- Hubei Key Laboratory of Animal Nutrition and Feed Science, School of Animal Science and Nutrition Engineering, Wuhan Polytechnic University, Wuhan, China
Preventative effects of Lactobacillus fermentum and Bacillus coagulans against Clostridium perfringens infection in broilers have been well-demonstrated. The present study was conducted to investigate the modulation of these two probiotics on intestinal immunity and microbiota of C. perfringens-challenged birds. The 336 one-day-old broilers were assigned to four groups with six replicates in each group. Birds in the control were unchallenged and fed a basal diet, and birds in the three challenged groups were dietary supplemented with nothing (Cp group), 1 × 109 CFU/kg of L. fermentum (Lf_Cp group), or 1 × 1010 CFU/kg of B. coagulans (Bc_Cp group). Challenge was performed from days 14 to 20, and samples were collected on days 21 and 28. Challenge upregulated interleukin (IL)-1β and transforming growth factor (TGF)-β4 mRNA expression in jejunum on day 21, which was downregulated by B. coagulans and L. fermentum, respectively (P < 0.05). Both probiotic groups upregulated jejunal IL-1β, interferon (IFN)-γ, IL-17, and TGF-β4 on day 28 as well as IFN-γ on day 21 (P < 0.05). The Bc_Cp group increased CD3+ T cell counts in the jejunal crypt on day 21 (P < 0.05). Challenge decreased the ileal ACE index on day 21 and cecal microbial richness on day 28, which were increased by probiotic treatments, and ileal bacterial richness decreased in the Bc_Cp group on day 28 (P < 0.05). Only ileal microbiota on day 21 was distinctly affected with an R-value at 0.3116 by ANOSIM analysis (P < 0.05). Compared with the control, ileal Firmicutes increased on day 21, and ileal Bacteroidetes and cecal Proteobacteria decreased on day 28 in challenged groups (P < 0.05). Challenge increased Romboutsia spp. in the ileum as well as unclassified f_Lachnospiraceae and Ruminococcus_torques group in the cecum, and decreased Lactobacillus spp. in the ileum on day 21, which were all conversely modulated by L. fermentum (P < 0.05). Challenge increased amino acid metabolism of ileal microbiota and membrane transport of cecal microbiota, and decreased amino acid metabolism of cecal microbiota on day 21, which were conversely regulated by both probiotics (P < 0.05). In conclusion, L. fermentum and B. coagulans attenuated the intestinal inflammation and microbial dysbiosis soon after C. perfringens challenge.
Introduction
Necrotic enteritis (NE) is one of the most important enteric diseases in poultry and causes huge economic losses to the industry worldwide (1). Clinical NE is characterized by diarrhea, intestinal necrotic lesions, severe morbidity and mortality, and lower growth rate and feed efficiency, while the subclinical form presents as poor growth performance without mortality (2, 3). Clostridium perfringens, the etiological agent of NE, is a spore-forming Gram-positive bacterium and commonly exists in environments such as soil, sewage, litter, feces of chickens, and the intestines of humans and animals (3). The C. perfringens population is normally < 102 to 104 colony-forming units (CFU) per gram of intestinal contents in the small intestine of healthy chickens compared to 107 to 109 CFU/g in NE birds (4).
Traditional NE mitigation strategies included antibiotic growth promoters and ionophores. With the increasing pressure to reduce antibiotic use in poultry production, the incidence of NE has been increasing in recent years (5) and alternative nutritional strategies are in urgent need. Probiotics, such as lactic acid bacteria, Bacillus, Bifidobacteria, and Enterococcus, are potentially alternative strategies to prevent or mitigate NE in birds (6). Probiotics exhibit protective effects by producing antimicrobial compounds, competitive excluding, enhancing the intestinal barrier, and stimulating host immunity (7–9). It has been reported that daily oral administration of Lactobacillus fermentum 1.2029 reduced the severity of NE lesions and ameliorated inflammation by upregulating interleukin (IL)-10 mRNA expression and downregulating interferon (IFN)-γ and toll-like receptor (TLR) 2 expression in the ileum of C. perfringens-challenged broiler chickens (10). Dietary supplementation with Bacillus coagulans decreased gut lesion scores as well as cecal population and hepatic translocation of C. perfringens, and increased mRNA expression of fowlicidin-2, an antimicrobial peptide, in the jejunum of NE birds (11).
In recent years, high-throughput sequencing technologies have been used to analyze the bacterial communities in different intestinal segments of chickens (12). Using this technology, Li et al. (13) reported that dietary addition with L. acidophilus decreased the Shannon index of ileal microbiota and restored the intestinal microbial community in C. perfringens-challenged broiler chickens by increasing relative abundance of beneficial bacteria such as Lactobacillus and decreasing abundance of pathogens such as Escherichia-Shigella. The NE birds' diet supplemented with Bacillus subtilis DSM 32315 had an intestinal microbiota with diversity and composition more similar to that of healthy counterparts reflected by the reduction of opportunistic pathogens and protein-fermentative bacteria (14, 15). However, the effects of L. fermentum and B. coagulans on the intestinal microbial profiles of NE birds are largely unknown. Therefore, the present study was conducted to investigate the influence of L. fermentum and B. coagulans on the intestinal immunity and microbiota of broiler chickens infected with C. perfringens.
Materials and Methods
Experimental Design and Bird Management
All animal procedures used in the present study were performed in compliance with Hubei Provincial Regulations for Laboratory Animals (011043145-029-2013-000009), and were approved by the Institutional Animal Care and Use Committee of Wuhan Polytechnic University. A total of 336 one-day-old sex-mixed broiler chicks were randomly assigned to four groups, each with six replicates of 14 birds. Birds in the control group were fed a corn-soybean meal basal diet and unchallenged by C. perfringens. Birds infected with C. perfringens were considered as positive control (Cp group). Challenged birds in the Lf_Cp and Bc_Cp groups were fed basal diets supplemented with 1 × 109 CFU/kg of L. fermentum and 1 × 1010 CFU/kg of B. coagulans, respectively. The L. fermentum (CGMCC No. 1.2029) was obtained from China General Microbiological Culture Collection Center (CGMCC, Beijing, China) and cultured in Mann-Rogosa-Sharpe broth. The overnight culture of L. fermentum was absorbed by nano silicon dioxide and probiotic concentration was 1 × 109 CFU/g of powder. The product of B. coagulans (1 × 1010 CFU/g) was obtained from Hubei Horwath Biotechnology Co. Ltd. (Wuhan, China). Both L. fermentum and B. coagulans powder was mixed in the diet at a dose of 0.1%. The basal diet was formulated according to the nutritional requirements of the National Research Council (1994). The composition and nutrient levels of the basal diet are presented in Supplementary Table 1. The trial lasted for 28 days. Birds were raised in wire cages in an environmentally controlled room with 23 h of light and were allowed ad libitum access to water and mashed diets throughout the trial.
Clostridium perfringens Challenge
The avian C. perfringens type A field strain (CVCC2030) was obtained from China Veterinary Culture Collection Center (Beijing, China). The NE model was established as Du et al. (16) described with modification. The C. perfringens was cultured in cooked meat medium and incubated anaerobically overnight at 37°C. The organism was counted using tryptose-sulfite-cycloserine agar plates. From days 14 to 20, challenged birds were orally gavaged with 1 mL of actively growing culture of C. perfringens (1 × 108 CFU/mL) once per day. Birds in the control group were gavaged with equal volumes of sterile meat medium.
Sample Collection
On days 21 and 28, two birds per replicate (12 birds per group) were randomly selected and euthanized by cervical dislocation. Jejunal mucosa was scraped from about 10 cm of the jejunal segment and stored at −80°C for total RNA isolation. Approximately 1 cm of the mid-jejunum was sampled and immediately fixed in 4% paraformaldehyde for immunohistochemical analysis. Digesta from the ileum and cecum was collected and stored at −80°C for DNA extraction.
RNA Isolation and Quantitative Real-Time PCR
Total RNA of jejunal samples was isolated using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol. The concentration and purity of total RNA was quantified by measuring its optical density at 260 and 280 nm with a NanoDrop® ND-2000 UV-VIS spectrophotometer (Thermo Scientific, Wilmington, DE, USA). RNA integrity was verified by agarose gel electrophoresis. One microgram of total RNA was reverse transcribed by the PrimeScript® RT reagent Kit with gDNA Eraser [Takara Biotechnology (Dalian) Co., Ltd., Dalian, China] according to the manufacturer's instructions. The quantitative real-time PCR assay was performed with the 7500 fluorescence detection system (Applied Biosystems, Foster City, CA, USA) according to optimized PCR protocols using the SYBR Premix Ex TaqTM kit [Takara Biotechnology (Dalian) Co., Ltd.]. The primer pairs for the amplification of genes encoding IL-1β, INF-γ, IL-13, IL-17, and transforming growth factor (TGF)-β4 are presented in Supplementary Table 2. β-actin served as an endogenous reference gene. The PCR conditions were an initial denaturation step at 95°C for 30 s, 40 cycles at 95°C for 5 s, annealing and extension temperature at 60°C for 34 s. Each biological sample was run in triplicate. To confirm amplification specificity, the PCR products from each primer pair were subjected to a melting curve analysis and subsequent agarose gel electrophoresis. Gene expression was quantified using the comparative threshold cycle method and the data were expressed as the relative values to the control group.
