
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 11 August 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.634286
This article is part of the Research Topic Antibiotic Resistance and its Continuity in the Environmental Niche View all 19 articles
Candida krusei (C. krusei) has been recently recognized as an important pathogen involved in mycotic mastitis of cows. The phenotypic and molecular characteristics of 15 C. krusei clinical isolates collected from cows with clinical mastitis in three herds of Yinchuan, Ningxia, were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry. In addition to sequencing analysis, the ERG11 gene that encodes 14α-demethylases, the expression of the ERG11 gene, and efflux transporters ABC1 and ABC2 in itraconazole-susceptible (S), itraconazole-susceptible dose dependent (SDD), and itraconazole-resistant (R) C. krusei isolates was also quantified by a quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) assay. Sequencing analysis revealed three synonymous codon substitutions of the ERG11 gene including T939C, A756T, and T642C in these C. krusei clinical isolates. Among them, T642C and T939C mutations were detected in itraconazole-resistant and -susceptible C. krusei isolates, but the A756T substitution was found only in itraconazole-resistant isolates. Importantly, the expression of the ERG11 gene in itraconazole-resistant isolates was significantly higher compared with itraconazole-SDD and itraconazole-susceptible isolates (p = 0.052 and p = 0.012, respectively), as determined by the qRT-PCR assay. Interestingly, the expression of the ABC2 gene was also significantly higher in itraconazole-resistant isolates relative to the itraconazole-SDD and itraconazole-susceptible strains. Notably, the expression of ERG11 was positively associated with resistance to itraconazole (p = 0.4177 in SDD compared with S, p = 0.0107 in SDD with R, and p = 0.0035 in S with R, respectively). These data demonstrated that mutations of the ERG11 gene were involved in drug resistance in C. krusei. The A756T synonymous codon substitution of the ERG11 gene was correlated with an increased expression of drug-resistant genes including ERG11 and ABC2 in itraconazole-resistant C. krusei isolates examined in this study.
Cow mastitis has a major negative impact on dairy industries, causing significant economic losses to farmers. Pathogenically, a wide variety of microorganisms have been identified as causative agents of cow mastitis, mainly bacteria and fungi (1). In cases of fungal infection of the mammary gland, yeasts of Candida genus are the most reported fungal pathogens in cow mastitis (2).
Historically, the mycotic mastitis caused by fungi of the Candida genus was first described by Fleischer as early as 1930 (3). Although the infection of C. albicans was considered as the most common cause of mastitis, cases of cow mastitis caused by the infection of non-albicans Candida spp. (NAC), such as C. krusei, C. parapsilosis, C. glabrata, and C. tropicalis, have increased significantly during the last decade (4). Among these NACs, C. krusei ranked as the fifth most common cause of cow mycotic mastitis (5, 6). C. krusei was reported as the causative agent of bovine mastitis and bronchopneumonia in Canada, Mexico, Japan, the United Kingdom, Turkey, Poland, and Algeria (7, 8). Our previous investigation also suggested that C. krusei was one of the most important pathogens in mycotic mastitis in dairy farms of the Yinchuan region in Ningxia, China. This result was in accordance with the report of Erbaş et al. (9).
Candida krusei has been regarded as a multidrug-resistant fungal pathogen because of its intrinsic resistance to fluconazole (FLC) (10, 11), with more than 96% of human clinical and veterinary isolates being fluconazole-resistant (12). Azole is one of the most common antifungal drugs in agricultural practices (13), including itraconazole, ketoconazole, and tetraconazole. Although it has been reported that multiple mechanisms are involved in drug resistance in Candida spp., mechanisms involved in alterations of target enzymes and upregulation of multidrug resistance (MDR)-related proteins are the common mechanisms of Candida resistant to azoles. In this regard, 14α-lanosterol demethylase (14-DM) is the target enzyme of azoles, which is responsible for the production of an ergosterol precursor and is encoded by the ERG11 gene. In C. albicans and C. parapsilosis, the efflux pump genes CDR1, CDR2, and MDR1 are also associated with azole resistance (14). Nowadays, although transporter genes ABC1 and ABC2 were involved in drug resistance in C. krusei (15, 16), increased lines of evidence suggested that changes in the expression of activity of target enzyme and upregulation of MDR were the main mechanisms of drug resistance in C. krusei (11, 16). To date, the study on mechanisms of Candida in azole resistance has mainly focused on C. albicans, C. glabrata, and C. tropicalis, but studies on azole resistance mechanisms in C. krusei, especially the C. krusei isolates from cow mastitis resistant to itraconazole, are limited, and the involvement of ERG11, ABC1, and ABC2 genes in the drug resistance of C. krusei has not been determined (17).
In the present study, we evaluated the profile of the susceptibility of C. krusei to itraconazole and investigated the potential alterations of the ERG11 gene and the differential expression of ERG11, ABC1, and ABC2 genes of 15 clinical C. krusei isolates that were isolated from cow mastitis in Yinchuan, Ningxia, China.
This study was submitted to and approved by the Ethic Committee of Animal Study in Ningxia University. A total of 465 quarter-milk samples were collected from the cows with clinical or subclinical mastitis, which originated from three herds in Yinchuan, Ningxia, China. Clinical or subclinical mastitis was defined by swelling, reduced milk flow, and abnormal milk appearance (watery to viscous with clots varying from gray-white to yellowish). Additionally, other signs of infection such as fever, inappetence, ataxia, and depression were also considered. These cows have been treated with antibiotics before sample collection. The isolates of C. krusei were identified by using Candida chromogenic medium (CHROMagar, France) (18) and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) with score values of >2.000 (VITEK® MS, BioMerieux, France) (19) and stored in liquid nitrogen.
