
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 06 January 2021
Sec. Parasitology
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.613718
This article is part of the Research Topic Zoonotic Diseases: Their Hosts and Vectors View all 18 articles
Salmonella spp. is widely considered one of the most important zoonotic pathogens worldwide. The close contact between reptiles and their owners provides favourable conditions for the transmission of zoonotic pathogen infections, and ~6% of human salmonellosis cases are acquired after direct or indirect contact with reptiles. Moreover, antimicrobial resistance is one of the most important health threats of the twenty-first century and has been reported in Salmonella strains isolated from pet reptiles, which could entail therapeutic consequences for their owners and breeders. The aim of this study was to assess Salmonella carriage by pet reptiles in pet shops and households, and their role in the transmission of antimicrobial resistance, to inform the owners about the possible risks factors. During the period between January 2019 and December 2019, 54 reptiles from pet shops and 69 reptiles from households were sampled in the Valencian Region (Eastern Spain). Three different sample types were collected from each reptile: oral cavity, skin, and cloacal swabs. Salmonella identification was based on ISO 6579-1:2017 (Annex D), serotyped in accordance with Kauffman-White-Le-Minor technique, and antibiotic susceptibility was assessed according to Decision 2013/652. The results of this study showed that 48% of the pet reptiles examined from households and pet shops carry Salmonella spp. All the strains isolated presented resistance to at least one antibiotic, and 72% were multidrug-resistant strains, the most frequently observed resistance patterns being gentamicin-colistin and gentamicin-colistin-ampicillin. The present study demonstrates that pet reptiles could be a source of human multidrug-resistant Salmonella infection. In this context, the most optimal prevention of multidrug-resistant Salmonella infections necessarily involves strict control of the sanitary status of reptile pet shops and hygienic handling by the individual owners at home.
Salmonella is widely considered one of the most important zoonotic pathogens worldwide. This pathogen has become an important public health concern with a significant economic impact, which has been estimated at 3.6 billion dollars annually (1, 2). In Europe, salmonellosis was responsible for 94,203 human cases, of which 9.3% corresponded to Spain (3). The infection usually causes self-limited diarrhoeal illness, although severe illness and death may occur, especially in children, elderly or immunocompromised adults (4). However, the overall epidemiological pattern of human salmonellosis cases is related to Salmonella-contaminated food from animal origin, especially eggs and poultry meat, and ~6% of human salmonellosis cases are acquired after direct or indirect contact with reptiles (3, 4).
In the last few years, exotic reptiles have risen in popularity as pets, with a population of over 7 million in European households (5). This increase in “living presents for children” is resulting in a trade of non-conventional species around the world, with Europe as the leading reptile importer (6, 7). The close contact between reptiles and their owners provides favourable conditions for the transmission of zoonotic pathogens infections, constituting a public health concern, as these pets have been considered as potential Salmonella carriers (7–11). Reptiles are natural reservoirs of Salmonella, which can hold a wide variety of serovars simultaneously without symptoms (12–14). However, reptile-associated salmonellosis seems to be responsible for more serious complications, with invasive disease and hospitalisation, especially in children (14, 15). From a public health standpoint, pet reptiles represent a persistent source of salmonellosis in households (16–19).
In addition, Salmonella multi-resistant strains emerge as a potential concern for public health safety, with implications of increased disease severity, longer hospitalisations and higher cost rates (20, 21). In this context, the World Health Organisation deemed antimicrobial resistance (AMR) one of the most important health threats, which could cause 10 million deaths a year by 2050, ahead of other diseases such as cancer (22, 23). In this sense, Salmonella has been included in the World Health Organisation priority list of twelve antibiotic-resistant bacteria (24). Interest in the role of reptiles as an antibiotic-resistant Salmonella reservoir has increased in recent years (7, 25, 26). Moreover, AMR had been reported in Salmonella isolated from captive reptiles, and their release could entail therapeutic consequences for their owners and breeders (16, 27). Moreover, the widespread use of antibiotics against Salmonella has been described in the international trade of pet reptiles, in order to prevent economical loses, as well as in animal welfare in crowded farms and long-distance transport (28–30). Therefore, more information on AMR in pet reptiles is needed in view of One Health (31).
In this context, the objective of the present study is to assess Salmonella carriage by pet reptiles in pet stores and households in Eastern Spain (Valencia Region) and gain more in-depth knowledge of their role in AMR transmission, in order to inform the owners about the possible risk factors.
All animals were handled according to the principles of animal care published by Spanish Royal Decree 53/2013 (32).
During the period between January 2019 and December 2019, a total of 349 samples from 123 different reptile species from households and pet shops reptiles were taken. Previously, the owners were contacted by advertising the project through the University community (Universidad Cardenal Herrera-CEU, and Universidad Politécnica de Valencia) and veterinary clinics of the Valencian Region (Eastern Spain).
A total of 37 species were identified from the 123 of the reptiles sampled (Table 1). From these species, 12 were classified as chelonians (order Chelonia), 16 as lizards (suborder Sauria) and 9 as snakes (suborder Ofidia) (Table 1). According to the individuals sampled from each group, 43.9% (54/123), 39.0% (48/123), and 17.1% (21/123) were chelonians, lizards and snakes, respectively (Table 1).
For each individual, whenever possible, samples from oral cavity (n = 114), skin (n = 123), and cloaca (n = 112) were taken using sterile cotton swabs (Cary Blair sterile transport swabs, DELTALAB®) (33). All individuals sampled were healthy and none of them presented clinical symptoms such as diarrhoea at the moment of sampling. In addition, an epidemiological questionnaire was filled in. The questionnaire contained information related to species, diet and the number of reptiles that cohabit in the same terrarium. The diet was classified as food from animal origin (including live prey, fresh meat and frozen meat), food of vegetable origin (including fruit and vegetables) and processed (including commercially manufactured reptile food). Moreover, the number of reptiles coexisting in the same terrarium was recorded as reptiles that inhabit alone, or reptiles that cohabit with two or more reptiles.
