- 1Department of Molecular Biology and Genetics, Faculty of Arts and Sciences, Ordu University, Ordu, Turkey
- 2Department of Comparative Biomedical Sciences, College of Veterinary Medicine, Mississippi State University, Mississippi State, MS, United States
Edwardsiella ictaluri is a Gram-negative facultative intracellular pathogen causing enteric septicemia of catfish (ESC), a devastating disease resulting in significant economic losses in the U.S. catfish industry. Bacterial secretion systems are involved in many bacteria's virulence, and Type VI Secretion System (T6SS) is a critical apparatus utilized by several pathogenic Gram-negative bacteria. E. ictaluri strain 93–146 genome has a complete T6SS operon with 16 genes, but the roles of these genes are still not explored. In this research, we aimed to understand the roles of two hemolysin co-regulated family proteins, Hcp1 (EvpC) and Hcp2. To achieve this goal, single and double E. ictaluri mutants (EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2) were generated and characterized. Catfish peritoneal macrophages were able to kill EiΔhcp2 better than EiΔevpC, EiΔevpCΔhcp2, and E. ictaluri wild-type (EiWT). The attachment of EiΔhcp2 and EiΔevpCΔhcp2 to ovary cells significantly decreased compared to EiWT whereas the cell invasion rates of these mutants were the same as that of EiWT. Mutants exposed to normal catfish serum in vitro showed serum resistance. The fish challenges demonstrated that EiΔevpC and EiΔevpCΔhcp2 were attenuated completely and provided excellent protection against EiWT infection in catfish fingerlings. Interestingly, EiΔhcp2 caused higher mortality than that of EiWT in catfish fingerlings, and severe clinical signs were observed. Although fry were more susceptible to vaccination with EiΔevpC and EiΔevpCΔhcp2, their attenuation and protection were significantly higher compared to EiWT and sham groups, respectively. Taken together, our data indicated that evpC (hcp1) is involved in E. ictaluri virulence in catfish while hcp2 is involved in adhesion to epithelial cells and survival inside catfish macrophages.
Introduction
Edwardsiella ictaluri (E. ictaluri) is the causative agent of enteric septicemia of catfish (ESC) (1). Although E. ictaluri is well-adapted to catfish, it can also infect other freshwater fish species (2–4). At the early stages of host invasion, E. ictaluri encounters the host immune system (5, 6). However, E. ictaluri is capable of surviving and replicating inside catfish professional phagocytic cells, macrophages, and neutrophils (7). To replicate successfully inside the host cells, E. ictaluri must resist and overcome the bacterial killing mechanisms present in the host macrophages and neutrophils, such as oxidative and nitrosative stress (8–10).
The survival of E. ictaluri highly depends on its resistance to host stress factors and modulating the host environment. Edwardsiella ictaluri encodes urease that is activated in acidic phagosomes of macrophage to cope with low pH (11, 12). Low pH and low phosphate concentration inside the phagosome can trigger the expression level of genes in both Type III and Type VI secretion systems (T3SS and T6SS), which assists E. ictaluri survival inside the host immune cells (13). It was shown that the effector proteins secreted via T3SS had an important role in virulence of E. ictaluri, and mutation of these genes caused decreased intracellular replication inside catfish head kidney-derived macrophages (14, 15).
Hemolysin co-regulated family proteins (Hcp) are involved in bacteria-host interaction. Particularly, they are involved in adhesion and invasion, intracellular survival of bacteria, bacterial cytotoxicity, and virulence (16). In E. ictaluri, T6SS proteins, Eip19 (evpE), Eip18 (evpC), Eip55 (evpB), Eip20 (evpA), have been first identified during the catfish host-pathogen interaction (17). The secretion of evpC is transcriptionally controlled by two-component system regulatory protein esrC in low-pH and phosphate conditions in E. ictaluri (13). Fish pathogen E. piscicida also possesses T6SS, which is required for virulence (18). In E. tarda, evpC plays a dual role as a chaperone and T6SS-dependent secreted protein (19). evpC belongs to Hcp family proteins and can bind to T6SS-dependent effector proteins in bacterial cytoplasm and guide effector proteins through the T6SS needle (20). Due to their role as a chaperone protein, evpC interacts with the T6SS-dependent effector proteins such as evpP in E. tarda (21). A recent study showed that evpP effector protein secreted via evpC could target the macrophages' inflammasome activation (22).
E. ictaluri genome has a complete T6SS operon with evpC (hcp1) while hcp2 is located outside of the T6SS operon. In this study, we evaluated the role of hcp genes in E. ictaluri-catfish interaction. Our study revealed roles of evpC and hcp2 in adhesion and invasion of catfish epithelial cells, survival and replication inside catfish peritoneal macrophages, adaptation to the stress factors, and virulence and efficacy in catfish.
Materials and Methods
Bacteria, Plasmids, and Media
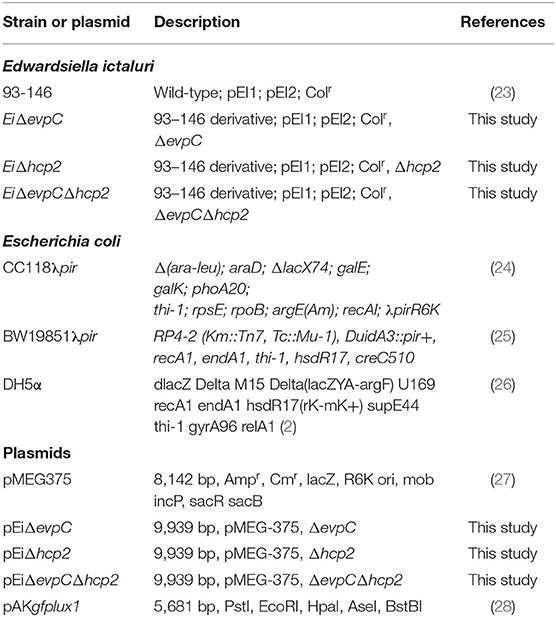
Bacterial strains and plasmids used in this work were listed in Table 1. Edwardsiella ictaluri strain 93–146 (EiWT) and isogenic hcp mutants were grown at 30°C in Brain Heart Infusion (BHI) broth or agar. Escherichia coli (E. coli) CC118λpir, BW19851 (ΔuidA3::pir), and DH5α strains were cultured on Luria–Bertani (LB) agar or broth and incubated at 37°C. Antibiotics were added to the culture medium at the following concentrations: gentamicin (10 μg/ml), ampicillin (100 μg/ml), and colistin (12.5 μg/ml).
In-frame Deletion of evpC and hcp2
The nucleotide sequences of evpC (NT01EI_RS11900) and hcp2 (NT01EI_RS14960) were obtained from the E. ictaluri 93–146 genome (GenBank accession: 95 CP001600) (29). The overlap extension PCR method was used to generate evpC and hcp2 in-frame deletion fragments. Briefly, external and internal primers were designed to amplify the regions for upstream and downstream of each gene (Table 2). Two amplified fragments were combined through splicing by overlap extension (SOEing) (30). The overlap PCR product and the pMEG375 suicide plasmid were digested with the same restriction enzymes, and the mutated insert was ligated into the pMEG375. After electroporation and selection of the correct plasmid in CC118, the plasmid was transferred to E. coli BW19851 by electroporation, which was then used to transfer the plasmid into E. ictaluri strain 93–146 by conjugation. Two-step selection was used to obtain in-frame deletion mutants. At the first step, ampicillin-resistant E. ictaluri colonies were inoculated into BHI broth containing ampicillin and colistin. In the second step, positive colonies were streaked on the BHI agar containing colistin only. These colonies were re-streaked on the BHI agar with 5% sucrose, 0.35% D-mannitol, and colistin. Ampicillin-sensitive colonies with the mutant band were in-frame deletion colonies. The deletion of each gene was confirmed by PCR and sequencing. For the construction of double mutant, E. coli BW19851 carrying pMEG375 with overlap hcp2 and EiΔevpC were conjugated, and two-step selection yielded EiΔevpCΔhcp2. PCR and sequencing confirmed the deletion of hcp2 in the double mutant.