Immunohistochemical Analysis
The number of CD3+ intraepithelial T cells in the jejunal villus and crypt was determined by immunohistochemical (IHC) analysis as Röhe et al. (17) described with modifications. Briefly, the fixed jejunal tissue was dehydrated and embedded with paraffin wax. The paraffin blocks were consecutively cut at 5 μm and the obtained sections were mounted on glass slides. The indirect immunohistochemical method was used for IHC analysis following the manufacturer's instructions (Boster Biological Technology Co., Ltd., Wuhan, China). After blocking, the jejunal sections were incubated with primary antibody (1:100 dilution) against CD3 T cell receptors overnight at 4°C using humidity chambers. A rat anti-human CD3 antibody, which cross-reacts with the chicken CD3 complex, was used as the primary antibody. Then, tissue sections were incubated with secondary antibody conjugated with horseradish peroxidase (HRP) and CD3+ T cells were visualized using a chromogenic dye. Additionally, slides were counterstained with hematoxylin. One of the images of stained slides is presented in Supplementary Figure 1. Finally, the CD3+ T cells were quantified using a light microscope (Olympus, Tokyo, Japan), which was equipped with a digital camera (Olympus, Tokyo, Japan) and an image analysis program (ProRes CapturePro software, Jenoptik, Jena, Germany). The data were presented as the number of CD3+ T cells per 1,000 μm2 in the villus and crypt regions, respectively.
DNA Extraction
Bacterial DNA from the ileal and cecal digesta was extracted using a QIAamp DNA Stool Mini Kit (Qiagen Inc., Valencia, CA, USA). The DNA concentration was measured with a NanoDrop® ND-2000 UV-VIS spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and the DNA quality was determined by agarose gel electrophoresis. Some samples were pooled together before DNA extraction. Therefore, eight and seven DNA samples in each group were enrolled for high-throughput sequencing on days 21 and 28, respectively.
Amplification and 16S rRNA Gene Sequencing
The DNA extracted from samples collected on day 21 was amplified and sequenced as Wu et al. (18) described. Briefly, the V3-V4 region of the 16S rRNA gene was amplified by two-stage PCR. The first stage of PCR was performed using the HiFi HotStart ReadyMix kit (Kapa Biosystems, Wilmington, MA, USA) with the forward primer (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and the reverse primer (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCACTACHVGGGTATCTAATCC-3′). The second stage of index PCR was processed using the Nexteta XT Index kit (Illumina, San Diego, CA, USA), in which the dual indices and Illumina sequencing adapters were attached. Then, PCR products were purified with the Agencourt AMPure XP purification system (Beckman, Brea, CA, USA) and quantified by the SpectraMax i3X Multi-mode detection platform (Molecular Devices, San Jose, CA, USA). The purified amplicons were sequenced using the Illumina MiSeq platform (San Diego, CA, USA).
Due to technical issues, the DNA amplification and bacterial 16S rRNA gene sequencing for samples collected on day 28 was performed by Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The V3-V4 variable region of 16S RNA gene was amplified with barcoded primer pair 338F/806R (338F: 5′-ACTCCTACGGGAGGCAGCAG-3′; 806R: GGACTACHVGGGTWTCTAAT-3′). The amplicons were then extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified by a QuantusTM Fluorometer (Promega Corporation, Madison, WI, USA). Purified amplicons were pooled in equimolar amounts and pair-end sequenced (2 × 300) on an Illumina MiSeq platform (San Diego, CA, USA).
Bioinformatics Analysis
Bioinformatics analysis was performed using the Majorbio Cloud Platform (http://www.majorbio.com). Raw sequencing reads were demultiplexed and quality-filtered by Trimmomatic software (version 0.36) and then merged by FLASH software (version 1.2.7). Operational taxonomic units (OTUs) were clustered with the 97% similarity cut-off using UPARSE software (version 7.1). The taxonomy of each OTU representative sequence was analyzed using the RDP Classifier (http://rdp.cme.msu.edu/) against the SILVA rRNA database (http://www.arb-silva.de) with a confidence threshold of 70%. After the elimination of interference sequence, a rarefaction curve and Venn diagram were created using R software (version 2.15.3). Analysis of alpha diversity was performed using Mothur software (version 1.30.1). Beta diversity was evaluated by the unweighted Unifrac principal co-ordinates analysis (PCoA) and the significance of bacterial separation among groups was reflected by ANOSIM analysis. Both analyses were conducted using R software. The top 10 differentially abundant bacteria at the genus level in the control vs. Cp group, Cp vs. Lf_Cp group, and Cp vs. Bc_Cp group were analyzed with Student's t-test and a P value threshold of 0.05 was used. PICRUSt analysis was conducted to predict the potential function of microbiota. The OTUs were normalized by copy number and metagenome prediction was categorized into the Kyoto Encyclopedia of Genes and Genomes (KEGG) at level 2 (19).
Statistical Analysis
The data of intestinal immune indices, bacterial alpha diversity, top 4 phyla, and top 5 predicted KEGG pathways were analyzed by one-way ANOVA using SPSS version 21.0 (SPSS Inc., Chicago, IL, USA). When significant differences were observed, individual group means were compared using Duncan's multiple comparison. If data did not comply with normal distribution, which was analyzed by a Kolmogorov-Smirnov test, a non-parameter test and pairwise comparisons were used to analyze the data. Data in figures are presented as mean and SE, while data in tables are given as mean and pooled SEM. P < 0.05 was considered statistically significant.
Results
Jejunal Cytokine Gene Expression
The mRNA expression of cytokines in the jejunum is revealed in Figure 1. At 21 days of age, compared with the control group, the Cp group upregulated the mRNA expression of IL-1β and TGF-β4 (Figure 1A, P < 0.05). However, compared with the Cp group, the Bc_Cp and Lf_Cp groups downregulated the IL-1β and TGF-β4 mRNA expression, respectively (P < 0.05). The gene expression of IL-13 and IL-17 was not significantly affected (P < 0.05). At 28 days of age, compared with the control and Cp groups, the Lf_Cp and Bc_Cp groups significantly upregulated the mRNA expression of IL-1β, INF-γ, IL-17, and TGF-β4 in the jejunum (Figure 1B, P < 0.05). The mRNA level of IL-13 was not influenced by challenge and dietary treatments (P > 0.05).
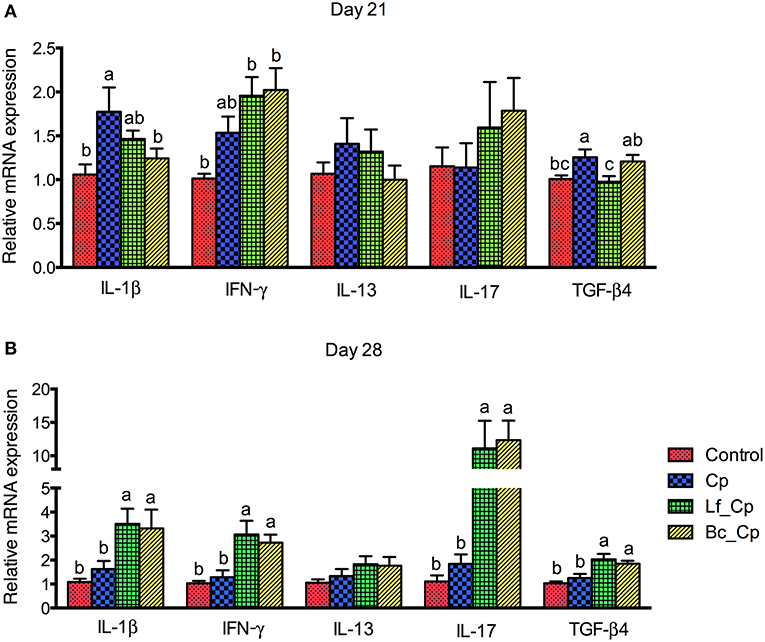
Figure 1. The cytokine mRNA expression in jejunal mucosa of Clostridium perfringens-challenged broiler chickens fed diets supplemented with Lactobacillus fermentum or Bacillus coagulans on days 21 (A) and 28 (B). Data were expressed as mean and SE from 12 chickens. Control, unchallenged group; Cp, C. perfringens-challenged group; Lf_Cp, challenged group with dietary supplementation of Lactobacillus fermentum; Bc_Cp, challenged group with dietary supplementation of Bacillus coagulans. IL, interleukin; IFN, interferon; TGF, transforming growth factor.
