- 1State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 2Animal Health Supervision Institute of Xinjiang, Ürümqi, China
- 3Jiangsu Co-innovation Center for the Prevention and Control of Important Animal Infectious Disease and Zoonose, Yangzhou University, Yangzhou, China
Bartonella are gram-negative intracellular bacteria; certain species of Bartonella can cause diseases in mammals and humans. Ticks play a major role in the transmission of Bartonella. Xinjiang is the largest province in China according to land area and has one-third of the tick species in China; the infection rate of Bartonella in ticks in the Xinjiang border areas has not been studied in detail. Therefore, this study investigated tick infections by Bartonella in Xinjiang border areas, and the purpose of the study was to fill in gaps in information regarding the genetic diversity of tick infections by Bartonella in Xinjiang. We tested 1,549 tick samples from domestic animals (sheep and cattle) for Bartonella using ribC-PCR. Positive samples from the ribC-PCR assay for Bartonella spp. were further subjected to PCR assays targeting the ITS, rpoB and gltA genes followed by phylogenetic analyses. Bartonella DNA was detected in 2.19% (34/1,549) of tick samples, and the ITS, rpoB and gltA genes of ribC gene-positive samples were amplified to identify nine samples of Bartonella melophagi. In this study, molecular analysis was used to assess the presence and genetic diversity of B. melophagi in ticks collected from sheep and cattle from Xinjiang, China. This study provides new information on the presence and identity of B. melophagi in ticks from sheep and cattle.
Introduction
Bacteria of the genus Bartonella are obligate gram-negative intracellular bacteria that primarily infect mammalian erythrocytes, macrophages, monocytes, and endothelial and dendritic cells (1, 2). B. bacilliformis was the only named species of Bartonella before 1990. At present, more than 36 species have been discovered, 17 of which are related to human and animal diseases (3).
Ruminants have played a vital role in ecology, agriculture and the economy worldwide. They are widely used by humans for different purposes, such as ecological indicators, dairy products and meat. At present, there are 5 kinds of Bartonella related to ruminants: B. bovis, B. chomelii, B. schoenbuchensis, B. capreoli, and B. melophagi. An increasing number of Candidatus Bartonella species and different genotypes have been reported in ruminants (4–7). Previous studies have shown that Bartonella DNA is amplified in ruminant-related blood-eating arthropods, including Hippoboscidae flies from Europe and Algeria (8, 9), Stomoxys spp. and Haematobia spp. from California (10), Rhipicephalus microplus from Brazilian Cerrado and Taiwan (11, 12), Haematopinus tuberculatus from Brazilian Cerrado and H. quadripertusus from Israel (12, 13), Haemaphysalis bispinosa from Malaysia (14), H. flava and Ixodes persulcatus from Korean (15), H. longicornis from North Korea and Korean (15, 16), Hyalomma anatolicum from Pakistan (17), Ctenocephalides felis from Tunisia (18), sheep keds (Melophagus ovinus) from Central Europe and northeastern Algeria (19, 20), and deer keds (Lipoptena cervi) from Norway (6).
Bartonella uses humans, cats, dogs, mice, horses, cows, rabbits and other wild animals around the world as hosts (3, 21, 22). It can be transmitted by insect vectors, such as ticks, human body louse, cat fleas, and sand flies (3, 22, 23). Ticks are a widely distributed vector insect worldwide and can transmit and carry a variety of zoonotic pathogens, including viruses, bacteria, parasites, spirochetes and Rickettsia (24, 25).
As the largest province in China, Xinjiang has a vast territory, a diverse ecological environment, and rich species. At present, Xinjiang has one-third of the tick species found in China (26). Ticks in Xinjiang have been shown to transmit multiple pathogenic infections, such as Babesia, Anaplasma, Rickettsia, Theileria and Tularemia (27–30); however, there are only a few studies on Bartonella infections in ticks in Xinjiang. Previous studies confirmed that sheep keds (M. ovinus) in Xinjiang were infected with B. melophagi (31). Therefore, we aimed to investigate Bartonella infection in ticks collected from the border areas of Xinjiang and its genetic diversity to fill in the existing lacunae.
Materials and Methods
Specimen Collection and Morphological Identification of the Ticks
From 2018 to 2019, we randomly collected tick samples from cattle and sheep at 23 sampling sites in 10 regions on the Xinjiang border, China (Figure 1, Table 1). Sampling sites included Kashi, Gongliu, Xinyuan, Nilka, Qapqal, Yining, Huocheng, Wensu, Wushi, Aheqi, Atushi, Hoboksar, Tacheng, Yumin, Pishan, Karakax, Habahe, Qinghe, Burqin, Jeminay, Wenquan, Qitai, and Barkol Kazak. The collected samples were stored in 50 mL centrifuge tubes and delivered to the laboratory. The ticks were identified based on morphological criteria following the descriptions provided by Deng et al. (32). Tick samples were collected with permission from the farmers.