Hemolysis Assay
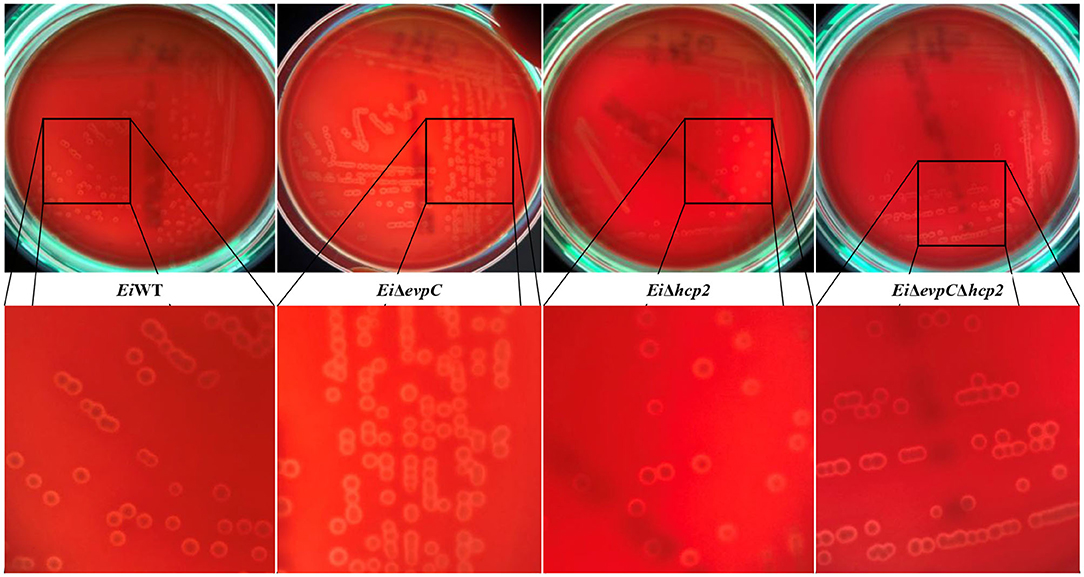
EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2, and EiWT were streaked on sheep blood agar plates (Fisher Scientific), which were incubated at 30°C for 48 h. Hemolytic activity of the mutants was visualized using a Stuart Colony Counter with sub-stage illumination (Cole-Parmer).
Construction of Bioluminescent Strains
pAKgfplux1 was used to construct bioluminescent EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 strains, as described previously (28). Briefly, E. coli SM10λpir carrying pAKgfplux1 and mutants were grown overnight and mixed at the ratio of 1:2 (donor: recipient). Mixture pellet was spotted on 0.45 μM filter paper placed on BHI agar and grown at 30°C for 24 h. Filter paper containing a mixture of bacteria was washed with BHI broth containing ampicillin and colistin, and serial dilutions were spread on selective BHI agar containing ampicillin and colistin. Ampicillin resistant mutant colonies carrying pAKgfplux1 appeared on the selective plates after 30°C for 24–48 h.
Serum Treatment
Bioluminescent EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 strains were exposed to catfish normal serum. Bioluminescent EiWT (positive control) and E. coli DH5α (negative control) were also included in each experiment. Catfish serum was collected as previously described (28). Then, 195 μl catfish serum was added to each well of a 96 black well-plate (Corning Costar). Next, 5 μl of the overnight bacterial culture [optical density at 600 nm (OD600) = 1.0] was mixed with the serum and inoculated for 4 h at 30°C. The experiment included four replicates, and bioluminescence was measured by using SpectraMax M5 Multi-Mode Microplate Reader (Molecular Devices). Bioluminescent images were taken using an IVIS Lumina XRMS In Vivo Imaging System Series III (PerkinElmer).
Bioluminescent Imaging
Sixteen specific-pathogen-free (SPF) catfish fingerlings (12.72 ± 1.00 cm, 24.95 ± 5.47 g) were obtained from the CVM hatchery and stocked into four tanks (4 fish/tank). Three tanks were assigned to EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 (treatments), and one tank for EiWT (positive control). After 1 week of acclimatization, the water level was reduced to 10 L, and 100 ml bacterial culture was added to each tank (final dose of 5 × 107 colony forming units, CFU, per ml of water). Following 1 h incubation, water flow was restored in each tank. Fish were anesthetized with 100 mg/L MS222, and bioluminescence emitted from the fish body was collected for one min by using IVIS Lumina XRMS In Vivo Imaging System Series III (PerkinElmer). Following bioluminescent imaging, fish were transferred to buckets with aerated water for recovery. Bioluminescent imaging was conducted at 0, 6, 12, and 24 h post-infection, and subsequent daily intervals until 14 days.
Bacterial Killing Assay
The bacterial killing assay was performed as previously described (12, 31, 32). Briefly, peritoneal macrophages were collected from a year-old channel catfish (250–300 g) injected with 1 ml squalene (Sigma). Following 4-day post-injection, peritoneal macrophages were harvested from five catfish by injecting 10 ml cold phosphate-buffered saline (1X, PBS) to the peritoneal cavity of catfish. Harvested cells were pooled and washed with PBS three times. The cells were resuspended in channel catfish macrophage medium (CCMM) including RPMI (RPMI 1640 sans phenol red & L-glutamine, Lonza) containing 1× glutamine substitute (GlutaMAX –I CTS, Invitrogen), 15 mM HEPES buffer (Invitrogen), in 0.18% sodium bicarbonate solution (Invitrogen), 0.05 mM 2-beta-mercaptoethanol (Sigma), and 5% heat-inactivated (HI) pooled channel catfish serum. Next, peritoneal macrophages (5 × 105 cells) were transferred into a 96-well plate (Evergreen Scientific), and bioluminescent E. ictaluri strains were added at a 1:1 ratio and mixed gently by pipetting up and down. The final volume of the cell-bacteria mixture was 200 μl in each well, and the plate included four replicate wells for each treatment and negative control (cell only). The plate was centrifuged at 1,500 rpm for 5 min at 24°C to compact the cells and bacteria at the bottom. The plate was then incubated for 1 h at 30°C to allow the invasion of catfish peritoneal macrophages by bioluminescent mutants and EiWT. Following the first incubation, the cell-bacteria mixture was centrifuged at 2,000 rpm for 7 min, and the media was removed. After this, CCMM containing 100 μg/ml gentamicin were added, and cells were incubated an additional 1 h at 30°C to kill non-phagocyted E. ictaluri. At the end of incubation, each well was washed three times with PBS, and peritoneal macrophages were suspended in CCMM with 10 μg/ml gentamicin. Finally, cells were transferred to black 96-well-plates (Fisher Scientific), and the plate was placed in Cytation 5 (BioTek) where the cells were incubated for 48 h under 5% CO2 at 30°C. Bioluminescence was captured every hour, and data were analyzed to determine the number of survived bioluminescent bacteria in catfish peritoneal macrophages.
Attachment and Invasion Assays
Attachment and invasion assays were performed by using channel catfish ovary (CCO) cell line, as described previously (33). Briefly, CCO cells were resuspended in DMEM medium (Sigma) supplemented with 10% fetal bovine serum and 4 mM L-glutamine at a final concentration of 1 x 107 cells ml−1. Bioluminescent mutants and EiWT were mixed with CCO cells at a 1:1 ratio and placed in a 24-well-plate. The cell-bacteria mixture's final volume was 1 ml in each well, and the plate included four replicate wells for each treatment and negative control (no bacteria). The plate was incubated 1 h at 28°C for the attachment of mutants and EiWT to CCO. After that, the cell suspensions were incubated in DMEM containing 100 μg/ml gentamicin for 1 h to kill the external bacteria. The plate was washed with PBS three times, and the invasion of E. ictaluri strains was determined by imaging IVIS Lumina XRMS In Vivo Imaging System Series III (PerkinElmer).
Stress Assays
The mutants' survival in oxidative stress in hydrogen peroxide (H2O2) (Sigma) and nitrosative stress in sodium nitroprusside (SNP) (Sigma) were tested in BHI (rich medium) and low phosphate minimal medium at pH 5.5 (MM19-P) (34). Bacteria were grown overnight, and OD600 adjusted to 0.5 for each culture. Five microliter of bacteria from each strain were inoculated into 195 μl of BHI and MM19-P broth containing 0.75 mM H2O2 (diluted from 30% stock solution) and 5 mM SNP. Each 96-well-black plate included three replicates for each mutant and EiWT as a positive control. The mean photon counts for each stress treatment were measured after 4, 8, 12, and 24 h incubation at 30°C by using IVIS Lumina XRMS In Vivo Imaging System Series III (PerkinElmer).
Virulence and Efficacy of Mutants in Catfish Fingerlings and Fry
Vaccination and efficacy were performed as previously described (35). Briefly, specific-pathogen-free (SPF) channel catfish fingerlings and fry were obtained from the MSU-CVM Hatchery. Catfish fingerlings (10.46 ± 0.86 cm, 14.03 ± 3.57 g) were stocked into 15 tanks at a rate of 25 fish/tank. Catfish fry were stocked in 12 tanks at a rate of 50 fish/tank. Fish were acclimated at 26–28°C for 1 week and fed twice a day. Chlorine, dissolved oxygen, and temperature were monitored daily. Treatments were randomly assigned to EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2 (vaccination), EiWT (positive control), and BHI (sham) groups. Each treatment had three replicates. Immersion vaccination was applied by lowering the water level in each tank to 10-L, and by adding 100 ml of bacterial culture (final dose of 2.4 × 107 CFU/ml water). After 1 h, water flow (1 liter/min) was restored to each tank. Mortalities were recorded daily for 21 days, and the percent mortalities were calculated for each group. To assess the protective capabilities of mutants, all fish that survived the EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 vaccination were re-challenged with EiWT (2.8 × 107 CFU/ml) 21 days post-vaccination as described above. Fish mortalities were recorded daily, and the experiment was terminated when no fish mortalities were observed for three consecutive days.
Statistical Analysis
The significance of the differences between treatment means was established by one-way ANOVA and two-way ANOVA procedures with Tukey's test in SAS for Windows 9.4 (SAS Institute, Inc., Cary, NC). The level of significance for all tests was set at p < 0.05.