The susceptibility assay was conducted using the Clinical and Laboratory Standard Institute (CLSI) broth microdilution (BMD) method. The CLSI BMD method was performed in a 96-well polystyrene microtiter plate in accordance with CLSI M27-A3 and M27-S4 guidelines (20, 21). The ranges of concentrations of tested drugs were as follows: 5-flucytosine (0.03–64 μg/ml), amphotericin B (0.008–16 μg/ml), fluconazole (0.03–64 μg/ml), itraconazole (0.03–64 μg/ml), and ketoconazole (0.03–64 μg/ml) (15, 22). The antifungal drugs were all purchased from Meilun Biotechnologies (Dalian, China). The C. krusei NCCLS reference strain ATCC 6258 served as quality control to ensure the test (3, 23); 1 ×103 CFU/ml working suspension of the C. krusei isolates was added into the each well. Results were recorded as resistant, susceptible dose dependent, and sensitive as shown in Table 1.
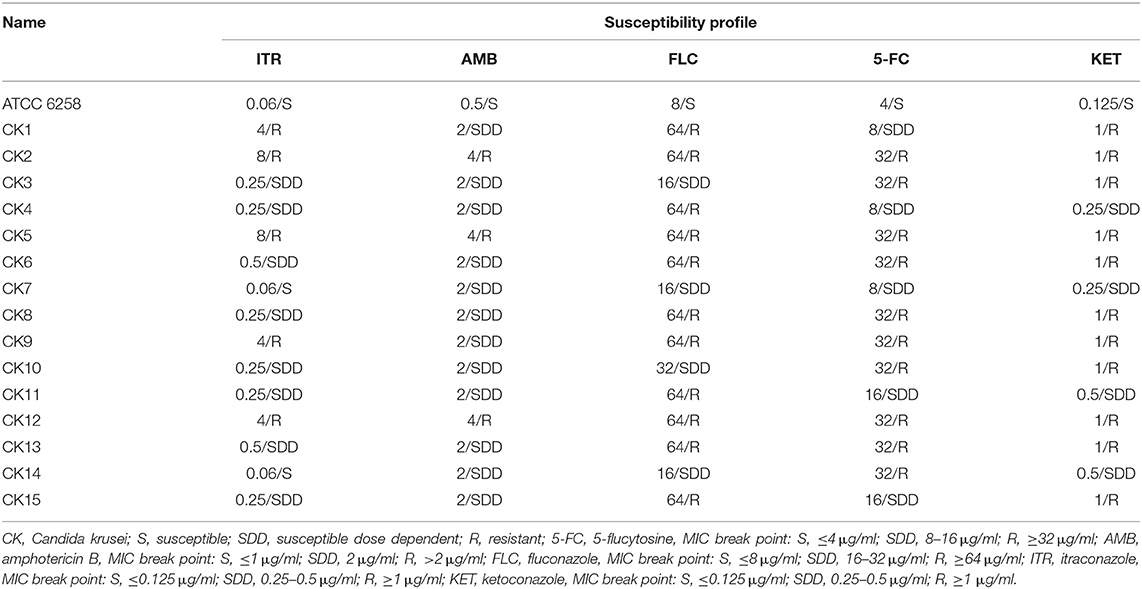
Table 1. Minimal inhibitory concentration (MIC) and susceptibility profile of C. krusei clinical isolates (n = 15).
Candida krusei isolates were subcultured twice on Sabouraud agar at 37°C for 18–24 h to revive and ensure the purity of cultures. A single colony was then transferred to 20 ml of liquid YPD (yeast extract 1%, dextrose 2%, and peptone 2%) broth and cultured at 35°C in a shaking incubator (120 rpm) exponential growth phase. The bacteria cells were collected by centrifugation at 3,000 rpm for 20 min, and the bacteria pellet was used for total genomic DNA preparation using a DNI5-A new Plant Genomic DNA Rapid Extraction Kit (Aidlab Biotechnologies, Beijing, China) according to the manufacturer's instruction. The isolated DNA was used as a template for amplification of the ERG11 gene. The primer set of ERG11 was designed by Primer 5.0 and synthesized at Sangon Biotech (Shanghai, China), based on the available sequence information of the C. krusei ERG11 gene (Gene accession number DQ903905) at the National Center for Biotechnology Information (NCBI) (Table 2). The PCR amplification was conducted in 25 μl volume containing 1 μl of genomic DNA (200 ng/μl), 0.5 μl of specific forward and reverse primers (50 μmol/L), and 12.5 μl of 2 × Phanta Max Master Mix (Vazyme Biotech, Nanjing, China). The PCR parameters were set as denaturation for 3 min at 95°C, followed by 35 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 1 min, and a final step of elongation (72°C for 5 min). The resultant PCR product was cleaned by gel purification in 1.5% agarose prior to being cloned into the pTOPO-TA Vector using a CV16-Zero Background pTOPO-Blunt Cloning Kit with Blue/white selection (Aidlab Biotechnologies, Beijing, China). The white colonies were analyzed for clones containing the DNA fragment of gene of interest. Eight to fifteen plasmids from clones generated from an identical PCR product were further sequenced for the ERG11 gene (Sangon Biotech, Shanghai, China). The sequences were then aligned with the online published sequence of the ERG11 gene of C. krusei strain (Gene Accession Number DQ903905) to determine gene mutation (17).
For quantitative real-time PCR (qRT-PCR) analysis, total RNA was extracted from C. krusei cultures with RNAiso Reagent (TaKaRa, Dalian, China) and reverse transcribed to cDNA with HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme Biotech, Nanjing, China) according to the manufacturer's instruction. For the ERG11 target gene and GAPDH reference gene, primer pairs were designed with the Primer 5.0 program and synthesized by Sangon, Shanghai, China (Table 2). qRT-PCR was conducted with a 20-μl volume containing the following reagents: 10 μl of 2 × ChamQ Universal SYBR qPCR Master Mix (Vazyme), 1 μl of total RNA sample, 0.5 μl of each primer pair at a concentration of 10 μmol/L, and 8 μl of distilled water. Each reaction was run in triplicate. Samples were subjected to an initial step at 95°C for 3 min, followed by 40 cycles, each of which consisted of 10 s at 95°C and 30 s at 60°C. Melting curves were recorded every 5 s. The fluorescence data were collected and analyzed with the QuantStudio Design Analysis Software 1.3.1. A 2−ΔΔCt algorithm was employed to analyze the relative expression levels of drug-resistant genes at resistant, susceptible dose-dependent, and sensitive strains.
