control in Tanzania. J Appl Ecol. (1996) 33:937. doi: 10.2307/2404675
22. Olayemi A, Fichet-Calvet E. Systematics, ecology, and host switching: attributes affecting emergence of the Lassa virus in rodents across Western Africa. Viruses. (2020) 12:312. doi: 10.3390/v12030312
23. Madhav NK, Wagoner KD, Douglass RJ, Mills JN. Delayed density-dependent prevalence of sin nombre virus antibody in montana deer mice (Peromyscus maniculatus) and implications for human disease risk. Vector-Borne Zoonotic Dis. (2007) 7:353–64. doi: 10.1089/vbz.2006.0605
24. Tkadlec E, Václavík T, Široký P. Rodent host abundance and climate variability as predictors of tickborne disease risk 1 year in advance. Emerg Infect Dis. (2019) 25:1738–41. doi: 10.3201/eid2509.190684
25. Andrianaivoarimanana V, Kreppel K, Elissa N, Duplantier J-M, Carniel E, Rajerison M, et al. Understanding the persistence of plague foci in Madagascar. PLoS Negl Trop Dis. (2013) 7:e2382. doi: 10.1371/journal.pntd.0002382
26. Childs JE, Mackenzie JS, Richt JA. Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission. Berlin, Heidelberg: Springer-Verlag (2007).
27. Jacob J, Tkadlec E. Rodent Outbreaks: Ecology and impacts in Europe dynamics and damage. In: Singleton G, Belmain S, Brown P, Hardy B, editors. Rodent Outbreaks: Ecology and Impacts, Los Baños, PA: International Rice Research Institute (2010).
28. Heroldová M, Bryja J, Zejda J, Tkadlec E. Structure and diversity of small mammal communities in agriculture landscape. Agric Ecosyst Environ. (2007) 120:206–10. doi: 10.1016/j.agee.2006.09.007
29. Jacob J, Manson P, Barfknecht R, Fredricks T. Common vole (Microtus arvalis) ecology and management: implications for risk assessment of plant protection products. Pest Manag Sci. (2014) 70:869–78. doi: 10.1002/ps.3695
30. Stenseth NC, Leirs H, Skonhoft A, Davis SA, Pech RP, Andreassen HP, et al. Mice, rats, and people: the bio-economics of agricultural rodent pests. Front Ecol Environ. (2003) 1:367. doi: 10.1890/1540-9295(2003)001[0367:MRAPTB]2.0.CO;2
31. Meerburg BG. Rodents are a risk factor for the spreading of pathogens on farms. Vet Microbiol. (2010) 142:464–5. doi: 10.1016/j.vetmic.2009.06.038
32. Daniels MJ, Hutchings MR, Greig A. The risk of disease transmission to livestock posed by contamination of farm stored feed by wildlife excreta. Epidemiol Infect. (2003) 130:561–68. doi: 10.1017/S0950268803008483
33. Luque-Larena JJ, Mougeot F, Arroyo B, Vidal MD, Rodríguez-Pastor R, Escudero R, et al. Irruptive mammal host populations shape tularemia epidemiology. PLOS Pathog. (2017) 13:e1006622. doi: 10.1371/journal.ppat.1006622
34. Lauret V, Delibes-Mateos M, Mougeot F, Arroyo-Lopez B. Understanding conservation conflicts associated with rodent outbreaks in farmland areas. Ambio. (2019) 49:1122–33. doi: 10.1007/s13280-019-01256-0
35. Führer H-P, Schneider R, Walochnik J, Auer H. Extraintestinal helminths of the common vole (Microtus arvalis) and the water vole (Arvicola terrestris) in Western Austria (Vorarlberg). Parasitol Res. (2010) 106:1001–4. doi: 10.1007/s00436-010-1753-x
36. Kallio-Kokko H, Uzcategui N, Vapalahti O, Vaheri A. Viral zoonoses in Europe. FEMS Microbiol Rev. (2005) 29:1051–77. doi: 10.1016/j.femsre.2005.04.012
37. Tokarz R, Markotić A, Margaletić J, Lipkin WI, Habuš J, Jain K, et al. Molecular survey of zoonotic agents in rodents and other small mammals in Croatia. Am J Trop Med Hyg. (2016) 94:466–73. doi: 10.4269/ajtmh.15-0517