Results
Hemolytic Activity of the Mutants
A beta-hemolysis with a narrow clear hemolytic zone around the colonies was observed, and hemolytic activity of EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 was similar to EiWT (Figure 1).
Survival of the Mutants Under Complement Stress
Channel catfish serum was used to evaluate the survival of mutants under complement stress. EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 were able to survive after 4 h of incubation in catfish serum (Figure 2A), and no significant differences have been detected between the mutants and EiWT (Figure 2B). However, significant differences in the intensity of bacterial bioluminescence were found between 0 and 4 h (p < 0.05; Figure 2C). These results indicate that EiWT and mutant strains were robust to complement killing and able to replicate in catfish serum.
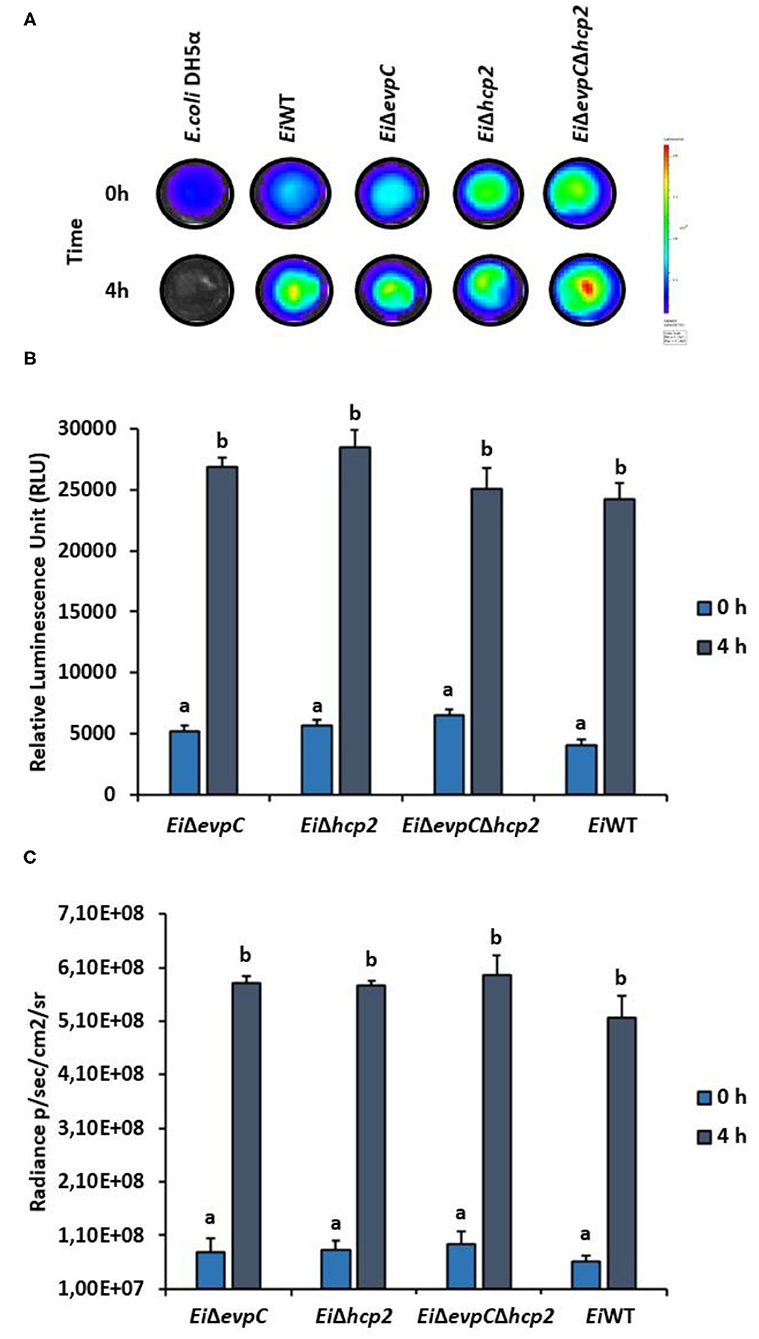
Figure 2. Treatment of EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2, and EiWT with normal catfish serum. (A) Bioluminescence of mutants, EiWT, and E. coli DH5α (negative control) treated with normal catfish serum at 0 and 4 h. Total photon emissions were collected by IVIS Lumina XRMS In Vivo Imaging System Series III at an exposure time of 1 min. (B) The bar graph indicates the relative luminescence unit (RLU) obtained by Cytation 5 from the mutants and EiWT treated with the catfish normal serum. (C) Total photon counts obtained from bioluminescent image in (A). Data represent the mean of each treatment ± SD. Letters (a and b) show significant differences between treatments at each time points (0 and 4 h) (p < 0.05).
Persistence of the Mutants in Catfish Fingerlings
The bioluminescent imaging was used to monitor the persistence of EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 in catfish fingerlings. EiWT was able to kill all catfish fingerlings in 5 days shortly after ESC clinical signs were observed (Figure 3A). Catfish fingerlings exposed to EiΔevpC and EiΔevpCΔhcp2 mutants survived, and clearance of mutants from the catfish fingerlings was observed. However, the immersion challenge of EiΔhcp2 showed severe mortality of all catfish fingerlings in 8 days (Figure 3A). The bioluminescent photon counts from fingerlings showed that the number of EiΔevpC and EiΔevpCΔhcp2 had peaked at the highest point at 48 h (Figure 3B). On the other hand, the bioluminescence of EiΔhcp2 was continued to increase after 48 h post-infection (Figure 3B). These findings demonstrated that EiΔevpC and EiΔevpCΔhcp2 were attenuated and cleared from catfish fingerlings while EiΔhcp2 was not attenuated.
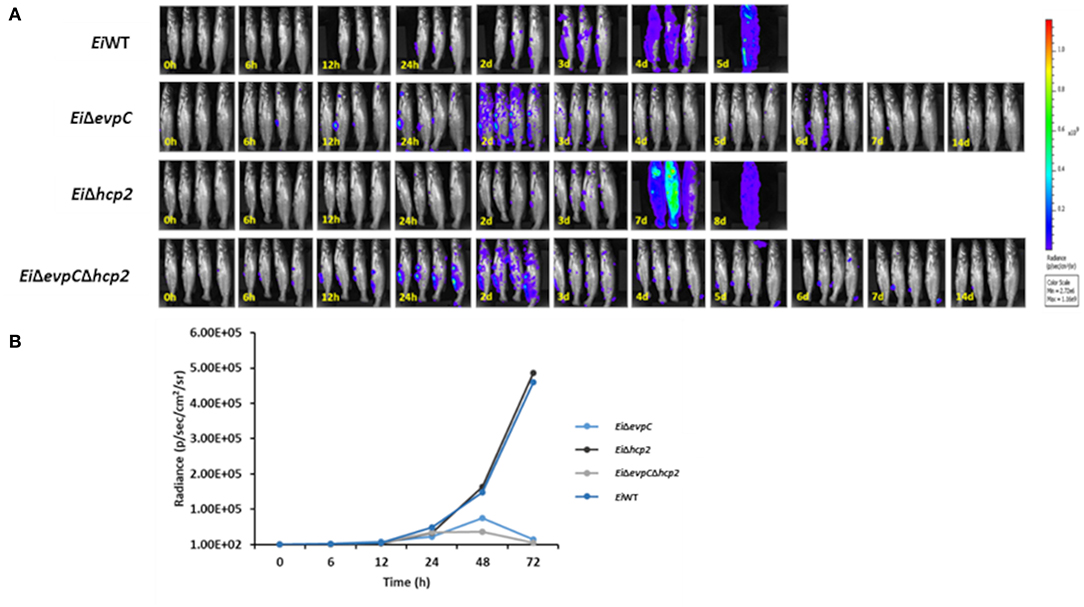
Figure 3. Bioluminescent imaging of live catfish fingerlings challenged with EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2, and EiWT. (A) The bioluminescent image was obtained from four fish at 0, 6, 12, 24 h, and subsequent daily intervals until 14 days. 0 h picture was taken after 1 h immersion challenge of catfish with mutants and EiWT. One of the four catfish challenged with EiWT died at 12 h post-infection. (B) Total photon emissions obtained from the images up to 72 h post-infection. Total photon emissions were collected by IVIS Lumina XRMS In Vivo Imaging System Series III at an exposure time of 1 min.
Bacterial Killing of the Mutants in Catfish Peritoneal Macrophages
EiWT and mutant strains were observed in phagosome/phagolysosome and cytoplasm of peritoneal macrophages by light microscopy (Figure 4A). The intensity of bacterial bioluminescence in catfish macrophages did not differ among the treatment groups at 0 h (Figure 4B). However, the luminescence of bacteria increased in all treatments at 6 h post-treatment. The intensity of luminescence from EiΔhcp2 was significantly lower than that of EiΔevpC at this time point (Figure 4B). Interestingly, bacterial luminescence decreased in all groups at 12 h post-treatment, and the luminescence of EiΔhcp2 was lower significantly compared to EiΔevpC at this time point. However, there were no significant differences in the intensity of luminescence between EiΔhcp2 and EiWT and EiΔevpCΔhcp2 at both 6 and 12 h post-treatment (Figure 4B). After 24 h, bacterial luminescence decreased in all treatment groups, and there were no significant differences between the treatments (Figure 4B). Our results suggest that EiWT and Hcp mutants are capable of surviving and replicating in catfish peritoneal macrophages up to 6 h post-treatment. However, peritoneal macrophages efficiently killed EiWT and Hcp mutant strains after 24 h of in vitro infection (Figure 4C).
