- 1Institute of Animal Science and Veterinary Medicine, Shandong Academy of Agricultural Sciences, Jinan, China
- 2College of Animal Science and Technology, Shandong Agricultural University, Taian, China
- 3Shandong Provincial Animal Disease Prevention and Control Center, Jinan, China
The present study was designed to explore the Salmonella prevalence and antimicrobial resistance characteristics in the context of chick mortality at hatching in China. Between December 2015 and August 2017, 1,288 dead-in-shell chicken embryos were collected from four breeder chicken hatcheries in Tai'an, Rizhao, Jining, and Heze, China. Salmonella isolates were successfully recovered from 6.7% of these embryos (86/1,288) and were evaluated for serotype, antimicrobial susceptibility, Class 1 integron prevalence, antimicrobial resistance gene expression, and multilocus sequence typing (MLST). Salmonella Thompson (37.2%), and Salmonella Infantis (32.6%) were the two most prevalent isolates in these chicken embryos, and 66.3% of isolates exhibited robust ampicillin resistance, while 55.8% of isolates exhibited multi-drug resistance (MDR). The majority of isolates harbored the blaTEM gene (74.4%), with the qnrS gene also being highly prevalent (50.0%). In contrast, just 27.9% of these isolates carried Class 1 integrons. These 86 isolates were separated into four sequence types (STs), whereby ST26 (32.2%) was the most prevalent. Overall, these results suggested that Salmonella infections may be an important cause of chicken embryo mortality in China, and that efforts to support the appropriate use of antibiotics in managing poultry populations are essential.
Introduction
Salmonella is an important foodborne pathogen that can cause serious illness in humans and animals (1). Over 2,600 Salmonella serovars have been detected to date (2), and these bacteria cause illness in roughly 1 million patients per year in the USA alone, leading to approximately $365 million in medical costs (3). Salmonellosis is also highly prevalent in China and is particularly common in elderly or immunocompromised individuals (4, 5). Most Salmonella infections occur as a consequence of the consumption of contaminated pork, poultry, or other foods, with poultry in particular being commonly identified as an important Salmonella reservoir species. A range of Salmonella serovars can infect poultry, causing significant morbidity and mortality and enabling horizontal transmission of these bacteria within flocks as well as vertical transmission of these bacteria to eggs, often resulting in embryonic mortality or death of newly hatched birds (6, 7). Prior work suggests that Salmonella infections are associated with 23.6% of dead breeder chicken embryos in Henan Province, China, with Salmonella Pullorum being the dominant serotype in this region (8). Similarly, Salmonella samples were isolated from 26.7% of dead-in-shell embryos in Jos, Central Nigeria, with Salmonella Hadar being dominant in this context (9). In order to control and prevent the spread of Salmonella through the food chain, it is vital that these sources of transmission and contamination be appropriately understood and managed.
Currently, Salmonella outbreaks are generally controlled via the application of antimicrobial agents. Widespread antibiotic application, however, has led to the emergence of antibiotic- and multidrug-resistant (MDR) Salmonella strains that can resist β-lactam and fluoroquinolone treatment, and that thus represent a major threat to global health (10–12). Such antimicrobial resistance can significantly increase treatment-related costs as well as rates of infection-related morbidity and mortality. The emergence of antibiotic-resistant Salmonella can occur in particular geographical regions and production sites, and may be confined to particular bacterial serotypes, emphasizing the importance of studying regional Salmonella epidemiology (13).
Bacterial genes associated with antibiotic resistance are commonly encoded by mobile genetic elements that can be transmitted between microbes, with DNA-based integrons being the primary mobile genetic elements responsible for the transmission of these genes via conjugation (14). The most common integrons in MDR Salmonella are class 1 integrons, which are also closely linked to resistance gene dissemination in a range of different pathogens (15).
Prior work has shown that the co-incubation of Salmonella-free and Salmonella-contaminated eggs can facilitate the horizontal transmission of these bacteria during hatching. After traversing the membrane, Salmonella can be extremely difficult to treat and generally further invades the egg whereupon it disrupts normal embryonic development (16, 17). Despite the critical importance of this pathogen, only a few studies have explored the prevalence of Salmonella in the context of chick mortality at hatching in Shandong, China (18). This study was therefore designed to assess Salmonella prevalence and antibiotic resistance characteristics in four breeder chicken hatcheries in this region in order to better understand the epidemiology of this foodborne pathogen.