Statistical analysis was performed with the GraphPad Prism program (GraphPad 8.0.1 Software Inc., San Diego, CA). The two-tailed Student's t-test was used to analyze significant differences between gene expression displayed by the distinct C. krusei strains; p < 0.05 was considered statistically significant.
A total of 15 C. krusei isolates (designated as CK1–CK15) were isolated from clinical samples from April 2018 to October 2019 in three herds at Yinchuan, Ningxia, China. Drug sensitivity testing was performed according to the broth microdilution method M27-A2 (NCCLS 2002), and the result showed that among 15 C. krusei isolates, 73.4, 73.4, and 66.7% were resistant to fluconazole (FLC), ketoconazole (KET), and 5-flucytosine (5-FC), respectively. However, 20 and 33.3% of the isolates were susceptible to amphotericin B (AMB) and itraconazole (ITR), respectively. Interestingly, isolates CK2, CK5, CK6, CK8, CK9, CK12, and CK13 showed resistance to both 5-FC and FLC. The rates of resistance to azole of these C. krusei isolates are listed in Table 3. Moreover, isolates CK2, CK5, and CK12 showed multidrug resistance to AMB, 5-FC, FLC, KET, and ITR. In contrast, the reference strain ATCC 6258 was susceptible to all of the five antifungal agents. Overall, among these 15 isolates, 5 were isolates resistant to ITR, 8 belonged to susceptible dose-dependent isolates, and 2 were isolates susceptible to ITR (Tables 1, 3).
The ERG11 gene fragment was amplified from ATCC 6258 and all 15 C. krusei isolates. The PCR product of the open frame of the ERG11 gene was 1,587 bp, which encodes 529 amino acids. Sequencing analysis identified four different mutations, three synonymous mutations (C642T, A756T, and T939C), and one missense mutation (C44T) in these 15 C. krusei isolates (Table 4). Synonymous mutations C642T and T939C were presented in all sequenced isolates, but the synonymous mutation A756T was found in the ERG11 gene of C. krusei isolates resistant to itraconazole (Table 4). One missense mutation was also found at 44 bp (C → T) of the ERG11 gene (Table 4), which resulted in an amino acid alteration from alanine to valine. However, such a missense mutation was also found in the reference strain ATCC 6258 and all C. krusei strains, which indicated that the C44T missense mutation might not be associated with drug resistance to azoles in C. krusei.
In order to examine whether an alteration of ERG11 gene expression was correlated with the drug resistance of C. krusei clinical isolates, the transcript of the ERG11 gene was accessed by a qRT-PCR assay. In comparison with the reference strain ATCC 6258, the relative ERG11 gene expression of field isolates in five itraconazole-resistant isolates was significantly upregulated (p < 0.01), while only two itraconazole-susceptible dose-dependent isolates showed a significantly upregulated ERG11 gene expression (p < 0.01, Figure 1A). The result showed that the transcript of the ERG11 gene in itraconazole-resistant isolates was significantly more abundant than itraconazole-susceptible strains (p = 0.0012, Figure 1B) and itraconazole-susceptible dose-dependent (SDD) strains (p = 0.0052, Figure 1B). However, there was no significant difference between itraconazole-susceptible dose-dependent isolates and itraconazole-susceptible isolates (p = 0.2562, Figure 1B).
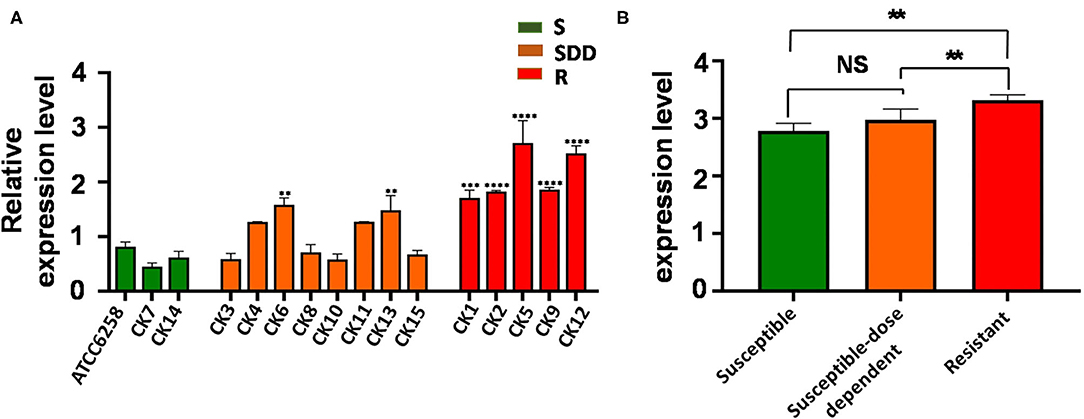
Figure 1. ERG11 relative gene expression levels in three groups of C. krusei clinical isolates. (A) Relative levels of ERG11 mRNA in all the C. krusei clinical isolates. ERG11 gene expression levels were quantified and normalized relative to the reference gene, GAPDH; S, itraconazole-susceptible; SDD, itraconazole-susceptible dose dependent; R, itraconazole-resistant. Asterisks indicate that the difference between field isolates with the reference strain ATCC 6258 is significant compared to the reference strain ATCC 6258: **p < 0.01 in CK6, CK13 with ATCC6258; ***p < 0.001 in CK1 with ATCC6258; and ****p < 0.0001 in CK2, CK5, CK9, CK12 with ATCC6258. (B) Log10+3 fold increase of gene expression levels in three groups (NS, no significance in SDD compared with S; *p < 0.05 in R with SDD; **p < 0.01 in R with S).
