
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 21 June 2021
Sec. Livestock Genomics
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.633898
This article is part of the Research Topic Marbling Fat in Livestock View all 13 articles
 Yaolu Zhao1
Yaolu Zhao1 Elke Albrecht1*
Elke Albrecht1* Zeyang Li2
Zeyang Li2 Johannes Schregel2
Johannes Schregel2 Quentin L. Sciascia2
Quentin L. Sciascia2 Cornelia C. Metges2
Cornelia C. Metges2 Steffen Maak1
Steffen Maak1Piglets with low birth weight (LBW) usually have reduced muscle mass and increased lipid deposition compared with their normal-birth-weight (NBW) littermates. Supplementation of piglets with amino acids during the first days of life may improve muscle growth and simultaneously alter the intramuscular lipid deposition. The aim of the current study was to investigate the influence of glutamine (Gln) supplementation during the early suckling period on lipid deposition in the longissimus muscle (MLD) and the role of different perilipin (PLIN) family members in this process. Four groups were generated consisting of 72 male LBW piglets and 72 NBW littermates. Piglets were supplemented with either 1 g Gln/kg body weight or an isonitrogenous amount of alanine (Ala) between days post natum (dpn) 1 and 12. Twelve piglets per group were slaughtered at 5, 12, and 26 dpn, and muscle tissue was collected. Perilipins were localized by immunohistochemistry in muscle sections. The mRNA and protein abundances of PLIN family members and related lipases were quantified by quantitative RT-PCR (qPCR) and western blots, respectively. While PLIN1 was localized around lipid droplets in mature and developing adipocytes, PLIN2 was localized at intramyocellular lipid droplets, PLIN3 and 4 at cell membranes of muscle fibers and adipocytes, and PLIN5 in the cytoplasm of undefined cells. The western blot results indicated higher protein abundances of PLIN2, 3, 4, and 5 in LBW piglets (p < 0.05) at 5 dpn compared with their NBW littermates independent of supplementation, while not directly reflecting the mRNA expression levels. The mRNA abundance of PLIN2 was lower while PLIN4 was higher in piglets at 26 dpn in comparison with piglets at 5 dpn (p < 0.01). Relative mRNA expression of LPL and CGI-58 was lowest in piglets at 5 dpn (p < 0.001). However, ATGL mRNA was not influenced by birth weight or supplementation, but the Spearman correlation coefficient analysis revealed close correlations with PLIN2, 4, and 5 mRNA at 5 and 26 dpn (r > 0.5, p < 0.001). The results indicated the importance of birth weight and age for intramuscular lipid deposition and different roles of PLIN family members in this process, but no clear modulating effect of Gln supplementation.
Low-birth-weight (LBW) piglets normally have a higher mortality rate and growth retardation of muscles compared with their normal-birth-weight (NBW) littermates, which in turn causes a delay in whole body growth (1). Furthermore, LBW piglets normally have greater fat deposition (2) and exhibit increased fatness at slaughter age (1). In the current project, we attempted to ameliorate the disadvantages of the LBW piglets with adapted nutrition, in particular with glutamine (Gln) supplementation, which was assumed insufficient in maternal milk especially for LBW piglets (3, 4). In our previous investigation (5), we observed fewer muscle fibers and more intramyocellular lipid droplets at 5 days post natum (dpn) in LBW piglets. Moreover, Gln supplementation was shown to increase intramuscular availability of free Gln in skeletal muscle in a short term and to influence muscle fiber size, but had no influence on further muscle morphology traits and a minor influence on the abundance of myosin heavy chain isoforms. The current study focused on the process of lipid deposition and redistribution between muscle fibers and developing adipocytes as part of the muscle development and how it was influenced by birth weight (BiW) and Gln supplementation.
Skeletal muscle development includes myogenesis, adipogenesis, as well as fibrogenesis (6). During the whole lifetime of pigs, lipogenesis interplays and competes with muscle growth (7, 8). However, the lipid content in skeletal muscle is not just a storage of nutritional energy; it is of great importance for meat quality (9). Appropriate intramuscular lipids in pork promote flavor, tenderness, and juiciness (10), and consequently, intramuscular lipid development has become increasingly importantly raised in pork production. Intramuscular fat in pig represents only a small part of total body fat (6). The lipids within skeletal muscle are either stored in intramuscular adipocytes or within muscle fibers as intramyocellular lipid droplets (5). These intramuscular lipid droplets, also named lipid or fat bodies, play a vital role in lipid homeostasis in skeletal muscle (11). They function in storing lipids and supplying energy (11–13), as well as providing lipids for membrane synthesis (14) and protecting the cells from lipotoxicity (15). Moreover, the intramuscular lipid droplets were found to be formed de novo (16), and their formation and physical functions are controlled by a series of proteins, such as synaptosomal-associated protein 23 (SNAP23) (17), caveolins (18), PAT family (19), and others. Among these proteins, PAT family proteins, named after perilipin, adipophilin, or adipose differentiation-related protein (ADRP), and the tail-interacting protein of 47 kDa (TIP47) (20) have drawn increasing attention in recent years. There are two new members in this family, S3–12 (21) and myocardial lipid droplet protein—MLDP (22), also known as OXPAT (23) or lipid storage droplet protein 5 (LSDP5) (24). These five proteins, also known as PLIN1–5 (25), participate in the process of lipid droplet formation, stabilization, and lipolysis in different cells (26). The encoding genes can serve as suitable biomarkers when studying lipid deposition and adipocyte development within skeletal muscle in piglets.
Our hypothesis was that differences in intramuscular lipid deposition between LBW and NBW piglets are modulated by Gln supplementation, and different PLIN family members are involved in this process. Therefore, the objective of the current study was to investigate the expression pattern of PLINs in LBW and NBW piglets during the first weeks after birth on mRNA and protein level and their modulation by Gln supplementation.
The study involved German Landrace male piglets with 72 LBW and 72 NBW littermates, nursed and supplemented with Gln or an isonitrogenous amount of alanine (Ala) as described in detail in our former publication (5). Twelve piglets per group were sacrificed at 5, 12, or 26 dpn. The four treatment groups were named LBW-GLN, LBW-ALA, NBW-GLN, and NBW-ALA. Experimental procedures and animal care were carried out strictly according to the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes (2010/63/EU) and were approved by the responsible State Office for Agriculture, Food Safety and Fishing Mecklenburg Western Pomerania, Germany (permission no. 7221.3-1-026/16). Tissue of musculus longissimus dorsi (MLD) and musculus semitendinosus (MST) was collected immediately after slaughter, snap frozen in liquid nitrogen, and subsequently stored at −80°C until analysis.
Muscle serial sections of MLD and MST were cut 10-μm thick using a cryostat microtome (CM3050 S, Leica, Bensheim, Germany). The muscle sections were stained with hematoxylin and eosin (H/E, hematoxylin: Dako, Glostrup, Denmark; eosin: Chroma Gesellschaft, Münster, Germany) or Oil Red O according to standard protocols. The H/E-stained images were used for measurement, and the Oil Red O-stained images were used to verify the selected adipocytes. Images were taken with an Olympus BX43 microscope (Olympus, Hamburg, Germany) equipped with a UC30 color camera and analyzed with Cell∧D imaging software (OSIS, Münster, Germany). At least 200 intramuscular adipocytes in both muscles were measured for each piglet using the interactive measurement module of the Cell∧D software. If <200 adipocytes could be found, as in many of 5 dpn piglets, all available adipocytes were measured.
Muscle RNA was isolated from MLD pieces (70–90 mg) using the RNeasy Fibrous Tissue Mini Kit (Qiagen, Hilden, Germany), following the manufacturer's instructions, and stored at −80°C. All RNA concentrations were measured with a NanoDrop ND-1000 spectrophotometer (Peqlab, Erlangen, Germany), and RNA integrity was determined using the Experion Automated Electrophoresis System and the RNA StdSens analysis chip (Bio-Rad, Munich, Germany). Then, first-strand cDNA was synthesized in a 20-μl reaction volume from 150 ng RNA with an iScript cDNA Synthesis Kit (Bio-Rad). Primers of reference and target genes were designed with Primer 3 web version 4.1.0 (http://primer3.ut.ee/) or adapted from published papers [tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta, YWHAZ (27); peptidylprolyl isomerase A, PPIA (28)], as shown in Table 1. All primers were synthesized by a commercial company (Sigma-Aldrich, Darmstadt, Germany). The annealing temperature of all primers was 60°C. To test these primers, qualitative polymerase chain reaction (PCR) was performed, and the products were subjected to 3% agarose gel electrophoresis and sequenced as described by Liu et al. (29). The quantitative RT-PCR (qPCR) was performed in duplicate as described by Schering et al. (30) with FastStart Essential DNA Green Master using a LightCycler® 96 real-time qPCR system (Roche, Basel, Switzerland). The mRNA expression values of target genes were normalized to two stable reference genes: YWHAZ and PPIA. The quantitation cycle (Cq) value was analyzed by the LightCycler® 96 system software. Efficiencies of amplifications were calculated by standard curves, which were calculated from serial dilutions (1:1, 1:10, 1:50, 1:100, and 1:200) using the formula and were within 1.8–2.2. The mRNA abundances were calculated as normalized relative quantities (NRQ) (31).