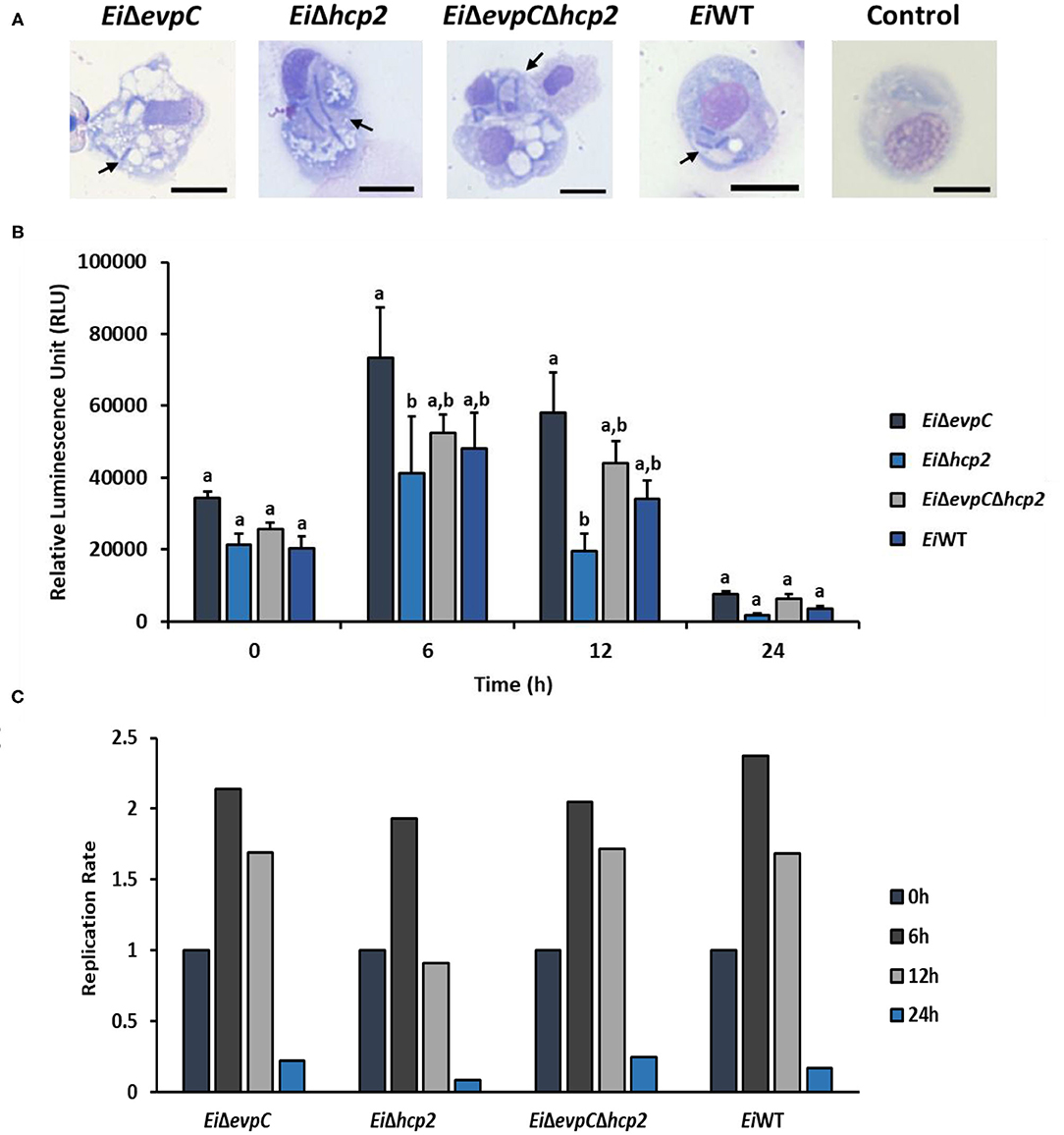
Figure 4. Bacterial killing assay of EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2, and EiWT. (A) Active uptake of mutants and EiWT in catfish peritoneal macrophages at 4 h post-treatment (100X). Black arrows mark engulfed bacteria in the cytoplasm and phagosomes and/or phagolysosomes of peritoneal macrophages. Scale bar = 10 μm. (B) The bar graph indicates mean relative luminescence unit (RLU) of four biological replicas obtained by Cytation 5 at an exposure time of 1 min. The graph represents the mean bioluminescence of each treatment ± SD. Letters above bars show the significant differences between treatments (p < 0.05). (C) Replication rate of mutants and EiWT in catfish peritoneal macrophages, which is calculated from RLU data in B by dividing RLU at 6, 12, and 24 h to RLU at 0 h.
Attachment and Invasion Capabilities of the Mutants in CCO Cells
CCO cell line was used to assess the attachment and invasion capabilities of EiWT and mutants, EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 (Figure 5A). The attachment ability of EiΔhcp2 and EiΔevpCΔhcp2 significantly declined compared to EiWT (p < 0.05; Figure 5B). However, no significant differences were recorded between EiΔevpC and EiWT (p < 0.05). In addition to the attachment, invasion of all mutants was reduced, but there were no significant differences compared to EiWT (p < 0.05; Figure 5C). These results indicate that Hcp mutants resulted in low attachment and invasion capabilities.
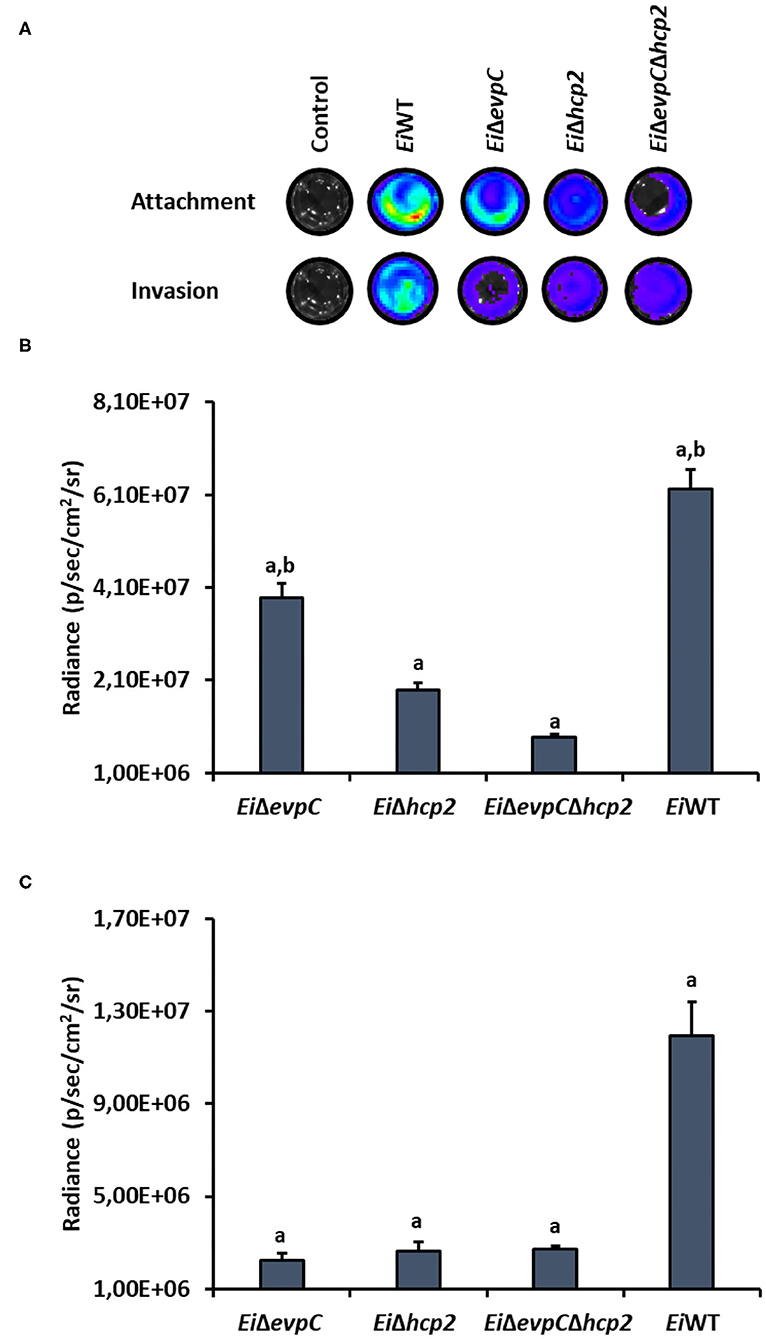
Figure 5. Attachment and invasion of bioluminescent EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2, and EiWT in CCO cells. (A) Attachment and invasion of E. ictaluri strains in CCO cells. Total photon emissions from the CCO cells were collected by IVIS Lumina XRMS In Vivo Imaging System Series III at an exposure time of 1 min. (B) Attachment: The bar graph indicates the mean photon emissions from four biological replicates after 1 h post-infection. Letters show the significant differences between treatments (p < 0.05). (C) Invasion: The bar graph indicates the mean photon emissions from four biological replicates after 1 h incubation in media containing gentamycin.