The collected samples were analysed within 24 h of collection according to ISO 6579-1:2017 (Annex D) recommendations (34). Samples were pre-enriched in 1:10 vol/vol Buffered Peptone Water 2.5% (BPW; Scharlau, Barcelona, Spain), and then incubated at 37 ± 1°C for 18 ± 2 h. The pre-enriched samples were transferred onto Semi-Solid Modification Rappaport Vassiliadis (MSRV; Difco, Valencia, Spain), and incubated at 41.5 ± 1°C for 24–48 h. For the positive plates, the cultures obtained in MSRV were inoculated onto two specific agar plates for Salmonella spp. detection: Xylose Lysine Deoxycholate Agar (XLD; Liofilchem, Valencia, Spain) and a selective chromogenic agar medium specific for detection of C8-esterase activity (ASAP, bioMérieux, Marcy l'Étoile, France), then incubated at 37 ± 1°C for 24 h. After incubation, one typical colony was collected and inoculated into a pre-dried nutrient agar plate (Scharlau, Barcelona, Spain), then incubated at 37 ± 1°C for 24 h. Finally, an API (API-20-E; bioMérieux, Madrid, Spain) biochemical test was performed to confirm Salmonella spp. The Salmonella isolates were stored at −80°C for further serotyping and antimicrobial susceptibility testing.
From each individual, the serotyping was performed from a cloacal strain, and when not present, a strain from the skin or oral cavity was analysed. Thus, all the strains were unfrozen and revived (ASAP) and the selected isolates were serotyped at the National Reference Laboratory for Animal Health (Algete, Madrid, Spain). The method used for serotyping was antigenic agglutination with specific antisera according to the White-Kauffmann-Le Minor scheme (35).
From all strains, Salmonella antimicrobial susceptibility was tested according to the European Committee on Antimicrobial Susceptibility Testing guidelines (36). Salmonella strains were inoculated into Mueller-Hinton agar (Scharlab, S.L.) to form a bacterial lawn and were allowed to dry for 30 min at ambient (25°C) temperature; then, the antibiotic discs were applied and plates were incubated at 37°C for 24 h. The antimicrobial agents selected were those set out in Decision 2013/652 (37), including three b-lactams: ampicillin (AMP, 10 μg), cefotaxime (CTX, 30 μg) and ceftazidime (CAZ, 30 μg); two quinolones: ciprofloxacin (CIP, 5 μg) and nalidixic acid (NA, 30 μg); one phenicol: chloramphenicol (CHL, 5 μg); one potentiated sulfonamide: trimethoprim-sulfamethoxazole (SXT, 1.25/23.75 μg); one polymyxin: colistin (COL, 10 μg); one macrolide: azithromycin (AZM, 15 μg); one glycylcycline: tigecycline (TGC, 15 μg); one aminoglycoside: gentamicin (GM, 10 μg); and one pyrimidine: trimethoprim (TM, 5 μg). The source for zone diameters used for interpretation of the test and plates after incubation at 37°C for 24 h was the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/clinical_breakpoints/), and where this was not possible, according to Clinical and Laboratory Standards Institute (CLSI) indications (https://clsi.org/media/2663/m100ed29_sample.pdf) (38). The isolate strains were categorised as susceptible (S) or resistant (R), based on EUCAST imperative criteria (39). Multidrug resistance (MDR) was defined as acquired resistance to at least one agent in two or more antimicrobial classes (40).
A Generalised Linear Model, which assumed a binomial distribution for Salmonella shedding, AMR and MDR, was fitted to the data to determine whether there was an association with the categorical variables (species and order or suborder of reptile, the habitat of the reptile, sample type, diet and number of reptiles that cohabit in the same terrarium). A reptile was considered Salmonella positive if one or more samples collected (oral cavity, skin and/or cloacal) tested positive. A P ≤ 0.05 was considered to indicate a statistically significant difference. Data are presented as least squares means ± standard error of the least squares means. In addition, a descriptive analysis has been done to assess the subespecies isolated in this study. Analyses were carried out using a commercially available software application (SPSS 24.0 software package; SPSS Inc., Chicago, IL, 2002).
From all samples collected during this study, 25.2 ± 2.3% (88/349) tested positive for Salmonella. The type of sample taken was significantly associated with Salmonella carriage (P = 0.000), with higher positive samples from cloaca (38.0 ± 4.6%, 43/112) than from skin (22.0 ± 3.7%, 27/123) and oral cavity (16.0 ± 3.4%, 18/114).
Salmonella spp. was detected in 48.0 ± 4.5% (59/123) of individuals sampled, with significant differences between snakes (76.0 ± 9.3%, 16/21) and lizards (69.0 ± 6.7%, 33/48), compared to chelonians (19.0 ± 5.3%, 10/54) (P = 0.000).
The reptiles sampled in this study inhabited households with private owners (56.1%, 69/123), as well as pet shops (43.9%, 54/123) in the Valencian Region (Eastern Spain). Significant differences for Salmonella isolation were found among the different reptile habitats (owners vs. pet shops) (P = 0.000), being higher in pet shop reptiles (67.0 ± 6.4%, 36/54) than in household pets (33.0 ± 5.7%, 23/69).