Materials and Methods
Sample Collection
Between December 2015 and August 2017, 1,288 dead-in-shell chicken embryos were collected from four breeder chicken hatcheries in Tai'an, Rizhao, Jining, and Heze. The lungs, heart, liver, and trachea were taken from each embryo and pooled, transported on ice, and analyzed within 6 h of collection in a laboratory. A bacterial culture was conducted as discussed previously (19). Briefly, 100 mL of buffered peptone water (BPW; Hopebiol, Qingdao, China) was combined with samples, followed by an 18-h incubation at 37°C. Next, 1 mL of pre-enrichment culture was combined with 10 mL of selenite cysteine (SC; Hopebiol, Qingdao, China) broth for 24 h at 42°C. A loop was then used to streak a sample of this SC broth culture on xylose lysine tergitol 4 (XLT4; Hopebiol, Qingdao, China) agar plates, followed by incubation for 24 h at 37°C. A bacterial genome extraction kit (QIAGEN, Mississayga, Ontario, Canada) was then used based on provided directions to isolate bacterial DNA, and polymerase chain reaction (PCR) amplification of the invA gene was used to confirm the identity of presumed Salmonella colonies (20).
Salmonella Serotyping
Salmonella isolate serotypes were established via the Kauffmann-White approach through slide agglutination using O and H antigen-specific sera (Tianrun Bio-Pharmaceutical, Ningbo, China) (21).
Antimicrobial Susceptibility Testing
A Kirby-Bauer disk diffusion approach was used to evaluate Salmonella sensitivity to treatment with 12 different common antibiotics, as per the protocols of the Clinical and Laboratory Standards Institute (22). Antibiotics used for these tests included ampicillin (AMP), cephalosporin/acid (CAC), cefazolin (CFZ), chloramphenicol (CHL), ciprofloxacin (CIP), nalidixic acid (NA), polymyxin B (PB), fosfomycin (FFN), gentamicin (GEN), tetracycline (TET), streptomycin (STR), and sulfamethoxazole (SXT). As a control, the ATCC 25922 and ATCC 35218 Escherichia coli strains were utilized and purchased from Beina Biotechnology Co., Ltd. All Salmonella isolates that were found to resist more than three antibiotic classes were defined as being MDR strains.
Class 1 Integrons and Antimicrobial Resistance Gene Detection
A bacterial genome extraction kit (QIAGEN) was used to isolate bacterial DNA, after which the qnrA, qnrB, qnrC, qnrD, qnrS, and aac(6')Ib-cr quinolone resistance genes were detected via PCR as detailed previously (23). Genes encoding β-lactamases, such as blaTEM, blaPSE, blaCMY−2, blaSHV, blaDHA−1, blaOXA, and blaCTX−M, were detected via PCR, as detailed previously (24, 25). Sequencing of all PCR products was then conducted. Class 1 integron gene cassettes were identified using primers and protocols discussed previously (26).
MLST
Seven housekeeping genes were used for multilocus sequence typing (MLST) profiling (aroC, dnaN, hemD, hisD, purE, sucA, and thrA), as defined by the University of College Cork (http://mlst.ucc.ie/). The Salmonella enterica MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Senterica) was used to assign STs to analyzed isolates.
Sequence-level relationships between Salmonella isolates were assessed by constructing an evolutionary phylogeny using MEGA6 via a maximum composite likelihood approach, with the topology of this phylogenetic tree being validated using 1,000 bootstrap replicates (27). To analyze resistance phenotypes and the relatedness of resistance gene expression profiles within this phylogenetic tree, the EvolView software package (http://www.evolgenius.info/evolview/#login) was used.