Tissue sections of MLD (thickness 8 μm) were cut with a CM3050 S cryostat microtome (Leica, Bensheim, Germany) and stained with antibodies against PLIN1–5 that were purchased from Novus Biologicals (NB110-40760, NB110-40878, NB110-40765, NBP2-13776, and NB110-60509; Wiesbaden-Nordenstadt, Germany). Fixation of muscle sections was done for 15 min in 4% paraformaldehyde. After washing 3 × 5 min with phosphate-buffered saline (PBS), the slides were incubated with 10% normal goat serum (NGS) in PBS for 15 min to block non-specific binding of the secondary antibody. Primary antibody (dilution 1:200 for PLIN1 and 4 and 1:100 for PLIN2, 3, and 5) incubation was performed for 1 h. Then, the slides were rinsed briefly, washed 3 × 10 min with PBS and subsequently incubated in the dark with secondary antibody (Alexa Fluor 488 Goat-Anti-Rabbit IgG, 1:1,000; Life Technologies, Darmstadt, Germany) for 45 min. Slides were washed 3 × 10 min with PBS, and the nuclei were stained with Hoechst 33258 for 5 min. After being washed 2 × with PBS and 1 × with distilled water for 5 min each, the slides were covered with ProLong Antifade (Thermo Fisher Scientific, Schwerte, Germany). All incubations were performed at room temperature in a humidity chamber. Two types of negative controls were generated to detect non-specific antibody binding, either blocking the primary antibodies with respective blocking peptides or omitting the primary antibody. No non-specific secondary antibody binding was detected, but weak non-specific binding of the antibodies against PLIN1 and 5 was observed (Supplementary Figure 1). Fluorescence signals were observed with a Nikon Microphot SA fluorescence microscope (Nikon, Düsseldorf, Germany) and a CC-12 color camera and recorded with Cell∧F image analysis software (OSIS, Münster, Germany).
Protein was isolated from all 144 MLD samples as described in details by Liu et al. (29) using CelLytic MT lyses reagent (Sigma-Aldrich, Munich, Germany) and protease inhibitor. Protein mixed with loading buffer was denatured at 95°C for 5 min and then loaded on Criterion TGX 12% gels (Bio-Rad) together with a molecular weight marker (PageRuler, Thermo Scientific, Schwerte, Germany). After electrophoresis, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Trans-Blot Turbo Transfer Pack, Bio-Rad) with a semi dry blotter (Trans-Blot, Bio-Rad). The Smart Protein Layers (SPL) Western Kit Red (PR913-R, NH DyeAGNOSTICS, Halle, Germany) was used to enable reliable protein quantification. Membranes were blocked with 5% non-fat dry milk or 10% ROTI Block (Carl Roth, Karlsruhe, Germany) in Tris-buffered saline (TBS) for 1 h at room temperature. All membranes were incubated with the primary antibodies, which were also used for immunohistochemistry (PLIN1 and 2 diluted at 1:15,000 and PLIN3, 4, and 5 diluted at 1:5,000) and a HRP-conjugated secondary antibody (Rabbit TrueBlot HRP, Rockland Immunochemicals, Limerick, PA, USA; diluted at 1:25,000). The antibody label was detected with highly sensitive chemiluminescence substrate (SuperSignal West Femto, Thermo Scientific). Chemiluminescence and fluorescence of calibrators and total protein were recorded with a ChemoCam HR-16 imager (INTAS, Göttingen, Germany) and quantified with LabImage 1D software (Kapelan Bio-Imaging, Leipzig, Germany). Target protein band volume was normalized to total protein abundance, and the SPL method enabled comparability among blots. All 144 samples were measured at least twice on separate blots (see example full blots in Supplementary Figure 2). Negative controls were generated to discriminate specific and non-specific antibody binding. Separate blots were processed that were incubated with the antibodies alone or with the antibodies together with respective blocking peptides, pre-incubated for 30 min (Supplementary Figure 3).
Statistical analysis was performed with the analysis of variance (ANOVA) model using the MIXED procedure of SAS statistical software (version 9.4, SAS Inst., Cary, USA). The fixed factors were BiW (LBW or NBW), supplementation (ALA or GLN), and age (5, 12, or 26), and respective interactions and sow were included as random factors. The used model was as follows:
Yijksl = μ + BiWi + Supj + Agek + (BiW × Sup)ij + (BiW × Age)ik + (Sup × Age)jk + (BiW × Sup × Age)ijk + Sows + eijksl,
where Yijksl = dependent variable (animal l from Sow s in BiW group i, Sup group j, and Age group k); μ = overall mean; BiWi = effect of birth weight i (i = 1–2); Supj = effect of supplementation j (j = 1–2); Agek = effect of age k (k = 1–3); (BiW × Sup)ij = effect of interaction; (BiW × Age)ik = effect of interaction; (Sup × Age)jk = effect of interaction; (BiW × Sup × Age)ijk = effect of interaction; Sows = random effect of sow s (s = 49); and eijksl = random residual error.
Least-square means (LSmeans) and standard errors (SE) were calculated for each fixed effect, and pairwise differences were tested by the Tukey–Kramer test. The SLICE statement of the MIXED procedure was used for partitioned analysis of the LSmeans for the interaction between BiW and supplementation within ages. Differences were considered significant if Tukey–Kramer adjusted p < 0.05 and a trend if 0.1 > p ≥ 0.05. The CORR procedure of SAS was used to calculate Spearman correlation coefficients. Correlations were regarded as low (0.3 < r < 0.5), moderate (0.5 < r < 0.7), high (0.7 < r < 0.9), and very high (r > 0.9) as described by Mukaka et al. (32).
Results of total lipid droplet area in muscle fibers and in adipocytes were reported in our former publication (5). We observed more lipid droplets in LBW piglets than in their NBW littermates at 5 dpn. Differences disappeared in older animals, and an influence of Gln supplementation was not detected. In the current study, we measured the size of individual intramuscular adipocytes. Adipocyte size distributions in MLD and MST are presented in Figure 1. The adipocyte size was mainly influenced by age and BiW, but no significant interaction effect among BiW, supplementation, and age was observed (p > 0.05). In both muscles, the mean adipocyte diameter was larger in piglets at 12 (p ≤ 0.001) or 26 dpn (p < 0.001) compared with animals at 5 dpn (Figure 1A). Size histograms showed that piglets at 26 dpn had more large adipocytes than at 5 or 12 dpn (Figures 1B,C). Comparing the adipocyte size distribution of MLD revealed more small-diameter adipocytes (20–30 μm, p = 0.03) at 5 dpn in NBW piglets and more large-diameter adipocytes (60–70 μm, p = 0.016 and 80–90 μm, p < 0.001) at 26 dpn in LBW piglets (Figure 1B). In addition, we found more medium-diameter fat cells (40–50 μm) in Gln-supplemented piglets at 26 dpn than in Ala-supplemented animals, independent of BiW groups. In MST, the mean diameter of adipocytes of LBW piglets was greater than that of their NBW littermates at 5 (p = 0.038) and 26 dpn (p = 0.03), which was not observed in MLD. Furthermore, NBW piglets had more small-size adipocytes (<20 μm, p < 0.05), whereas LBW animals had more medium-size adipocytes (20–30 μm, p = 0.012 and 30–40 μm, p = 0.077) at 5 dpn. At 26 dpn, LBW piglets had more large-diameter fat cells (50–80 μm, p < 0.05) in MST than NBW piglets. Fewer adipocytes with a diameter smaller than 10 μm were observed in GLN-supplemented piglets in comparison with Ala-supplemented animals (p = 0.037), regardless of BiW group.