Moreover, the number of reptiles cohabiting the same terrarium were known for 111 of the 123 reptiles analysed, 49 for pet shops and 66 for households. In pet shops, significant differences were found between the number of reptiles present in the same terrarium and Salmonella shedding (P = 0.008). Thus, 89 ± 7.4% of reptiles that cohabit in terrariums with two or more reptiles were positive for Salmonella (16/18), while 58 ± 8.9% of reptiles that inhabit terrariums alone were positive for the bacterium (18/31). In contrast, for private owners' reptiles, no significant differences were observed between reptiles that cohabit in terrariums with two or more reptiles or alone and Salmonella shedding (P = 0.064), 21.0 ± 7.8% (16/38) and 42.0 ± 8.0% (6/28), respectively.
The diet was significantly associated with Salmonella carriage (P = 0.000), with higher frequency in reptiles that were fed with food from animal origin (65.0 ± 5.6%, 47/72), in contrast to reptiles that were fed with food from vegetable origin, and processed (24.0 ± 6.6%, 10/42, and 22.0 ± 13.9%, 2/9, respectively).
From the 59 strains selected for serotyping, 51 were viable after culture and were serotyped. All Salmonella isolates were classified as Salmonella enterica. The most represented subspecies were S. enterica (56.9%, 29/51), S. houtenae (19.6%, 10/51), S. diarizonae (11.8%,6/51), S. salamae (9.8%, 5/51) and S. arizonae (2.0%, 1/51). Fifteen different serovars of S. enterica subspecies were identified (Table 2). From all the strains serotyped, one Salmonella enterica serovar was indeterminate.
Seventy-five out of 88 Salmonella strains isolated were viable after culture and included in the antimicrobial susceptibility study. All strains analysed were resistant to at least one out of the twelve antibiotics tested (n = 75/75). The highest percentages of AMR were found to COL (97.3%, n = 73), followed by GM (84.0%, n = 63), AMP (46.7%, n = 35) and TGC (42.7%, n = 32), AZM (26.7%, n = 20), NAL (12.0%, n = 9), CHL (9.3%, n = 7), SXT and TM (8.0%, n = 6, both), and finally CAZ (6.7%, n =5), CTX (4.0%, n = 3), and CIP (1.3%, n = 1) (P = 0.000). Antimicrobial resistance of the different Salmonella enterica serovars was summarised in Table 3.
Furthermore, a total of 72.0% (54/75) Salmonella isolates were resistant to two or more antimicrobials. No significant differences in MDR rates were shown between lizards (78.0%, 32/41), chelonians (73.3%, 11/15) and snakes (57.9%, 11/19) (P = 0.206). Although the type of sample collected was not significantly associated with MDR carriage, oral cavity (75.0%, 12/16), skin (77.3%, 17/22) and cloacal samples (67,6%, 25/37) (P = 0.692), significant differences were found between the type of sample and the different types of reptiles, except for lizards (Table 4, P < 0.05). Moreover, no significant differences were found between the habitat (pet shop and household), diet (food from animal origin, vegetable origin, and processed) and MDR Salmonella carriage (P = 0.065 and P = 0.432, respectively)
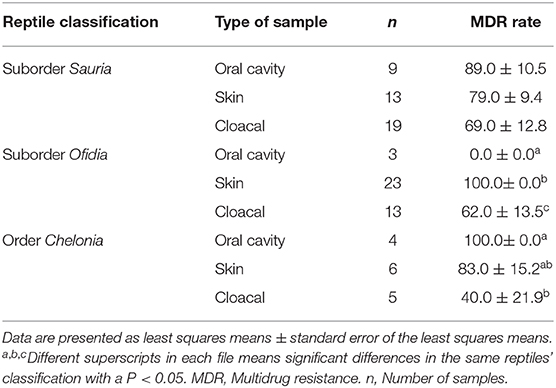
Table 4. Multidrug-resistant Salmonella isolated according to the type of sample collected in the different type of reptiles.
Overall, 25 different resistance patterns were observed. The combination of GM-COL (18.7%, 14/75) was the most frequently observed pattern, followed by GM-COL-AMP and GM-COL-TGC (10.7%, 8/75, both), COL alone (9.3%, 7/75) and GM-COL-AMP-TGC (8.0%, 6/75).
The present study demonstrates that 48% of the pet reptiles examined from households and pet shops carry Salmonella spp. All the strains isolated showed resistance to at least one antibiotic and 72% were multidrug-resistant strains. To our knowledge, this is the first study in the literature evaluating the prevalence and the antimicrobial resistance of this zoonotic pathogen from a considerable sample size in pet reptiles of Eastern Spain (Valencia Region).
Reptiles have been known to be important carriers of Salmonella spp. worldwide, which may pose a health hazard as a source of human infection, particularly in children (4, 16, 41–43). However, there is a lack of consensus regarding the role of reptile shops on MDR Salmonella strains spreading. The results of this study showed that Salmonella strains isolated in reptiles from shops were twice as high as those from private owners (67 vs. 33%) (44, 45). This may be due to poor hygienic management of terrariums, especially in pet shops where they are usually occupied and ensuring a proper cleaning and disinfection procedure is not easy (45). This fact could facilitate that MDR strains remain in the shop environment among different reptile batches. In addition, the reptiles' stress related to cohabiting with individuals of different ages and origins could result in an increase in the bacterial infection, shedding in the terrarium and reptile-to-reptile transmission (45, 46). Conversely, reptiles from private owners are exposed to better hygiene practises and less stressful environments, leading to lower Salmonella shedding (45).