Results
Salmonella Prevalence
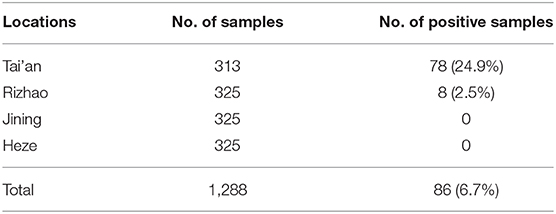
In total, we recovered Salmonella isolates from 6.7% of analyzed embryos (86/1,288), including 78 isolates from Tai'an (numbers 1–78) and 8 isolates from Rizhao (numbers 79–86). No isolate was recovered from Jining or Heze (Table 1).
Serotyping revealed four serotypes, including S. Thompson (n = 32), S. Infantis (n = 28), S. Enteritidis (n = 25), and S. Manhattan (S. Manhattan) (n = 1). S. Thompson (37.2%) and S. Infantis (32.6%) accounted for the majority of these isolates.
Antimicrobial Susceptibility Testing
These 86 Salmonella isolates were tested for resistance to 12 common antibiotics, revealing resistance rates as follows: ampicillin (66.3%), nalidixic acid (59.3%), tetracycline (47.7%), chloramphenicol (40.7%), sulfamethoxazole (38.4%), streptomycin (29.1%), and fosfomycin (2.3%). All Salmonella strains exhibited susceptibility or intermediate susceptibility to other tested antibiotics. Of these 86 isolates, 55.8% were classified as MDR isolates (Figure 1).
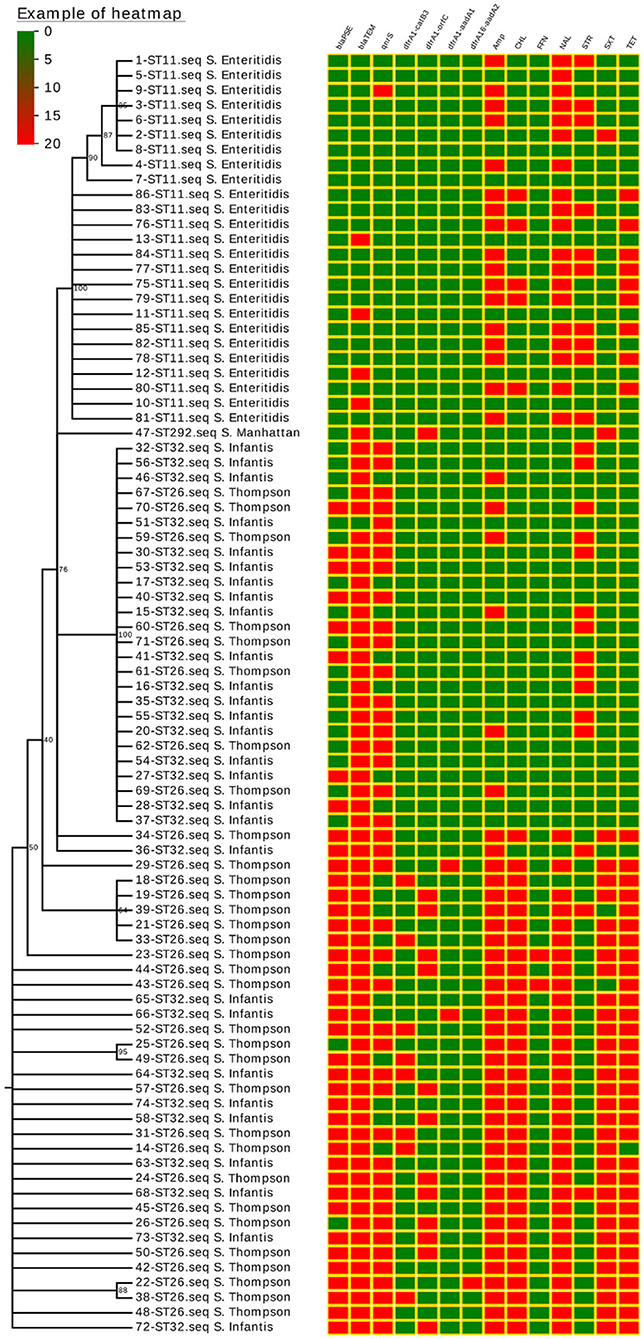
Figure 1. Salmonella isolate phylogenetic relationships, drug-resistance gene expression (blaPSE, blaTEM, and qnrS), class 1 integron structure (dfrA1-catB3, dfrA1-orfC, dfrA1-aadA1, and dfrA1-aadA2), and antimicrobial resistance (AMP, CHL, FFN, NAL, STR, SXT, and TET). Isolated strain numbers are given with ST designations and serotypes. Red and green squares correspond to the presence or absence of a given gene, respectively, or to the resistance or lack of resistance to a particular antibiotic, respectively.