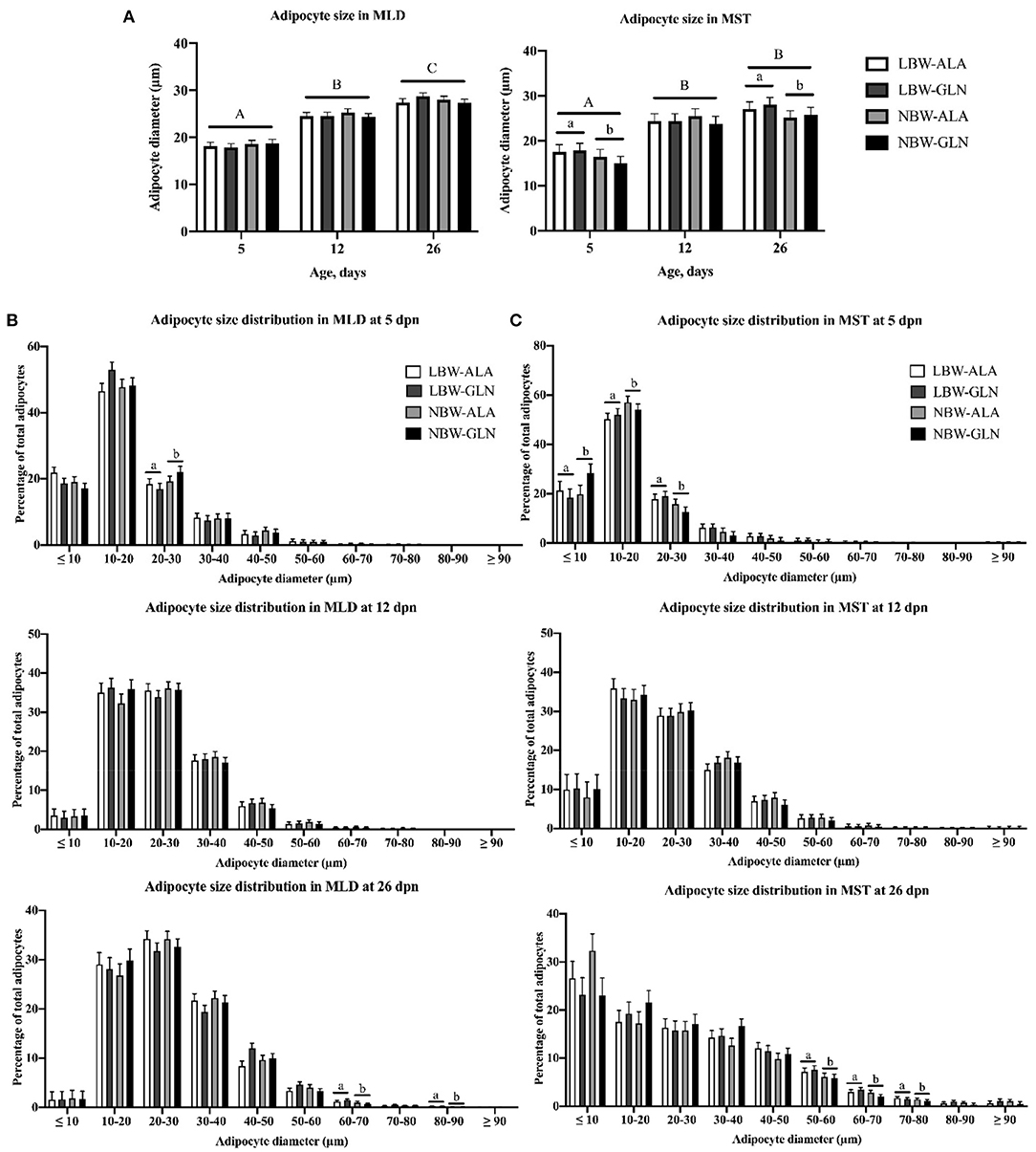
Figure 1. Intramuscular adipocyte diameter and adipocyte size frequency distribution within M. longissimus (MLD) and M. semitendinosus (MST) of low- (LBW) and normal-birth-weight (NBW) piglets at 5, 12, and 26 dpn, supplemented with glutamine (GLN) or alanine (ALA) (n = 12 per group). (A) Intramuscular adipocyte diameter within MLD and MST; intramuscular adipocyte size distribution within (B) MLD and (C) MST. Values represent LSmeans and SE in error bars. a,bSignificant differences between LBW and NBW piglets; A−CSignificant differences among ages (p < 0.05, Tukey–Kramer test).
The abundance of PLIN1–5 and interacting lipolysis-related genes was quantified with RT-qPCR (Figure 2) to elucidate their contribution to the observed differences in lipid deposition between LBW and NBW piglets. The statistical analyses revealed no interaction effect among BiW, supplementation, and age for the mRNA abundances of the investigated genes (p > 0.05). Age and BiW were also the main influencing factors for the mRNA abundances of the investigated genes. The PLIN1 mRNA level was overall higher in LBW than in NBW piglets at 5 dpn (p = 0.033), and in particular, it was higher in LBW-GLN piglets than in NBW-GLN piglets (p = 0.017, Figure 2A). However, no significant effects of supplementation (p = 0.53) or age (p = 0.127) were observed. Relative mRNA expression of PLIN2 was higher in piglets at 5 dpn in comparison with animals at 12 (p < 0.001) or 26 dpn (p = 0.04, Figure 2B). Furthermore, LBW-ALA piglets had more PLIN2 mRNA compared with their NBW-ALA littermates at 5 dpn (p = 0.008). Independent of supplementation, PLIN2 mRNA abundance was greater in LBW piglets than in NBW piglets at 5 dpn (p = 0.001). However, this effect was not observed at 12 (p = 0.811) or 26 dpn (p = 0. 949), and no supplementation effects were detected (p = 0.462) independent of ages. The relative mRNA abundance of PLIN3 was too low to be reliably detected in MLD of piglets in this study. The abundance of PLIN4 mRNA was lower in piglets at 5 dpn (p = 0.005, Figure 2C) and 12 dpn (p = 0.014) compared with piglets at 26 dpn and was not influenced by BiW (p = 0.570) or supplementation (p = 0.172). The abundance of PLIN5 mRNA in piglets at 12 dpn tended to be lower than in piglets at 5 dpn (p = 0.062, Figure 2D) and was lower than in animals at 26 dpn (p < 0.001). Furthermore, LBW-ALA piglets tended to have higher PLIN5 mRNA levels than NBW-ALA piglets at 5 dpn (p = 0.074). Overall, independent of Gln supplementation, LBW piglets had more PLIN5 mRNA compared with NBW at 5 dpn (p = 0.042), but not at 12 (p = 0.874) or 26 dpn (p = 0.538). The results did not show an effect of Gln supplementation on PLIN5 mRNA abundance (p = 0.851).
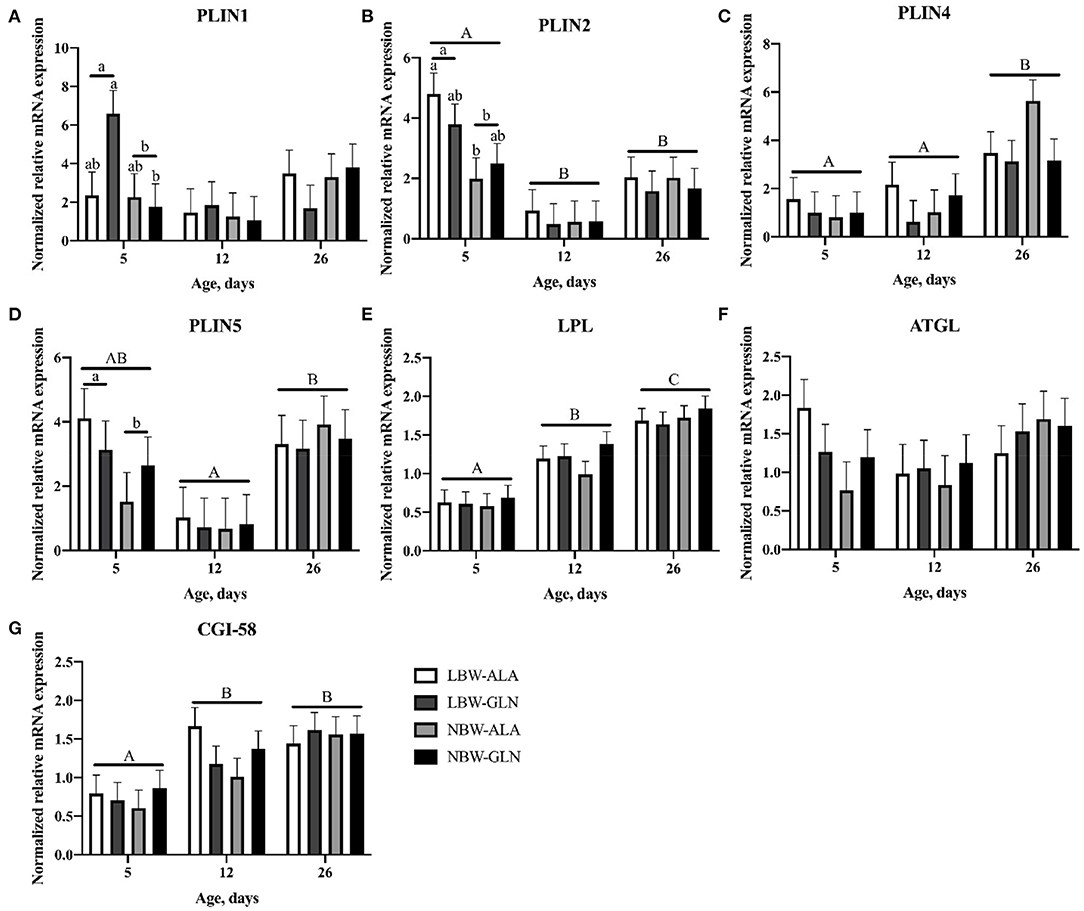
Figure 2. Normalized relative mRNA expression of perilipins and related lipases in M. longissimus of low- (LBW) and normal-birth-weight (NBW) piglets at 5, 12, and 26 dpn, supplemented with glutamine (GLN) or alanine (ALA) (n = 12 per group). Values represent LSmeans and SE in error bars of PLIN1 (A), PLIN2 (B), PLIN4 (C), PLIN5 (D), LPL (E), ATGL (F), and CGI-58 (G) mRNA abundance, normalized to YWHAZ and PPIA. a,bSignificant differences between LBW and NBW piglets at the same age; A−CSignificant differences among ages (p < 0.05, Tukey–Kramer test).