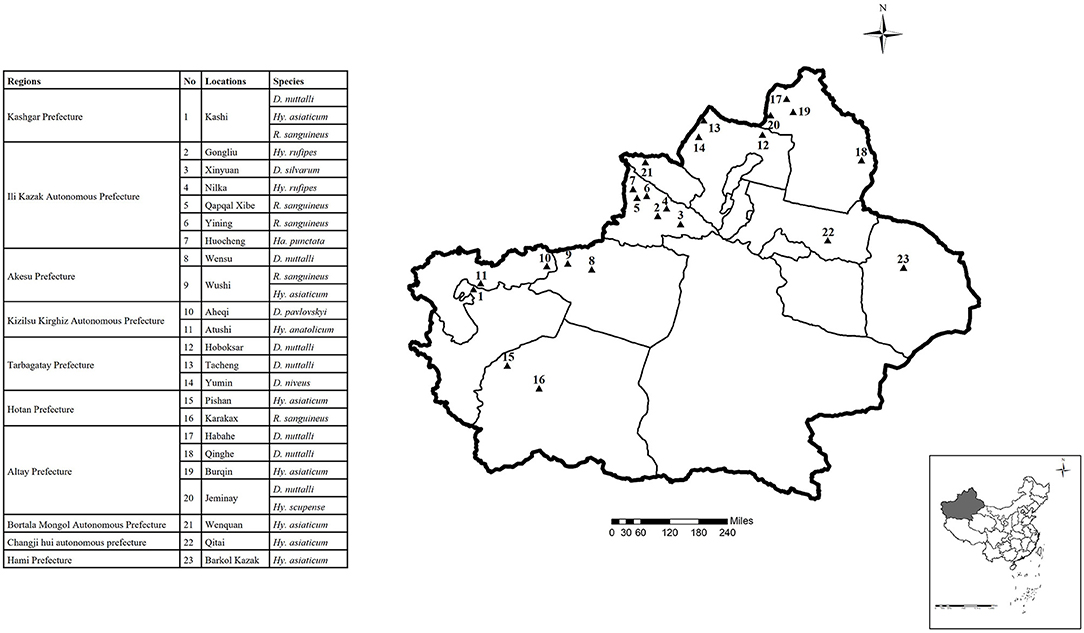
Figure 1. Locations of the sample sites for tick collection in the border areas of Xinjiang (different locations are coded by number; 1-23 indicate the sampling points). The map was made by ArcMap 10.2 (https://developers.arcgis.com/).
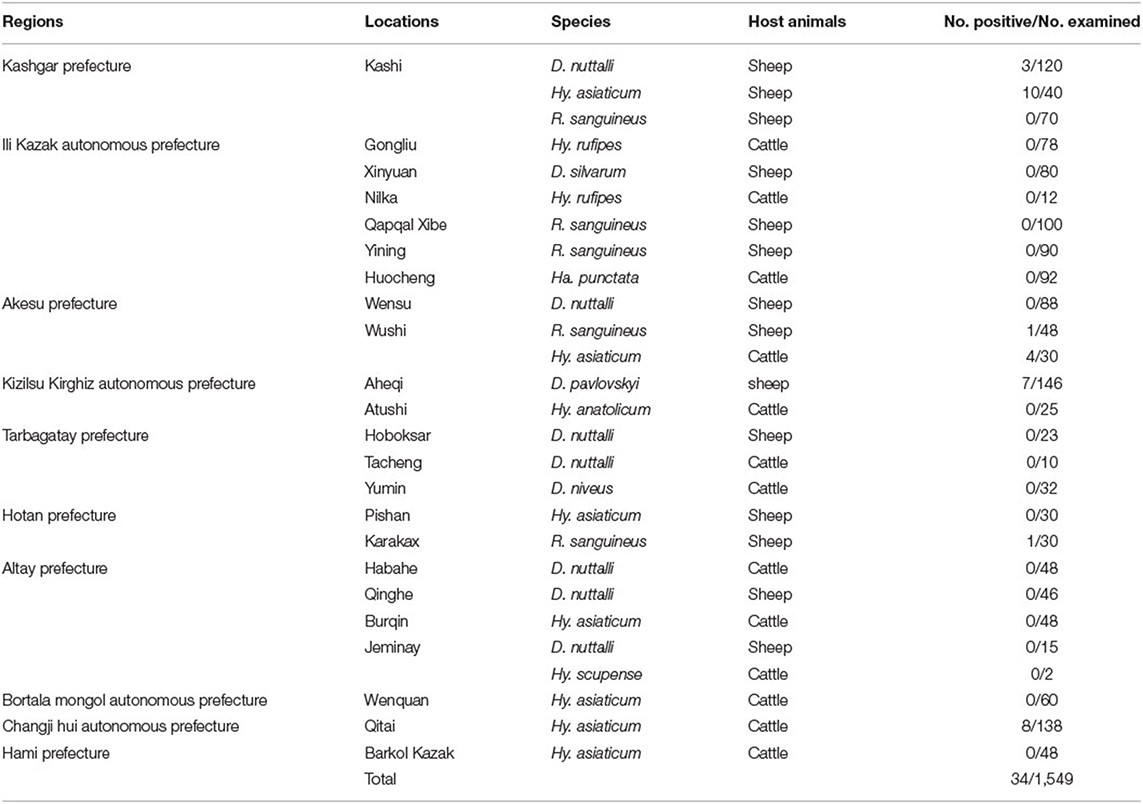
Table 1. Bartonella spp. detection in ticks collected from different localities of the Xinjiang border.
DNA Extraction and Molecular Analysis
Tick samples were placed in 50 mL sterilized centrifuge tubes and washed individually twice with 75% ethanol followed by rinsing with double-distilled water (ddH2O) until the liquid was clear. DNA was extracted from each sample using a QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol, and the extracted DNA was stored at −20°C. Initial screening of Bartonella DNA was performed using ribC-PCR. To further characterize the positive samples in this study, positive samples by ribC-PCR assay for Bartonella spp. were further subjected to PCR assays targeting the ITS, rpoB and gltA genes, followed by phylogenetic analyses (33–36). A negative control was prepared with ddH2O; a positive control was prepared in our laboratory using DNA extracted from Hy. asiaticum infected by B. melophagi. The PCR products of the partial gltA, rpoB, ribC and ITS genes were purified using an E.Z.N.A.® gel extraction kit (Omega, USA) and cloned into the pGEM-T Easy vector (Promega, USA).