In reptiles, Salmonella is spread by faecal-oral route with an asymptomatic natural colonisation of the enteric tract, so in this study cloacal swabs collected were more sensitive for Salmonella isolation than other samples collected, such as skin or oral cavity. However, it is important to highlight that because Salmonella is excreted through faeces, it could contaminate the reptile's skin, oral cavity and the environment, being a source of infection for humans who handle the reptile or who are exposed to the reptile's environment (11, 46–50). Moreover, the Salmonella serovars most frequently detected in this study have been cited previously in reptile studies (51, 52), as well as in human outbreaks (3, 51–55). In addition, it has been reported that cold-blooded animals could be the major reservoir for the subspecies houtenae, diarizonae, salamae, and arizonae (56, 57).
The results of this study showed higher Salmonella prevalence among snakes and lizards compared to chelonians, in accordance with previous research (11, 31, 52, 58, 59). Particular attention has recently been given to snakes and lizards, as human interaction with these reptiles has become increasingly common in domestic environments (11, 60). In this sense, it is important to highlight that these reptiles are mainly fed with food from animal origin, which represents an important source of Salmonella (49, 61, 62). Previous studies carried out in the United Kingdom reported the important role of commercial feeder rodents in bacterial transmission among reptiles, and even their owners (63, 64). Thus, handling Salmonella-contaminated feeder rodents, as well as cross-contamination in the kitchen due to the rodents being kept in the freezer and thawed in microwaves, also in contact with food for human consumption, have been linked to human Salmonella outbreaks (63, 65). In this context, to avoid Salmonella infection of reptiles, control of food products of animal origin has to be mandatory for the food suppliers (64).
On the other hand, special attention must be given to chelonians, due to their popularity as a pet for children. In this context, several countries, such as the US, have implemented strict bans in an attempt to curtail chelonian-associated salmonellosis; however, in Europe there are not many regulations to control its prevalence (53, 66). In the present study, Salmonella has been isolated from 18.5% of the chelonians tested. Seasonal effects, such as hibernation or season of sampling, have been speculated by previous studies to explain the low isolation rate of Salmonella in chelonians compared to other reptiles (31). Moreover, the diet may also have an important role (43, 49) because, as reported above, a large proportion of the chelonians are fed with food from vegetable origin or processed, and not from animal origin, frequently related with Salmonella outbreaks (11, 31, 67).
The increase in MDR Salmonella strains is of worldwide interest because it enhances the risk of therapeutic failure in cases of life-threatening salmonellosis in human and veterinary medicine (68, 69). In fact, it has been estimated that AMR could be the main cause of human mortality in 2050 (22). One of the most relevant outcomes in this study was the level of MDR isolated from pet reptiles, the most frequently observed resistance patterns being GM-COL and GM-COL-AMP. The high resistance against GM could be explained due to the indiscriminate use of aminoglycosides in pet reptile breeders, especially in the chelonian industry (28, 70). The use of GM as prophylactic Salmonella treatment in eggs to ensure sanitary conditions is a common practise in the US, the main supplier country of live reptiles for the EU (28, 71). Indeed, this practise has contributed to the finding of high-level plasmid-mediated gentamicin resistance in Salmonella isolated in its breeder farms (28).
Polymyxins have been widely used against Gramme-negative infections in animals, especially in animal production, the origin of several products involved in reptile feeding (72, 73). Currently, polymyxins such as COL represent the last line of defence against severe resistant infections in humans (72). Thus, it is highly restricted for animal infection treatments, and it is expected that resistance to this antibiotic will decrease in the coming years (74–76).
AMP was the third most frequent resistance shown in this study of Salmonella reptile strains, in line with a previous study conducted on species of gecko in Italy (10). This antibiotic is the most widely used in human medicine in Spain (76) and could thus be implicated in possible transmission of resistance from humans to reptiles, as a consequence of the direct and indirect contact between reptiles and their owners (75, 77, 78).
On the other hand, the level of resistance to NAL, CHL, SXT, TM, CAZ, CTX, and CIP in reptile Salmonella strains was relatively low. Resistance to CAZ contrasts with a survey conducted on geckos by Russo et al. (10), who showed a high resistance of Salmonella species to this antimicrobial, that is used in reptiles to treat infections or prophylactically after a traumatic injury (79). Moreover, it is important to note that fluoroquinolones (e.g., CIP) are the drugs of choice for invasive salmonellosis infections in humans (adults) and cephalosporins (e.g., CTX and CAZ) in children (21), although both are implicated in a reduced effectiveness of Salmonella treatment (77).
Should be essential to inform pet reptile owners about the risks of wrongly handling these animals (60). Proper hygienic management measures should be taken, such as the use of gloves when cleaning the reptile, and even during cleaning and disinfection of the surfaces that come in contact with the pet reptile (13). Moreover, it is important to thorough hand washing after handling the reptiles, especially the wounds due to bites or scratch (13). Reptiles and its feed should keep away from the kitchen and areas where the owners prepare their own food (64). Besides, before introducing a new reptile in the household, microbiological exams should be carried out to avoid cross-infection (13). Finally, it highlights the importance of extreme caution with young children and immunocompromised patients, because they are especially susceptible to Salmonella spp. infections (60).
The present study clearly demonstrates that pet reptiles could be a source of human MDR Salmonella infection. The problem of MDR in reptiles could start with the shops, where Salmonella presence is extremely high, and seems to be linked with the origin of reptile food. In this context, the most optimal prevention of MDR Salmonella infections involves strict control of the sanitary status of reptile pet shops and hygienic handling of the individuals in the household. Nevertheless, it is important to highlight that the number of included samples is relatively small, which may restrict the interpretation of our results to Eastern Spain. Further studies are needed to validate our results in a larger study sample.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethical review and approval was not required for the animal study because All animals were handled according to the principles of animal care published by Spanish Royal Decree 53/2013 (32). Written informed consent was obtained from the owners for the participation of their animals in this study.