38. Schmidt S, Essbauer SS, Mayer-Scholl A, Poppert S, Schmidt-Chanasit J, Klempa B, et al. Multiple infections of rodents with zoonotic pathogens in Austria. Vector-Borne Zoonotic Dis. (2014) 14:467–75. doi: 10.1089/vbz.2013.1504
39. Pawelczyk A, Bajer A, Behnke JM, Gilbert FS, Sinski E. Factors affecting the component community structure of haemoparasites in common voles (Microtus arvalis) from the Mazury Lake District region of Poland. Parasitol Res. (2004) 92:270–84. doi: 10.1007/s00436-003-1040-1
40. Fuehrer H-P, Blöschl I, Siehs C, Hassl A. Detection of Toxoplasma gondii, Neospora caninum, and Encephalitozoon cuniculi in the brains of common voles (Microtus arvalis) and water voles (Arvicola terrestris) by gene amplification techniques in western Austria (Vorarlberg). Parasitol Res. (2010) 107:469–73. doi: 10.1007/s00436-010-1905-z
41. Meerburg BG, Singleton GR, Kijlstra A. Rodent-borne diseases and their risks for public health. Crit Rev Microbiol. (2009) 35:221–70. doi: 10.1080/10408410902989837
42. Grzybek M, Antolová D, Tołkacz K, Alsarraf M, Behnke-Borowczyk J, Nowicka J, et al. Seroprevalence of Toxoplasma gondii among sylvatic rodents in Poland. Animals. (2021) 11:1048. doi: 10.3390/ani11041048
43. Beerli O, Guerra D, Baltrunaite L, Deplazes P, Hegglin D. Microtus arvalis and Arvicola scherman: key players in the Echinococcus multilocularis life cycle. Front Vet Sci. (2017) 4:216. doi: 10.3389/fvets.2017.00216
44. Telfer S, Lambin X, Birtles RJ, Beldomenico PM, Burthe SJ, Paterson S, et al. Species interactions in a parasite community drive infection risk in a wildlife population. Science. (2010) 330:243–6. doi: 10.1126/science.1190333
45. Luque-Larena JJ, Mougeot F, Viñuela J, Jareño D, Arroyo L, Lambin X, et al. Recent large-scale range expansion and outbreaks of the common vole (Microtus arvalis) in NW Spain. Basic Appl Ecol. (2013) 14:432–41. doi: 10.1016/j.baae.2013.04.006
46. Mougeot F, Lambin X, Rodríguez-Pastor R, Romairone J, Luque-Larena JJ. Numerical response of a mammalian specialist predator to multiple prey dynamics in Mediterranean farmlands. Ecology. (2019) 100:e02776. doi: 10.1002/ecy.2776
47. Rodríguez-Pastor R. Ecology of rodent outbreaks and zoonotic diseases: common voles in the farmland of Northwest Spain (thesis). Department of Agroforestry Sciences, University of Valladolid, Valladolid, Spain (2018)
48. Rodríguez-Pastor R, Mougeot F, Vidal MD, Jado I, González-Martín-Niño RM, Escudero R, et al. Zoonotic bacteria in fleas parasitizing common voles, Northwestern Spain. Emerg Infect Dis. (2019) 25:1423–5. doi: 10.3201/eid2507.181646
49. Rodríguez-Pastor R, Escudero R, Lambin X, Vidal MD, Gil H, Jado I, et al. Zoonotic pathogens in fluctuating common vole (Microtus arvalis) populations: occurrence and dynamics. Parasitology. (2019) 146:389–98. doi: 10.1017/S0031182018001543
50. Vidal MD, Alzaga V, Luque-Larena JJ, Mateo R, Arroyo L, Viñuela J. Possible interaction between a rodenticide treatment and a pathogen in common vole (Microtus arvalis) during a population peak. Sci Total Environ. (2009) 408:267–71. doi: 10.1016/j.scitotenv.2009.10.001
51. Vidal MD, Mougeot F, Rodríguez-Pastor R, Jubete F, Arroyo B, González Martín-Niño R, et al. Molecular evidence of Francisella tularensis in nature, 10 years after a large outbreak of tularemia in Spain. In: 9th International Conference on Tularemia. Montréal, QC (2018).
52. Lopes de Carvalho I, Pinto M, Rodríguez-Pastor R, Isidro J, Nunes C, Mougeot F, et al. Genome-scale comparison of Francisella tularensis strains isolated in an endemic region of Spain. In 9th International Conference on Tularemia. Montréal, QC (2018).