Class 1 Integron and Antimicrobial Resistance Gene Detection
A single quinolone resistance gene (qnrS) was detected in these 86 Salmonella isolates, with this gene being carried by 50.0% of analyzed strains. Two different β-lactamase genes were found to be expressed in these isolates, with blaTEM being the more common of the two (74.4%), followed by blaPSE (46.5%). No isolates harbored the blaCTX−M, blaCMY−2, blaSHV, blaOXA, or blaDHA−1 genes (Figure 1).
Integrons were detected in 24 of these 86 Salmonella isolates (27.9%), and all Salmonella isolates harboring integrons were classified as MDR isolates with the exception of one isolate that was only resistant to two antibiotics. All detected Salmonella integrons encoded resistance gene cassettes, including the dfrA1-orfC (n = 13), dfrA1-catB3 (n = 8), dfrA1-aadA1 (n = 2), and dfrA16-aadA2 (n = 1) cassettes (Figure 1).
MLST
In an MLST analysis, these 86 Salmonella isolates were classified into four ST (Sequence Type) types, with ST26 being the most dominant (37.2%), followed by ST32 (32.6%), ST11 (29.1%), and ST292 (1.2%). These four STs were consistent with the observed serovars identified in this study, with ST26 corresponding to S. Thompson, ST32 to S. Infantis, ST11 to S. Enteritidis, and ST292 to S. Manhattan.
The majority of these ST strains were successfully grouped into a phylogenetic tree, which revealed no significant differences in drug-resistance gene expression patterns or drug-resistance spectra among strains within each ST (Figure 1).
Discussion
In this study, we collected Salmonella isolates from 6.7% of analyzed chicken embryos, and the positive rate was similar to the eggs collected from poultry farms in Yangling (6.6%) (28) but lower than that from commercial chicken farms in China (25, 29). These differences in Salmonella isolation rates may be attributable to regional or seasonal differences, or to variations between studies with respect to the techniques used to collect samples. The relatively low isolation rate of Salmonella suggests that Salmonella is not the main cause of chicken embryo death, but it may be caused by other reasons, and further research is needed.
Serotyping is an effective approach to evaluating modes of transmission to develop strategies for preventing disease spread within poultry facilities (30). We found that S. Thompson, which is a member of Salmonella serogroup C1 commonly associated with disease in humans (31) and isolated in poultry and poultry eggs (8, 28, 32), was the most prevalent isolate in the present study. This is in contrast to studies in Shanghai and Sichuan that had identified S. Enteritidis as the most common serovar in commercial chicken farms (25, 33), while S. Indiana was found to be dominant in Shandong (34), and S. Weltevreden was dominant in Central Vietnam (35). Our study is the first to have reported the presence of S. Manhattan in Shandong, which is also found from broiler chickens in Kagoshima, Japan (36). Given that we observed clear overlap between the Salmonella serotypes isolated from chicken embryos and the strains known to cause human disease, this underscores the fact that Salmonella can be transmitted to humans through the consumption of contaminated food products (37).
Herein, we found that ampicillin and nalidixic acid were the most commonly resisted antibiotics, in line with findings from several other studies (38, 39), indicating that the use of these drugs may be widespread in laying hens. We also detected high rates of tetracycline resistance, consistent with the fact that this antibiotic is commonly used in the context of animal production (40). We found that the Salmonella isolates in the present study were largely sensitive to cephalosporin/acid, cefazolin, ciprofloxacin, polymyxin B, fosfomycin, and gentamicin, likely owing to the limited use of these antibiotics in the study area. We frequently detected MDR Salmonella isolates from dead-in-shell chicken embryos in the present study, consistent with findings from commercial chicken farms in China (41). In addition, S. Thompson showed a high MDR rate (24/32, 75.0%) in this study, which was different from other study that most of S. Indiana showed MDR (34). These MDR Salmonella isolates are of particular concern because they represent a major threat to public health if transmitted to humans through the food chain (42).