ABC transporters are involved in drug resistance; next, we therefore sought to examine the alteration of ABC transporters in C. krusei isolates. Interestingly, unlike the ERG11 gene, none of field isolates showed an upregulated ABC1 gene expression as compared with that of the reference strain ATCC 6258 (Figure 2A). The results of qRT-PCR for ABC1 genes showed that the expression of ABC1 gene mRNA was not significantly different between itraconazole-resistant isolates, itraconazole-susceptible dose-dependent isolates, and itraconazole-susceptible strains (p = 0.3844, p = 0.9997, and p = 0.2996, respectively, Figure 2B). Similar to that seen in the ERG11 gene, the relative ABC2 gene expression was extremely significantly upregulated in four itraconazole-resistant isolates (p < 0.01) and significant in one itraconazole-resistant isolate (p < 0.05), as compared with the reference strain ATCC 6258, whereas only two itraconazole-susceptible dose-dependent isolates showed an extremely significant upregulation of ABC2 gene expression compared to the reference strain ATCC 6258 (p < 0.01, Figure 3A). Intriguingly, the transcript of the ABC2 gene in itraconazole-resistant isolates was more abundant relative to itraconazole-susceptible strains (p = 0.0035) and itraconazole-susceptible dose-dependent strains (p = 0.0107, Figure 3B). No significant difference was found in the expression of the ABC2 gene was determined between the itraconazole-susceptible dose-dependent group and itraconazole-susceptible C. krusei isolates (p = 0.4177, Figure 3B).
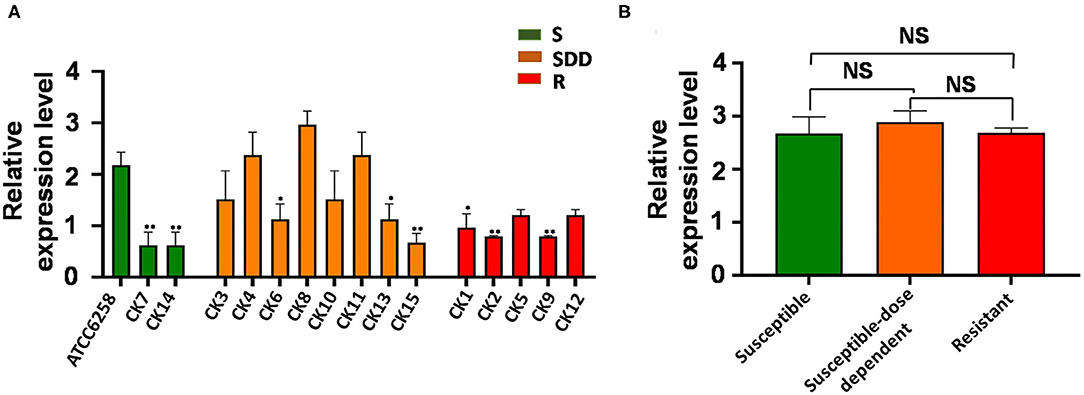
Figure 2. ABC1 relative gene expression levels in three groups of C. krusei clinical isolates. (A) Relative levels of ABC1 mRNA in all the C. krusei clinical isolates. ABC1 gene expression levels were quantified and normalized relative to the reference gene, GAPDH; S, itraconazole-susceptible; SDD, itraconazole-susceptible dose dependent; R, itraconazole-resistant. Asterisks indicate that the difference between field isolates with the reference strain ATCC 6258 is significant compared to the reference strain ATCC 6258: *p < 0.05 in CK1, CK6, CK13 with ATCC6258 and **p < 0.01 in CK2, CK7, CK9, CK14, CK15 with ATCC6258. (B) Log10+3 fold increase of gene expression levels in three groups (NS, no significance in SDD compared with S; *p < 0.05 in R with SDD; **p < 0.01 in R with S).
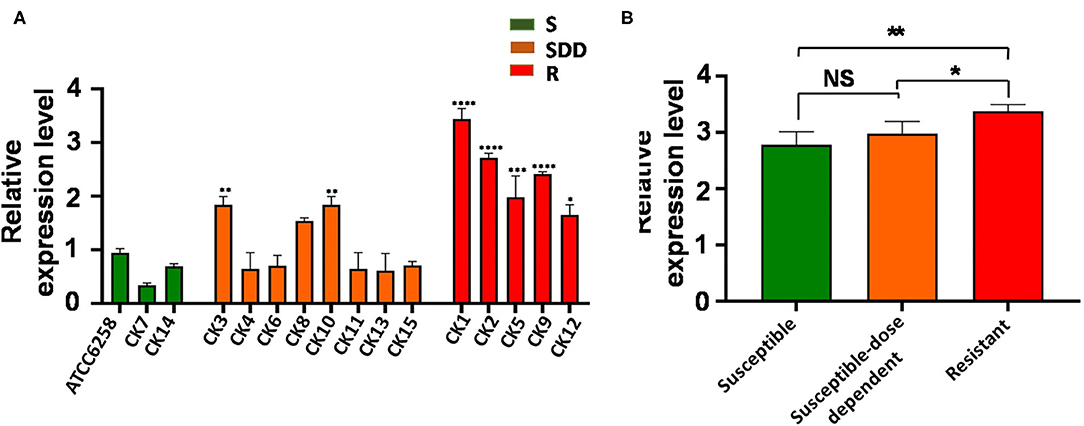
Figure 3. ABC2 relative gene expression levels in three groups of C. krusei clinical isolates. (A) Relative levels of ABC2 mRNA in all the C. krusei clinical isolates. ABC2 gene expression levels were quantified and normalized relative to the reference gene, GAPDH; S, itraconazole-susceptible; SDD, itraconazole-susceptible dose dependent; R, itraconazole-resistant. Asterisks indicate that the difference between field isolates with the reference strain ATCC 6258 is significant compared to the reference strain ATCC 6258: *p < 0.05 in CK12 with ATCC6258; **p < 0.01 in CK3, CK10 with ATCC6258; ***p < 0.001 in CK5 with ATCC6258; and ****p < 0.0001 in CK1, CK2, CK9 with ATCC6258. (B) Log10+3 fold increase of gene expression levels in three groups (NS, no significance in SDD compared with S; *p < 0.05 in R with SDD; **p < 0.01 in R with S).