53. González-Barrio D, Jado I, Viñuela J, García JT, Olea PP, Arce F, et al. Investigating the role of micromammals in the ecology of Coxiella burnetii in Spain. Animals. (2021) 11:654. doi: 10.3390/ani11030654
54. Luque-Larena JJ, Mougeot F, Vidal Roig D, Lambin X, Rodríguez-Pastor R, Rodríguez-Valín E, et al. Tularemia outbreaks and common vole (Microtus arvalis) irruptive population dynamics in Northwestern Spain, 1997–2014. Vector-Borne Zoonotic Dis. (2015) 15:568–70. doi: 10.1089/vbz.2015.1770
55. Jareño D, Viñuela J, Luque-Larena JJ, Arroyo L, Arroyo B, Mougeot F. Factors associated with the colonization of agricultural areas by common voles Microtus arvalis in NW Spain. Biol Invasions. (2015) 17:2315–27. doi: 10.1007/s10530-015-0877-4
56. Rodríguez-Pastor R, Luque-Larena JJ, Lambin X, Mougeot F. “Living on the edge”: the role of field margins for common vole (Microtus arvalis) populations in recently colonised Mediterranean farmland. Agric Ecosyst Environ. (2016) 231:206–17. doi: 10.1016/j.agee.2016.06.041
57. Romairone J, Jiménez J, Luque-Larena JJ, Mougeot F. Spatial capture-recapture design and modelling for the study of small mammals. PLoS ONE. (2018) 13:e0198766. doi: 10.1371/journal.pone.0198766
58. Rodríguez-Pastor R, Escudero R, Vidal MD, Mougeot F, Arroyo B, Lambin X, et al. Density-dependent prevalence of Francisella tularensis in fluctuating vole populations, Northwestern Spain. Emerg Infect Dis. (2017) 23:1377–9. doi: 10.3201/eid2308.161194
59. Luque-Larena JJ, Mougeot F, Arroyo B, Lambin X. “Got rats?” Global environmental costs of thirst for milk include acute biodiversity impacts linked to dairy feed production. Glob Chang Biol. (2018) 24:2752–4. doi: 10.1111/gcb.14170
60. Fargallo JA, Martínez-Padilla J, Viñuela J, Blanco G, Torre I, Vergara P, et al. Kestrel-prey dynamic in a Mediterranean region: The effect of generalist predation and climatic factors. PLoS ONE (2009) 4:e4311. doi: 10.1371/journal.pone.0004311
61. Kman NE, Nelson RN. Infectious agents of bioterrorism: a review for emergency physicians. Emerg Med Clin North Am. (2008) 26:517–47. doi: 10.1016/j.emc.2008.01.006
62. World Health Organization. WHO Guidelines on Tularaemia. Geneva: World Health Organization (2007).
63. Bossi P, Tegnell A, Baka A, Loock V, Hendriks J, Werner A, et al. Bichat guidelines for the clinical management of tularaemia and bioterrorism-related tularaemia. Euro Surveill. (2004) 9:27–8. doi: 10.2807/esm.09.12.00503-en
64. Maurin M, Gyuranecz M. Tularaemia: clinical aspects in Europe. Lancet Infect Dis. (2016) 16:113–24. doi: 10.1016/S1473-3099(15)00355-2
65. Abd H, Johansson T, Golovliov I, Sandström G, Forsman M. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl Environ Microbiol. (2003) 69:600–6. doi: 10.1128/AEM.69.1.600-606.2003
66. Telford SR III, Goethert HK. Ecology of Francisella tularensis. Annu Rev Entomol. (2020) 65:351–72. doi: 10.1146/annurev-ento-011019-025134
67. Carvalho CL, Lopes de Carvalho I, Zé-Zé L, Núncio MS, Duarte EL. Tularaemia: a challenging zoonosis. Comp Immunol Microbiol Infect Dis. (2014) 37:85–96. doi: 10.1016/j.cimid.2014.01.002
68. Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. (2007) 1105:30–66. doi: 10.1196/annals.1409.011
69. Anda P, Segura del Pozo J, García JMD, Escudero R, Peña FJG, Velasco MCL, et al. Waterborne outbreak of tularemia associated with crayfish fishing. Emerg Infect Dis. (2001) 7:575–82. doi: 10.3201/eid0707.017740
70. Hestvik G, Warns-Petit E, Smith LA, Fox NJ, Uhlhorn H, Artois M, et al. The status of tularemia in Europe in a one-health context: a review. Epidemiol Infect. (2015) 143:2137–60. doi: 10.1017/S0950268814002398
71. European Centre for Disease Prevention and Control. Surveillance Atlas of Infectious Diseases. (2020) Available online at: https://atlas.ecdc.europa.eu/public/index.aspx?Dataset=27&HealthTopic=55 (accessed June 3, 2021).