Birth weight and Gln supplementation had no effect on mRNA abundance of LPL (p = 0.641 and p = 0.29, respectively). There was only an age effect detected; thus, more LPL mRNA was measured in piglets at 26 dpn compared with piglets at 5 (p < 0.001, Figure 2E) and 12 dpn (p < 0.001). Similarly, the mRNA level of CGI-58 was higher in piglets at 12 (p = 0.026, Figure 2F) and 26 dpn (p < 0.001) compared with animals at 5 dpn. No BiW (p = 0.561) or Gln supplementation (p = 0.774) effects were observed. There was a trend for higher level of ATGL mRNA in LBW-ALA compared with NBW-ALA piglets at 5 dpn (p = 0.054). Moreover, ATGL mRNA tended to be higher in LBW piglets than in NBW animals at 5 dpn independent of supplementation (p = 0.054), but it was not affected by Gln supplementation (p = 0.738) or age (p = 0.335).
The proteins encoded by the five PLIN genes were detected with immunohistochemistry in the MLD of piglets. Different localizations and staining patterns were observed for the individual PLIN proteins. A strong signal was observed for PLIN1 around fat vacuoles in mature and developing adipocytes (Figures 3A–C). Additional weak signals in muscle tissue were identified as unspecific (see Supplementary Figure 1) by blocking specific antibody bindings with the respective blocking peptide. In contrast, PLIN2 was localized exclusively at intramyocelluar lipid droplets (Figures 3D–F). The staining decreased with age according to the reduction of lipid droplets within muscle fibers. Similar staining patterns were observed for PLIN3 and PLIN4, both located at the periphery of muscle fibers and large adipocytes (Figures 4A–D), whereas PLIN5 was detected in the cytosol of many small, undefined cells between muscle fibers (Figures 4E,F).
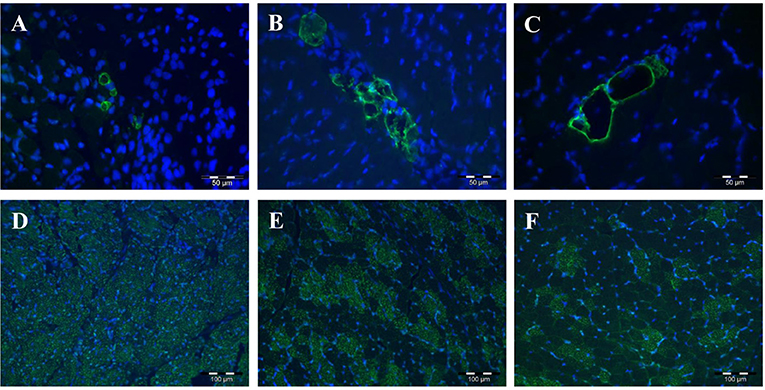
Figure 3. Immunohistochemical localization of PLIN1 (A–C) and PLIN2 (D–F) in porcine M. longissimus at 5, 12, and 26 dpn, respectively. PLIN1 and 2 were stained green. Nuclei were counterstained blue with Hoechst 33258.
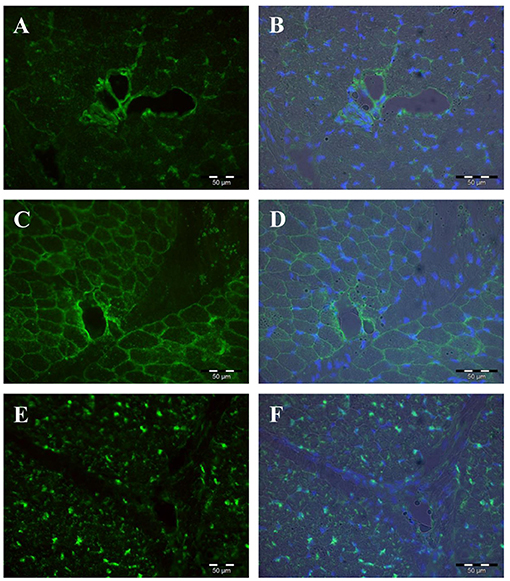
Figure 4. Immunohistochemical localization of PLIN3 (A,B), PLIN4 (C,D), and PLIN5 (E,F) in porcine M. longissimus at 12 dpn. Nuclei were counterstained blue with Hoechst 33258. The right panel shows merged images of fluorescence (PLIN and Hoechst 33258) and bright field.
Normalized protein abundances of PLIN1–5 within MLD were determined with western blots and are shown in Figure 5. A significant interaction effect among BiW, supplementation, and age was observed for the protein abundance of PLIN3 (p = 0.017), but for no other protein (p > 0.05). Age and BiW were the main factors influencing protein abundances of perilipins in MLD of piglets. The protein abundances of all five perilipins (Figures 5A–E) were higher in piglets at 5 dpn compared with animals at 12 dpn (p < 0.01) and 26 dpn (p < 0.01). The protein content of PLIN1 was not influenced by BiW, whereas PLIN3 and 4 protein abundances (Figures 5C,D) were higher in LBW-GLN compared with NBW-GLN piglets at 5 dpn (p < 0.001 and p = 0.021, respectively). Regardless of supplementation, protein abundances of PLIN2–5 (Figures 5B–E) were higher in LBW piglets compared with their NBW littermates at 5 dpn (p = 0.015, p = 0.009, p = 0.012, and p = 0.016, respectively). However, an influence of Gln supplementation on protein abundances of PLIN1–5 was not observed (p = 0.642, p = 0.522, p = 0.578, p = 0.460, and p = 0.68, respectively).
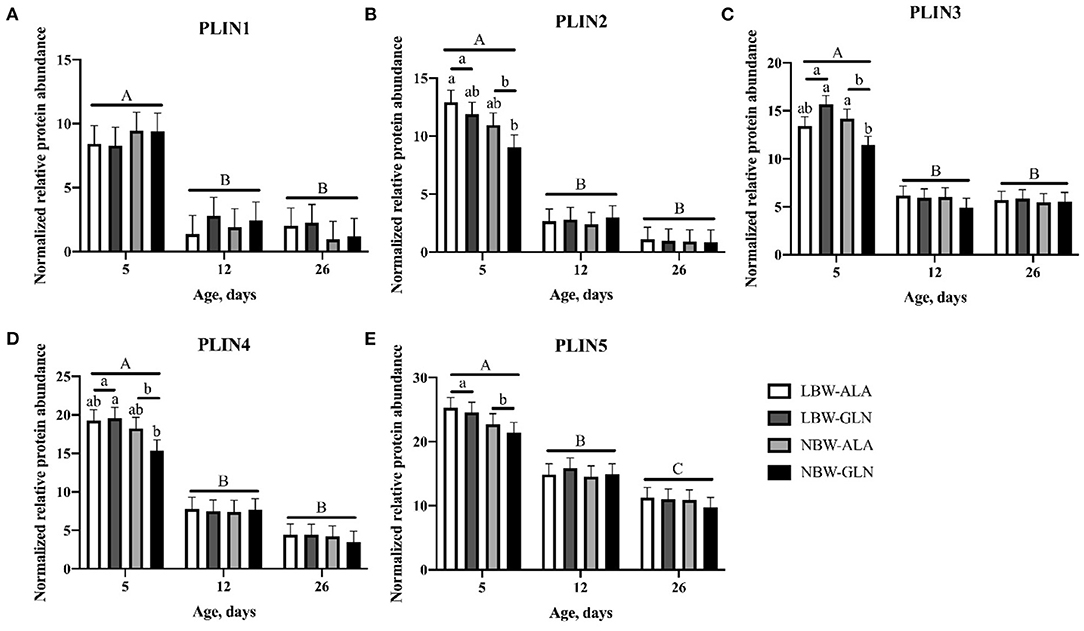
Figure 5. Normalized relative protein abundance of PLIN1 (A), PLIN2 (B), PLIN3 (C), PLIN4 (D), and PLIN5 (E) in M. longissimus of low- (LBW) and normal-birth-weight (NBW) piglets at 5, 12, and 26 dpn, supplemented with glutamine (GLN) or alanine (ALA) (n = 12 per group). Values represent LSmeans and SE in error bars of protein abundances normalized to total protein. a,bSignificant differences between piglets at the same age; A−CSignificant differences among ages (p < 0.05, Tukey–Kramer test).