Our previous study demonstrated that C. krusei was a predominant pathogen isolated from mycotic mastitis of cows in Yinchuan, Ningxia, China (12), suggesting that it may be an important fungal pathogen of mycotic mastitis of cows in this area. The crucial roles of drug-resistant genes ERG11, ABC1, and ABC2 in FLC-resistant clinical isolates of C. krusei from human have been well-established (15, 24). However, the pathogenic molecular mechanism of C. krusei isolated from cow mastitis remains unclear. In the present report, we evaluated the susceptibility profiles and mutations in the ERG11 gene in 15 clinical C. krusei isolates. The expression of drug-resistant genes ERG11, ABC1, and ABC2 between isolates susceptible, susceptible dose-dependent, and resistant to ITR was also analyzed. We identified three synonymous mutations and one missense mutation in the ERG11 gene in these clinical C. krusei isolates, as previously described in human C. krusei. Furthermore, the A756T was only presented in ITR-resistant strains, suggesting that it might be correlated with drug resistance in C. krusei, while mutations T642C and T939C were presented in all these 15 C. krusei isolates. The expression of drug-resistant ERG11 and ABC2 was also significantly higher in ITR-resistant C. krusei isolates compared to ITR-susceptible and susceptible dose-dependent isolates, suggesting a correlation of mutations of the ERG11 gene with the resistance to antifungal agents in C. krusei.
In the present study, based on the CLSI BMD method's susceptibility to ITR, the 15 C. krusei clinical isolates from cows with clinical mastitis could be divided into three groups: ITR-susceptible (2 isolates), ITR-susceptible dose-dependent (8 isolates), and ITR-resistant (5 isolates). The antifungal testing showed that 13.3, 53.3, and 33.3% were susceptible, susceptible dose-dependent, and resistant to itraconazole among these C. krusei clinical isolates, respectively. Notably, 13 of the 15 C. krusei isolates (73.4%) were also resistant to FLC and KET, 10 of the 15 C. krusei isolates (66.7%) were resistant to flucytosine, and 3 of the 15 isolates (20%) were resistant to amphotericin B. This finding was consistent with a study by Namvar et al. (25), but was different from reports by others (8, 9, 26). Of interest, the rate of resistance to antifungal agents in these C. krusei isolates was lower than our previous findings (12), which might be attributed to the reduction in the use of antifungal drugs during breeding. Consistent with our previous studies, C. krusei isolates were double-resistant and multidrug-resistant to antifungal drugs. It is strongly recommended that ketoconazole and other azole antifungal agents should not be used in the treatment of C. krusei infection in dairy cows in Ningxia, China, owing to high drug resistance.
It is worth noting that CK2, CK5, and CK12 showed resistance to amphotericin B, but such cases are rare (27). However, several lines of studies evidenced an increased minimum inhibitory concentration (MIC) of amphotericin B in C. krusei isolates. In Candida spp., resistance to amphotericin B was found to be associated with a decreased ergosterol content of cell membrane (28–30). In addition, an inactivation of ERG3 could substitute 14α-methylfecosterol with ergosterol, thus reducing ergosterol levels, which, in turn, resulted in deficient ergosterol to counter the function of amphotericin B (31). Moreover, several other mutations in the ergosterol biosynthetic genes such as ERG2, ERG5, ERG6, and ERG24 can also result in C. albicans and C. glabrata resistant to amphotericin B (32–34). However, such mutations have not been reported in C. krusei and further studies are needed in future studies.
A number of studies have focused on ERG11 gene mutation in Candida species (35–37). In order to fully understand ERG11 gene mutation of C. krusei, the whole open reading frame of the ERG11 gene was amplified for sequencing analysis, and three synonymous mutations and one missense mutation were identified in this study. C44T has one missense mutation and was found in all 15 isolates and the reference strain, suggesting that it had no impact on the itraconazole resistance of C. krusei. Molecularly, the C44T mutation resulted in the alteration of alanine to valine in the 15th amino acid of 14α-lanosterol demethylase (14-DM); this mutation might occur outside the active site of the ERG11 gene, which might not affect the mutual interaction of azole and 14-DM, or a single missense mutation might not be sufficient to change the affinity of the 14-DM to azole (15). Moreover, synonymous mutations (C642T, A756T, and T939C) in the 15 isolates were consistent with a previous report (24). Synonymous mutations can affect transcription, splicing, mRNA transport, and translation, any of which could change phenotype, rendering the synonymous mutation non-silent (38). He et al. (15) also reported that C642T, T1389C, and G1536C mutations occurred in all the experimental strains. In addition, the T1389C mutation was also reported by Ricardo et al. (16). Tavakoli et al. (35) revealed a heterozygous polymorphism at position T939C in the ERG11 coding region and speculated that this polymorphism might play a key role in the transcriptional regulation of genes and be involved in the processes of ergosterol biosynthesis. Of note, several previously reported mutations, including the T418C missense mutation (16), and C51T, T1389C, and G1536C synonymous mutations (15), have not been found in this study; thus, limited clinical isolates (15) were analyzed. Mechanistically, previous studies on C. albicans and C. tropicalis have demonstrated that the missense mutation was associated with resistance to azole, which was partly through changing the conformation of the target enzyme 14-DM, which, in turn, decreased its drug affinity and influenced the enzyme's function in ergosterol biosynthesis (39, 40). The resistance mechanism of these resistant strains may be due to one or multiple mutations in these genes, which needs further investigation.