72. Gutiérrez MP, Bratos MA, Garrote JI, Dueñas A, Almaraz A, Alamo R, et al. Serologic evidence of human infection by Francisella tularensis in the population of Castilla y León (Spain) prior to 1997. FEMS Immunol Med Microbiol. (2003) 35:165–9. doi: 10.1016/S0928-8244(03)00002-6
73. Gyuranecz M. Tularaemia. Infectious Diseases of Wild Mammals and Birds in Europe. Oxford, UK: Wiley-Blackwell (2012). p. 303–9.
74. Rossow H, Forbes KM, Tarkka E, Kinnunen PM, Hemmilä H, Huitu O, et al. Experimental infection of voles with Francisella tularensis indicates their amplification role in tularemia outbreaks. PLoS ONE.(2014) 9:e108864. doi: 10.1371/journal.pone.0108864
75. Samia NI, Kausrud KL, Heesterbeek H, Ageyev VS, Begon M, Chan K-S, et al. Dynamics of the plague-wildlife-human system in Central Asia are controlled by two epidemiological thresholds. Proc Natl Acad Sci. (2011) 108:14527–32. doi: 10.1073/pnas.1015946108
76. Rossow H, Ollgren J, Hytönen J, Rissanen H, Huitu O, Henttonen H, et al. Incidence and seroprevalence of tularaemia in Finland, 1995 to 2013: regional epidemics with cyclic pattern. Eurosurveillance. (2015) 20:21209. doi: 10.2807/1560-7917.ES2015.20.33.21209
77. Rodríguez-Ferri E-F. Tularemia: una aproximación a su estudio integral en Castilla y León. León: Academia Ciencias Veterinarias de Castilla y León (2017).
78. González-Barrio D, Viñuela J, Santamaría AE, Olea PP, García JT, Ruiz-Fons F. Prevalence of zoonotic pathogens in common vole (Microtus arvalis) during a population outbreak. In: Proceedings of the 15th Rodens et Spatium. Olomouc (2016).
79. Herrero-Cófreces S, Flechoso MF, Rodríguez-Pastor R, Luque-Larena JJ, Mougeot F. Patterns of flea infestation in rodents and insectivores from intensified agro-ecosystems, Northwest Spain. Parasit Vectors. (2021) 14:16. doi: 10.1186/s13071-020-04492-6
80. Gutiérrez R, Krasnov BR, Morick D, Gottlieb Y, Khokhlova IS, Harrus S. Bartonella infection in rodents and their flea ectoparasites: an overview. Vector-Borne Zoonotic Dis. (2015) 15:27–39. doi: 10.1089/vbz.2014.1606
81. Shah HA, Huxley P, Elmes J, Murray KA. Agricultural land-uses consistently exacerbate infectious disease risks in Southeast Asia. Nat Commun. (2019) 10:4299. doi: 10.1038/s41467-019-12333-z
82. Cleri DJ, Ricketti AJ, Vernaleo JR. Fever of unknown origin due to zoonoses. Infect Dis Clin North Am. (2007) 21:963–96. doi: 10.1016/j.idc.2007.08.009
83. Crump JA, Morrissey AB, Nicholson WL, Massung RF, Stoddard RA, Galloway RL, et al. Etiology of severe non-malaria febrile illness in Northern Tanzania: a prospective cohort study. PLoS Negl Trop Dis. (2013) 7:e2324. doi: 10.1371/journal.pntd.0002324
84. Ariza-Miguel J, Johansson A, Fernández-Natal MI, Martínez-Nistal C, Orduña A, Rodríguez-Ferri EF, et al. Molecular investigation of tularemia outbreaks, Spain, 1997–2008. Emerg Infect Dis. (2014) 20:754–61. doi: 10.3201/eid2005.130654
85. Telford SR, Goethert HK. Toward an understanding of the perpetuation of the agent of tularemia. Front Microbiol. (2011) 1:150. doi: 10.3389/fmicb.2010.00150
86. González-Esteban J, Villate I. Microtus arvalis (Pallas, 1778). In: Palomo LJ, Gisbert J, Blanco JC, editors. Atlas y Libro Rojo de los Mamíferos Terrestres de España. Madrid: Dirección General para la Biodiversidad-SECEM-SECEMU (2007). p. 128–30.