Sequencing and Phylogenetic Analysis
The nucleotide sequences were confirmed by bidirectional sequencing at TSINGKE Biotech, China. The nucleotide sequences were compared with the reference sequences of GenBank (www.ncbi.nlm.nih.gov/nuccore/) by SeqMan (www.dnastar.com/); the forward and reverse primers of the sequences after bidirectional sequencing were selected using SeqMan; extra sequences at the ends of the forward and reverse primers were removed, and corrected calibration was performed after manual adjustment as needed. The sequences were aligned with sequences downloaded in GenBank using MAFFT (37). ModelFinder (38) was used to select the best model using the BIC criterion. Phylogenetic inference was founded on maximum likelihood (ML) analysis through the IQ-TREE (39) program of PhyloSuite V1.2.2 (40). The trees were edited in Figtree v1.4.3 (https://github.com/rambaut/figtree/releases). The confidence values for each branch of the phylogenetic tree were determined by using 1,000 repeat analyses.
Results
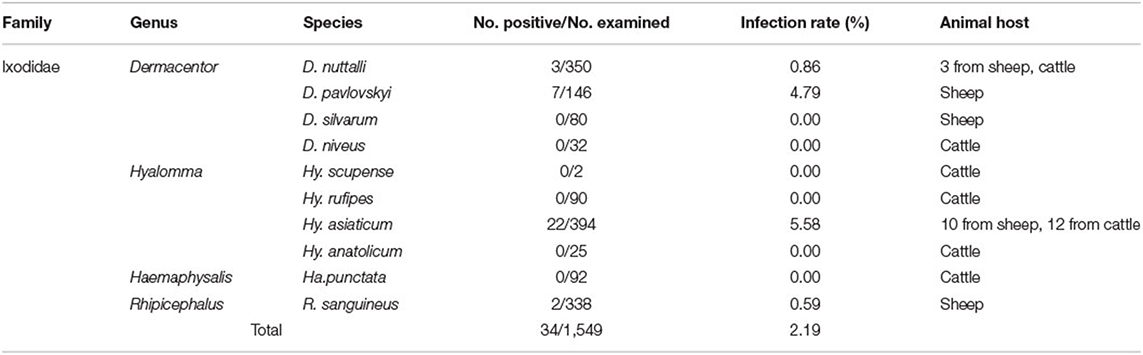
A total of 1,549 tick samples were collected from livestock in various regions of the Xinjiang border (Table 1). All tick samples represented a single family [Ixodidae], four genera [Dermacentor (n = 608), Hyalomma (n = 511), Rhipicephalus (n = 338), and Haemaphysalis (n = 92)], and ten species [D. nuttalli (n = 350), D. pavlovskyi (n = 146), D. silvarum (n = 80), D. niveus (n = 32), Hy. rufipes (n = 90), Hy. scupense (n = 2), Hy. anatolicum (n = 25), Hy. asiaticum (n = 394), R. sanguineus (n = 338), and Ha. punctata (n = 92)] (Table 2).
In this study, we assayed Bartonella in 1,549 tick samples from livestock (cattle and sheep) using ribC-PCR and detected Bartonella DNA in 2.19% (34/1,549) of tick samples. The positive samples had the following proportions: D. nuttalli 3 (n = 350; 0.86%), D. pavlovskyi 7 (n = 146; 4.79%), Hy. asiaticum 22 (n = 394; 5.58%), and R. sanguineus 2 (n = 338; 0.59%). Meanwhile, the gltA, ITS and rpoB genes were amplified by PCR and sequencing. A total of 17 Bartonella ribC (three from D. nuttalli, seven from Hy. asiaticum, five from D. pavlovskyi, and two from R. sanguineus), nine gltA (two from D. nuttalli, four from Hy. asiaticum, two from D. pavlovskyi, and one from R. sanguineus), 12 ITS (three from D. nuttalli, six from Hy. asiaticum, two from D. pavlovskyi, and one from R. sanguineus) and eight rpoB (one from D. nuttalli, four from Hy. asiaticum, two from D. pavlovskyi and one from R. sanguineus) sequences were obtained.
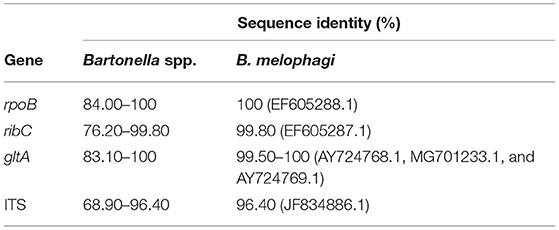
The results of sequence analysis showed that there were no basic differences among the rpoB (n = 8), gltA (n = 9), ribC (n = 17), and ITS (n = 12) sequences obtained in this study. The sequence identity between the examined B. melophagi and other known Bartonella species was 84–100% for rpoB, 76.20–99.80% for ribC, 83.10–100% for gltA, and 68.90–96.40% for ITS. The sequence identity between the examined B. melophagi and known B. melophagi was 100% for rpoB (EF605288.1), 99.80% for ribC (99.80), 99.50–100% for gltA (AY724768.1, MG701233.1, and AY724769.1), and 96.40% for ITS (JF834886.1) (Table 3).
In the phylogenetic inference based on gltA and rpoB genes, the gltA and rpoB sequences obtained in this study were clustered into a branch of Bartonella associated with ruminants and supported by 98 and 100% bootstrap analysis. The gltA and rpoB sequences of B. melophagi in this study were closest to AY724768.1 and EF605288.1, respectively (Figures 2, 3).