In addition, the expression of ABC2 and ERG11 genes was significantly upregulated in C. krusei veterinary clinical isolates resistant to itraconazole. Previous studies demonstrated that resistance to azole was also due to the increased expression of ERG11. This results in insufficient azole activity owing to the overproduction of the target enzyme (41). Although ERG11 overexpression has been reported in C. krusei, the mechanism behind the overexpression remains unclear (15, 35). Another mechanism of resistance to azole is via the decreased intracellular accumulation of azole. This can be due to efflux pump activity or changes in the cell membrane. In this regard, drug efflux pumps belong to either the ATP-binding cassette (ABC) family of transporters or the major facilitator super family (MFS) class. These proteins can pump out fungicidal compounds across the cell membrane, and their overexpression results in multidrug resistance phenotype in pathogenic fungus (17). In contrast to members of the MFS class that are actuated by electrochemical proton-motive force, ABC family members depend on the hydrolysis of ATP for energy (42, 43). Indeed, along with the decrease of susceptibility to itraconazole, the ABC2 gene mRNA expression in the isolates appeared to increase. Although it was considered that Abc1p played an important role in the innate FLC resistance of C. krusei (16, 17), the ABC2 gene could be activated slower than the ABC1 gene in the presence of voriconazole (16). Given voriconazole tolerance, the Abc1p efflux pump is supposed to be more efficient in expelling drug and plays a late role in the development of resistance, and the accumulation of itraconazole in drug-susceptible C. krusei was higher than that of resistant strains (44). Interestingly, Venkateswarlu et al. found that two isolates highly resistant to fluconazole showed a different sensitivity to itraconazole, suggesting that itraconazole and fluconazole in C. krusei may have different resistance mechanisms (44). The authors stated that C. krusei resistant to itraconazole was due to decreased drug accumulation in cells and speculated that there might exist more efflux pumps contributing to itraconazole resistance of C. krusei, which could be well-explained in this study. The overexpression of both ERG11 and ABC2 has been reported to be involved in itraconazole resistance in C. krusei (15, 35). However, an unusual transient or stable resistance of C. krusei to voriconazole has also emerged. Overexpression of ABC2 and ERG11, as observed in itraconazole resistance, imparts a transient resistance to voriconazole, while a more stable resistance was observed due to the overexpression of ABC1 and point mutation in ERG11 (16). Taking this into account, we speculate that the resistance mechanisms of itraconazole and voriconazole in C. krusei clinical isolates may be different. Moreover, other genes encoding ATP-dependent efflux transporters may occur in C. krusei, such as a CgSNQ2 homologous gene that was verified as an azole-associated resistance gene in C. glabrata (45). Although the C. krusei genome has been sequenced, it is not completely annotated yet; thus, other transporter genes were not assessed. Our mRNA expression data showed that Abc2p may play a more important role in itraconazole resistance of C. krusei, instead of Abc1p.
This study has enriched our knowledge in the veterinary clinical C. krusei resistance gene expression and mutation data by comparing the difference between the veterinary clinical and the human clinical C. krusei, and further deepened our understanding of the resistance mechanism of C. krusei in veterinary clinics. The limitation of this study is that the sample size was small and no drug susceptibility test and resistance mechanism research related to echinocandin has been included, which require further investigations.
In conclusion, in this study, we found that C. krusei veterinary clinical isolates exhibited a different susceptibility to antifungal agents. Mechanistically, the A756T mutation in the ERG11 gene resulted in an upregulation of drug-resistant genes ERG11 and ABC2, substantially enhancing the resistance to itraconazole of C. krusei. Although we have identified four point mutations in the ERG11 gene associated with itraconazole resistance and have already described their role on the itraconazole resistance of C. krusei, it is necessary to confirm the effect of these mutations by site-directed mutagenesis of the C. krusei strain in the future. Nevertheless, this study may thus provide an insight into the mechanism of the resistance of C. krusei to antifungal agents, which warrants for further investigation.
The datasets generated for this study are available on request to the corresponding author.
The animal study was reviewed and approved by Ethics Committee of Animal Research of Ningxia University.
JD, YW, and XZ conceived and designed the experiments. JD and JF analyzed the data and drafted the manuscript. JD, WM, JF, and XL performed experiments and acquired data. XL and XZ interpreted data and critically revised the manuscript. All authors read and approved the final version of the manuscript.
This study was supported by a grant from the National Natural Science Foundation of China (No. 32060816), a grant from the Key Research and Development Plan Project of Ningxia Hui Autonomous Region (No. 2017BN04), and a grant from Natural Science Foundation of Ningxia (2019AAC03006). These funds play no role in the design of the study; collection, analysis, and interpretation of data; and the writing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank all the study participants, farm owners, veterinarians, and personnel of the Center of Disease Control and Veterinary Institute in Ningxia Hui Autonomous Region of China, who helped in the realization of this study.
1. Krukowski H, Lisowski A, Rózański P, Skórka A. Yeasts and algae isolated from cows with mastitis in the south-eastern part of Poland. Pol J Vet Sci. (2006) 9:181–4.