CM, OL, and SV: conceptualisation. OL, JV-G, CM, and SV: data curation. LL-R, CM, OL, and JV-G: methodology. CM and SV: investigation, writing–review and editing, and funding acquisition. CM and LL-R: writing–original draft preparation. SV: project administration. All authors: have read and agreed to the published version of the manuscript.
This work was funded by Universidad Cardenal Herrera-CEU (IDOC 19/15, and INDI 20-21).
LL-R was supported by a research grant from the Generalitat Valenciana-Fondo Social Europeo (ACIF/2020/376).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank the reptile owners, veterinary clinics and pet shops for allowing access to their reptiles to enable this work to be carried out. Also, the authors wish to thank the “Improvement of Production System-related Food Safety and End Products” research group (Veterinary Faculty, University Cardenal Herrera-CEU) for the technical support. The English text version was revised by N. Macowan English Language Service.
1. USDA (United States Department of Agriculture). USDA ERS—Cost Estimates of Foodborne Illnesses. (2014). Available online at: https://www.ers.usda.gov/data-products/cost-estimates-of-foodborne-illnesses.aspx (accessed April 24, 2020).
2. WHO (World Health Organization). Interventions for the Control of Non-typhoidal Salmonella spp. 30th ed. L. Thorgeir Rome: Microbiological Risk Assessment Series (2016).
3. EFSA and ECDC. The European Union One Health 2018 zoonoses report. EFSA J. (2019) 17:1–276. doi: 10.2903/j.efsa.2019.5926
4. Mermin J, Hutwagner L, Vugia D, Shallow S, Daily P, Bender J, et al. Reptiles, amphibians, and human Salmonella infection: a population-based, case-control study. Clin Infect Dis. (2004) 38:S253–61. doi: 10.1086/381594
6. Bush ER, Baker SE, Macdonald DW. Global trade in exotic pets 2006-2012. Conserv Biol. (2014) 28:663–76. doi: 10.1111/cobi.12240
7. Bertelloni F, Chemaly M, Cerri D, Le Gall F, Ebani VV. Salmonella infection in healthy pet reptiles: bacteriological isolation and study of some pathogenic characters. Acta Microbiol Immunol Hung. (2016) 63:203–16. doi: 10.1556/030.63.2016.2.5
8. Stockman JA. Multistate outbreak of Salmonella infections associated with small turtle exposure, 2007–2008. Yearb Pediatr. (2011) 2011:288–9. doi: 10.1016/S0084-3954(10)79733-5
9. Jiménez RR, Barquero-Calvo E, Abarca JG, Porras LP. Salmonella isolates in the introduced asian house gecko (Hemidactylus frenatus) with emphasis on Salmonella Weltevreden, in two regions in Costa Rica. Vector-Borne Zoonotic Dis. (2015) 15:550–5. doi: 10.1089/vbz.2015.1785
10. Russo TP, Varriale L, Borrelli L, Pace A, Latronico M, Menna LF, et al. Salmonella serotypes isolated in geckos kept in seven collections in southern Italy. J Small Anim Pract. (2018) 59:294–7. doi: 10.1111/jsap.12808
11. Ramos CP, Santana JA, Morcatti Coura F, Xavier RGC, Leal CAG, Oliveira Junior CA, et al. Identification and characterization of Escherichia coli, Salmonella spp., Clostridium perfringens, and C. difficile isolates from reptiles in brazil. Biomed Res Int. (2019) 2019:1–9. doi: 10.1155/2019/9530732
12. Wikström VO, Fernström LL, Melin L, Boqvist S. Salmonella isolated from individual reptiles and environmental samples from terraria in private households in Sweden. Acta Vet Scand. (2014) 56:7. doi: 10.1186/1751-0147-56-7
13. Ebani VV. Domestic reptiles as source of zoonotic bacteria: a mini review. Asian Pac J Trop Med. (2017) 10:723–8. doi: 10.1016/j.apjtm.2017.07.020
14. Dudek B, Ksiazczyk M, Krzyzewska E, Rogala K, Kuczkowski M, Wozniak-Biel A, et al. Comparison of the phylogenetic analysis of PFGE profiles and the characteristic of virulence genes in clinical and reptile associated Salmonella strains. BMC Vet Res. (2019) 15:1–12. doi: 10.1186/s12917-019-2019-1
15. Walters MS, Simmons L, Anderson TC, De Ment J, Van Zile K, Matthias LP, et al. Outbreaks of salmonellosis from small turtles. Pediatrics. (2016) 137:1–9. doi: 10.1542/peds.2015-1735
16. Seepersadsingh N, Adesiyun AA. Prevalence and antimicrobial resistance of Salmonella spp. in pet mammals, reptiles, fish aquarium water, and birds in Trinidad. J Vet Med Ser B Infect Dis Vet Public Heal. (2003) 50:488–93. doi: 10.1046/j.0931-1793.2003.00710.x
17. Lowther SA, Medus C, Scheftel J, Leano F, Jawahir S, Smith K. Foodborne outbreak of Salmonella subspecies IV infections associated with contamination from bearded dragons. Zoonoses Public Health. (2011) 58:560–6. doi: 10.1111/j.1863-2378.2011.01403.x
18. Gay N, Le Hello S, Weill FX, de Thoisy B, Berger F. Salmonella serotypes in reptiles and humans, French Guiana. Vet Microbiol. (2014) 170:167–71. doi: 10.1016/j.vetmic.2014.01.024
19. Kuroki T, Ishihara T, Nakajima N, Furukawa I, Une Y. Prevalence of Salmonella enterica subspecies enterica in red-eared sliders trachemys scripta elegans retailed in pet shops in Japan. Jpn J Infect Dis. (2019) 72:38–43. doi: 10.7883/yoken.JJID.2018.140
20. O'Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. In: O'Neil, editor. The Review on Antimicrobial Resistance Chaired. London (2014).