We found that the qnrS gene was expressed by the majority of isolates in the present study, in contrast to a prior study of commercial chicken farms in Shandong where this gene was not detected (44). We found that 22 Salmonella isolates harboring the qnrS gene were resistant to nalidixic acid, indicating that these strains may exhibit a chromosomal quinolone resistance-determining region point mutation. The aac(6')Ib-cr gene is a key mediator of bacterial resistance to ciprofloxacin and norfloxacin treatment (43). However, no bacteria harboring this aac(6')Ib-cr gene were detected in the present study, in contrast to the findings of a prior analysis of commercial Chinese chicken farms, which found this gene to be present in >90% of Salmonella isolates (44). The high prevalence of PMQR genes in Salmonella isolates underscores the importance of prudently utilizing fluoroquinolones in order to minimize the development of high-level fluoroquinolone resistance.
We found that blaTEM was the most common β-lactamase gene expressed among isolates in the present study, followed by blaPSE, consistent with a similar report from commercial chicken farms in Shandong (34), although these results were inconsistent with those from a study of slaughterhouses and retail meat products in Sichuan, where the blaOXA was the most common such gene, followed by blaTEM, blaPSE, and blaCMY−2 (25). We found that the majority of analyzed Salmonella isolates harboring blaTEM and blaPSE exhibited ampicillin resistance, indicating that β-lactamases may be the main mechanism in Gram-negative bacteria to overcome penicillin-derived antibiotics.
We found that 27.9% of our Salmonella isolates harbored integrons, consistent with a prior report from farm animals in Shandong (45), although this rate was higher than that reported for Salmonella isolates in the Netherlands (46). All but one of the Salmonella isolates harboring these integrons in the present study were classified as MDR isolates, consistent with a model wherein class 1 integrons are linked to the emergence of MDR in Salmonella. We additionally identified both S. Thompson and S. Infantis strains harboring these integrons.
An MLST approach led to the classification of these 86 Salmonella isolates into four STs, all of which have been previously linked to the incidence of human salmonellosis (31, 39). ST26 was the most prevalent ST in the present study, in contrast to the results of a prior study of breeder chicken flocks in nine Chinese provinces, which found ST92 to be the most prevalent in sample sites (18). We also observed a close relationship between STs and serovars. When a phylogenetic tree was used to assess relationships between ST genotypes and antibiotic resistance profiles, we observed marked similarities in drug-resistance characteristics for Salmonella isolates within each of these STs.
Conclusion
In summary, we explored the characteristics of Salmonella infections in the context of chicken mortality at hatching in Shandong, China. We found the clinically important S. Thompson and S. Infantis serovars to be dominant among isolates recovered in the present analysis, and the majority of other isolates were also related to salmonellosis in humans. Overall, our data emphasize the importance of conducting antibiotic susceptibility testing when choosing appropriate antibiotics to treat Salmonella infections in order to minimize the risk of further facilitating the spread of drug-resistant strains of these dangerous bacteria.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
XiaonZ, SS, and XiaomZ: data curation. SS and XiaomZ: formal analysis. SS: funding acquisition. XiaonZ, ZJ, and HT: investigation. ZJ, JY, GW, and FW: methodology. XiaonZ: writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National key R&D project (2016YFD050 1608); Shandong Agricultural Major Applied Technology Innovation Program (SD2019XM009). The High-Level Talents and Innovative Team Recruitment Program of the Shandong Academy of Agricultural Sciences, China (CXGC2018E10).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Layton AN, Galyov EE. Salmonella-induced enteritis: molecular pathogenesis and therapeutic implications. Expert Rev Mol Med. (2007) 9:1–17. doi: 10.1017/S1462399407000373