2. Dworecka-Kaszak B, Krutkiewicz A, Szopa D, Kleczkowski M, Biegańska M. High prevalence of Candida yeast in milk samples from cows suffering from mastitis in Poland. Sci World J. (2012) 2012:1–5. doi: 10.1100/2012/196347
3. Costa EO, Gandra CR, Pires MF, Coutinho SD, Castilho W, Teixeira CM. Survey of bovine mycotic mastitis in dairy herds in the State of São Paulo, Brazil. Mycopathologia. (1993) 124:13–7. doi: 10.1007/bf01103051
4. de Casia dos Santos R, Marin JM. Isolation of Candida spp. from mastitic bovine milk in Brazil. Mycopathologia. (2005) 159:251–3. doi: 10.1007/s11046-004-2229-2
5. Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Nagy E, Dobiasova S, et al. Candida krusei, a multidrug-resistant opportunistic fungal pathogen: geographic and temporal trends from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J Clin Microbiol. (2008) 46:515–21. doi: 10.1128/jcm.01915-07
6. Douglass AP, Offei B, Braun-Galleani S, Coughlan AY, Martos AAR, Ortiz-Merino RA, et al. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: one species, four names. PLoS Pathog. (2018) 14:e1007138. doi: 10.1371/journal.ppat.1007138
7. Ksouri S, Djebir S, Hadef Y, Benakhla A. Survey of bovine mycotic mastitis in different mammary gland statuses in two North-Eastern Regions of Algeria. Mycopathologia. (2015) 179:327–31. doi: 10.1007/s11046-014-9845-2
8. Sonmez M, Erbas G. Isolation and identification of Candida spp. from mastitis cattle milk and determination of antifungal susceptibilities. Int J Vet Sci. (2017) 6:104–7.
9. Erbaş G, Parin U, Kirkan S, Savaşan S, ÖZavci MV, Yüksel HT. Identification of Candida strains with nested PCR in bovinemastitis and determination of antifungal susceptibilities. Turkish J Vet Anim Sci. (2017) 41:757–63. doi: 10.3906/vet-1704-39
10. Gómez-Gaviria M, Mora-Montes HM. Current aspects in the biology, pathogeny, and treatment of Candida krusei, a neglected fungal pathogen. Infect Drug Resist. (2020) 13:1673–89. doi: 10.2147/idr.s247944
11. Jamiu AT, Albertyn J, Sebolai OM, Pohl CH. Update on Candida krusei, a potential multidrug-resistant pathogen. Med Mycol. (2021) 59:14–30. doi: 10.1093/mmy/myaa031
12. Du J, Wang X, Luo H, Wang Y, Liu X, Zhou X. Epidemiological investigation of non-albicans Candida species recovered from mycotic mastitis of cows in Yinchuan, Ningxia of China. BMC Vet Res. (2018) 14:251. doi: 10.1186/s12917-018-1564-3
13. Rocha MF, Alencar LP, Paiva MA, Melo LM, Bandeira SP, Ponte YB, et al. Cross-resistance to fluconazole induced by exposure to the agricultural azole tetraconazole: an environmental resistance school? Mycoses. (2016) 59:281–90. doi: 10.1111/myc.12457
14. Chen LM, Xu YH, Zhou CL, Zhao J, Li CY, Wang R. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Med Res. (2010) 38:536–45. doi: 10.1177/147323001003800216
15. He X, Zhao M, Chen J, Wu R, Zhang J, Cui R, et al. Overexpression of both ERG11 and ABC2 genes might be responsible for itraconazole resistance in clinical isolates of Candida krusei. PLoS ONE. (2015) 10:e0136185. doi: 10.1371/journal.pone.0136185
16. Ricardo E, Miranda IM, Faria-Ramos I, Silva RM, Rodrigues AG, Pina-Vaz C. In vivo and in vitro acquisition of resistance to voriconazole by Candida krusei. Antimicrob Agents Chemother. (2014) 58:4604–11. doi: 10.1128/aac.02603-14
17. Lamping E, Ranchod A, Nakamura K, Tyndall JD, Niimi K, Holmes AR, et al. Abc1p is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei. Antimicrob Agents Chemother. (2009) 53:354–69. doi: 10.1128/aac.01095-08
18. Hulimane S, Maluvadi-Krishnappa R, Mulki S, Rai H, Dayakar A, Kabbinahalli M. Speciation of Candida using CHROMagar in cases with oral epithelial dysplasia and squamous cell carcinoma. J Clin Exp Dent. (2018) 10:e657–60. doi: 10.4317/jced.54737
19. Maldonado I, Cataldi S, Garbasz C, Relloso S, Striebeck P, Guelfand L, et al. [Identification of Candida yeasts: conventional methods and MALDI-TOF MS]. Rev Iberoam Micol. (2018) 35:151–4. doi: 10.1016/j.riam.2018.02.002
20. Xiao M, Chen SC, Kong F, Xu XL, Yan L, Kong HS, et al. Distribution and antifungal susceptibility of Candida species causing candidemia in China: an update from the CHIF-NET study. J Infect Dis. (2020) 221(Suppl 2):S139–47. doi: 10.1093/infdis/jiz573
21. Aslani N, Abastabar M, Hedayati MT, Shokohi T, Aghili SR, Diba K, et al. Molecular identification and antifungal susceptibility testing of Candida species isolated from dental plaques. J Mycol Med. (2018) 28:433–6. doi: 10.1016/j.mycmed.2018.05.006
22. Barry AL, Pfaller MA, Brown SD, Espinel-Ingroff A, Ghannoum MA, Knapp C, et al. Quality control limits for broth microdilution susceptibility tests of ten antifungal agents. J Clin Microbiol. (2000) 38:3457–9. doi: 10.1128/jcm.38.9.3457-3459.2000
23. Fadda ME, Pisano MB, Scaccabarozzi L, Mossa V, Deplano M, Moroni P, et al. Use of PCR-restriction fragment length polymorphism analysis for identification of yeast species isolated from bovine intramammary infection. J Dairy Sci. (2013) 96:7692–7. doi: 10.3168/jds.2013-6996