87. Biggins DE, Kosoy MY. Influences of introduced plague on North American mammals: implications from ecology of plague in Asia. J Mammal. (2001) 82:906–16. doi: 10.1644/1545-1542(2001)082<0906:IOIPON>2.0.CO;2
88. Chantrey J, Dale TD, Read JM, White S, Whitfield F, Jones D, et al. European red squirrel population dynamics driven by squirrelpox at a gray squirrel invasion interface. Ecol Evol. (2014) 4:3788–99. doi: 10.1002/ece3.1216
89. Kreizinger Z, Erdélyi K, Felde O, Fabbi M, Sulyok KM, Magyar T, et al. Comparison of virulence of Francisella tularensis ssp. holarctica genotypes B.12 and B.FTNF002-00. BMC Vet Res. (2016) 13:46. doi: 10.1186/s12917-017-0968-9
90. Bell JF, Stewart SJ. Chronic shedding tularemia nephritis in rodents: possible relation to occurrence of Francisella tularensis in lotic waters. J Wildl Dis. (1975) 11:421–30. doi: 10.7589/0090-3558-11.3.421
91. Turner AK, Beldomenico PM, Bown KJ, Burthe SJ, Jackson JAA, Lambin X, et al. Host–parasite biology in the real world: the field voles of Kielder. Parasitology. (2014) 141:997–1017. doi: 10.1017/S0031182014000171
92. Kuehn A, Schulze C, Kutzer P, Probst C, Hlinak A, Ochs A, et al. Tularaemia seroprevalence of captured and wild animals in Germany: the fox (Vulpes vulpes) as a biological indicator. Epidemiol Infect. (2013) 141:833–40. doi: 10.1017/S0950268812001008
93. Origgi FC, Wu N, Pilo P. Francisella tularensis infection in a stone marten (Martes foina) without classic pathological lesions consistent with tularemia. J Vet Diagnostic Investig. (2013) 25:519–21. doi: 10.1177/1040638713489124
94. Petersen JM, Mead PS, Schriefer ME. Francisella tularensis: an arthropod-borne pathogen. Vet Res. (2009) 40:07. doi: 10.1051/vetres:2008045
95. Kittl S, Francey T, Brodard I, Origgi FC, Borel S, Ryser-Degiorgis M-P, et al. First European report of Francisella tularensis subsp. holarctica isolation from a domestic cat. Vet Res. (2020) 51:109. doi: 10.1186/s13567-020-00834-5
96. Lopes de Carvalho I, Zé-Zé L, Alves AS, Pardal S, Lopes RJ, Mendes L, et al. Borrelia garinii and Francisella tularensis subsp. holarctica detected in migratory shorebirds in Portugal. Eur J Wildl Res. (2012) 58:857–61. doi: 10.1007/s10344-012-0617-3
97. Mani RJ, Morton RJ, Clinkenbeard KD. Ecology of tularemia in central US endemic region. Curr Trop Med Rep. (2016) 3:75–9. doi: 10.1007/s40475-016-0075-1
Keywords: colonizing rodent, infectious pathogens, intensive agriculture, land-use changes, Francisella tularensis, Microtus arvalis
Citation: Herrero-Cófreces S, Mougeot F, Lambin X and Luque-Larena JJ (2021) Linking Zoonosis Emergence to Farmland Invasion by Fluctuating Herbivores: Common Vole Populations and Tularemia Outbreaks in NW Spain. Front. Vet. Sci. 8:698454. doi: 10.3389/fvets.2021.698454
Received: 21 April 2021; Accepted: 14 July 2021;
Published: 12 August 2021.
Edited by:
Alexis Ribas Salvador, University of Barcelona, SpainReviewed by:
Luca Villa, University of Milan, ItalyCopyright © 2021 Herrero-Cófreces, Mougeot, Lambin and Luque-Larena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silvia Herrero-Cófreces, c2lsdmlhLmhlcnJlcm8uY29mcmVjZXNAdXZhLmVz; François Mougeot, ZnJhbmNvaXMubW91Z2VvdEB1Y2xtLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.