Since most investigated traits were age dependent, Spearman correlation coefficients were calculated for each age group separately. We detected relationships among investigated genes and between gene expression and phenotypic traits (Supplementary Table 1). The relative mRNA abundances of PLIN2, 4, and 5 were moderately or highly correlated among each other in all age groups (r > 0.50, p < 0.001). The PLIN1 mRNA was moderately correlated with mRNA of PLIN2, 4, and 5 at 12 and 26 dpn (r > 0.6, p < 0.001). Furthermore, ATGL mRNA was closely correlated with CGI-58 and PLIN2, 4, and 5 mRNA at 5 dpn (r > 0.4, p < 0.001), whereas LPL mRNA was correlated with ATGL and CGI-58 only in piglets at 5 dpn (r > 0.6, p < 0.001). The protein abundances of the five investigated PLINs were not as closely correlated as mRNA, with only weak-to-high correlation among PLIN2 and PLIN4 and 5 in all age groups (r > 0.3, p < 0.001). There was no clear relationship between gene expression and intramuscular adipocyte area, except a weak negative correlation between PLIN1, 4, and 5 and adipocyte area at 12 dpn (r < −0.3, p < 0.05). However, we observed weak or moderate correlations between intramyocelluar lipid droplet area and protein abundances of PLIN2–5 (r > 0.3, p < 0.05) at 12 dpn and mRNA of PLIN1, 2, 4, and 5 as well as protein abundance of PLIN2 and 5 at 26 dpn (r = 0.357, p < 0.05). The mean adipocyte diameter showed a low negative correlation with intramuscular adipocyte area (r = −0.355, p < 0.05) at 12 dpn and with PLIN4 mRNA (r = −0.337, p < 0.05) at 26 dpn.
Our previous study indicated a clear difference between LBW and NBW piglets in the abundance of lipid droplets within muscle fibers at 5 dpn (5). The area percentage of intramyocellular lipid droplets decreased during the first days of life by more than 80%. The difference between LBW and NBW piglets disappeared with age. At the same time, intramuscular adipocytes started to develop with high individual variation. The current study was conducted to elucidate the process of lipid deposition and redistribution between muscle fibers and developing adipocytes as part of the muscle development and its modulation by Gln. Intramyocellular lipid droplets are vital organelles sequestering lipids, regulating intramyocelluar lipid homeostasis by preventing from lipotoxicity (11, 15). Within skeletal muscle, the perilipin family of proteins plays a vital role in lipid droplet metabolism (33). It is not known whether Gln can directly interact with and modulate perilipin family members, but PLINs are biomarkers for lipid deposition and adipocyte development within skeletal muscle in pigs (25, 26, 34). Therefore, we investigated the expression pattern of the five known members of the perilipin family and their related lipases in skeletal muscle of LBW piglets and their NBW littermates, which were orally supplemented with Gln or Ala during the first 12 days of life.
Myogenesis and adipogenesis are competitive processes during muscle development (7, 8). Thus, when the muscle growth is delayed, as reported in LBW piglets (5, 10), nutrients are available for the development of intramuscular adipocytes at the same time. Gondret et al. (10) observed a greater adipocyte diameter in the MST of LBW pigs at 112 kg compared with high birth weight pigs. In agreement with Gondret et al. (10), we observed larger adipocytes in the MST, but not MLD, of LBW compared with NBW piglets during the first weeks of life. In addition, LBW piglets had more large-size adipocytes in both muscles at 26 dpn, whereas NBW animals had more small-size adipocytes at 5 dpn. Together with the result of our previous study that LBW had more intramyocelluar lipid droplets compared with NBW piglets (5), this suggests that LBW piglets stored the lipids in intramyocellular lipid droplets rather than in adipocytes during the first days of life. Then, with increasing age, the lipids were transferred to intramuscular adipocytes. Overall, LBW piglets had more intramuscular lipid deposition during the suckling period than their NBW littermates.
All five members of the perilipin family of proteins were detected in MLD of piglets in our study with mainly distinct localizations. The first member, PLIN1, was originally discovered in adipocytes (35) surrounding lipid droplets (36). Other PLINs were consecutively detected. The five members of the perilipin family are proteins associated with the surface of lipid droplets and function in the formation, stabilization, and utilization of lipid droplets (26). Wolins et al. (37) elucidated the process how PLINs mediate lipid droplet formation in cultured 3T3-L1 adipocytes. According to their studies, nascent lipid droplets are coated first by PLIN3 and 4, then PLIN2 is recruited to the surface of growing lipid droplets. Afterwards, PLIN1 starts to bind the mature lipid droplets, stabilizing and protecting them from lipolysis. However, PLIN1 starts to promote triglyceride hydrolysis when it is phosphorylated by protein kinase A (38). In previous experiments, PLIN1 was discovered to be around large adipocytes (35, 39) and was thought to be restricted within white and brown adipose tissue or steroidogenic cells (40, 41). Our results indicate that PLIN1 can also be detected within skeletal muscle at the intramuscular adipocytes, in line with the findings of Gandolfi et al. (34). Furthermore, Skinner et al. (42) reported that PLIN1 can also be detected in the cytosol when it moves from smooth endoplasmic reticulum to mature adipocytes. While PLIN1 is a suitable marker for intramuscular fat deposition in pigs, PLIN2 was traditionally considered a marker for lipid droplets within skeletal muscle as it is located around small lipid droplets (34, 43). MacPherson et al., however, argued this point as the distribution of PLIN2 was only 60–80% parallel to intramuscular lipid droplets in some studies (44–46). Nevertheless, in most investigations, PLIN2 was found ubiquitously, appearing as the most abundant protein of the perilipin family in skeletal muscle (46, 47), which could not be concluded from our study. It shares a similar function and homology with PLIN1 and is able to replace PLIN1 when it is lacking (47, 48). The study of Xu et al. revealed that PLIN2 is competing with PLIN1 for binding at the surface of lipid droplets in adipocytes (49). There was no co-localization of PLIN1 and PLIN2 observed in MLD of piglets in our study. While PLIN1 and PLIN2 share similar functions, PLIN3 and PLIN4, both interacting with nascent lipid droplets, were localized at the periphery of muscle fibers and intramuscular adipocytes in our study. Previous studies have indicated that PLIN3 and 4 were also located cytosolically (21, 33, 50). However, in this study, we did not observe a clear association of PLIN3 and 4 with lipid droplets beside a weak cytosolic signal, which could indicate binding of smaller nascent lipid droplets that could not be detected under the microscope. The last member of this family, PLIN5, was localized within undefined cells between the muscle fibers in our study. It could not be clarified what kind of cells were stained and whether this was a specific binding of the PLIN5 antibody, although blocking the epitope with the respective peptide completely prevented antibody binding and staining. Previous studies found that PLIN5 could be recruited to the mitochondria to supply energy (33, 51); thus, it could be important during skeletal muscle growth. Altogether, the members of the perilipin family have different localizations within skeletal muscle tissue according to their distinct roles in lipid storage and metabolism.
The expression of PLINs and lipolysis-related genes was measured to elucidate whether the observed differences in intramuscular lipid deposition between LBW and NBW piglets are regulated by different PLIN family members and whether the availability of free Gln in the muscle was involved. There are many examples in the literature that PLIN1 mRNA expression is correlated with fat deposition in different species, e.g., pigs (52), cattle (53), and humans (54). Our results indicated a higher PLIN1 mRNA level, but not a higher protein level, in LBW piglets at 5 dpn compared with their NBW littermates. The lower protein abundance of PLIN1 in piglets at 26 dpn in comparison with animals at 5 dpn was in contrast to the increasing appearance of adipocytes with age (5). However, possible explanations for this expression pattern of PLIN1 are as follows: (i) the size of adipocytes is smaller in younger piglets; hence, more PLIN1 protein is required to enclose lipid droplets of small adipocytes as they have a larger summarized surface. (ii) The intramuscular fat was not evenly distributed in the muscle (5); thus, the results may vary within the same muscle, and different parts were used for histology and mRNA or protein extraction. (iii) The relative protein abundance was normalized to the total protein abundance, which is increasingly dominated by structural muscle proteins (55). This applies also for other PLIN proteins; thus, the age effect will not be discussed in the following.