Survival and Stress Resistance of the Mutants in BHI and MM19-P
The survival and resistance of the EiWT and mutants to nitrite oxide and hydrogen peroxide were evaluated. The exposure of mutants and EiWT to SNP and H2O2 in BHI and MM19-P showed a variation in the growth rate of bacteria (Figures 6A,B). Their resistance was increased in MM19-P compared to BHI up to 12 h (Figures 6C,E). Due to the low pH (5.5) in MM19-P, the resistance of mutants and EiWT was enhanced in 0 and 4 h. The mutants and EiWT strains grew exponentially up to 24 h in BHI whereas their growth was restricted in MM19-P at 24 h (Figures 6D,F). EiΔevpCΔhcp2 double mutant had more resistance to SNP and H2O2 stress in BHI and MM19-P up to 12 h. However, EiΔevpC and EiΔhcp2 showed a similar growth rate in BHI and MM19-P.
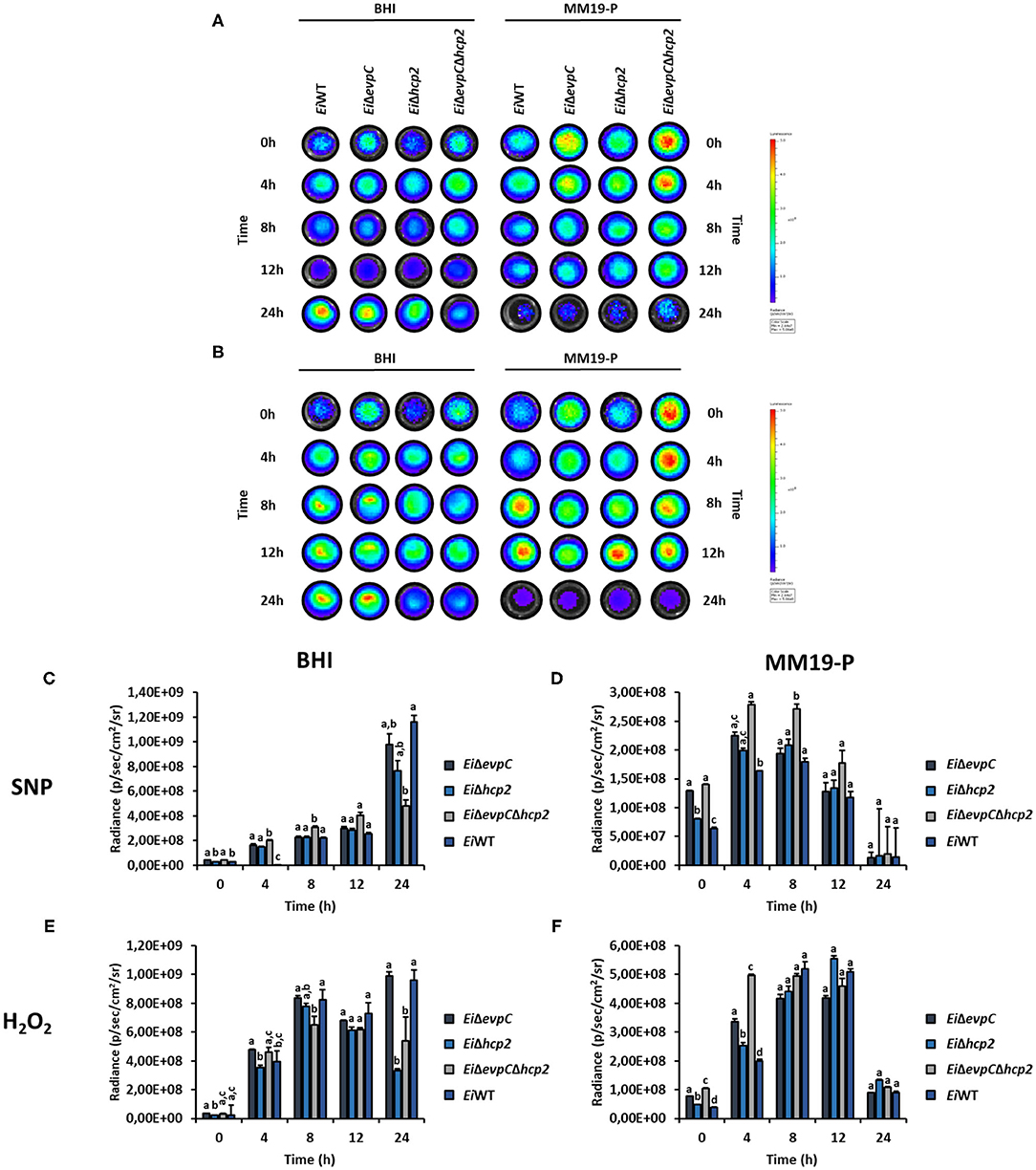
Figure 6. Survival and stress resistance of EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2, and EiWT in BHI and MM19. Bioluminescent imaging of mutants and EiWT exposed to H2O2 (A) and SNP (B) in BHI and MM19 for 24 h. The picture represents one of three biological replicates. Total photon emissions from E. ictaluri under stress were collected by IVIS Lumina XRMS In Vivo Imaging System Series III at an exposure time of 1 min. (C,D) Total photon counts obtained from bioluminescent mutants and EiWT exposed to SNP in BHI and MM19 for 24 h. (E,F) Total photon counts obtained from bioluminescent mutants and EiWT exposed to H2O2 in BHI and MM19 for 24 h. Data represent the mean of three biological replicates ± SD. Letters show the significant differences between treatments at each time point (p < 0.05).
Assessment of Virulence and Efficacy of the Mutants in Catfish
The immersion challenge was used to evaluate mutants' virulence and efficacy in catfish fingerlings and fry (Figure 7). EiΔevpC (0% mortality) and EiΔevpCΔhcp2 (0% mortality) were completely attenuated in catfish fingerlings in comparison to EiWT (67.53% mortality) (p < 0.05). Interestingly, EiΔhcp2 caused a severe and rapid death (93.94% mortality) in catfish fingerlings (Figure 7A). The protection of EiΔevpC (0 % mortality) and EiΔevpCΔhcp2 (0% mortality) were significantly better than EiWT (Figure 7B). In catfish fry challenge, virulence of EiΔevpC (18.72% mortality) and EiΔevpCΔhcp2 (35.90% mortality) decreased significantly in comparison to EiWT (100% mortality) (Figure 7C; p < 0.05). These mutants showed protection in fry compared to the sham-vaccinated group (Figure 7D). The protection of EiΔevpC (33.93% mortality) was better than that of EiΔevpCΔhcp2 (58.42% mortality).
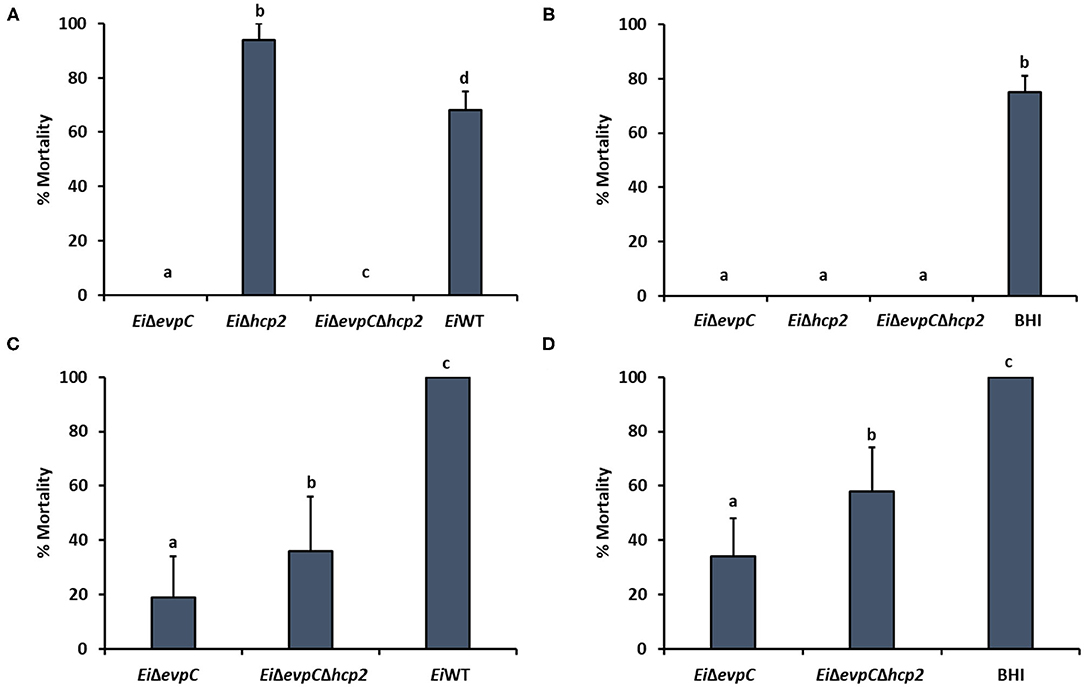
Figure 7. Virulence and vaccine efficacy of EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2, and EiWT in catfish. Percent mortality in virulence (A) and efficacy (B) of EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2 mutants, and EiWT in catfish fingerlings. Fish mortalities were recorded for 21 days. Percent mortality in virulence (C) and efficacy (D) of mutants and EiWT in catfish fry. Fish mortalities were recorded for 21 days.