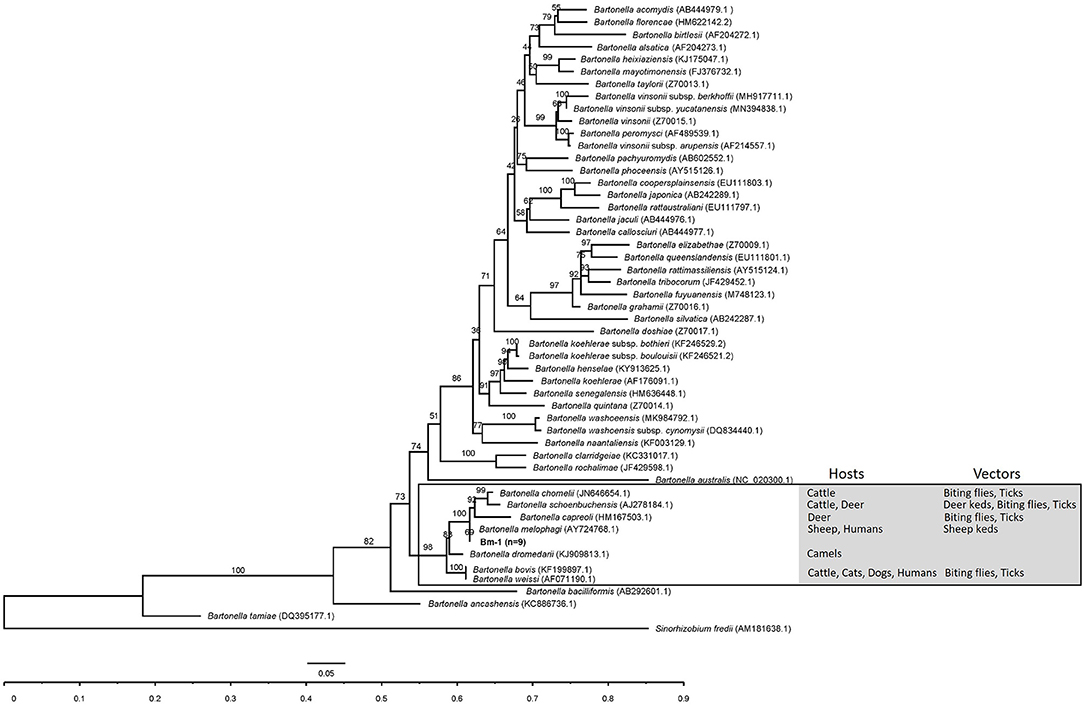
Figure 2. The phylogenetic tree of Bartonella gltA sequences was constructed by using maximum likelihood (ML) and GTR+I+G4+F as evolutionary models. The sequences detected in this study are shown in bold, and the numbers at the nodes correspond to bootstrap values. Sinorhizobium fredii was used as the out group.
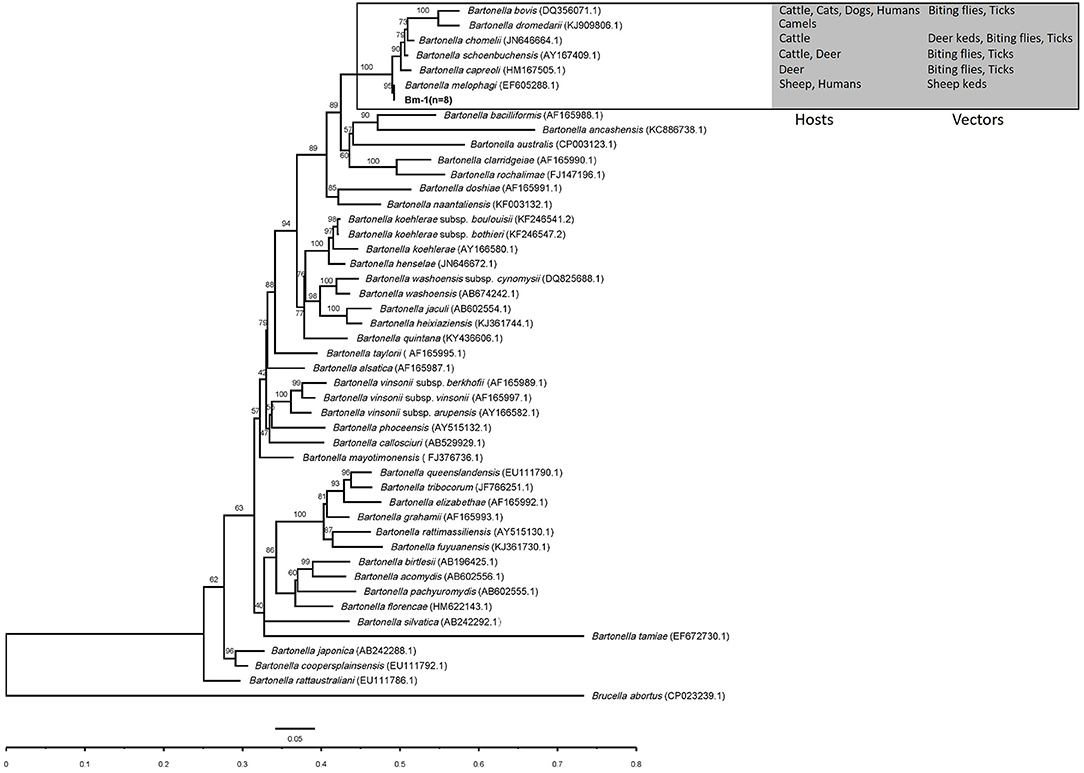
Figure 3. The phylogenetic tree of Bartonella rpoB sequences was constructed by using maximum likelihood (ML) and HKY+I+G4+F as evolutionary models. The sequences detected in this study are shown in bold, and the numbers at the nodes correspond to bootstrap values. Brucella abortus was used as the out group.