Jejunal CD3+ T Cell Counts in Jejunum
The CD3+ T cell counts in the villus and crypts of the jejunum are presented in Figure 2. At 21 days of age, the Lf_Cp group tended to increase the villus CD3+ T cell counts in contrast to the Cp group (Figure 2A, P = 0.059). The Bc_Cp group had higher crypt CD3+ T cell counts than that of the other three groups (P < 0.05), among which the cell count did not significantly differ (P > 0.05). At 28 days of age, the jejunal CD3+ cell counts were not significantly affected by treatments (Figure 2B, P > 0.05). The Lf_Cp group tended to have a higher CD3+ T cell number in the crypt of the jejunum than that of the Cp and Bc_Cp groups (P = 0.087).
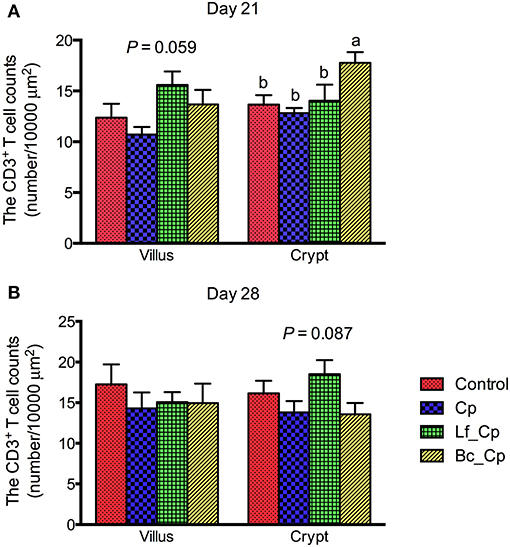
Figure 2. The CD3+ T cell counts in villus and crypt of jejunum of Clostridium perfringens-challenged broiler chickens fed diets supplemented with Lactobacillus fermentum or Bacillus coagulans on days 21 (A) and 28 (B). Data were expressed as mean and SE from 12 chickens. Control, unchallenged group; Cp, C. perfringens-challenged group; Lf_Cp, challenged group with dietary supplementation of Lactobacillus fermentum; Bc_Cp, challenged group with dietary supplementation of Bacillus coagulans.
Quality of Sequencing Data and Venn Diagram
The sequencing of ileal and cecal samples collected on days 21 and 28 generated a total of 3,215,602, 4,590,839, 1,482,180, and 1,624,227 effective sequences, respectively (Supplementary Tables 3, 4). The median read length for these samples was 451, 449, 433, and 433 base pairs, respectively. The Good's coverage (Supplementary Tables 3, 4) and rarefaction curves (Supplementary Figure 2) showed that sufficient sequencing coverage for all samples was achieved. All treatments shared 270 and 400 OTUs in ileal and cecal microbial communities at 21 days of age, respectively (Supplementary Figures 3A,C). At 28 days of age, there were 226 and 406 common OTUs in ileal and cecal microbiota of the four groups, respectively (Supplementary Figures 3B,D).
Alpha Diversity of Microbiota in Ileum and Cecum
As seen in Table 1, at 21 days of age, compared with the control group, the Cp group tended to decrease Sobs (P = 0.087) and Chao1 (P = 0.096) indices and significantly reduced the ACE index of ileal microbiota (P < 0.05). Meanwhile, the Bc_Cp group had comparable Sobs and ACE indices to the control. The alpha diversity of cecal microbiota was not significantly affected by treatments on day 21. At 28 days of age, the Bc_Cp group had lower richness indices (Sobs, Chao1, and ACE) than those of the other three groups in ileal microbiota (P < 0.05). Compared with the Cp group, the Lf_Cp group tended to increase the Shannon index (P = 0.055) and decrease the Simpson index of ileal microbiota (P = 0.087). In the cecum, the Cp group decreased in richness indices (Chao1 and ACE) in contrast to the control and Bc_Cp groups (P < 0.05). The Cp group tended to have lower Sobs (P = 0.075) and Shannon (P = 0.058) indices of cecal microbiota than those in the other three groups.
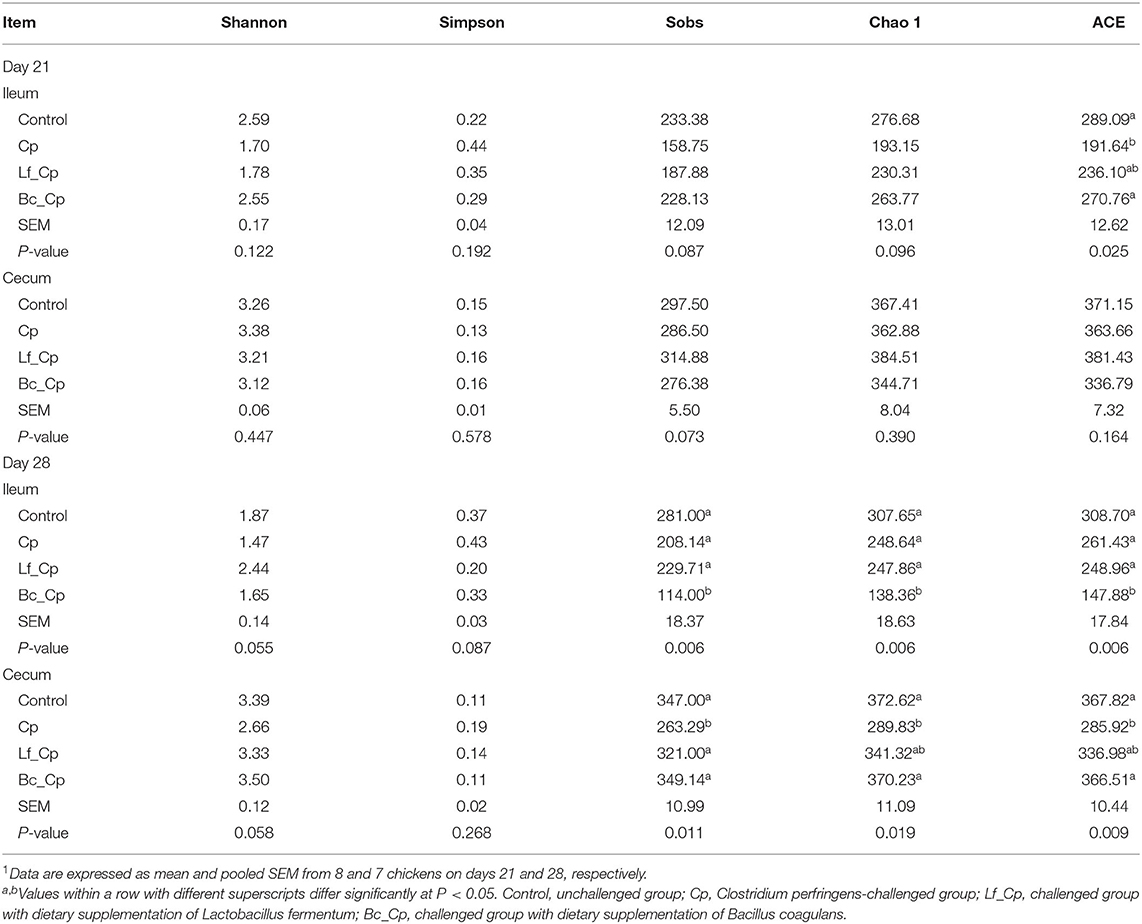
Table 1. The alpha diversity of ileal and cecal microbiota in broiler chickens1.
Beta Diversity of Microbiota in Ileum and Cecum
Beta diversity illustrated via PCoA is presented in Figure 3. At 21 days of age, the Lf_Cp group had distinct ileal microbiota compared with the other three groups, with a slight similarity with the control (Figure 3A). Meanwhile, ileal microbiota on day 21 in the Cp group was much similar to that in the Bc_Cp group. Cecal microbiota on day 21 as well as ileal and cecal microbiota on day 28 was not notably differentiated by treatments. However, the positive R-values in ANOSIM analysis suggested that the inter-group difference of microbial communities in both the ileum and cecum at two time points was greater than the intra-group difference (Table 2, P < 0.05).