PLIN2 was previously reported to stimulate uptake of fatty acids in transfected COS-7 cells (56) and promote lipid droplet enlargement in murine fibroblasts (57). Consistent with our published data of intramyocellular lipid droplets (5), we observed higher PLIN2 mRNA and protein abundance in LBW compared with NBW piglets at 5 dpn in the current study. Thus, this variance of intramuscular lipid droplet content in piglets with different birth weight might result from the different expression pattern of PLIN2. Although, we observed weak-to-moderate positive correlations between intramyocellular lipid droplet area and PLIN2 protein abundance only at 12 and 26 dpn.
The protein abundance of PLIN3 and 4 was higher in LBW piglets at 5 dpn in comparison with NBW piglets, and both proteins had similar localizations in muscle tissue. We observed staining of small particles within particular muscle fibers, but different from the PLIN2 staining pattern. This is in accordance with previous studies indicating that PLIN3 functions in the biogenesis of small, nascent lipid droplets together with PLIN4 (21, 37, 58, 59). However, in PLIN3 blots, which were incubated with blocking peptide, the 47-kDa (theoretical size of PLIN3) band was only partially blocked, whereas a 63-kDa (similar to PLIN1) band was completely blocked. Therefore, the PLIN3 antibody possibly non-specifically binds to other members of the PLIN family. Additionally, the PLIN3 protein results should be considered with caution because of the very low mRNA abundance. Interestingly, PLIN4 blocking experiments indicated the size of PLIN4 protein at about 80 kDa (Supplementary Figure 3), which was smaller than the theoretical protein size (164 kDa). Whether it is an unknown isoform needs further clarification. Among the perilipin family, PLIN4 is the only protein that does not share the same PAT domain and a four-helix bundle with other PLINs and cannot mediate lipolysis (21, 26). Pourteymour et al. reported PLIN4 was more expressed in slow-twitch than in fast-twitch fibers in humans (60). Even though we did not investigate this specifically, it may also be similar in pigs, since most lipid droplets are located in slow-twitch fibers and are associated with PLINs.
Previous studies have suggested that PLIN5 is correlated with oxidative activities in skeletal muscle. Bosma et al. reported that overexpression of PLIN5 in the tibialis anterior muscle of rats increased oxidative gene expression and intramyocellular lipid content (61). Furthermore, this group reported higher interactivity of lipid droplets and mitochondria in human vastus lateralis muscle, rat tibialis anterior muscle, or cultured HEK293 cells upon overexpression of PLIN5 (62). In our study, LBW piglets had a higher level of PLIN5 mRNA and protein than NBW piglets at 5 dpn, regardless of supplementation. This suggests more oxidative activities in skeletal muscle of LBW piglets in the first days of life, probably to generate energy for muscle growth. Furthermore, PLIN5 protein was negatively correlated with adipocyte diameter in piglets at 26 dpn, suggesting PLIN5 is not involved in adipocyte growth and utilization of the lipids stored in intramyocellular lipid droplets.
Altogether, PLIN2–5 were more abundant in muscle tissue of LBW piglets than in NBW animals during their first days of life. A possible explanation for this observation may be that LBW piglets expend fewer fatty acids for their muscle and body growth than their NBW littermates, and the excess fatty acids are synthesized to triglycerides and stored in intramyocellular lipid droplets, preventing lipotoxicity. However, with increasing age, body growth or lipid deposition in adipocytes requires more energy, and the excess lipids in lipid droplets are then utilized. There is no indication from the current study that Gln supplementation alters the expression level of PLINs in porcine skeletal muscle.
The uptake of fatty acids from the blood is an important energy source for skeletal muscle metabolism, and LPL plays an important role in triacylglycerol hydrolysis from lipoproteins (63, 64). It is a rate-limiting lipase during this process and is mainly synthesized in skeletal muscle (64, 65). In a previous study, a tissue-specific over-expression of LPL was reported to increase the fatty acid content in mouse skeletal muscle (66, 67). Therefore, in the current study, we hypothesized that LPL expression was positively related to intramuscular lipid droplets, which are synthesized de novo from fatty acids (16). In the present study, relative LPL mRNA expression was significantly higher in piglets at 26 dpn compared with younger animals at 5 and 12 dpn, indicating an increasing demand of fatty acids in porcine skeletal muscle during the early postnatal period. The mRNA level of LPL was not influenced by BiW, suggesting that the higher content of intramyocellular lipid droplets in LBW piglets may not result from increased fatty acid uptake.
In addition to promoting lipid droplet formation and stabilization, PLINs were reported to modulate hydrolysis of the main content of lipid droplets, the triglycerides, together with ATGL and its coactivator CGI-58 (44). Moreover, ATGL is a vital lipase in skeletal muscle (41). During lipolysis within adipocytes, PLIN1 is phosphorylated first, then CGI-58 combined with PLIN1 is released from the surface of lipid droplets and activates ATGL (33). Subsequently, ATGL interacts with triacylglycerols of the lipid droplet cores and metabolizes them to diacylglycerols and fatty acids (33). In non-adipose tissue, e.g., myocytes, where PLIN1 is lacking, ATGL and CGI-58 could possibly mediate lipolysis with PLIN2 without being phosphorylated (44, 68, 69). Perilipin 3 and 5 could be both phosphorylated with stimulation to promote lipolysis in skeletal muscle (69). However, few studies have investigated the interaction of PLIN3 and lipases. Additionally, PLIN5 is able to bind CGI-58 or ATGL separately and is the only member of the PLIN family that binds ATGL directly (41, 70, 71). However, the interaction of PLIN5 and ATGL prevents lipolysis unless phosphorylation of PLIN5 (41). Our results indicated that mRNA expression of CGI-58 was not influenced by BiW or Gln supplementation despite the difference in lipid droplet distribution between LBW and NBW piglets at 5 dpn. Moreover, CGI-58, as a regulator of ATGL activity (72), exhibited higher mRNA expression in piglets at 12 and 26 dpn in comparison with piglets at 5 dpn, indicating increased lipolysis activity with increasing age. However, the lipolysis rate of triglycerides in skeletal muscle is controlled by ATGL (69), but its relative mRNA expression in these animals was not influenced by BiW, supplementation, or age. Whether post translationally the regulation of ATGL played a role needs further clarification.
Relative mRNA expressions of PLIN2, 4, and 5 were closely correlated among each other within skeletal muscle of piglets in this study, suggesting their similar or interactive roles in intramuscular lipid droplet mobilization during the first weeks of life. Moreover, although LPL was not influenced by BiW, supplementation, or age in this study, its mRNA expression showed moderate correlations with the mRNA abundance of PLIN2, PLIN5, ATGL, and CGI-58 in piglets at 5 dpn. This could be a sign of LPL's function in intramuscular lipid droplet formation together with these genes. In addition, as ATGL-mediated lipolysis requires interactions with PLIN family members (41), close correlations among ATGL mRNA and PLINs in animals at 5 and 26 dpn were observed in this study. Consistent with that, the study of MacPherson et al. (44, 69) revealed distinct roles of PLINs in mediating lipolysis. Overexpression of PLIN5 correlated with an increase of ATGL expression to maintain lipid homeostasis in the study of Bosma et al. (61). All these data revealed that PLIN family members are closely correlated with the important lipases in skeletal muscle.
Perilipins have their distinct roles in regulating the lipid droplet formation, distribution, and mobilization in porcine skeletal muscle. Their expression and function are closely related as indicated by high correlations among PLIN2–5 at both mRNA and protein level. The results revealed BiW as an important factor influencing their expression pattern, with a higher expression of most PLINs in LBW piglets at 5 dpn. Furthermore, the close interactive correlations among PLIN family and related lipases suggested the PLINs play important roles in lipolysis together with LPL, ATGL, and CGI-58 within porcine skeletal muscle in the early postnatal phase. Oral supplementation with Gln had only negligible effects on the expression level of PLINs and related lipases.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by State Office for Agriculture, Food Safety and Fishing Mecklenburg Western Pomerania, Germany (permission No. 7221.3-1-026/16).