Discussion
Our study reports the prevalence of Bartonella in ticks in the border area of Xinjiang, China. A total of 1,549 tick samples were collected in the current study, and 2.19% (340/1,549) contained Bartonella DNA assayed by ribC-PCR. The gltA, rpoB and ITS genes of the ribC gene-positive samples were selectively amplified, and BLASTn analysis showed that the gltA, rpoB, ITS and ribC genes were all B. melophagi. This may be due to the potential pitfall in this study, because the positive control we selected is Bartonella melophagi. In view of this, more follow-up studies are needed, such as using another Bartonella as a positive control. Previous studies on Bartonella in ticks showed that the positive rates were 2.40% (7/292) in China (41), 2.48% (23/929) in 16 states of the USA (42), 2.32% (26/1,119) in California (43), and 2.2% (5/133) in Korea (15). Our results are similar to those reported previously. Another investigation of Bartonella in ticks from different countries reported positive rates of 1.05% (2/191) in Lithuania (44), 6.86% (19/277) in Algeria (9), 10.31% (13/126) in Brazil (12), 18.04% (57/316) in Poland (45), 10.4% (13/125) and 15.7% (40/254) in the Brazilian Cerrado and Taiwan (11, 12), 4.0% (8/200) in Malaysia (14), 5.8% (5/85) and 2.5% (1/40) in Korea (15), 8.56% (25/292) in North Korea (16), and 0.4% (1/234) in Pakistan (17). Previous studies on Bartonella in other ruminant-related blood-eating arthropods showed that the positive rates were 78.79% (26/33) in Hippoboscidae flies from Algeria (9) 4.21% (4/95) and 84.62% (11/13) in H. tuberculatus from the Brazilian Cerrado and H. quadripertusus from Israel (12, 13), 2.93% (16/546) in C. felis from Tunisia (18), 100% (133/133) and 36.87% (104/282) in M. ovinus from Central Europe and northeastern Algeria (19, 20), and 84.75% (50/59) and 93.75% (45/48) in L. cervi from Norway and Europe (6, 8). There were differences in the positive rates of Bartonella in ticks from different countries and regions, which may be related to the location, number, detection methods and ecological environment of the collected samples. Interestingly, the positive rate of Bartonella is also different in different arthropods. Because Bartonella species are mainly transmitted through vectors, it is speculated that the prevalence and richness of specific arthropods play a vital role.
Xinjiang is the largest province in China and is rich in tick resources. Forty-two species of ticks belonging to nine genera were reported in Xinjiang, accounting for more than 1/3 of the total number of tick species in China (26, 46, 47); the wide distribution of ticks has a significant impact on the development of animal husbandry and on public health. In recent years, B. melophagi was detected in M. ovinus from northeastern Algeria (20), China (31), central Europe (19), the western United States (48), and northern Oromia, Ethiopia (12). B. melophagi was detected in white-tailed deer (49) and human blood (50) in the United States. Xinjiang is located in the hinterland of Eurasia and has a land border of more than 5,600 km bordering Russia, Kazakhstan, Kyrgyzstan, Tajikistan, Pakistan, Mongolia, India, and Afghanistan. With the proposal of the Belt and Road Initiative of China, import and export trade becomes more frequent, and Bartonella may spread among different countries and regions with the import and export of trade products.
In the phylogenetic inference, the gltA and rpoB sequences obtained in this study were clustered into a branch with Bartonella related to ruminants. In the cluster, the hosts of Bartonella were mainly ruminants, such as cattle, sheep, camel, and deer, and the vectors of Bartonella mainly included deer keds, sheep keds, biting flies, and ticks (Figures 2, 3). However, ticks can be invoked as vectors for several ruminant-related Bartonella. This suggests that certain vectors, especially ticks, may play a key role in the spread of Bartonella. At the same time, ticks have a wide range of hosts, which makes it possible for Bartonella to spread across species through ticks. Ruminant-related Bartonella clustered into one branch, which may be explained by the relationship between Bartonella and the host. A host-specific association between B. washoensis and squirrel was revealed by using multisite sequence analysis. The results showed that B. washoensis may have co-speciated with the host squirrel. The phylogenetic relationships showed that the B. washoensis strain of Spermophilus is mainly related to the host genus rather than to its geographic origin (51). There may be a symbiotic relationship between M. ovinus and B. melophagi (8), and B. melophagi may transfer between M. ovinus and sheep (48). Interestingly, we also detected B. melophagi in ticks collected from cattle (Table 3). The coparasitism of ticks and M. ovinus in sheep may be considered a route of B. melophagi transmission. At present, there are no reports on associations between B. melophagi and sheep disease. B. melophagi was isolated from two women with a history of animal contact (50), which may suggest that B. melophagi is a possible human pathogen. In brief, our study provides evidence for the presence of B. melophagi DNA in ticks collected from cattle and sheep; however, further studies are needed to demonstrate that ticks are the vectors of B. melophagi. Additionally, we need to systematically investigate the prevalence of Bartonella in ticks, domestic mammals and people with a history of animal contact in Xinjiang, China. In general, this study is an important step in the control and public health safety of tick-borne Bartonella disease in mainland China and its neighboring countries.
Conclusions
In this study, molecular analysis was used to assess the presence and genetic diversity of B. melophagi in ticks collected from sheep and cattle from Xinjiang, China. This study provides new information on the presence and identity of B. melophagi in ticks from sheep and cattle. Ticks are used as the transmission medium of Bartonella, and it is necessary to investigate the prevalence of Bartonella in ticks, animals and people with animal contact.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MT536936.1; https://www.ncbi.nlm.nih.gov/genbank/, MT549891.1; https://www.ncbi.nlm.nih.gov/genbank/, MT549892.1; https://www.ncbi.nlm.nih.gov/genbank/, MT549893.1; https://www.ncbi.nlm.nih.gov/genbank/, MT549894.1; https://www.ncbi.nlm.nih.gov/genbank/, MT549895.1; https://www.ncbi.nlm.nih.gov/genbank/, MT635398.1; https://www.ncbi.nlm.nih.gov/genbank/, MT534396.1.