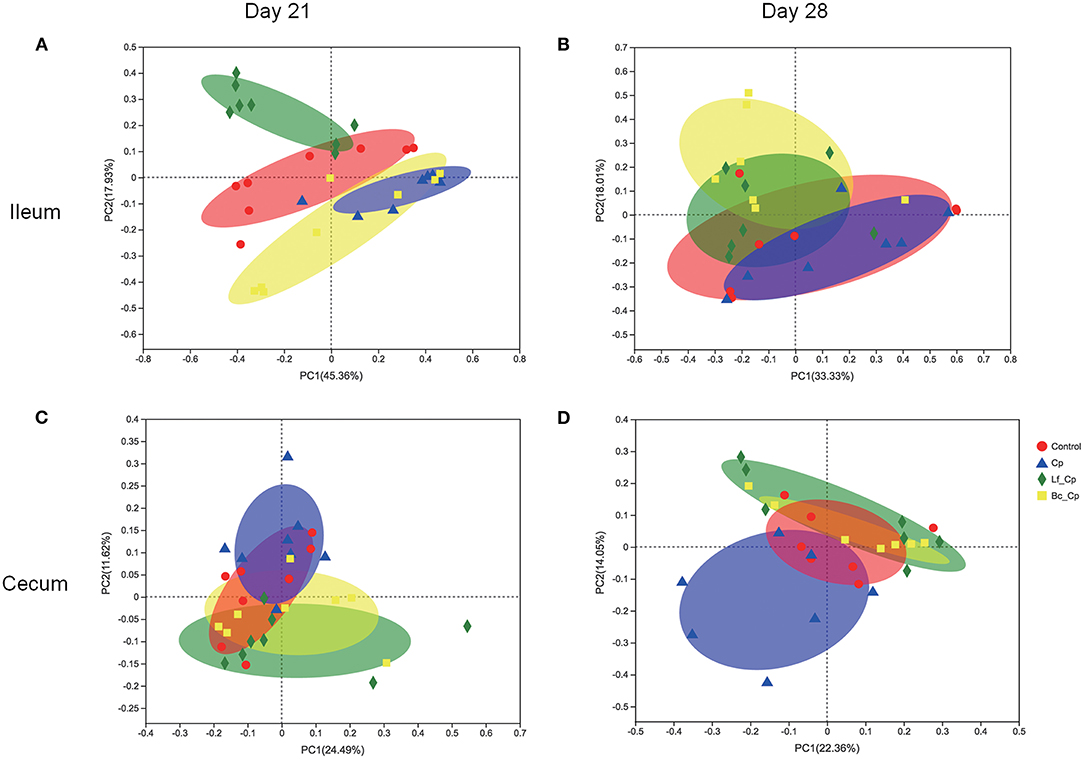
Figure 3. The PCoA plots of intestinal microbiota of Clostridium perfringens-challenged broiler chickens fed diets supplemented with Lactobacillus fermentum or Bacillus coagulans on days 21 (A,C) and 28 (B,D). Data were derived from 8 and 7 chickens on days 21 and 28, respectively. Control, unchallenged group; Cp, C. perfringens-challenged group; Lf_Cp, challenged group with dietary supplementation of Lactobacillus fermentum; Bc_Cp, challenged group with dietary supplementation of Bacillus coagulans.
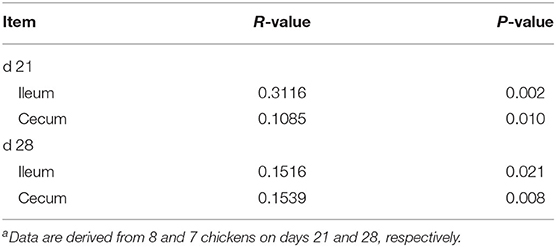
Table 2. Similarities of bacterial composition among groups by ANOSIM analysisa.
Bacterial Composition at Phylum Level in Ileum and Cecum
The ileal and cecal bacterial composition at the phylum level is shown in Supplementary Figure 4. The top four phyla in each intestinal segment and at each time point are presented in Table 3. At 21 days of age, compared with the control, the Cp and Lf_Cp groups increased the relative abundance of Firmicutes in the ileum (P < 0.05). The bacterial compositions at the phylum level in the cecum were not significantly affected by treatments (P > 0.05). At 28 days of age, compared with the control, the Cp and Bc_Cp groups decreased in ileal Bacteroidetes abundance, and all other groups reduced in cecal Proteobacteria abundance (P < 0.05).
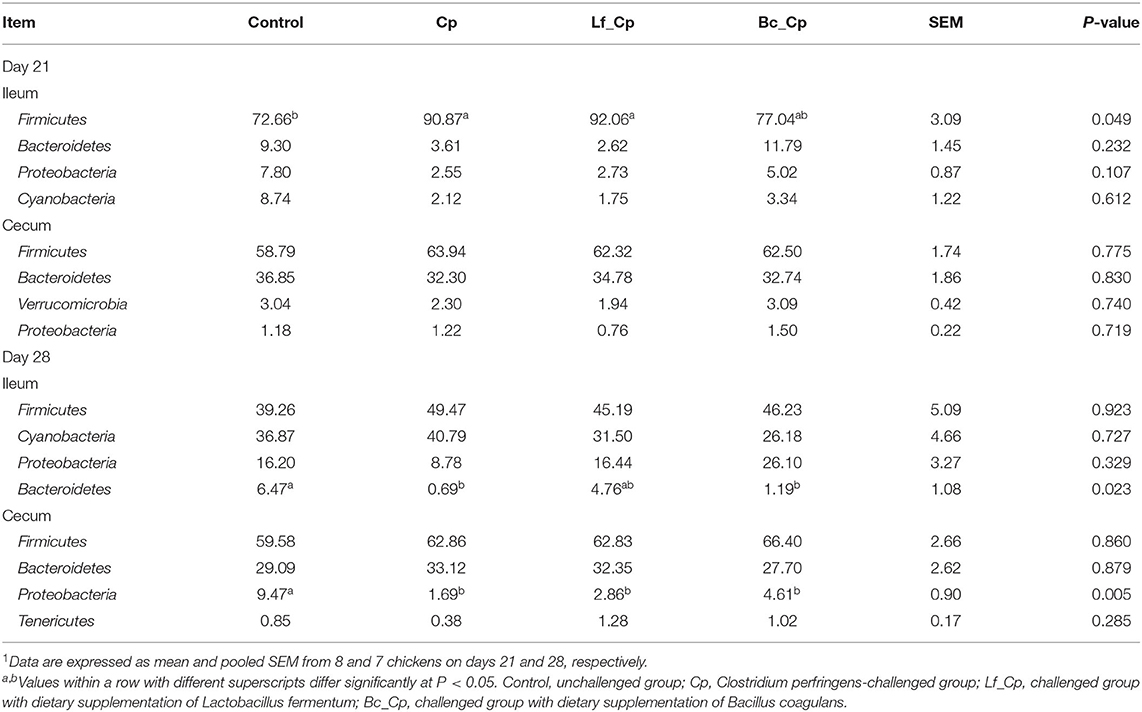
Table 3. The top 4 phyla in the ileal and cecal microbiota of broiler chickens1.
Top 10 Differentially Abundant Bacteria at Genus level in Ileum and Cecum
The top 10 differentially abundant genera in selected groups are seen in Figures 4–6.
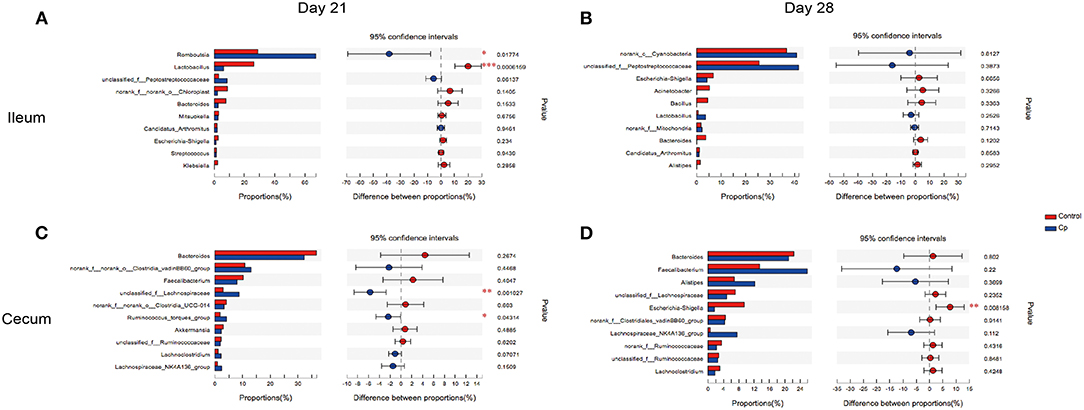
Figure 4. The top 10 differentially abundant bacteria at genus level in control vs. Cp groups. Ileal and cecal microflora on days 21 (A,C) and 28 (B,D) was analyzed. Data were derived from 8 and 7 chickens on days 21 and 28, respectively. Control, unchallenged group; Cp, C. perfringens-challenged group. *P < 0.05, **P < 0.05, ***P < 0.01.
At 21 days of age, compared with the control, the Cp group increased the relative abundance of Romboutsia spp. and decreased in Lactobacillus spp. abundance in the ileum (Figure 4A, P < 0.05). Compared with the Cp group, the Lf_Cp group decreased the relative contents of Romboutsia spp. and unclassified f_Peptostreptococcaceae, and increased the Lactobacillus spp. abundance in the ileum (Figure 5A, P < 0.05). The Bc_Cp group had a higher ileal Bacteroides spp. content than that of the Cp group (Figure 6A, P < 0.05). In contrast to the control, the Cp group increased unclassified f_Lachnospiraceae and Ruminococcus_torques group abundance in the cecum (Figure 4C, P < 0.05), both of which was decreased in the Lf_Cp group (Figure 5C, P < 0.05). Compared with the Cp group, the Bc_Cp group decreased cecal unclassified f_Lachnospiraceae content (Figure 6C, P < 0.05).