2. Son IZ, Jie CE, Keys S, Zhao J, Brown EW. Analysis of pulsed field gel electrophoresis profiles using multiple enzymes for predicting potential source reservoirs for strains of Salmonella enteritidis and Salmonella typhimurium isolated from humans. Infect Genet Evol. (2013) 16:226–33. doi: 10.1016/j.meegid.2013.01.020
3. Centers for Disease Control and Prevention. Salmonella serotype Tennessee in powdered milk products and infant formula Canada and the United States. Morb Mortal Wkly Rep. (2014) 42:516–7. doi: 10.1001/jama.1993.03510040036011
4. Lee LA, Puhr ND, Maloney EK, Bean NH, Tauxe RV. Increase in antimicrobial-resistant Salmonella infections in the United States, 1989-1990. J Infect Dis. (1994) 170:128–34. doi: 10.1093/infdis/170.1.128
5. Liang Z, Ke BX, Deng XL, Liang JH, Ran LL, Lu G, et al. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009-2012. BMC Infect Dis. (2015) 15:53. doi: 10.1186/s12879-015-0784-4
6. Vo AT, Van DE, Fluit AC, Heck ME, Verbruggen A, Maas HM, et al. Distribution of Salmonella enterica serovars from humans, livestock and meat in Vietnam and the dominance of Salmonella Typhimurium phage type 90. Vet Microb. (2006) 113:153–8. doi: 10.1016/j.vetmic.2005.10.034
7. Yang J, Gao S, Chang Y, Su M, Xie Y, Sun S. Occurrence and characterization of Salmonella isolated from large-scale breeder farms in Shandong Province, China. Biomed Res Int. (2019) 5:8159567. doi: 10.1155/2019/8159567
8. Xu YH, Zhou X, Jiang ZH, Qi Y, Abdelaziz E, Yue M. Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder hicken hatcheries in Henan, China. Front Cell Infect Microbiol. (2020) 15:00597. doi: 10.3389/fcimb.2020.00497
9. Maryam M, Lawal M, Abdul-Ganiyu A, Aliyu M, Samuel A, Lisa B. Prevalence of Salmonella associated with chick mortality at hatching and their susceptibility to antimicrobial agents. Vet Microbiol. (2010) 140:131–5. doi: 10.1016/j.vetmic.2009.07.009
10. Gong J, Xu M, Zhu C, Miao J, Liu X, Xu B, et al. Antimicrobial resistance, presence of integrons and biofilm formation of Salmonella Pullorum isolates from eastern China (1962-2010). Avian Pathol. (2013) 42:290–4. doi: 10.1080/03079457.2013.788129
11. Lai J, Wu CM, Wu CB, Qi J, Wang Y, Wang HY, et al. Serotype distribution and antibiotic resistance of Salmonella in food-producing animals in Shandong province of China, 2009 and 2012. Int J Food Microbiol. (2014) 180:30–8. doi: 10.1016/j.ijfoodmicro.2014.03.030
12. Song Y, Yu L, Zhang Y, Dai Y, Wang P, Feng C, et al. Prevalence and characteristics of multidrug-resistant mcr-1-positive Escherichia coli isolates from broiler chickens in Tai'an, China. Poultry Sci. (2020) 99:1117–23. doi: 10.1016/j.psj.2019.10.044
13. Fei X, Yin K, Yin C, Hu Y, Li J, Zhou Z, et al. Analyses of prevalence and molecular typing reveal the spread of antimicrobial-resistant Salmonella infection across two breeder chicken farms. Poultry Sci. (2018) 97:4374–83. doi: 10.3382/ps/pey305
14. Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. (2006) 4:608–20. doi: 10.1038/nrmicro1462
15. Wright GD. Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol. (2010) 13:589–94. doi: 10.1016/j.mib.2010.08.005
16. Cason JA, Cox NA, Bailey JS. Transmission of Salmonella typhimurium during hatching of broiler chicks. Avian Dis. (1994) 38:583–8. doi: 10.2307/1592082
17. Cox NA, Berrang ME, Cason JE. Salmonella penetration of egg shells and proliferation in broiler hatching eggs: a review. Poultry Sci. (2000) 79:1571–4. doi: 10.1093/ps/79.11.1571
18. Song Y, Wang F, Liu Y, Song Y, Zhang L, Zhang F, et al. Occurrence and characterization of Salmonella isolated from chicken breeder flocks in nine Chinese provinces. Front Vet Sci. (2020) 7:479. doi: 10.3389/fvets.2020.00479
19. Yan H, Li L, Alam MJ, Shinoda S, Miyoshi S, Shi L. Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int J Food Microbiol. (2010) 143:230–4. doi: 10.1016/j.ijfoodmicro.2010.07.034
20. Malorny B, Hoorfar J, Hugas M, Heuvelink A, Fach P, Ellerbroek L, et al. Interlaboratory diagnostic accuracy of a Salmonella specific PCR-based method. Int J Food Microb. (2003) 89:241–9. doi: 10.1016/S0168-1605(03)00154-5
21. Grimont PAD, Weill FX. Antigenic Formulae of the Salmonella Serovars. 9th ed. Paris: WHO collaborating Centre for Reference and Research on Salmonella, Institut Pasteur. 13 p.
22. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 29th Informational Supplement, M100-S29. Pennsylvania, USA: CLSI (2019).
23. Jiang HX, Song L, Liu J, Zhang XH, Ren YN, Zhang WH, et al. Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int J Antimicrob Agents. (2014) 43:242–7. doi: 10.1016/j.ijantimicag.2013.12.005
24. Batchelor M, Hopkins K, Threlfall EJ, Clifton-Hadley FA, Stallwood AD, Davies RH, et al. bla(CTX-M) genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob Agents Chemother. (2005) 49:1319–22. doi: 10.1128/AAC.49.4.1319-1322.2005
25. Li RC, Lai J, Wang Y, Liu SL, Li Y, Liu KY, et al. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int J Food Microbiol. (2013) 163:14–8. doi: 10.1016/j.ijfoodmicro.2013.01.020
26. Kerrn MB, Klemmensen T, Frimodt-Moller N, Espersen F. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J Antimicrob Chemother. (2002) 50:513–6. doi: 10.1093/jac/dkf164
27. Alves VF, Nino-Arias FC, Pitondo-Silva A, Araújo F, Oliverira G, Toubas L, et al. Molecular characterisation of Staphylococcus aureus from some artisanal Brazilian dairies. Int Dairy J. (2018) 85:247–53. doi: 10.1016/j.idairyj.2018.06.008
28. Li W, Li H, Zheng S, Wang Z, Sheng H, Shi C, et al. Prevalence, serotype, antibiotic susceptibility, and genotype of Salmonella in eggs from poultry farms and marketplaces in Yangling, Shaanxi province, China. Front Microbiol. (2020) 11:1482. doi: 10.3389/fmicb.2020.01482
29. Cui MQ, Xie MY, Qu ZN, Zhao SJ, Wang JW, Wang Y, et al. Prevalence and antimicrobial resistance of Salmonella isolated from an integrated broiler chicken supply chain in Qingdao. China Food Control. (2016) 62:270–6. doi: 10.1016/j.foodcont.2015.10.036
30. Paudyal N, Pan H, Elbediwi M, Zhou X, Peng X, Li X, et al. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC microbiol. (2019) 19:226. doi: 10.1186/s12866-019-1598-0
31. Gaulin C, Fiset M, Duchesne C, Ramsay D, Savard N, Urbanek A, et al. Salmonella Thompson outbreak associated with consumption of chicken shawarma and the usefulness of genome sequencing in the investigation. Can Commun Dis Rep. (2017) 43:186–92. doi: 10.14745/ccdr.v43i09a05
32. Shah DH, Paul NC, Sischo WC, Crespo R, Guard J. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci. (2017) 96:687–702. doi: 10.3382/ps/pew342
33. Liu WB, Chen J, Huang YY, Liu B, Shi XM. Serotype, genotype, and antimicrobial susceptibility profiles of Salmonella from chicken farms in Shanghai. J Food Protect. (2010) 73:562–7. doi: 10.4315/0362-028X-73.3.562
34. Lu Y, Wu CM, Wu GJ, Zhao HY, He T, Cao XY, et al. Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog Dis. (2011) 8:1. doi: 10.1089/fpd.2010.0605
35. Lettini AA, Than TV, Marafin E, Longo A, Antonello K, Zavagnin P, et al. Distribution of Salmonella serovars and antimicrobial susceptibility from poultry and swine farms in Central Vietnam. Zoonoses Public Health. (2016) 63:569–76. doi: 10.1111/zph.12265
36. Duc VM, Nakamoto Y, Fujiwara A, Toyofuku H, Obi T, Chuma T, et al. Prevalence of Salmonella in broiler chickens in Kagoshima, Japan in 2009 to 2012 and the relationship between serovars changing and antimicrobial resistance. BMC Vet Res. (2019) 15:108. doi: 10.1186/s12917-019-1836-6