Author Contributions
JN and QR performed experiments. HL, MA, YLu, and ZM participated in sample collection. GL, ZC, JinL, and QR identification of tick samples. JN, JG, WL, XX, ZQ, ZW, and YT performed data analysis. JW, YLi, GG, JiaL, HY, and GL revised the manuscript. All authors read and approved the final manuscript.
Funding
The Central Public-interest Scientific Institution Basal Research Fund (Y2019YJ07-04 and Y2018PT76), the National Key Research, Development Programme of China (2019YFC1200502, 2019YFC1200500, 2017YFD0501206, and 2017YFD0501200), NPRC-2019-194-30 supported the design of the study and sample collection, and NSFC (1572511, 1702229, and 1471967) supported the writing of the manuscript and supported the analysis. ASTIP (CAAS-ASTIP-2016-LVRI), NBCITS (CARS-37), and the Jiangsu Co-innovation Center Programme for the Prevention and Control of Important Animal Infectious Disease and Zoonoses supported the interpretation of data in this study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank HL, MA, YLu, and ZM for their assistance in sample collection and ZC and GL for their contribution to sample identification. We are also grateful to YL, GG, JiaL, and HY for their revision of the manuscript.
References
1. Eicher SC, Dehio C. Bartonella entry mechanisms into mammalian host cells. Cell Microbiol. (2012) 14:1166–73. doi: 10.1111/j.1462-5822.2012.01806.x
2. Musso T, Badolato R, Ravarino D, Stornello S, Panzanelli P, Merlino C, et al. Interaction of Bartonella henselae with the murine macrophage cell line J774: infection and proinflammatory response. Infect Immun. (2001) 69:5974–80. doi: 10.1128/IAI.69.10.5974-5980.2001
3. Breitschwerdt EB. Bartonellosis, one health and all creatures great and small. Vet Dermatol. (2017) 28:96. doi: 10.1111/vde.12413
4. Dahmani M, Sambou M, Scandola P, Raoult D, Fenollar F, Mediannikov O. Bartonella bovis and candidatus Bartonella davousti in cattle from senegal. Comp Immunol Microbiol Infect Dis. (2017) 50:63–9. doi: 10.1016/j.cimid.2016.11.010
5. Raya AP, Jaffe DA, Chomel BB, Ota MS, Tsou PM, Davis AZ, et al. Detection of Bartonella species, including candidatus Bartonella ovis sp. nov, in ruminants from mexico and lack of evidence of Bartonella DNA in saliva of common vampire bats (Desmodus rotundus) predating on them. Vet Microbiol. (2018) 222:69–74. doi: 10.1016/j.vetmic.2018.06.018
6. Razanske I, Rosef O, Radzijevskaja J, Klepeckiene K, Lipatova I, Paulauskas A. Infections with Bartonella spp. in free-ranging cervids and deer keds (Lipoptena cervi) in Norway. Comp Immunol Microbiol Infect Dis. (2018) 58:26–30. doi: 10.1016/j.cimid.2018.06.003
7. Gonçalves LR, Teixeira MMG, Rodrigues AC, Mendes NS, Matos CA, Pereira CL, et al. Molecular detection of Bartonella species and Haemoplasmas in wild African buffalo (Syncerus caffer) in Mozambique, Africa. Parasitol Open. (2018) 4:10. doi: 10.1017/pao.2018.10
8. Halos L, Jamal T, Maillard R, Girard B, Guillot J, Chomel B, et al. Role of hippoboscidae flies as potential vectors of Bartonella spp. infecting wild and domestic ruminants. Appl Environ Microbiol. (2004) 70:6302–5. doi: 10.1128/AEM.70.10.6302-6305.2004
9. Boularias G, Azzag N, Gandoin C, Bouillin C, Chomel B, Haddad N, et al. Bartonella bovis and Bartonella chomelii infection in dairy cattle and their ectoparasites in Algeria. Comp Immunol Microbiol Infect Dis. (2020) 70:101450. doi: 10.1016/j.cimid.2020.101450
10. Chung CY, Kasten RW, Paff SM, Van Horn BA, Vayssier-Taussat M, Boulouis HJ, et al. Bartonella spp. DNA associated with biting flies from California. Emerg Infect Dis. (2004) 10:1311–3. doi: 10.3201/eid1007.030896
11. Tsai YL, Chomel BB, Chang CC, Kass PH, Conrad PA, Chuang ST. Bartonella and Babesia infections in cattle and their ticks in Taiwan. Comp Immunol Microbiol Infect Dis. (2011) 34:179–87. doi: 10.1016/j.cimid.2010.11.003
12. Goncalves LR, Harrus S, Gutierrez R, Herrera HM, de Souza Ramos IA, Porfirio GEO, et al. Molecular detection and genetic diversity of Bartonella species in large ruminants and associated ectoparasites from the Brazilian Cerrado. Transbound Emerg Dis. (2020) 67:1888–97. doi: 10.1111/tbed.13517
13. Gutierrez R, Cohen L, Morick D, Mumcuoglu KY, Harrus S, Gottlieb Y. Identification of different Bartonella species in the cattle tail louse (Haematopinus quadripertusus) and in cattle blood. Appl Environ Microbiol. (2014) 80:5477–83. doi: 10.1128/AEM.01409-14