21. EFSA (European Food Safety Authority). The European union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. (2019) 17:1–278. doi: 10.2903/j.efsa.2019.5598
22. O'Neill J. Tackling drug-resistant infections globally. Rev Antimicrob Resist. (2016) 7:110. doi: 10.4103/2045-080X.186181
23. WHO. Ten Threats to Global Health in 2019. (2019). Available online at: https://www.who.int/vietnam/news/feature-stories/detail/ten-threats-to-global-health-in-2019 (accessed May 12, 2020).
24. Tacconelli E, Carrara E, Savoldi A, Kattula D, Burkert F. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. (2017). Available online at: http://www.cdc.gov/drugresistance/threat-report-2013/ (accessed April 24, 2020).
25. Gorski L, Jay-Russell MT, Liang AS, Walker S, Bengson Y, Govoni J, et al. Diversity of pulsed-field gel electrophoresis pulsotypes, serovars, and antibiotic resistance among Salmonella. isolates from wild amphibians and reptiles in the California Central Coast. Foodborne Pathog Dis. (2013) 10:540–8. doi: 10.1089/fpd.2012.1372
26. Zhang J, Kuang D, Wang F, Meng J, Jin H, Yang X, et al. Turtles as a possible reservoir of nontyphoidal Salmonella in Shanghai, China. Foodborne Pathog Dis. (2016) 13:428–33. doi: 10.1089/fpd.2015.2107
27. Wei Z, Xu X, Yan M, Chang H, Li Y, Kan B, et al. Salmonella Typhimurium and Salmonella Enteritidis infections in sporadic diarrhea in children: source tracing and resistance to third-generation cephalosporins and ciprofloxacin. Foodborne Pathog Dis. (2019) 16:244–55. doi: 10.1089/fpd.2018.2557
28. Díaz MA, Cooper RK, Cloeckaert A, Siebeling RJ. Plasmid-mediated high-level gentamicin resistance among enteric bacteria isolated from pet turtles in Louisiana. Appl Environ Microbiol. (2006) 72:306–12. doi: 10.1128/AEM.72.1.306-312.2006
29. Giacopello C, Foti M, Fisichella V, Latella G, Aleo A, Mammina C. Antibiotic resistance in Salmonella isolated from tegus (Tupinambis spp.). J Exot Pet Med. (2012) 21:328–31. doi: 10.1053/j.jepm.2012.09.008
30. Goławska O, Zajac M, Maluta A, Pristas P, Hamarová L, Wasyl D. Complex bacterial flora of imported pet tortoises deceased during quarantine: another zoonotic threat? Comp Immunol Microbiol Infect Dis. (2019) 65:154–9. doi: 10.1016/j.cimid.2019.05.007
31. Chen CY, Chen WC, Chin SC, Lai YH, Tung KC, Chiou CS, et al. Prevalence and antimicrobial susceptibility of salmonellae isolates from reptiles in Taiwan. J Vet Diagnostic Investig. (2010) 22:44–50. doi: 10.1177/104063871002200107
32. Spain. Royal Degree 53/2013, 1st of Febrary, por el que se establecen las normas básicas aplicables para la protección de los animales utilizados en experimentación y otros fines científicos, incluyendo la docencia. Boletín Oficial del Estado. (2013) 34:11370–421.
33. Pees M, Rabsch W, Plenz B, Fruth A, Prager R, Simon S, et al. Evidence for the transmission of Salmonella from reptiles to children in Germany, July 2010 to October 2011. Eurosurveillance. (2013) 18:1–10. doi: 10.2807/1560-7917.ES2013.18.46.20634
34. ISO. Microbiology of Food and Animal Feeding Stuffs. Horizontal Method for the Detection of Salmonella spp. 6579-1:2017. Genève: International Organization for Standardization (2017).
35. Grimont PAD, Weill FX. Antigenic Formulae of the Salmonella Serovars. 9th ed. Paris: WHO Collaborating Centre for Reference and Research on Salmonella (2007).
36. Matuschek E, Brown DFJ, Kahlmeter G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect. (2014) 20:O255–66. doi: 10.1111/1469-0691.12373
37. European Commission. Commission Implementing Decision of 12 November 2013 on the Monitoring and Reporting of Antimicrobial Resistance in Zoonotic and Commensal Bacteria (Notified Under Document C(2013) 7145). (2013). Available online at: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32013D0652 (accessed May 5, 2020).
38. CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Twenty—fourth Informational Supplement. CLSI document M100—S24. Wayne, PA: s.n (2014).
39. EUCAST. New Definitions of S, I and R from 2019. (2019). Available online at: https://www.eucast.org/newsiandr/ (accessed May 1, 2020).
40. EFSA and ECDC. (European Food Safety Authority and European Centre for Disease Prevention and Control). EU Protocol for Harmonised Monitoring of Antimicrobial Resistance in Human Salmonella and Campylobacter Isolates. Stockholm: EFSA; ECDC (2016).