YZ performed most experiments, analyzed the data, and drafted the manuscript. EA designed the study, performed parts of the experiments and analyses, and helped with writing the manuscript. CM designed and supervised the animal experiment, provided resources, acquired funding, and revised the manuscript. QS, ZL, and JS performed the animal experiment, provided resources, and reviewed and edited the manuscript. SM supervised and administered the project, provided resources, and reviewed and edited the manuscript. All authors approved the final submitted manuscript.
YZ was funded by a China Scholarship Council (CSC) grant. The project was partly funded by Deutsche Forschungsgemeinschaft (DFG), Bonn, Germany (grant number ME 1420/10-1). The publication of this article was funded by the Open Access Fund of the FBN.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors acknowledge the excellent technical assistance of E. Schwitulla, S. Foß, F. Feldt, and K. Gürtler; Dr. A. Tuchscherer for statistical advice; Dr. R. Pfuhl and the staff of the experimental slaughterhouse; as well as the EAS team.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.633898/full#supplementary-material
1. D'Inca R, Gras-Le Guen C, Che LQ, Sangild PT, Le Huerou-Luron I. Intrauterine growth restriction delays feeding-induced gut adaptation in term newborn pigs. Neonatology. (2011) 99:208–16. doi: 10.1159/000314919
2. Jamin A, Seve B, Thibault JN, Floc'h N. Accelerated growth rate induced by neonatal high-protein milk formula is not supported by increased tissue protein synthesis in low-birth-weight piglets. J Nutr Metab. (2012) 2012:545341. doi: 10.1155/2012/545341
3. Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. (2009) 37:1–17. doi: 10.1007/s00726-009-0269-0
4. Wu G, Bazer FW, Johnson GA, Knabe DA, Burghardt RC, Spencer TE, et al. Triennial growth symposium: important roles for L-glutamine in swine nutrition and production. J Anim Sci. (2011) 89:2017–30. doi: 10.2527/jas.2010-3614
5. Zhao Y, Albrecht E, Sciascia QL, Li Z, Görs S, Schregel J, et al. Effects of oral glutamine supplementation on early postnatal muscle morphology in low and normal birth weight piglets. Animals (Basel). (2020) 10:1976. doi: 10.3390/ani10111976
6. Hausman GJ, Basu U, Du M, Fernyhough-Culver M, Dodson MV. Intermuscular and intramuscular adipose tissues: bad vs. good adipose tissues. Adipocyte. (2014) 3:242–55. doi: 10.4161/adip.28546
7. Hocquette JF, Gondret F, Baeza E, Medale F, Jurie C, Pethick DW. Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers. Animal. (2010) 4:303–19. doi: 10.1017/S1751731109991091
8. Yan X, Zhu MJ, Dodson MV, Du M. Developmental programming of fetal skeletal muscle and adipose tissue development. J Genomics. (2013) 1:29–38. doi: 10.7150/jgen.3930
9. Zhao SM, Ren LJ, Chen L, Zhang X, Cheng ML, Li WZ, et al. Differential expression of lipid metabolism related genes in porcine muscle tissue leading to different intramuscular fat deposition. Lipids. (2009) 44:1029–37. doi: 10.1007/s11745-009-3356-9
10. Gondret F, Lefaucheur L, Juin H, Louveau I, Lebret B. Low birth weight is associated with enlarged muscle fiber area and impaired meat tenderness of the longissimus muscle in pigs. J Anim Sci. (2006) 84:93–103. doi: 10.2527/2006.84193x
11. Walther TC, Farese RV Jr. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. (2012) 81:687–714. doi: 10.1146/annurev-biochem-061009-102430
12. Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. (1999) 24:109–15. doi: 10.1016/S0968-0004(98)01349-8
13. Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem. (2012) 287:2273–9. doi: 10.1074/jbc.R111.309088
14. Thiam AR, Farese RV Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. (2013) 14:775–86. doi: 10.1038/nrm3699
15. Kusminski CM, Shetty S, Orci L, Unger RH, Scherer PE. Diabetes and apoptosis: lipotoxicity. Apoptosis. (2009) 14:1484–95. doi: 10.1007/s10495-009-0352-8
16. Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. (2008) 20:378–85. doi: 10.1016/j.ceb.2008.05.009
17. Olofsson SO, Bostrom P, Andersson L, Rutberg M, Levin M, Perman J, et al. Triglyceride containing lipid droplets and lipid droplet-associated proteins. Curr Opin Lipidol. (2008) 19:441–7. doi: 10.1097/MOL.0b013e32830dd09b
18. Pol A, Martin S, Fernandez MA, Ingelmo-Torres M, Ferguson C, Enrich C, et al. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the golgi complex and between the cell surface and lipid bodies. Mol Biol Cell. (2005) 16:2091–105. doi: 10.1091/mbc.e04-08-0737
19. Lu XY, Gruia-Gray J, Copeland NG, Gilbert DJ, Jenkins NA, Londos C, et al. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mammalian Genome. (2001) 12:741–9. doi: 10.1007/s00335-01-2055-5
20. Londos C, Brasaemle DL, Schultz CJ, Segrest JP, Kimmel AR. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin Cell Dev Biol. (1999) 10:51–8. doi: 10.1006/scdb.1998.0275
21. Wolins NE, Skinner JR, Schoenfish MJ, Tzekov A, Bensch KG, Bickel PE. Adipocyte protein S3-12 coats nascent lipid droplets. J Biol Chem. (2003) 278:37713–21. doi: 10.1074/jbc.M304025200
22. Yamaguchi T, Matsushita S, Motojima K, Hirose F, Osumi T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J Biol Chem. (2006) 281:14232–40. doi: 10.1074/jbc.M601682200
23. Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Croce MA, Gropler MC, et al. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. (2006) 55:3418–28. doi: 10.2337/db06-0399
24. Dalen KT, Dahl T, Holter E, Arntsen B, Londos C, Sztalryd C, et al. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim Biophys Acta. (2007) 1771:210–27. doi: 10.1016/j.bbalip.2006.11.011
25. Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. (2010) 51:468–71. doi: 10.1194/jlr.R000034
26. Itabe H, Yamaguchi T, Nimura S, Sasabe N. Perilipins: a diversity of intracellular lipid droplet proteins. Lipids Health Dis. (2017) 16:83. doi: 10.1186/s12944-017-0473-y
27. Erkens T, Van Poucke M, Vandesompele J, Goossens K, Van Zeveren A, Peelman LJ. Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol. (2006) 6:41. doi: 10.1186/1472-6750-6-41
28. Uddin MJ, Cinar MU, Tesfaye D, Looft C, Tholen E, Schellander K. Age-related changes in relative expression stability of commonly used housekeeping genes in selected porcine tissues. BMC Res Notes. (2011) 4:441. doi: 10.1186/1756-0500-4-441
29. Liu Y, Albrecht E, Schering L, Kuehn C, Yang R, Zhao Z, et al. Agouti signaling protein and its receptors as potential molecular markers for intramuscular and body fat deposition in cattle. Front Physiol. (2018) 9:172. doi: 10.3389/fphys.2018.00172
30. Schering L, Albrecht E, Komolka K, Kuhn C, Maak S. Increased expression of thyroid hormone responsive protein (THRSP) is the result but not the cause of higher intramuscular fat content in cattle. Int J Biol Sci. (2017) 13:532–44. doi: 10.7150/ijbs.18775
31. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. (2007) 8:R19. doi: 10.1186/gb-2007-8-2-r19
32. Mukaka MM. Statistics Corner: a guide to appropriate use of correlation coefficient in medical research. Malawi Med J. (2012) 24:69–71.