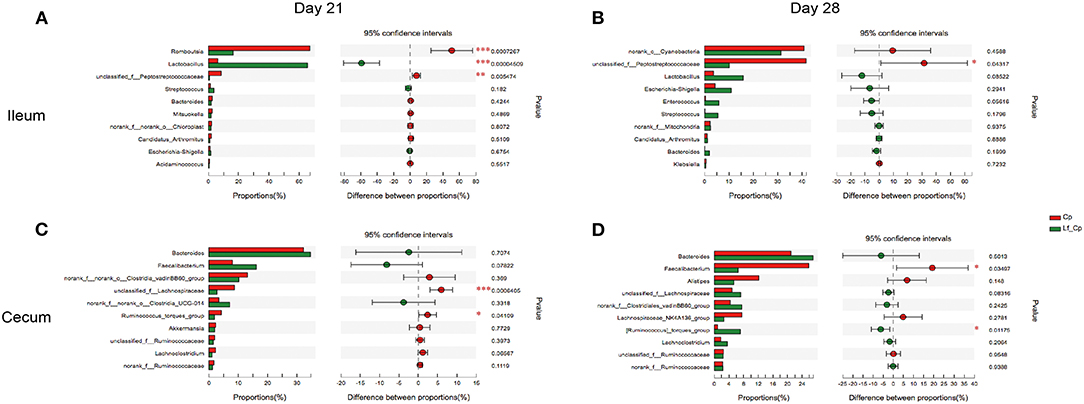
Figure 5. The top 10 differentially abundant bacteria at genus level in Cp vs. Lf_Cp groups. Ileal and cecal microflora on days 21 (A,C) and 28 (B,D) was analyzed. Data were derived from 8 and 7 chickens on days 21 and 28, respectively. Cp, C. perfringens-challenged group; Lf_Cp, challenged group with dietary supplementation of Lactobacillus fermentum. *P < 0.05, **P < 0.05, ***P < 0.01.
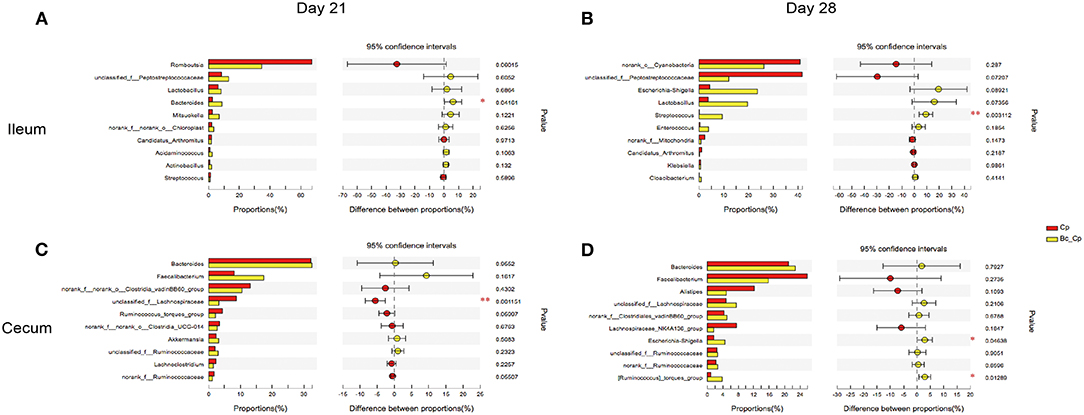
Figure 6. The top 10 differentially abundant bacteria at genus level in Cp vs. Bc_Cp groups. Ileal and cecal microflora on days 21 (A,C) and 28 (B,D) was analyzed. Data were derived from 8 and 7 chickens on days 21 and 28, respectively. Cp, C. perfringens-challenged group; Bc_Cp, challenged group with dietary supplementation of Bacillus coagulans. *P < 0.05, **P < 0.05.
At 28 days of age, C. perfringens challenge did not induce significant changes in ileal microbiota (Figure 4B, P > 0.05). Compared with the Cp group, the Lf_Cp group decreased in the relative abundance of unclassified f_Peptostreptococcaceae in the ileum (Figure 5B, P < 0.05). Meanwhile, the Bc_Cp group increased ileal Streptococcus spp. content (Figure 6B, P < 0.05). In the cecum, C. perfringens challenge decreased the relative abundance of Escherichia-Shigella (Figure 4D, P < 0.05). Compared with the Cp group, the Lf_Cp group reduced the relative content of Faecalibacterium spp. and increased Ruminococcus_torques group content in the cecum (Figure 5D, P < 0.05). The Bc_Cp group increased the relative abundance of both Escherichia-Shigella and Ruminococcus_torques group in the cecum (Figure 6D, P < 0.05).
Predicted Functional Potentials of OTUs in Ileum and Cecum
The top five presumptive functions of microbiota at level 2 of the KEGG pathways are illustrated in Table 4. At 21 days of age, the Lf_Cp group had a higher abundance of the replication and repair pathway and translation pathway than that of the other three groups in the ileum (P < 0.05). Compared with the control, the Cp group increased ileal abundance of amino acid metabolism, but the Lf_Cp group decreased this pathway abundance (P < 0.05). In contrast to the control, the Cp group increased the abundance of the membrane transport pathway, but decreased amino acid metabolism in the cecum (P < 0.05). Both probiotic-supplemented groups exhibited opposite changes to the Cp group (P < 0.05) and did not significantly differ from the control (P > 0.05). The Lf_Cp group had higher abundance of the replication and repair pathway than that of the control in the cecum (P < 0.05). At 28 days of age, compared with the control and Cp groups, the Bc_Cp and Lf_Cp groups increased the abundance of ileal membrane transport and cecal carbohydrate metabolism, respectively (P < 0.05).
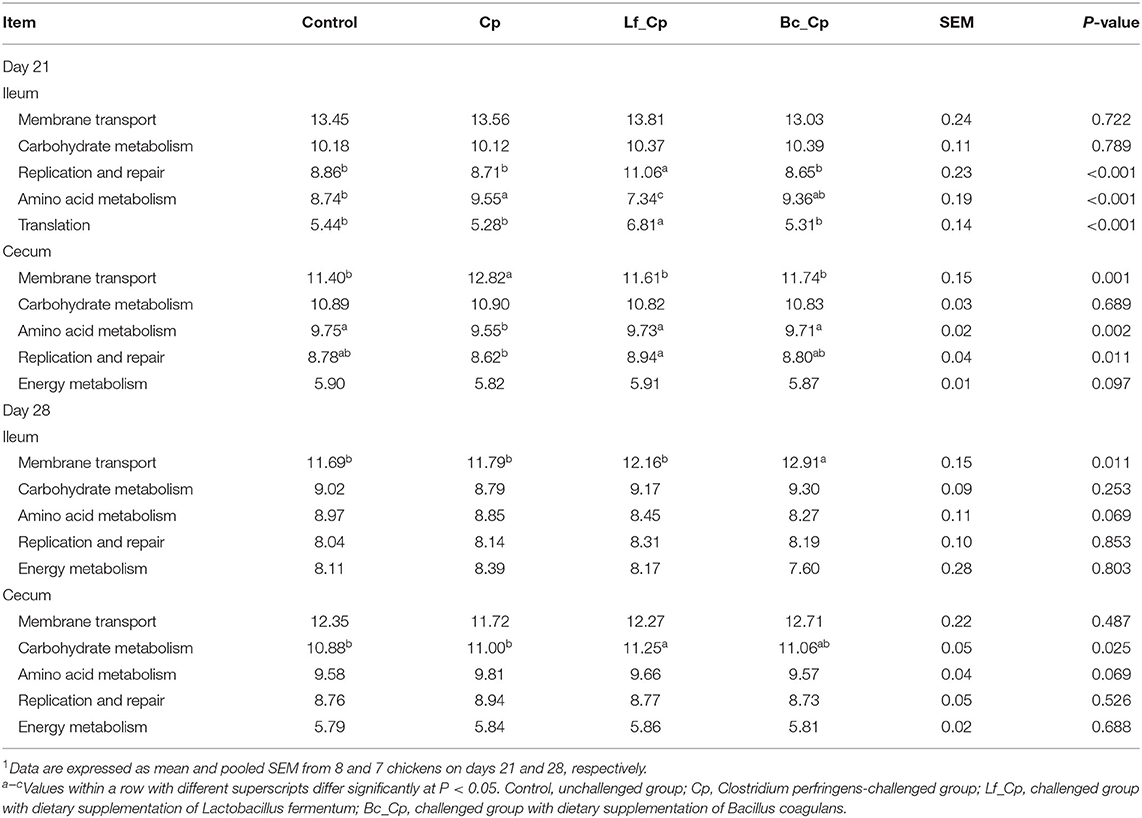
Table 4. The top 5 abundant microbial pathway grouped into level 2 functional categories using PICRUSt1.