Discussion
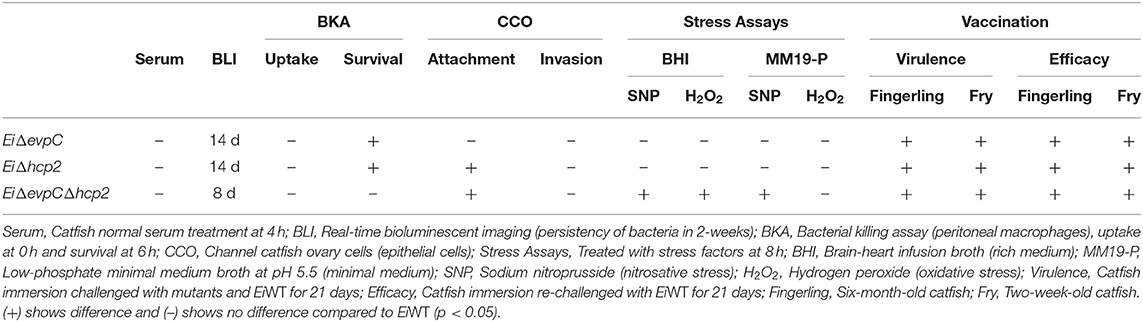
This research aimed to determine the potential roles of hcp1 (evpC) and hcp2 of T6SS in E. ictaluri virulence in catfish. To achieve this, EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 mutants were constructed and persistence in catfish, survival and replication inside catfish peritoneal macrophages, attachment and invasion capabilities in catfish epithelial cells, adaptation and survival to stress factors, and virulence and efficacy in catfish were investigated (Table 3).
Almost all strains of E. ictaluri show beta-type homolysis and hemolytic zone is narrow (36). Hemolytic activity can vary between strains and there is no clear correlation between hemolytic activity and virulence (37, 38). Our study indicated that hemolytic activities of EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 were similar to EiWT, and deletion of evpC and hcp2 genes did not have any effect on E. ictaluri hemolytic activity. Hemolysin co-regulated family proteins (Hcp) are involved in adhesion and invasion, intracellular survival of bacteria, bacterial cytotoxicity, and virulence (16).
Edwardsiella ictaluri can evade the complement system in catfish serum and establish a systemic infection. Edwardsiella ictaluri can differentially regulate its proteins in catfish serum (39). Our study revealed that EiΔevpC, EiΔhcp2, and EiΔevpCΔhcp2 were resistant to complement killing in catfish blood. Mutation in evpC, hcp2, and evpC-hcp2 did not affect E. ictaluri's resistance to complement killing, which indicates that Hcp family proteins of T6SS are not essential for E. ictaluri to survive in catfish serum.
The real-time bioluminescent imaging is a quantification method that allows detection E. ictaluri infection and persistence of mutants in catfish (40, 41). The bioluminescence from EiΔevpC (7.5 × 104 photons−1 cm−2 steradian−1) and EiΔevpCΔhcp2 (3.6 × 104 photons−1 cm−2 steradian−1) was reached the peak at 48 h post-infection, after which bacterial clearance from catfish was observed. However, bioluminescence from EiΔhcp2 (1.4 × 105 photons−1 cm−2 steradian−1) and EiWT (1.6 × 105 photons−1 cm−2 steradian−1) was gradually increased, even after 48 h post-infection, until the fish dies. Our result showed that EiΔevpC and EiΔevpCΔhcp2 had no mortalities for 14 days, although the bioluminescence quantity of EiΔevpC and EiΔevpCΔhcp2 started to increase earlier than EiΔhcp2 and EiWT. Our bioluminescent imaging data were corroborated with our virulence and efficacy study showing that persistence and replication of EiΔevpC and EiΔevpCΔhcp2 in catfish up to 48 h post-infection and decrease afterward may stimulate catfish an immune response, hence the survival of catfish.
Hcp family proteins are secreted inside host macrophages and required for intracellular survival in host macrophages (42, 43). Lack of a functional hcp reduced survival of Burkholderia pseudomallei in macrophages (44, 45). In E. tarda, the deletion of evpC caused a lower replication rate in gourami phagocytes (46). In this study, we found that the mutation in hcp2 displayed a lower replication rate for the intracellular growth of E. ictaluri inside the catfish peritoneal macrophages. The numbers of macrophages with intracellular EiΔevpC, EiΔhcp2, EiΔevpCΔhcp2, and EiWT bacteria were similar at 0 and 24 h post-infection. At 6 and 12 h post-infection, the number of cells with intracellular EiΔevpC and EiWT were similar, but intracellular replication of EiΔhcp2 in macrophages was significantly impaired. This result suggests that hcp2 may be necessary for E. ictaluri replication within catfish macrophages. This may depend on lack of hcp2 or a hcp2-dependent effector protein, which warrant further investigation.
Hcp family proteins are involved in adherence and invasion of the host epithelial tissues. Disruption in hcp genes could cause different results in Hcp-mediated cell adhesion and invasion activity of pathogenic bacteria. Hcp mutants displayed reduced adhesion and invasion of epithelial cells in Campylobacter jejuni, E. coli, and Vibrio parahaemolyticus (47–49). However, the deletion of hcp caused increased adhesion and invasion of MODE-K cell line in Helicobacter hepaticus (50). We demonstrated that evpC and hcp2 mutants' adhesion capabilities were dissimilar while their invasion capabilities were similar in CCO cells. Mutation in evpC did not decrease adherence to CCO cells while a mutation in hcp2 and both in evpC and hcp2 did. This suggests that hcp2 is required for epithelial cell attachment of E. ictaluri whereas both evpC and hcp2 are not essential for epithelial cell invasion of E. ictaluri.
T6SS facilitates the uptake of important metals under stress conditions by releasing proteinaceous metallophores into the host environment (51). The role of T6SS in manganese scavenging under oxidative stress has been revealed in Burkholderia thailandensis (52). Intracellular compartmentalization of Salmonella typhimurium inside macrophages initiates stress conditions, including nitrosative and oxidative stress, to suppress the replication of bacteria (53). T6SS effectors are involved in bacterial survival in oxidative stress (54, 55). In E. ictaluri and E. piscicida, the T6SS effector EvpP enhanced resistance to oxidative stress (56, 57). To investigate the role of Hcp family proteins of T6SS in E. ictaluri stress resistance, we applied nitrosative and oxidative stress with SNP and H2O2 in BHI and in MM19-P to imitate stressful phagosome conditions. Our results indicated that mutants and EiWT were able to grow in nutrient rich media, but SNP and H2O2 stress reduced survival of EiΔhcp2 and EiΔevpCΔhcp2 at 24 h. It seems that hcp2 is more critical for E. ictaluri to cope with SNP and H2O2 stress in presence of nutrients. In nutrient restricted media, mutants and EiWT were able to grow up to 4 h, but SNP and H2O2 stress suppressed EiWT growth more than mutants, which may indicate both evpC and hcp2 are not critical in phagosome conditions.
In vivo and in vitro infection models indicated that Hcp family proteins were associated with bacterial virulence and host colonization. In Aeromonas hydrophila, E. coli, and B. pseudomallei, hcp is required for virulence because hcp mutants were less virulent than wild-type (58, 59). Additionally, evpC was essential for the virulence of E. tarda (18). Here, we showed that evpC contributed to the pathogenicity of E. ictaluri in catfish. Vaccination of catfish fingerlings with EiΔevpC and EiΔevpCΔhcp2 provided complete protection against ESC in catfish fingerlings. However, EiΔhcp2 showed a hypervirulent phenotype causing higher mortality with severe symptoms in catfish fingerlings and was not tested in catfish fry. The mortality rates of EiΔevpC and EiΔevpCΔhcp2 in catfish fry immersion challenge indicated that EiΔevpC showed significantly less mortality and better protection compared to EiΔevpCΔhcp2.
Although evpC is located in the T6SS operon of E. ictaluri, hcp2 is founded ~67 kilobases (kb) away from evpC (Figure 8). It is possible that putative hcp2 might be an effector protein in E. ictaluri. It is worth noting that the protein sequence alignment of evpC and hcp2 had no significant match (data not shown). Thus, these two proteins classified in the Hcp protein family may have a different role in E. ictaluri.