41. Gambino-Shirley K, Stevenson L, Wargo K, Burnworth L, Roberts J, Garrett N, et al. Notes from the field: four multistate outbreaks of human Salmonella infections linked to small turtle exposure—United States, 2015. Morb Mortal Wkly Rep. (2016) 65:655–6. doi: 10.15585/mmwr.mm6525a3
42. Kiebler CA, Bottichio L, Simmons L, Basler C, Klos R, Gurfield N, et al. Outbreak of human infections with uncommon Salmonella serotypes linked to pet bearded dragons, 2012–2014. Zoonoses Public Health. (2020) 67:425–34. doi: 10.1111/zph.12701
43. Sodagari HR, Habib I, Shahabi MP, Dybing NA, Wang P, Bruce M. Veterinary sciences a review of the public health challenges of salmonella and turtles. Vet Sci. (2020) 7:1–12. doi: 10.3390/vetsci7020056
44. Nakadai A, Kuroki T, Kato Y, Suzuki R, Yamai S, Yaginuma C, et al. Prevalence of Salmonella spp. in pet reptiles in Japan. J Vet Med Sci. (2005) 67:97–101. doi: 10.1292/jvms.67.97
45. Marin C, Vega S, Marco-Jiménez F. Tiny turtles purchased at pet stores are a potential high risk for Salmonella human infection in the Valencian Region, Eastern Spain. Vector-Borne Zoonotic Dis. (2016) 16:455–60. doi: 10.1089/vbz.2016.1950
46. Lukac M, Pedersen K, Prukner-Radovcic E. Prevalence of Salmonella in captive reptiles from Croatia. J Zoo Wildl Med. (2015) 46:234–40. doi: 10.1638/2014-0098R1.1
47. Ebani VV, Cerri D, Fratini F, Meille N, Valentini P, Andreani E. Salmonella enterica isolates from faeces of domestic reptiles and a study of their antimicrobial in vitro sensitivity. Res Vet Sci. (2005) 78:117–21. doi: 10.1016/j.rvsc.2004.08.002
48. Marin C, Ingresa-Capaccioni S, González-Bodi S, Marco-Jiménez F, Vega S. Free-living turtles are a reservoir for Salmonella but not for campylobacter. PLoS ONE. (2013) 8:e72350. doi: 10.1371/journal.pone.0072350
49. Clancy MM, Davis M, Valitutto MT, Nelson K, Sykes JM. Salmonella infection and carriage in reptiles in a zoological collection. J Am Vet Med Assoc. (2016) 248:1050–9. doi: 10.2460/javma.248.9.1050
50. Guyomard-Rabenirina S, Weill FX, Le Hello S, Bastian S, Berger F, Ferdinand S, et al. Reptiles in Guadeloupe (French West Indies) are a reservoir of major human Salmonella enterica serovars. PLoS ONE. (2019) 14:e0220145. doi: 10.1371/journal.pone.0220145
51. Ricard C, Mellentin J, Ben Abdallah Chabchoub R, Kingbede P, Heuclin T, Ramdame A, et al. Méningite à Salmonelle chez un nourrisson due à une tortue domestique. Arch Pediatr. (2015) 22:605–7. doi: 10.1016/j.arcped.2013.09.019
52. Bjelland AM, Sandvik LM, Skarstein MM, Svendal L, Debenham JJ. Prevalence of Salmonella serovars isolated from reptiles in Norwegian zoos. Acta Vet Scand. (2020) 62:3. doi: 10.1186/s13028-020-0502-0
53. De Jong B, Andersson Y, Ekdahl K. Effect of regulation and education on reptile-associated salmonellosis. Emerg Infect Dis. (2005) 11:398–403. doi: 10.3201/eid1103.040694
54. Jones SB, Spalloni W, Ferreccio F, Postigo J, Fernández A, Porte L, et al. Salmonella spp. Gastroenteritis associated to pet turtles in three infants. Rev Chil Infectol. (2015) 32:334–8. doi: 10.4067/S0716-10182015000400013
55. Bosch S, Tauxe RV, Behravesh CB. Turtle-associated salmonellosis, United States, 2006-2014. Emerg Infect Dis. (2016) 22:1149–55. doi: 10.3201/eid2207.150685
56. Briones V, Téllez S, Goyache J, Ballesteros C, Del Pilar Lanzarot M, Domínguez L, et al. Salmonella diversity associated with wild reptiles and amphibians in Spain. Environ Microbiol. (2004) 6:868–71. doi: 10.1111/j.1462-2920.2004.00631.x
57. Bauwens L, Vercammen F, Bertrand S, Collard JM, De Ceuster S. Isolation of Salmonella from environmental samples collected in the reptile department of Antwerp Zoo using different selective methods. J Appl Microbiol. (2006) 101:284–9. doi: 10.1111/j.1365-2672.2006.02977.x
58. Abalem De Sá IV, Solari CA. Salmonella in Brazilian and imported pet reptiles. Brazilian J Microbiol. (2001) 32:293–7. doi: 10.1590/S1517-83822001000400007
59. Geue L, Löschner U. Salmonella enterica in reptiles of German and Austrian origin. Vet Microbiol. (2002) 84:79–91. doi: 10.1016/S0378-1135(01)00437-0
60. Whiley H, Gardner MG, Ross K. A review of Salmonella and squamates (Lizards, snakes and amphisbians): implications for public health. Pathogens. (2017) 6:1–15. doi: 10.3390/pathogens6010001
61. Fuller CC, Jawahir SL, Leano FT, Bidol SA, Signs K, Davis C, et al. A multi-state Salmonella Typhimurium outbreak associated with frozen vacuum-packed rodents used to feed snakes. Zoonoses Public Health. (2008) 55:481–7. doi: 10.1111/j.1863-2378.2008.01118.x
62. Vrbova L, Sivanantharajah S, Walton R, Whitfield Y, Lee C, Picard I, et al. Outbreak of Salmonella Typhimurium associated with feeder rodents. Zoonoses Public Health. (2018) 65:386–94. doi: 10.1111/zph.12442
63. Kanagarajah S, Waldram A, Dolan G, Jenkins C, Ashton PM, Carrion Martin AI, et al. Whole genome sequencing reveals an outbreak of Salmonella Enteritidis associated with reptile feeder mice in the United Kingdom, 2012-2015. Food Microbiol. (2018) 71:32–8. doi: 10.1016/j.fm.2017.04.005
64. Marin C, Martelli F, Rabie A, Davies R. Commercial frozen mice used by owners to feed reptiles are highly externally contaminated with Salmonella Enteritidis PT8. Vector-Borne Zoonotic Dis. (2018) 18:453–7. doi: 10.1089/vbz.2018.2295
65. Cartwright EJ, Nguyen T, Melluso C, Ayers T, Lane C, Hodges A, et al. A multistate investigation of antibiotic-resistant Salmonella enterica serotype I 4,[5],12:i:-Infections as part of an International Outbreak Associated with Frozen Feeder Rodents. Zoonoses Public Health. (2016) 63:62–71. doi: 10.1111/zph.12205
66. Cohen ML, Potter M, Pollard R, Feldman RA. Turtle-associated salmonellosis in the United States. Effect of Public Health Action, 1970 to 1976. JAMA. (1980) 243:1247–9. doi: 10.1001/jama.1980.03300380027016