Discussion
The previously reported results from the current study showed that the C. perfringens challenge-induced NE model was successful, evidenced by decreased averaged feed intake and increased jejunal lesion score as well as elevated C. perfringens counts in the ileum and cecum (20). Furthermore, both L. fermentum and B. coagulans addition inhibited intestinal pathogen colonization and B. coagulans exhibited superior effects in alleviating intestinal injury (20). Therefore, the intestinal immunity and microbiota of C. perfringens-challenged birds fed L. fermentum and B. coagulans-supplemented diets were further investigated in the present study.
The in vivo and in vitro studies have demonstrated that C. perfringens challenge evoked intense intestinal inflammatory responses via TLRs or nucleotide-binding oligomerization domains triggered the nuclear factor kappa B signaling pathway in broilers (9, 21). This signaling cascade resulted in the activation of macrophages and/or dendritic cells, which in turn led to the differentiation of naïve T helper (Th) cells into mature effector Th1, Th2, Th17, and Treg cells, which characteristically produced IL-1, INF-γ, IL-13, IL-17, and TGF-β, respectively (22, 23). Fasina and Lillehoj (24) reported that C. perfringens infection induced intestinal inflammation via the activation of Th2 and Th17 cells and inhibition of Treg cells, proven by the upregulation of IL-13 and IL-17 and downregulation of TGF-β4. In the present study, C. perfringens challenge upregulated jejunal IL-1β and TGF-β4 expression, which was downregulated by B. coagulans and L. fermentum, respectively, soon after the infection (day 21). However, both probiotics increased the mRNA expression of IL-1β, INF-γ, IL-13, IL-17, and TGF-β in the jejunum at seven days post infection (day 28). This suggested that probiotics showed inhibitory and stimulatory effects on cytokine expression of Th cells in the acute and recovery phase of C. perfringens challenge, respectively. Meanwhile, they mainly modulated the functions of Th1, Th17, and Treg cells. However, Emami et al. (25) reported that Bacillus licheniformis alone or combined with prebiotic and essential oil downregulated the IFN-γ, IL-10, and IL-17 mRNA abundance in the jejunum of broilers with subclinical NE, indicating a inhibition of Th2, Th17, and Treg cell function.
Intraepithelial lymphocytes (IEL), consisting of natural killer cells, T cells, and B cells, are an important component in gut-associated lymphoid tissue, which is one of the major immunological systems in chickens (26). Increase of the general T cell (CD3+) population within the intestinal epithelium indicates a strong local immune system with increased resistance to enteric diseases (27). In the present study, L. fermentum tended to increase CD3+ T cell density in the villus on day 21 and in the crypt on day 28, and B. coagulans significantly elevated crypt CD3+ T cell population in the jejunum of C. perfringens-challenged broilers. This suggested that L. fermentum and B. coagulans addition might induce intestinal T cell responses to fight against C. perfringens infection in broiler chickens. Consistently, Bai et al. (28) observed that L. fermentum JS combined with Saccharomyces cerevisiae stimulated the intestinal T cell immune system by increasing the subpopulations of IEL CD3+, CD4+, and CD8+ T cells in the jejunum of broiler chickens. It was reported that B. subtilis spores as adjuvants with inactivated H9N2 enhanced CD4+ and CD8+ T cell responses in White Leghorn chickens against avian influenza H9N2 (29).
It has been demonstrated that the diversity of microbial community contributed to the homeostasis of intestinal microbiota and the resistance to pathogens (30). In the present study, C. perfringens challenge decreased the richness (Sobs, Chao1, and ACE indices) and diversity (Shannon index) of cecal microbiota at 28 days of age, and both probiotic groups had comparable alpha diversity to the control. The same change of ACE index was seen in the ileal microbiota at 21 days of age. This indicated that the impaired intestinal microbial richness and diversity was restored by probiotic addition. Partly consistent with our findings, Li et al. (13) reported that C. perfringens infection decreased the richness of ileal bacterial community of broiler chickens and dietary L. acidophilus addition reduced its diversity, but both challenge and probiotic supplementation increased the richness and diversity of cecal microbiota. Furthermore, we found that B. coagulans significantly decreased ileal bacterial richness at 28 days of age. In a non-challenged model, broilers fed B. coagulans TBC169 had higher diversity of jejunal microbiota than that of the control on day 21, but lower diversity on day 42, although both of which did not achieve statistical significance (31).
In the current study, beta diversity analysis indicated only ileal microbiota at 21 days of age was markedly altered by treatments with the highest R-value (0.3116) by ANOSIM analysis. The unchallenged and challenged birds without probiotic addition had distinct microbiota only in the ileum on day 21. Compared with the C. perfringens-challenged group, L. fermentum alone and both probiotics altered intestinal microbiota on days 21 and 28, respectively. Necrotic enteritis in broilers was caused by C. perfringens proliferation and manifested by lesions in the small intestine (1). Moreover, the exogenous C. perfringens administration could not totally colonize in the intestine and they might be gradually excreted after challenge. This was evidenced by our previous observation that the relative abundance of C. perfringens in ileal and cecal contents quantified by qPCR in the challenged group without probiotic supplementation was much higher than that in the control on day 21 and no significant C. perfringens abundance was seen on day 28 (20). The influence of probiotic addition on the intestinal microbiota of birds challenged by C. perfringens might be disclosed in the following description.
It has been reported that Firmicutes, Cyanobacteria, Proteobacteria, and Bacteroidetes were the four predominant bacterial phyla in ileal microbiota (19, 32), and Firmicutes, Proteobacteria, Bacteroidetes, Tenericutes, and Verrucomicrobia were the predominant phyla in cecal microbiota (13, 33). This was consistent with our findings. Meanwhile, challenged birds in the Cp and Lf_Cp groups had higher relative abundance of ileal Firmicutes than that of birds in the control group on day 21, and challenged birds had lower ileal Bacteroidetes and cecal Proteobacteria abundance on day 28. Contrary to our observation, Xu et al. (34) demonstrated that NE severity was related to a decrease of Firmicutes and an increase of Bacteroidetes and Proteobacteria. Moreover, Zhang et al. (19) observed that C. perfringens challenge significantly decreased ileal Firmicutes abundance in broilers. The increase of Firmicutes population could suppress C. perfringens and restore intestinal homeostasis (35). It was speculated that the increase of Firmicutes abundance in the Cp and Lf_Cp groups might be a strategy for the host to maintain ileal microbial homeostasis. The increase of Bacteroidetes was associated with a decrease of nutrient absorption (36). Proteobacteria contain a wide variety of pathogens such as Escherichia coli, Salmonella, and Shigella, which can colonize in the intestine of chickens (37). The decrease of Proteobacteria and Bacteroidetes in challenged birds on day 28 might indicate that a healthy bacterial community was achieved in the recovery phase post challenge.
At 21 days of age, C. perfringens challenge increased Romboutsia spp. in the ileum as well as unclassified f_Lachnospiraceae and Ruminococcus_torques group in the cecum, and decreased Lactobacillus spp. in the ileum, which were all conversely modulated by dietary L. fermentum addition. Romboutsia spp. was reported to be associated with less severe immune responses accompanied with decreasing levels of pro-inflammatory cytokines in plasma (38). It was predominant in the feces of nursing home residents with Clostridium difficile colonization (39). Therefore, the decrease of Romboutsia spp. abundance in challenged birds fed L. fermentum-supplemented diet might benefit microbial homeostasis and alleviate inflammation. In a NE challenge model of broilers, B. subtilis DSM 32315 decreased the Lachnospiraceae members in the cecum (15), this was consistent with our findings that both L. fermentum and B. coagulans reduced the cecal Lachnospiraceae abundance. Ruminococcus spp. could produce lantibiotics that enhanced sterilization activity against some clostridia and bifidobacteria species (40), and the increase of Ruminococcus spp. was associated with mucin glycans (41). Xu et al. (34) reported that higher Ruminococcus spp. and Bacteroides spp. abundance was observed in the ileum of NE birds, which was decreased by B. licheniformis supplementation. Dietary L. fermentum decreased Ruminococcus spp. in the cecum, but B. coagulans increased Bacteroides spp. in the ileum of C. perfringens-challenged birds. Bacteroides spp. can evolve into a pathogenic form and will increase when the gut is pathologically impaired (42). In the current study, L. fermentum significantly increased Lactobacillus spp. in the ileum on day 21 and decreased Faecalibacterium spp. in the cecum on day 28. Lactobacillus spp. is a well-known beneficial genus in the gut and functions by maintaining intestinal bacterial community, improving immunity and gut integrity, and facilitating nutrition absorption (43). Complying with our findings, Li et al. (13) reported that L. acidophilus supplementation increased Lactobacillus spp. and suppressed Faecalibacterium spp., a potentially pathogenic flora.