14. Kho KL, Koh FX, Jaafar T, Nizam QN, Tay ST. Prevalence and molecular heterogeneity of Bartonella bovis in cattle and Haemaphysalis bispinosa ticks in peninsular Malaysia. BMC Vet Res. (2015) 11:153. doi: 10.1186/s12917-015-0470-1
15. Kang JG, Ko S, Kim HC, Chong ST, Klein TA, Chae JB, et al. Prevalence of Anaplasma and Bartonella spp. in ticks collected from Korean water deer (Hydropotes inermis argyropus). Korean J Parasitol. (2016) 54:87–91. doi: 10.3347/kjp.2016.54.1.87
16. Kang JG, Ko S, Smith WB, Kim HC, Lee IY, Chae JS. Prevalence of Anaplasma, Bartonella and Borrelia species in Haemaphysalis longicornis collected from goats in North Korea. J Vet Sci. (2016) 17:207–16. doi: 10.4142/jvs.2016.17.2.207
17. Ghafar A, Cabezas-Cruz A, Galon C, Obregon D, Gasser RB, Moutailler S, et al. Bovine ticks harbour a diverse array of microorganisms in Pakistan. Parasit Vectors. (2020) 13:1. doi: 10.1186/s13071-019-3862-4
18. Zouari S, Khrouf F, M'Ghirbi Y, Bouattour A. First molecular detection and characterization of zoonotic Bartonella species in fleas infesting domestic animals in Tunisia. Parasit Vectors. (2017) 10:436. doi: 10.1186/s13071-017-2372-5
19. Rudolf I, Betasova L, Bischof V, Venclikova K, Blazejova H, Mendel J, et al. Molecular survey of arthropod-borne pathogens in sheep keds (Melophagus ovinus), central Europe. Parasitol Res. (2016) 115:3679–82. doi: 10.1007/s00436-016-5175-2
20. Boucheikhchoukh M, Mechouk N, Benakhla A, Raoult D, Parola P. Molecular evidence of bacteria in Melophagus ovinus sheep keds and Hippobosca equina forest flies collected from sheep and horses in northeastern Algeria. Comp Immunol Microbiol Infect Dis. (2019) 65:103–109. doi: 10.1016/j.cimid.2019.05.010
21. Iannino F, Salucci S, Di Provvido A, Paolini A, Ruggieri E. Bartonella infections in humans dogs and cats. Veterinaria Italiana. (2018) 54:63–72. doi: 10.12834/VetIt.398.1883.2
22. Ben-Tekaya H, Gorvel JP, Dehio C. Bartonella and Brucella–weapons and strategies for stealth attack. Cold Spring Harb Perspect Med. (2013) 3:a010231. doi: 10.1101/cshperspect.a010231
23. Schouls LM, Van de Pol I, Rijpkema SGT schot CS. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in dutch Ixodes ricinus ticks. J Clin Microbiol. (1999) 37:2215–22. doi: 10.1128/JCM.37.7.2215-2222.1999
24. Wu XB, Na RH, Wei SS, Zhu JS, Peng HJ. Distribution of tick-borne diseases in China. Parasites Vector. (2013) 6:1–8. doi: 10.1186/1756-3305-6-119
25. Yu ZJ, Wang H, Wang TH, Sun WY, Yang XL, Liu JZ. Tick-borne pathogens and the vector potential of ticks in China. Parasites Vectors. (2015) 8:1–8. doi: 10.1186/s13071-014-0628-x
26. Chen Z, Yang XJ, Bu FJ, Yang XH, Yang XL, Liu JZ. Ticks (acari: ixodoidea: argasidae, ixodidae) of China. Experimental Appl Acarol. (2010) 51:393–404. doi: 10.1007/s10493-010-9335-2
27. Yu PF, Liu ZJ, Niu QL, Yang JF, Abdallah MO, Chen Z, et al. Molecular evidence of tick-borne pathogens in Hyalomma anatolicum ticks infesting cattle in Xinjiang uygur autonomous region, Northwestern China. Experi Appl Acarol. (2017) 73:269–81. doi: 10.1007/s10493-017-0162-6
28. Song SN, Chen CF, Yang MH, Zhao SS, Wang BJ, Hornok S, et al. Diversity of Rickettsia species in border regions of northwestern China. Parasites Vector. (2018) 11:1–7. doi: 10.1186/s13071-018-3233-6
29. Song R, Wang Q, Guo F, Liu X, Song S, Chen C, et al. Detection of Babesia spp., Theileria spp. and Anaplasma ovis in border regions, northwestern China. Transboundary Emerg Dis. (2018) 65:1537–44. doi: 10.1111/tbed.12894
30. Wang YH, Mao LL, Sun YW, Wang ZJ, Zhang JY, Zhang JB, et al. A novel Trancisella-like endosymbiont in Haemaphysalis longicornis and Hyalomma asiaticum, China. Vector-Borne Zoonotic Dis. (2018) 18:669–76. doi: 10.1089/vbz.2017.2252
31. Liu YH, He B, Li F, Li KR, Zhang LY, Li XQ, et al. Molecular identification of Bartonella melophagi and Wolbachia supergroup f from sheep keds in Xinjiang, China. Korean J Parasitol. (2018) 56:365–70. doi: 10.3347/kjp.2018.56.4.365
33. Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by pcr-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. (1995) 33:1797–803. doi: 10.1128/JCM.33.7.1797-1803.1995
34. Rozental T, Ferreira MS, Guterres A, Mares-Guia MA, Teixeira BR, Goncalves J, et al. Zoonotic pathogens in atlantic forest wild rodents in Brazil: Bartonella and Coxiella infections. Acta Tropica. (2017) 168:64–73. doi: 10.1016/j.actatropica.2017.01.003
35. Johnson G, Ayers M, McClure SCC, Richardson SE, Tellier R. Detection and identification of Bartonella species pathogenic for humans by PCR amplification targeting the riboflavin synthase gene (ribC). J Clin Microbiol. (2003) 41:1069–72. doi: 10.1128/JCM.41.3.1069-1072.2003