33. Morales PE, Bucarey JL, Espinosa A. Muscle lipid metabolism: role of lipid droplets and perilipins. J Diabetes Res. (2017) 2017:1789395. doi: 10.1155/2017/1789395
34. Gandolfi G, Mazzoni M, Zambonelli P, Lalatta-Costerbosa G, Tronca A, Russo V, et al. Perilipin 1 and perilipin 2 protein localization and gene expression study in skeletal muscles of European cross-breed pigs with different intramuscular fat contents. Meat Sci. (2011) 88:631–7. doi: 10.1016/j.meatsci.2011.02.020
35. Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchettemackie EJ, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. (1991) 266:11341–6. doi: 10.1016/S0021-9258(18)99168-4
36. Blanchettemackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, et al. Perilipin is located on the surface-layer of intracellular lipid droplets in adipocytes. J Lipid Res. (1995) 36:1211–26. doi: 10.1016/S0022-2275(20)41129-0
37. Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel P. S3-12, adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem. (2005) 280:19146–55. doi: 10.1074/jbc.M500978200
38. Brasaemle DL, Rubin B, Harten IA, Gruia-Gray J, Kimmel AR, Londos C. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J Biol Chem. (2000) 275:38486–93. doi: 10.1074/jbc.M007322200
39. Yamaguchi T, Omatsu N, Matsushita S, Osumi T. CGI-58 interacts with perilipin and is localized to lipid droplets - possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J Biol Chem. (2004) 279:30490–7. doi: 10.1074/jbc.M403920200
40. Dalen KT, Schoonjans K, Ulven SM, Weedon-Fekjaer MS, Bentzen TG, Koutnikova H, et al. Adipose tissue expression of the lipid droplet-associating proteins S3-12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma. Diabetes. (2004) 53:1243–52. doi: 10.2337/diabetes.53.5.1243
41. Wang H, Bell M, Sreenivasan U, Hu H, Liu J, Dalen K, et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem. (2011) 286:15707–15. doi: 10.1074/jbc.M110.207779
42. Skinner JR, Harris LA, Shew TM, Abumrad NA, Wolins NE. Perilipin 1 moves between the fat droplet and the endoplasmic reticulum. Adipocyte. (2013) 2:80–6. doi: 10.4161/adip.22864
43. Listenberger LL, Ostermeyer-Fay AG, Goldberg EB, Brown WJ, Brown DA. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res. (2007) 48:2751–61. doi: 10.1194/jlr.M700359-JLR200
44. MacPherson RE, Peters SJ. Piecing together the puzzle of perilipin proteins and skeletal muscle lipolysis. Appl Physiol Nutr Metab. (2015) 40:641–51. doi: 10.1139/apnm-2014-0485
45. MacPherson RE, Herbst EA, Reynolds EJ, Vandenboom R, Roy BD, Peters SJ. Subcellular localization of skeletal muscle lipid droplets and PLIN family proteins OXPAT and ADRP at rest and following contraction in rat soleus muscle. Am J Physiol Regul Integr Comp Physiol. (2012) 302:R29–36. doi: 10.1152/ajpregu.00163.2011
46. Shaw CS, Sherlock M, Stewart PM, Wagenmakers AJ. Adipophilin distribution and colocalization with lipid droplets in skeletal muscle. Histochem Cell Biol. (2009) 131:575–81. doi: 10.1007/s00418-009-0558-4
47. Brasaemle DL, Barber T, Wolins NE, Serrero G, BlanchetteMackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. (1997) 38:2249–63. doi: 10.1016/S0022-2275(20)34939-7
48. Jiang HP, Serrero G. Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc Natl Acad Sci USA. (1992) 89:7856–60. doi: 10.1073/pnas.89.17.7856
49. Xu G, Sztalryd C, Lu X, Tansey JT, Gan J, Dorward H, et al. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J Biol Chem. (2005) 280:42841–7. doi: 10.1074/jbc.M506569200
50. Wolins NE, Rubin B, Brasaemle DL. TIP47 associates with lipid droplets. J Biol Chem. (2001) 276:5101–8. doi: 10.1074/jbc.M006775200
51. Wang H, Sreenivasan U, Hu H, Saladino A, Polster BM, Lund LM, et al. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res. (2011) 52:2159–68. doi: 10.1194/jlr.M017939
52. Li B, Weng Q, Dong C, Zhang Z, Li R, Liu J, et al. A key gene, PLIN1, can affect porcine intramuscular fat content based on transcriptome analysis. Genes. (2018) 9:194. doi: 10.3390/genes9040194
53. Shirouchi B, Albrecht E, Nuernberg G, Maak S, Olavanh S, Nakamura Y, et al. Fatty acid profiles and adipogenic gene expression of various fat depots in Japanese Black and Holstein steers. Meat Sci. (2014) 96:157–64. doi: 10.1016/j.meatsci.2013.06.027
54. Gjelstad IM, Haugen F, Gulseth HL, Norheim F, Jans A, Bakke SS, et al. Expression of perilipins in human skeletal muscle in vitro and in vivo in relation to diet, exercise and energy balance. Arch Physiol Biochem. (2012) 118:22–30. doi: 10.3109/13813455.2011.630009
55. Fröhlich T, Kemter E, Flenkenthaler F, Klymiuk N, Otte KA, Blutke A, et al. Progressive muscle proteome changes in a clinically relevant pig model of Duchenne muscular dystrophy. Sci Rep. (2016) 6:33362. doi: 10.1038/srep33362
56. Gao J, Serrero G. Adipose differentiation related protein (ADRP) expressed in transfected COS-7 cells selectively stimulates long chain fatty acid uptake. J Biol Chem. (1999) 274:16825–30. doi: 10.1074/jbc.274.24.16825
57. Imamura M, Inoguchi T, Ikuyama S, Taniguchi S, Kobayashi K, Nakashima N, et al. ADRP stimulates lipid accumulation and lipid droplet formation in murine fibroblasts. Am J Physiol-Endoc M. (2002) 283:E775–E83. doi: 10.1152/ajpendo.00040.2002
58. Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, et al. TIP47 functions in the biogenesis of lipid droplets. J Cell Biol. (2009) 185:641–55. doi: 10.1083/jcb.200812042
59. Skinner JR, Shew TM, Schwartz DM, Tzekov A, Lepus CM, Abumrad NA, et al. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem. (2009) 284:30941–8. doi: 10.1074/jbc.M109.013995
60. Pourteymour S, Lee S, Langleite TM, Eckardt K, Hjorth M, Bindesboll C, et al. Perilipin 4 in human skeletal muscle: localization and effect of physical activity. Physiol Rep. (2015) 3:12481. doi: 10.14814/phy2.12481
61. Bosma M, Sparks LM, Hooiveld GJ, Jorgensen JA, Houten SM, Schrauwen P, et al. Overexpression of PLIN5 in skeletal muscle promotes oxidative gene expression and intramyocellular lipid content without compromising insulin sensitivity. Bba-Mol Cell Biol L. (2013) 1831:844–52. doi: 10.1016/j.bbalip.2013.01.007
62. Bosma M, Minnaard R, Sparks LM, Schaart G, Losen M, de Baets MH, et al. The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem Cell Biol. (2012) 137:205–16. doi: 10.1007/s00418-011-0888-x
63. Tan MH, Sata T, Havel RJ. The significance of lipoprotein lipase in rat skeletal muscles. J Lipid Res. (1977) 18:363–70. doi: 10.1016/S0022-2275(20)41685-2
64. Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab. (2009) 297:E271–88. doi: 10.1152/ajpendo.90920.2008
65. Kersten S. Physiological regulation of lipoprotein lipase. Biochim Biophys Acta. (2014) 1841:919–33. doi: 10.1016/j.bbalip.2014.03.013
66. Levakfrank S, Radner H, Walsh A, Stolberger R, Knipping G, Hoefler G, et al. Muscle-specific overexpression of lipoprotein-lipase causes a severe myopathy characterized by proliferation of mitochondria and peroxisomes in transgenic mice. J Clin Invest. (1995) 96:976–86. doi: 10.1172/JCI118145
67. Kim JK, Fillmore JJ, Chen Y, Yu CL, Moore IK, Pypaert M, et al. Tissue-specific overexpression of lipoprotein lipase causes tissue-specific insulin resistance. P Natl Acad Sci USA. (2001) 98:7522–7. doi: 10.1073/pnas.121164498
68. Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. (2009) 50:3–21. doi: 10.1194/jlr.R800031-JLR200
69. MacPherson RE, Ramos SV, Vandenboom R, Roy BD, Peters SJ. Skeletal muscle PLIN proteins, ATGL and CGI-58, interactions at rest and following stimulated contraction. Am J Physiol Regul Integr Comp Physiol. (2013) 304:R644–50. doi: 10.1152/ajpregu.00418.2012
70. Granneman JG, Moore HP, Mottillo EP, Zhu Z. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J Biol Chem. (2009) 284:3049–57. doi: 10.1074/jbc.M808251200
71. Granneman JG, Moore HP, Mottillo EP, Zhu Z, Zhou L. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J Biol Chem. (2011) 286:5126–35. doi: 10.1074/jbc.M110.180711
Keywords: glutamine, low birth weight, lipid deposition, muscle gene expression, perilipins, pig
Citation: Zhao Y, Albrecht E, Li Z, Schregel J, Sciascia QL, Metges CC and Maak S (2021) Distinct Roles of Perilipins in the Intramuscular Deposition of Lipids in Glutamine-Supplemented, Low-, and Normal-Birth-Weight Piglets. Front. Vet. Sci. 8:633898. doi: 10.3389/fvets.2021.633898
Received: 26 November 2020; Accepted: 10 May 2021;
Published: 21 June 2021.
Edited by:
Xiao Li, Northwest A and F University, ChinaReviewed by:
Xin Wu, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Zhao, Albrecht, Li, Schregel, Sciascia, Metges and Maak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elke Albrecht, ZWxrZS5hbGJyZWNodEBmYm4tZHVtbWVyc3RvcmYuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.