Peptostreptococcaceae was one of the dominant families in the ileum of broilers (13, 32). In a broiler NE model established by Eimeria maxima and C. perfringens co-infection, challenged birds had higher ileal Peptostreptococcaceae and Streptococcus spp. abundance as compared to counterparts fed Bacillus direct-fed microbials (44). Similarly, L. fermentum decreased unclassified f_Peptostreptococcaceae in the ileum of C. perfringens-challenged birds on both days 21 and 28. However, B. coagulans increased Streptococcus spp. in the ileum on day 28. It has been indicated that there was a positive correlation between the relative abundance of Escherichia-Shigella with NE occurrence in broilers (45). Numerous studies evidenced that C. perfringens challenge increased the Escherichia-Shigella abundance in the ileum, and probiotic supplementation significantly or numerically decreased it (13, 25). However, in the present study, C. perfringens challenge decreased Escherichia-Shigella in the cecum on day 28, which was increased by B. coagulans. These data suggested that B. coagulans exerted a negative effect on intestinal microbiota at 7 days post challenge.
Membrane transport, carbohydrate metabolism, amino acid metabolism, replication and repair, and energy metabolism were the dominant functions of microbiota, which was confirmed by Zhang et al. (19). Membrane transport pathways are essential to cell viability and growth and are thereby crucial for the survival of bacteria in the gut ecosystem (46). Zhang et al. (19) reported that C. perfringens challenge decreased the abundance of the membrane transport function of ileal microbiota. In contrast, we found C. perfringens infection increased the membrane transport of cecal microbiota on day 21, which was decreased by probiotic addition. Meanwhile, challenged birds fed B. coagulans-supplemented diet had higher membrane transport abundance in the ileum on day 28. This indicated that the activities of ileal and cecal microbiota showed different responses to challenge and probiotic addition. It has been demonstrated that decreased carbohydrate metabolism and increased amino acid metabolism was observed in inflamed mucosal microbiota of ulcerative colitis patients (47). In the present study, C. perfringens challenge increased amino acid metabolism of ileal microbiota, but decreased it in cecal microbiota on day 21, which were conversely changed by probiotic addition. This might be attributed to the mucolytic characteristic and ileal colonization of C. perfringens (48). Dietary L. fermentum and B. coagulans affected the bacterial function by changing the microbial composition. Furthermore, challenged birds supplemented with L. fermentum had higher abundance of the replication and repair of ileal and cecal microbiota as well as translation of ileal microbiota on day 21. Consistently, Cui et al. (49) reported that dietary addition with B. subtilis and Clostridium butyricum increased the replication and repair of jejunal microbiota in broilers, which improved the renewal and vigorous vitality of bacteria.
Conclusion
C. perfringens challenge induced inflammatory responses and intestinal microbial dysbiosis soon after challenge and these impairments diminished at 7 days post challenge. Dietary L. fermentum and B. coagulans supplementation attenuated inflammation by modulating Th cell function and T cell responses, and restored intestinal microflora by enriching beneficial bacteria and suppressing harmful flora. L. fermentum exhibited superior microbial modulatory effects to B. coagulans.
Data Availability Statement
The sequencing data presented in the study are deposited in the Sequence Read Archive of the NCBI, accession number SPR304801.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Wuhan Polytechnic University.
Author Contributions
SG, YiX, and BD conceived and designed the experiment. SG, YiX, and TW performed the experiment. SG, YuX, TW, and DZ analyzed the data. SG, YuX, ZZ, and BD wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31702130), the Hubei Provincial Key Project for Scientific and Technical Innovation (2018ABA110), the Open Project of Hubei Key Laboratory of Animal Nutrition and Feed Science (DKXY2020014), and the Launching Research Fund from Wuhan Polytechnic University (2019RZ03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.680742/full#supplementary-material
References
1. Shojadoost B, Vince AR, Prescott JF. The successful experimental induction of necrotic enteritis in chickens by Clostridium perfringens: a critical review. Vet Res. (2012) 43:74. doi: 10.1186/1297-9716-43-74
2. Kaldhusdal M, Hofshagen M. Barley inclusion and avoparcin supplementation in broiler diets. 2. Clinical, pathological, and bacteriological findings in a mild form of necrotic enteritis. Poult Sci. (1992) 71:1145–53. doi: 10.3382/ps.0711145
3. Songer JG. Clostridial enteric diseases of domestic animals. Clin Microbiol Rev. (1996) 9:216–34. doi: 10.1128/CMR.9.2.216-234.1996
4. Kondo F. In vitro lecithinase activity and sensitivity to 22 antimicrobial agents of Clostridium perfringens isolated from necrotic enteritis of broiler chickens. Res Vet Sci. (1988) 45:337–40.
5. Cooper KK, Songer JG. Necrotic enteritis in chickens: a paradigm of enteric infection by Clostridium perfringens type A. Anaerobe. (2009) 15:55–60. doi: 10.1016/j.anaerobe.2009.01.006
6. Khalique A, Zeng D, Shoaib M, Wang H, Qing X, Rajput DS, et al. Probiotics mitigating subclinical necrotic enteritis (SNE) as potential alternatives to antibiotics in poultry. AMB Express. (2020) 10:50. doi: 10.1186/s13568-020-00989-6
7. La Ragione RM, Woodward MJ. Competitive exclusion by Bacillus subtilis spores of Salmonella enterica serotype Enteritidis and Clostridium perfringens in young chickens. Vet Microbiol. (2003) 94:245–56. doi: 10.1016/S0378-1135(03)00077-4
8. Guo S, Liu D, Zhang B, Li Z, Li Y, Ding B, et al. Two Lactobacillus species inhibit the growth and alpha-toxin production of Clostridium perfringens and induced proinflammatory factors in chicken intestinal epithelial cells in vitro. Front Microbiol. (2017) 8:2081. doi: 10.3389/fmicb.2017.02081
9. Wu Y, Zhen W, Geng Y, Wang Z, Guo Y. Pretreatment with probiotic Enterococcus faecium NCIMB 11181 ameliorates necrotic enteritis-induced intestinal barrier injury in broiler chickens. Sci Rep. (2019) 9:10256. doi: 10.1038/s41598-019-46578-x
10. Cao L, Yang XJ, Li ZJ, Sun FF, Wu XH, Yao JH. Reduced lesions in chickens with Clostridium perfringens-induced necrotic enteritis by Lactobacillus fermentum 1.2029. Poult Sci. (2012) 91:3065–71. doi: 10.3382/ps.2012-02548
11. Wu Y, Shao Y, Song B, Zhen W, Wang Z, Guo Y, et al. Effects of Bacillus coagulans supplementation on the growth performance and gut health of broiler chickens with Clostridium perfringens-induced necrotic enteritis. J Anim Sci Biotechnol. (2018) 9:9. doi: 10.1186/s40104-017-0220-2
12. Borda-Molina D, Seifert J, Camarinha-Silva A. Current perspectives of the chicken gastrointestinal tract and its microbiome. Comput Struct Biotechnol J. (2018) 16:131–9. doi: 10.1016/j.csbj.2018.03.002
13. Li Z, Wang W, Liu D, Guo Y. Effects of Lactobacillus acidophilus on gut microbiota composition in broilers challenged with Clostridium perfringens. PLoS ONE. (2017) 12:e0188634. doi: 10.1371/journal.pone.0188634
14. Bortoluzzi C, Serpa Vieira B, de Paula Dorigam JC, Menconi A, Sokale A, Doranalli K, et al. Bacillus subtilis DSM 32315 supplementation attenuates the effects of Clostridium perfringens challenge on the growth performance and intestinal microbiota of broiler chickens. Microorganisms. (2019) 7:71. doi: 10.3390/microorganisms7030071
15. Whelan RA, Doranalli K, Rinttilä T, Vienola K, Jurgens G, Apajalahti J. The impact of Bacillus subtilis DSM 32315 on the pathology, performance, and intestinal microbiome of broiler chickens in a necrotic enteritis challenge. Poult Sci. (2019) 98:3450–63. doi: 10.3382/ps/pey500
16. Du E, Gan L, Li Z, Wang W, Liu D, Guo Y. In vitro antibacterial activity of thymol and carvacrol and their effetcs on broiler chickens challenged with Clostridium perfringens. J Anim Sci Biotechnol. (2015) 6:58. doi: 10.1186/s40104-015-0055-7