36. Jensen WA, Fall MZ, Rooney J, Kordick DL, Breitschwerdt EB. Rapid identification and differentiation of Bartonella species using a single-step PCR assay. J Clin Microbiol. (2000) 38:1717–22. doi: 10.1128/JCM.38.5.1717-1722.2000
37. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/molbev/mst010
38. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. (2017) 14:587–89. doi: 10.1038/nmeth.4285
39. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. (2015) 32:268–74. doi: 10.1093/molbev/msu300
40. Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, et al. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. (2020) 20:348–55. doi: 10.1111/1755-0998.13096
41. Hou J, Ling F, Liu Y, Zhang R, Song X, Huang R, et al. A molecular survey of Anaplasma, Ehrlichia, Bartonella and Theileria in ticks collected from southeastern China. Exp Appl Acarol. (2019) 79:125–35. doi: 10.1007/s10493-019-00411-2
42. Maggi RG, Toliver M, Richardson T, Mather T, Breitschwerdt EB. Regional prevalences of Borrelia burgdorferi, Borrelia bissettiae, and Bartonella henselae in Ixodes affinis, Ixodes pacificus and Ixodes scapularis in the USA. Ticks Tick Borne Dis. (2019) 10:360–64. doi: 10.1016/j.ttbdis.2018.11.015
43. Chang CC, Hayashidani H, Pusterla N, Kasten RW, Madigan JE, Chomel BB. Investigation of Bartonella infection in ixodid ticks from California. Comp Immunol Microbiol Infect Dis. (2002) 25:229–36. doi: 10.1016/S0147-9571(02)00012-7
44. Lipatova I, Razanske I, Jurgelevicius V, Paulauskas A. Bartonella washoensis infection in red squirrels (Sciurus vulgaris) and their ectoparasites in Lithuania. Comp Immunol Microbiol Infect Dis. (2020) 68:101391. doi: 10.1016/j.cimid.2019.101391
45. Dwuznik D, Mierzejewska EJ, Drabik P, Kloch A, Alsarraf M, Behnke JM, et al. The role of juvenile Dermacentor reticulatus ticks as vectors of microorganisms and the problem of ‘meal contamination'. Exp Appl Acarol. (2019) 78:181–202. doi: 10.1007/s10493-019-00380-6
46. Fan MY, Wang JG, Jiang YX, Zong DG, Lenz B, Walker DH. Isolation of a spotted fever group rickettsia from a patient and related ecologic investigations in Xinjiang Uygur autonomous region of China. J Clin Microbiol. (1987) 25:628–32. doi: 10.1128/JCM.25.4.628-632.1987
47. Wang YZ, Mu LM, Zhang K, Yang MH, Zhang L, Du JY, et al. A broad-range survey of ticks from livestock in Northern Xinjiang: changes in tick distribution and the isolation of Borrelia burgdorferi sensu stricto. Parasit Vectors. (2015) 8:449. doi: 10.1186/s13071-015-1021-0
48. Kosoy M, Bai Y, Enscore R, Rizzo MR, Bender S, Popov V, et al. Bartonella melophagi in blood of domestic sheep (Ovis aries) and sheep keds (Melophagus ovinus) from the southwestern us: cultures, genetic characterization, and ecological connections. Vet Microbiol. (2016) 190:43–49. doi: 10.1016/j.vetmic.2016.05.009
49. Izenour K, Zikeli S, Kalalah A, Ditchkoff SD, Starkey LA, Wang C, et al. Diverse Bartonella spp. detected in white-tailed deer (Odocoileus virginanus) and associated keds (Lipoptena mazamae) in the southeastern United States. J Wild Dis. (2020) 56:505–11. doi: 10.7589/2019-08-196
50. Maggi RG, Kosoy M, Mintzer M, Breitschwerdt EB. Isolation of candidatus Bartonella melophagi from human blood. Emerg Infect Dis. (2009) 15:66–8. doi: 10.3201/eid1501.081080
Keywords: Bartonella melophagi, ticks, ixodidae, cattle, sheep, Xinjiang
Citation: Ni J, Ren Q, Lin H, Aizezi M, Luo J, Luo Y, Ma Z, Chen Z, Liu W, Guo J, Qu Z, Xu X, Wu Z, Tan Y, Wang J, Li Y, Guan G, Luo J, Yin H and Liu G (2021) Molecular Evidence of Bartonella melophagi in Ticks in Border Areas of Xinjiang, China. Front. Vet. Sci. 8:675457. doi: 10.3389/fvets.2021.675457
Received: 03 March 2021; Accepted: 25 May 2021;
Published: 22 June 2021.
Edited by:
Andrei Daniel Mihalca, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Muhammad Hammad Hussain, Ministry of Agriculture and Fisheries, OmanMichael Kosoy, KB One Health LLC, United States
Copyright © 2021 Ni, Ren, Lin, Aizezi, Luo, Luo, Ma, Chen, Liu, Guo, Qu, Xu, Wu, Tan, Wang, Li, Guan, Luo, Yin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangyuan Liu, bGl1Z3Vhbmd5dWFuQGNhYXMuY24=; Qiaoyun Ren, cmVucWlhb3l1bkBnbWFpbC5jb20=
 Jun Ni1
Jun Ni1 Jin Luo
Jin Luo Ze Chen
Ze Chen Wenge Liu
Wenge Liu Xiaofeng Xu
Xiaofeng Xu Jinming Wang
Jinming Wang Guiquan Guan
Guiquan Guan Jianxun Luo
Jianxun Luo