67. Wojdat E, Kwiatek K, Zasadny R. Microbiological quality of petfood in Poland. Pol J Vet Sci. (2004) 7:207–9.
68. Threlfall EJ, Fisher IS, Berghold C, Gerner-Smidt P, Tschäpe H, Cormican M, et al. Antimicrobial drug resistance in isolates of Salmonella enterica from cases of salmonellosis in humans in Europe in 2000: results of international multi-centre surveillance. Euro Surveill. (2003) 8:41–5. doi: 10.2807/esm.08.02.00400-en
69. Münch S, Braun P, Wernery U, Kinne J, Pees M, Flieger A, et al. Prevalence, serovars, phage types, and antibiotic susceptibilities of Salmonella strains isolated from animals in the United Arab Emirates from 1996 to 2009. Trop Anim Health Prod. (2012) 44:1725–38. doi: 10.1007/s11250-012-0130-4
70. Arnafia W, Ningrum SG, Adji RS, Lukman DW, Pasaribu FH, Wayan I, et al. Aislamiento de Salmonella en tiendas de mascotas de reptiles y su suceptibilidad a los antibioticos en Indonesia. Hum Vet Med Int J Bioflux Soc Res Artic. (2016) 8:177–88.
71. Auliya M, Altherr S, Ariano-Sanchez D, Baard EH, Brown C, Brown RM, et al. Trade in live reptiles, its impact on wild populations, and the role of the European market. Biol Conserv. (2016) 204:103–19. doi: 10.1016/j.biocon.2016.05.017
72. Sartelli M, Weber DG, Ruppé E, Bassetti M, Wright BJ, Ansaloni L, et al. Antimicrobials: a global alliance for optimizing their rational use in intra-abdominal infections (AGORA). World J Emerg Surg. (2016) 11:1–32. doi: 10.1186/s13017-016-0089-y
73. Elbediwi M, Li Y, Paudyal N, Pan H, Li X, Xie S, et al. Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980–2018). Microorganisms. (2019) 7:461. doi: 10.3390/microorganisms7100461
74. Aarestrup FM. The livestock reservoir for antimicrobial resistance: a personal view on changing patterns of risks, effects of interventions and the way forward. Philos Trans R Soc B Biol Sci. (2015) 370:20140085. doi: 10.1098/rstb.2014.0085
75. ECDC/EFSA/EMA (European Centre for Disease Prevention and Control/European Food Safety Authority/ European Medicines Agency). ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals: Joint Interagency Antimicrobial Consumption and Resistan. EFSA J. (2017) 15:1–135. doi: 10.2903/j.efsa.2017.4872
76. MAPA (Ministry of Agriculture, Fishing and Food). Informe JIACRA España. Primer análisis integrado del consumo de antibióticos y su relación con la aparición de resistencia. AEMPS. Plan Nac. Resist. Antibióticos. (2018). p. 1–165. Available online at: http://www.resistenciaantibioticos.es/es/system/files/field/files/informe_jiacra-espana.pdf?file=1andtype=nodeandid=410andforce=0 (accessed April 24, 2020).
77. Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev. (2015) 28:901–37. doi: 10.1128/CMR.00002-15
78. EMA (European Medicines Agency). Categorisation of Antibiotics in the European Union. (2019). Ema/Cvmp/Chmp/682198/2017 31. Available online at: https://www.ema.europa.eu/en/documents/report/categorisation-antibiotics-european-union-answer-request-european-commission-updating-scientific_en.pdf (accessed May 1, 2020).
Keywords: reptile-associated salmonellosis, multidrug-resistant Salmonella, pet reptiles, One Health, zoonosis, Salmonella
Citation: Marin C, Lorenzo-Rebenaque L, Laso O, Villora-Gonzalez J and Vega S (2021) Pet Reptiles: A Potential Source of Transmission of Multidrug-Resistant Salmonella. Front. Vet. Sci. 7:613718. doi: 10.3389/fvets.2020.613718
Received: 03 October 2020; Accepted: 08 December 2020;
Published: 06 January 2021.
Edited by:
Rodrigo Morchón García, University of Salamanca, SpainReviewed by:
Laura Rinaldi, University of Naples Federico II, ItalyCopyright © 2021 Marin, Lorenzo-Rebenaque, Laso, Villora-Gonzalez and Vega. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clara Marin, Y2xhcmEubWFyaW5AdWNoY2V1LmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.