24. Feng W, Yang J, Wang Y, Chen J, Xi Z, Qiao Z. ERG11 mutations and upregulation in clinical itraconazole-resistant isolates of Candida krusei. Can J Microbiol. (2016) 62:938–43. doi: 10.1139/cjm-2016-0055
25. Namvar Z, Sepahy AA, Tabatabaei RR, Rezaie S. Antifungal susceptibility of non-albicans Candida spp. isolated fromraw milk and human blood in Alborz and Tehran provinces Iran. J Microbiol. (2019) 11:520–6. doi: 10.18502/ijm.v11i6.2224
26. Xiao M, Fan X, Chen SC, Wang H, Sun ZY, Liao K, et al. Antifungal susceptibilities of Candida glabrata species complex, Candida krusei, Candida parapsilosis species complex and Candida tropicalis causing invasive candidiasis in China: 3 year national surveillance. J Antimicrob Chemother. (2015) 70:802–10. doi: 10.1093/jac/dku460
27. Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. (2013) 11:e1001692. doi: 10.1371/journal.pbio.1001692
28. Scorzoni L, de Paula ESAC, Marcos CM, Assato PA, de Melo WC, de Oliveira HC, et al. Antifungal therapy: new advances in the understanding and treatment of mycosis. Front Microbiol. (2017) 8:36. doi: 10.3389/fmicb.2017.00036
29. Anderson TM, Clay MC, Cioffi AG, Diaz KA, Hisao GS, Tuttle MD, et al. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol. (2014) 10:400–6. doi: 10.1038/nchembio.1496
30. Prasad R, Banerjee A, Shah Abdul H. Resistance to antifungal therapies. Essays Biochem. (2017) 61:157–66. doi: 10.1042/EBC20160067
31. Young LY, Hull CM, Heitman J. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob Agents Chemother. (2003) 47:2717–24. doi: 10.1128/aac.47.9.2717-2724.2003
32. Sanglard D. Emerging threats in antifungal-resistant fungal pathogens. Front Med. (2016) 3:11. doi: 10.3389/fmed.2016.00011
33. Hull CM, Bader O, Parker JE, Weig M, Gross U, Warrilow AG, et al. Two clinical isolates of Candida glabrata exhibiting reduced sensitivity to amphotericin B both harbor mutations in ERG2. Antimicrob Agents Chemother. (2012) 56:6417–21. doi: 10.1128/aac.01145-12
34. Jensen RH, Astvad KM, Silva LV, Sanglard D, Jørgensen R, Nielsen KF, et al. Stepwise emergence of azole, echinocandin and amphotericin B multidrug resistance in vivo in Candida albicans orchestrated by multiple genetic alterations. J Antimicrob Chemother. (2015) 70:2551–5. doi: 10.1093/jac/dkv140
35. Tavakoli M, Zaini F, Kordbacheh M, Safara M, Raoofian R, Heidari M. Upregulation of the ERG11 gene in Candida krusei by azoles. Daru. (2010) 18:276–80.
36. Jensen RH. Resistance in human pathogenic yeasts and filamentous fungi: prevalence, underlying molecular mechanisms and link to the use of antifungals in humans and the environment. Dan Med J. (2016) 63:B5288.
37. Sardari A, Zarrinfar H, Mohammadi R. Detection of ERG11 point mutations in Iranian fluconazole-resistant Candida albicans isolates. Curr Med Mycol. (2019) 5:7–14. doi: 10.18502/cmm.5.1.531
38. Goymer P. Synonymous mutations break their silence. Nat Rev Genet. (2007) 8:92–92. doi: 10.1038/nrg2056
39. Vandeputte P, Larcher G, Bergès T, Renier G, Chabasse D, Bouchara JP. Mechanisms of azole resistance in a clinical isolate of Candida tropicalis. Antimicrob Agents Chemother. (2005) 49:4608–15. doi: 10.1128/aac.49.11.4608-4615.2005
40. Strzelczyk JK, Slemp-Migiel A, Rother M, Gołabek K, Wiczkowski A. Nucleotide substitutions in the Candida albicans ERG11 gene of azole-susceptible and azole-resistant clinical isolates. Acta Biochim Pol. (2013) 60:547–52. doi: 10.18388/abp.2013_2019
41. MacPherson S, Akache B, Weber S, De Deken X, Raymond M, Turcotte B. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother. (2005) 49:1745–52. doi: 10.1128/aac.49.5.1745-1752.2005
42. Pao SS Paulsen IT Saier MH Jr. Major facilitator superfamily. Microbiol Mol Biol Rev. (1998) 62:1–34.
43. Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol. (2009) 10:218–27. doi: 10.1038/nrm2646
44. Venkateswarlu K, Denning DW, Manning NJ, Kelly SL. Reduced accumulation of drug in Candida krusei accounts for itraconazole resistance. Antimicrob Agents Chemother. (1996) 40:2443–6. doi: 10.1128/aac.40.11.2443
Keywords: Candida krusei, resistance, mycotic mastitis, cows, ERG11, ABC1, ABC2
Citation: Du J, Ma W, Fan J, Liu X, Wang Y and Zhou X (2021) The A756T Mutation of the ERG11 Gene Associated With Resistance to Itraconazole in Candida Krusei Isolated From Mycotic Mastitis of Cows. Front. Vet. Sci. 8:634286. doi: 10.3389/fvets.2021.634286
Received: 27 November 2020; Accepted: 14 July 2021;
Published: 11 August 2021.
Edited by:
Marina Spinu, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Veronica Risco-Castillo, INRA École Nationale Vétérinaire d'Alfort (ENVA), FranceCopyright © 2021 Du, Ma, Fan, Liu, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoming Liu, bHhtMTk2NkBueHUuZWR1LmNu; Yujiong Wang, d3lqQG54dS5lZHUuY24=; Xuezhang Zhou, emhvdXh1ZXpoYW5nQG54dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.