17. Röhe I, Gobel TW, Goodarzi Boroojeni F, Zentek J. Effect of feeding soybean meal and differently processed peas on the gut mucosal immune system of broilers. Poult Sci. (2017) 96:2064–73. doi: 10.3382/ps/pew491
18. Wu T, Lyu Y, Li X, Wu M, Yu K, Li S, et al. Impact of N-acetylcysteine on the gut microbiota in the piglets infected with porcine epidemic diarrhea virus. Front Vet Sci. (2021) 7:582338. doi: 10.3389/fvets.2020.582338
19. Zhang B, Lv Z, Li Z, Wang W, Li G, Guo Y. Dietary L-arginine supplementation alleviates the intestinal injury and modulates the gut microbiota in broiler chickens challenged by Clostridium perfringens. Front Microbiol. (2018) 9:1716. doi: 10.3389/fmicb.2018.01716
20. Xia Y, Zhang Y, Xu J, Guo S, Ding B. Effects of Lactobacillus fermentum and Bacillus coagulans on the growth performance and gut health of broilers challenged with Clostridium perfringens. China Anim Husbandry Vet Med. (2019) 46:2927–36. doi: 10.16431/j.cnki.1671-7236.2019.10.013 (Abstract in English)
21. Guo S, Li C, Liu D, Guo Y. Inflammatory responses to a Clostridium perfringens type A strain and alpha-toxin in primary intestinal epithelial cells of chicken embryos. Avian Pathol. (2015) 44:81–91. doi: 10.1080/03079457.2015.1005573
22. Cosmi L, Maggi L, Santarlasci V, Liotta F, Annunziato F. T helper cells plasticity in inflammation. Cytometry Part A. (2014) 85:36–42. doi: 10.1002/cyto.a.22348
23. Oh ST, Lillehoj HS. The role of host genetic factors and host immunity in necrotic enteritis. Avian Pathol. (2016) 45:313–6. doi: 10.1080/03079457.2016.1154503
24. Fasina YO, Lillehoj HS. Characterization of intestinal immune response to Clostridium perfringens infection in broiler chickens. Poult Sci. (2018) 98:188–98. doi: 10.3382/ps/pey390
25. Emami NK, Calik A, White MB, Kimminau EA, Dalloul RA. Effect of probiotics and multi-component feed additives on microbiota, gut barrier and immune responses in broiler chickens during subclinical necrotic enteritis. Front Vet Sci. (2020) 7:572142. doi: 10.3389/fvets.2020.572142
26. Yun CH, Lillehoj HS, Lillehoj EP. Intestinal immune responses to coccidiosis. Dev Comp Immunol. (2000) 24:303–24. doi: 10.1016/s0145-305x(99)00080-4
27. Dalloul RA, Lillehoj HS, Shellem TA, Doerr JA. Effect of vitamin A deficiency on host intestinal immune response to Eimeria acervulina in broiler chickens. Poult Sci. (2002) 81:1509–15. doi: 10.1093/ps/81.10.1509
28. Bai SP, Wu AM, Ding XM, Lei Y, Bai J, Zhang KY, et al. Effects of probiotic-supplemented diets on growth performance and intestinal immune characteristics of broiler chickens. Poult Sci. (2013) 92:663–70. doi: 10.3382/ps.2012-02813
29. Lee JE, Kye Y-C, Park S-M, Shim B-S, Yoo S, Hwang E, et al. Bacillus subtilis spores as adjuvants against avian influenza H9N2 induce antigen-specific antibody and T cell responses in White Leghorn chickens. Vet Res. (2020) 51:68. doi: 10.1186/s13567-020-00788-8
30. Konstantinov SR, Favier CF, Zhu WY, Williams BA, Klüss J, Souffrant WB, et al. Microbial diversity studies of the porcine gastrointestinal ecosystem during weaning transition. Anim Res. (2004) 53:317–24. doi: 10.1051/animres:2004019
31. Li C-L, Wang J, Zhang H-J, Wu S-G, Hui Q-R, Yang C-B, et al. Intestinal morphologic and microbiota responses to dietary Bacillus spp. in a broiler chicken model. Front Physiol. (2019) 9:1968. doi: 10.3389/fphys.2018.01968
32. Wang W, Li Z, Lv Z, Zhang B, Lv H, Guo Y. Effects of Kluyveromyces marxianus supplementation on immune responses, intestinal structure and microbiota in broiler chickens. PLoS ONE. (2017) 12:e0180884. doi: 10.1371/journal.pone.0180884
33. Proctor A, Phillips GJ. Differential effects of bacitracin methylene disalicylate (BMD) on the distal colon and cecal microbiota of young broiler chickens. Front Vet Sci. (2019) 6:114. doi: 10.3389/fvets.2019.00114
34. Xu S, Lin Y, Zeng D, Zhou M, Zeng Y, Wang H, et al. Bacillus licheniformis normalize the ileum microbiota of chickens infected with necrotic enteritis. Sci Rep. (2018) 8:1744. doi: 10.1038/s41598-018-20059-z
35. Fasina YO, Newman MM, Stough JM, Liles MR. Effect of Clostridium perfringens infection and antibiotic administration on microbiota in the small intestine of broiler chickens. Poult Sci. (2016) 95:247–60. doi: 10.3382/ps/pev329
36. Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. (2011) 94:58–65. doi: 10.3945/ajcn.110.010132
37. Mora A, Herrera A, Mamani R, López C, Alonso MP, Blanco JE, et al. Recent emergence of clonal group O25b:K1:H4-B2-ST131 ibeA strains among Escherichia coli poultry isolates, including CTX-M-9-producing strains, and comparison with clinical human isolates. Appl Environ Microbiol. (2010) 76:6991–7. doi: 10.1128/AEM.01112-10
38. Liang G, Malmuthuge N, Bao H, Stothard P, Griebel PJ, Guan LL. Transcriptome analysis reveals regional and temporal differences in mucosal immune system development in the small intestine of neonatal calves. BMC Genomics. (2016) 17:602. doi: 10.1186/s12864-016-2957-y
39. Araos R, Andreatos N, Ugalde J, Mitchell S, Mylonakis E, D'agata EMC. Fecal microbiome among nursing home residents with advanced dementia and Clostridium difficile. Digest Dis Sci. (2018) 63:1525–31. doi: 10.1007/s10620-018-5030-7
40. Dabard J, Bridonneau C, Phillipe C, Anglade P, Molle D, Nardi M, et al. Ruminococcin A, a new lantibiotic produced by a Ruminococcus gnavus strain isolated from human feces. Appl Environ Microbiol. (2001) 67:4111–8. doi: 10.1128/AEM.67.9.4111-4118.2001
41. Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE. (2013) 8:e76341. doi: 10.1371/journal.pone.0076341
42. Rocha ER, Smith CJ. Ferritin-like family proteins in the anaerobe Bacteroides fragilis: when an oxygen storm is coming, take your iron to the shelter. Biometals. (2013) 26:577–91. doi: 10.1007/s10534-013-9650-2
43. Bermudez-Brito M, Plaza-Díaz J, Muñoz-Quezada S, Gómez-Llorente C, Gil A. Probiotic mechanisms of action. Ann Nutr Metab. (2012) 61:160–74. doi: 10.1159/000342079
44. Hernandez-Patlan D, Solis-Cruz B, Pontin KP, Hernandez-Velasco X, Merino-Guzman R, Adhikari B, et al. Impact of a Bacillus direct-fed microbial on growth performance, intestinal barrier integrity, necrotic enteritis lesions, and ileal microbiota in broiler chickens using a laboratory challenge model. Front Vet Sci. (2019) 6:108. doi: 10.3389/fvets.2019.00108
45. Yang W-Y, Lee Y, Lu H, Chou C-H, Wang C. Analysis of gut microbiota and the effect of lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. PLoS ONE. (2019) 14:e0205784. doi: 10.1371/journal.pone.0205784
46. Lyons PP, Turnbull JF, Dawson KA, Crumlish M. Phylogenetic and functional characterization of the distal intestinal microbiome of rainbow trout Oncorhynchus mykissfrom both farm and aquarium settings. J Appl Microbiol. (2017) 122:347–63. doi: 10.1111/jam.13347
47. Davenport M, Poles J, Leung JM, Wolff MJ, Abidi WM, Ullman T, et al. Metabolic alterations to the mucosal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. (2014) 20:723–31. doi: 10.1097/MIB.0000000000000011
48. Liu D, Guo S, Guo Y. Xylanase supplementation to a wheat-based diet alleviated the intestinal mucosal barrier impairment of broiler chickens challenged by Clostridium perfringens. Avian Pathol. (2012) 41:291–8. doi: 10.1080/03079457.2012.684089
Keywords: Lactobacillus fermentum, Bacillus coagulans, Clostridium perfringens, immunity, gut microbiota, broiler chicken
Citation: Guo S, Xi Y, Xia Y, Wu T, Zhao D, Zhang Z and Ding B (2021) Dietary Lactobacillus fermentum and Bacillus coagulans Supplementation Modulates Intestinal Immunity and Microbiota of Broiler Chickens Challenged by Clostridium perfringens. Front. Vet. Sci. 8:680742. doi: 10.3389/fvets.2021.680742
Received: 18 March 2021; Accepted: 19 April 2021;
Published: 31 May 2021.
Edited by:
Amlan Kumar Patra, West Bengal University of Animal and Fishery Sciences, IndiaReviewed by:
Bingkun Zhang, China Agricultural University, ChinaShad Mahfuz, Sylhet Agricultural University, Bangladesh
Copyright © 2021 Guo, Xi, Xia, Wu, Zhao, Zhang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binying Ding, ZGJ5aW5nNzQ3MUAxMjYuY29t
†These authors have contributed equally to this work
 Shuangshuang Guo
Shuangshuang Guo Yu Xi†
Yu Xi† Tao Wu
Tao Wu