37. Mezal EH, Stefanova R, Khan AA. Isolation and molecular characterization of Salmonella enterica serovar Javiana from food, environmental and clinical samples. Int J Food Microbiol. (2013) 164:113–8. doi: 10.1016/j.ijfoodmicro.2013.03.021
38. Dessie HK, Bae DH, Lee YJ. Characterization of integrons and their cassettes in Escherichia coli and Salmonella isolates from poultry in Korea. Poultry Sci. (2013) 92:3036–43. doi: 10.3382/ps.2013-03312
39. Cai YQ, Tao J, Jiao Y, Fei X, Zhou L, Wang Y, et al. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int J Food Microbiol. (2016) 222:56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020
40. Miranda JM, Rodriguez JA, Galan-Vidal CA. Simultaneous determination of tetracyclines in poultry muscle by capillary zone electrophoresis. J Chromatogr A. (2009) 1216:3366–71. doi: 10.1016/j.chroma.2009.01.105
41. Kuang XH, Hao HH, Dai MH, Wang YL, Ahmad I, Liu ZL, et al. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front Microbiol. (2015) 6:602. doi: 10.3389/fmicb.2015.00602
42. Rosangela EZ, Camila L, Ana PP, Mallu JS, Ricardo APS, Cibeli V, et al. Multidrug resistance and ESBL-producing Salmonella spp. isolated from broiler processing plants. Braz J Microbiol. (2016) 47:191–5. doi: 10.1016/j.bjm.2015.11.021
43. Pribul BR, Festivo ML, Soares de Souza MM, Rodrigues DP. Characterization of quinolone resistance in Salmonella spp. isolates from food products and human samples in Brazil. Braz J Microbiol. (2016) 47:196–201. doi: 10.1016/j.bjm.2015.04.001
44. Lu Y, Zhao HY, Liu YQ, Zhou XP, Wang JY, Liu T, et al. Characterization of quinolone resistance in Salmonella enterica sevovar Indiana from chickens in China. Poultry Sci. (2015) 94:454–60. doi: 10.3382/ps/peu133
45. Zhao X, Yang J, Zhang B, Sun S, Chang W. Characterization of integrons and resistance genes in Salmonella isolates from farm animals in Shandong Province, China. Front Microbiol. (2017) 8:1300. doi: 10.3389/fmicb.2017.01300
Keywords: chicken embryos, class 1 integrons, antimicrobial resistance, MLST, Salmonella
Citation: Zhao X, Ju Z, Wang G, Yang J, Wang F, Tang H, Zhao X and Sun S (2021) Prevalence and Antimicrobial Resistance of Salmonella Isolated From Dead-in-Shell Chicken Embryos in Shandong, China. Front. Vet. Sci. 8:581946. doi: 10.3389/fvets.2021.581946
Received: 10 July 2020; Accepted: 27 January 2021;
Published: 16 March 2021.
Edited by:
Roswitha Merle, Freie Universität Berlin, GermanyReviewed by:
Olga Makarova, Freie Universität Berlin, GermanyAnna Schmitz, Freie Universität Berlin, Germany
Yantao Wu, Yangzhou University, China
Copyright © 2021 Zhao, Ju, Wang, Yang, Wang, Tang, Zhao and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuhong Sun, anF5YmZreWpzQDE2My5jb20=; Xiaomin Zhao, WE1aSEFPNjZAc2RhdS5lZHUuY24=
†These authors have contributed equally to this work
 Xiaonan Zhao
Xiaonan Zhao Zijing Ju2†
Zijing Ju2† Fangkun Wang
Fangkun Wang Shuhong Sun
Shuhong Sun