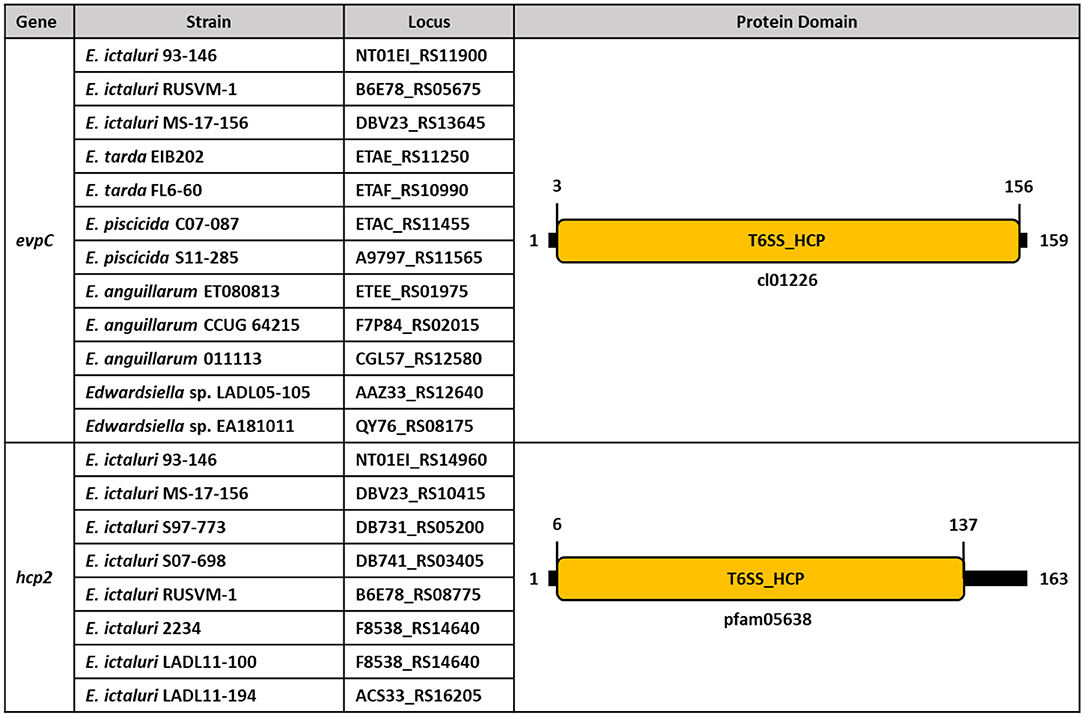
Figure 8. Conserved protein domains and families of evpC and hcp2 in Edwardsiella species. Schematic representation of putative hcp1 (evpC) and hcp2 of Hcp family proteins in E. ictaluri. Protein domains and families were identified by using NCBI CDD and colored. Protein length is represented by black bars, and numbers indicate protein and domain sizes. Protein family and domain names are inside and below the rectangles.
In conclusion, the two Hcp family proteins found in the E. ictaluri genome seems to have diverse roles in E. ictaluri pathogenesis. hcp2 is important in adherence to epithelial cells and replication within macrophages. However, evpC plays a crucial role in E. ictaluri virulence in catfish. Therefore, secretion of potential evpC and hcp2 dependent effector proteins via T6SS need more investigation.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee at Mississippi State University.
Author Contributions
SK, AOK, LP, and AK conceived and designed the experiments. SK, HA, and AOK performed the experiments. SK wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This project was supported by Agriculture and Food Research Initiative competitive Grant No. 2016-67015-24909 from the USDA National Institute of Food and Agriculture. Bioluminescence imaging was supported by USDA-ARS Biophotonics Initiative #58-6402-3-018. SK was supported by a fellowship from the Ministry of National Education of Turkey.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Laboratory Animal Resources and Care at the College of Veterinary Medicine for providing the SPF channel catfish and Dr. Larry A. Hanson for providing channel catfish ovary cell lines.
References
1. Hawke JP, Mcwhorter AC, Steigerwalt AG, Brenner DJ. Edwardsiella ictaluri sp. nov., the causative agent of enteric septicemia of catfish. Int Systemat Evol Microbiol. (1981) 31:396–400. doi: 10.1099/00207713-31-4-396
2. Hawke JP, Kent M, Rogge M, Baumgartner W, Wiles J, Shelley J, et al. Edwardsiellosis caused by Edwardsiella ictaluri in laboratory populations of zebrafish danio rerio. J Aquat Anim Health. (2013) 25:171–83. doi: 10.1080/08997659.2013.782226
3. Plumb JA, Sanchez DJ. Susceptibility of five species of fish to Edwardsiella ictaluri. J Fish Dis. (1983) 6:261–266. doi: 10.1111/j.1365-2761.1983.tb00075.x
4. Soto E, Griffin M, Arauz M, Riofrio A, Martinez A, Cabrejos ME. Edwardsiella ictaluri as the causative agent of mortality in cultured nile tilapia. J Aquat Anim Health. (2012) 24:81–90. doi: 10.1080/08997659.2012.675931
5. Miyazaki T, Plumb JA. Histopathology of Edwardsiella ictaluri in channel catfish, Ictalurus punctatus (Rafinesque). J Fish Dis. (1985) 8:389–92. doi: 10.1111/j.1365-2761.1985.tb00961.x
6. Shoemaker CA, Klesius PH, Plumb JA. Killing of Edwardsiella ictaluri by macrophages from channel catfish immune and susceptible to enteric septicemia of catfish. Vet Immunol Immunopathol. (1997) 58:181–90. doi: 10.1016/S0165-2427(97)00026-3
7. Booth NJ, Elkamel A, Thune RL. Intracellular replication of Edwardsiella ictaluri in channel catfish macrophages. J Aquat Anim Health. (2006) 18:101–8. doi: 10.1577/H05-025.1
8. Chen DX, Ainsworth AJ. Assessment of metabolic activation of channel catfish peripheral blood neutrophils. Dev Comp Immunol. (1991) 15:201–8. doi: 10.1016/0145-305X(91)90011-M
9. Yao J, Li C, Zhang J, Liu S, Feng J, Wang R, et al. Expression of nitric oxide synthase (NOS) genes in channel catfish is highly regulated and time dependent after bacterial challenges. Dev Comp Immunol. (2014) 45:74–86. doi: 10.1016/j.dci.2014.02.005
10. Yeh HY, Klesius PH. Changes of serum myeloperoxidase and nitric oxide in the early stage of Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus (Rafinesque). J Fish Dis. (2013) 36:441–6. doi: 10.1111/jfd.12038
11. Baumgartner WA, Dubytska L, Rogge ML, Mottram PJ, Thune RL. Modulation of vacuolar pH is required for replication of Edwardsiella ictaluri in channel catfish macrophages. Infect Immun. (2014) 82:2329–36. doi: 10.1128/IAI.01616-13
12. Booth NJ, Beekman JB, Thune RL. Edwardsiella ictaluri encodes an acid-activated urease that is required for intracellular replication in channel catfish (Ictalurus punctatus) macrophages. Appl Environ Microbiol. (2009) 75:6712–20. doi: 10.1128/AEM.01670-09
13. Rogge ML, Thune RL. Regulation of the Edwardsiella ictaluri type III secretion system by pH and phosphate concentration through EsrA, EsrB, and EsrC. Appl Environ Microbiol. (2011) 77:4293–302. doi: 10.1128/AEM.00195-11
14. Dubytska LP, Rogge ML, Thune RL. Identification and characterization of putative translocated effector proteins of the Edwardsiella ictaluri type III secretion system. mSphere. (2016) 1:1–14. doi: 10.1128/mSphere.00039-16
15. Zhao LJ, Lu JF, Nie P, Li AH, Xiong BX, Xie HX. Roles of plasmid-encoded proteins, EseH, EseI and EscD in invasion, replication and virulence of Edwardsiella ictaluri. Vet Microbiol. (2013) 166:233–41. doi: 10.1016/j.vetmic.2013.05.023
16. Hachani A, Wood TE, Filloux A. Type VI secretion and anti-host effectors. Curr Opin Microbiol. (2016) 29:81–93. doi: 10.1016/j.mib.2015.11.006
17. Moore MM, Fernandez DL, Thune RL. Cloning and characterization of Edwardsiella ictaluri proteins expressed and recognized by the channel catfish Ictalurus punctatus immune response during infection. Dis Aquat Organ. (2002) 52:93–107. doi: 10.3354/dao052093
18. Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. (2007) 66:1192–206. doi: 10.1111/j.1365-2958.2007.05993.x
19. Jobichen C, Chakraborty S, Li M, Zheng J, Joseph L, Mok YK, et al. Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS ONE. (2010) 5:e12910. doi: 10.1371/journal.pone.0012910
20. Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. (2007) 104:15508–13. doi: 10.1073/pnas.0706532104
21. Hu W, Anand G, Sivaraman J, Leung KY, Mok YK. A disordered region in the EvpP protein from the type VI secretion system of Edwardsiella tarda is essential for EvpC binding. PLoS ONE. (2014) 9:e110810. doi: 10.1371/journal.pone.0110810
22. Chen H, Yang D, Han F, Tan J, Zhang L, Xiao J, et al. The bacterial T6SS effector EvpP prevents NLRP3 inflammasome activation by inhibiting the Ca2+-dependent MAPK-Jnk pathway. Cell Host Microbe. (2017) 21:47–58. doi: 10.1016/j.chom.2016.12.004
23. Lawrence ML, Cooper RK, Thune RL. Attenuation, persistence, and vaccine potential of an Edwardsiella ictaluri purA mutant. Infect Immun. (1997) 65:4642–51. doi: 10.1128/IAI.65.11.4642-4651.1997
24. Herrero M, de Lorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J Bacteriol. (1990) 172:6557–67. doi: 10.1128/JB.172.11.6557-6567.1990
25. Metcalf WW, Jiang W, Wanner BL. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene. (1994) 138:1–7. doi: 10.1016/0378-1119(94)90776-5
26. Taylor RG, Walker DC, McInnes RR. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. (1993) 21:1677–78. doi: 10.1093/nar/21.7.1677
27. Dozois CM, Daigle F, Curtiss R III. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA. (2003) 100:247–52. doi: 10.1073/pnas.232686799
28. Karsi A, Lawrence ML. Broad host range fluorescence and bioluminescence expression vectors for Gram-negative bacteria. Plasmid. (2007) 57:286–95. doi: 10.1016/j.plasmid.2006.11.002
29. Williams ML, Gillaspy AF, Dyer DW, Thune RL, Waldbieser GC, Schuster SC, et al. Genome sequence of Edwardsiella ictaluri 93-146, a strain associated with a natural channel catfish outbreak of enteric septicemia of catfish. J Bacteriol. (2012) 194:740–1. doi: 10.1128/JB.06522-11
30. Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. (1989) 77:61–8. doi: 10.1016/0378-1119(89)90359-4
31. Kordon AO, Abdelhamed H, Ahmed H, Park JY, Karsi A, Pinchuk LM. Phagocytic and bactericidal properties of channel catfish peritoneal macrophages exposed to Edwardsiella ictaluri live attenuated vaccine and wild-type strains. Front Microbiol. (2018) 8:2638. doi: 10.3389/fmicb.2017.02638
32. Russo R, Shoemaker CA, Panangala VS, Klesius PH. In vitro and in vivo interaction of macrophages from vaccinated and non-vaccinated channel catfish (Ictalurus punctatus) to Edwardsiella ictaluri. Fish Shellfish Immunol. (2009) 26:543–52. doi: 10.1016/j.fsi.2009.02.011
33. Santander J, Martin T, Loh A, Pohlenz C, GatlinDM III, Curtiss R III. Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology. (2013) 159:1471–86. doi: 10.1099/mic.0.066639-0
34. Collins LA, Thune RL. Development of a defined minimal medium for the growth of Edwardsiella ictaluri. Appl Environ Microbiol. (1996) 62:848–52. doi: 10.1128/AEM.62.3.848-852.1996
35. Karsi A, Gulsoy N, Corb E, Dumpala PR, Lawrence ML. High-throughput bioluminescence-based mutant screening strategy for identification of bacterial virulence genes. Appl Environ Microbiol. (2009) 75:2166–75. doi: 10.1128/AEM.02449-08
36. Waltman WD, Shotts EB, Hsu TC. Biochemical characteristics of Edwardsiella ictaluri. Appl Environ Microbiol. (1986) 51:101–4. doi: 10.1128/AEM.51.1.101-104.1986
37. Stanley LA, Hudson JS, Schwedler RE, Hayasaka SS. Extracellular products associated with virulent and avirulent strains of Edwardsiella ictaluri from channel catfish. J Aquat Anim Health. (1994) 6:36–43. doi: 10.1577/1548-8667(1994)006<0036:EPAWVA>2.3.CO;2
38. Williams ML, Lawrence ML. Identification and characterization of a two-component hemolysin from Edwardsiella ictaluri. Vet Microbiol. (2005) 108:281–9. doi: 10.1016/j.vetmic.2005.04.017
39. Dumpala PR, Lawrence ML, Karsi A. Identification of differentially regulated Edwardsiella ictaluri proteins during catfish serum treatment. J Aquat Anim Health. (2018) 30:50–6. doi: 10.1002/aah.10007
40. Dahal N, Abdelhamed H, Lu J, Karsi A, Lawrence ML. Tricarboxylic acid cycle and one-carbon metabolism pathways are important in Edwardsiella ictaluri virulence. PLoS ONE. (2013) 8:e65973. doi: 10.1371/journal.pone.0065973
41. Karsi A, Menanteau-Ledouble S, Lawrence ML. Development of bioluminescent Edwardsiella ictaluri for noninvasive disease monitoring. FEMS Microbiol Lett. (2006) 260:216–23. doi: 10.1111/j.1574-6968.2006.00310.x
42. Burtnick MN, DeShazer D, Nair V, Gherardini FC, Brett PJ. Burkholderia mallei cluster 1 type VI secretion mutants exhibit growth and actin polymerization defects in RAW 264.7 murine macrophages. Infect Immun. (2010) 78:88–99. doi: 10.1128/IAI.00985-09
43. Peng Y, Wang X, Shou J, Zong B, Zhang Y, Tan J, et al. Roles of Hcp family proteins in the pathogenesis of the porcine extraintestinal pathogenic Escherichia coli type VI secretion system. Sci Rep. (2016) 6:26816. doi: 10.1038/srep26816
44. Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, et al. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun. (2011) 79:1512–25. doi: 10.1128/IAI.01218-10
45. Hopf V, Gohler A, Eske-Pogodda K, Bast A, Steinmetz I, Breitbach K. BPSS1504, a cluster 1 type VI secretion gene, is involved in intracellular survival and virulence of Burkholderia pseudomallei. Infect Immun. (2014) 82:2006–15. doi: 10.1128/IAI.01544-14
46. Srinivasa Rao PS, Yamada Y, Tan YP, Leung KY. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol. (2004) 53:573–86. doi: 10.1111/j.1365-2958.2004.04123.x
47. Lertpiriyapong K, Gamazon ER, Feng Y, Park DS, Pang J, Botka G, et al. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS ONE. (2012) 7:e42842. doi: 10.1371/journal.pone.0042842
48. Yu Y, Yang H, Li J, Zhang P, Wu B, Zhu B, et al. Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch Microbiol. (2012) 194:827–35. doi: 10.1007/s00203-012-0816-z
49. Zhou Y, Tao J, Yu H, Ni J, Zeng L, Teng Q, et al. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect Immun. (2012) 80:1243–51. doi: 10.1128/IAI.05994-11
50. Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. (2010) 7:265–76. doi: 10.1016/j.chom.2010.03.004
51. Chen WJ, Kuo TY, Hsieh FC, Chen PY, Wang CS, Shih YL, et al. Involvement of type VI secretion system in secretion of iron chelator pyoverdine in Pseudomonas taiwanensis. Sci Rep. (2016) 6:32950. doi: 10.1038/srep32950
52. Si M, Zhao C, Burkinshaw B, Zhang B, Wei D, Wang Y, et al. Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc Natl Acad Sci USA. (2017) 114:E2233–42. doi: 10.1073/pnas.1614902114
53. Burton NA, Schurmann N, Casse O, Steeb AK, Claudi B, Zankl J, et al. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe. (2014) 15:72–83. doi: 10.1016/j.chom.2013.12.006
54. Wan B, Zhang Q, Ni J, Li S, Wen D, Li J, et al. Type VI secretion system contributes to enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS). PLoS Pathog. (2017) 13:e1006246. doi: 10.1371/journal.ppat.1006246
55. Wang T, Si M, Song Y, Zhu W, Gao F, Wang Y, et al. Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. (2015) 11:e1005020. doi: 10.1371/journal.ppat.1005020
56. Kalindamar S, Kordon AO, Abdelhamed H, Tan W, Pinchuk LM, Karsi A. Edwardsiella ictaluri evpP is required for colonisation of channel catfish ovary cells and necrosis in anterior kidney macrophages. Cell Microbiol. (2020) 22:e13135. doi: 10.1111/cmi.13135
57. Qin L, Wang X, Gao Y, Bi K, Wang W. Roles of EvpP in Edwardsiella piscicida-macrophage interactions. Front Cell Infect Microbiol. (2020) 10:53. doi: 10.3389/fcimb.2020.00053
58. de Pace F, Nakazato G, Pacheco A, de Paiva JB, Sperandio V, da Silveira W. D. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect Immun. (2010) 78:4990–8. doi: 10.1128/IAI.00531-10
Keywords: T6SS, EvpC, Hcp, macrophages, CCO cells, virulence, Ictalurus punctatus
Citation: Kalindamar S, Abdelhamed H, Kordon AO, Pinchuk LM and Karsi A (2021) Hemolysin Co-regulated Family Proteins Hcp1 and Hcp2 Contribute to Edwardsiella ictaluri Pathogenesis. Front. Vet. Sci. 8:681609. doi: 10.3389/fvets.2021.681609
Received: 20 March 2021; Accepted: 07 May 2021;
Published: 02 June 2021.
Edited by:
Changyong Cheng, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Javier Santander, Memorial University of Newfoundland, CanadaLingzhi Zhang, East China University of Science and Technology, China
Copyright © 2021 Kalindamar, Abdelhamed, Kordon, Pinchuk and Karsi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Attila Karsi, a2Fyc2lAY3ZtLm1zc3RhdGUuZWR1
 Safak Kalindamar
Safak Kalindamar Hossam Abdelhamed
Hossam Abdelhamed Adef O. Kordon
Adef O. Kordon Lesya M. Pinchuk
Lesya M. Pinchuk Attila Karsi
Attila Karsi