- 1College of Animal Medical Science, Henan University of Animal Husbandry and Economy, Zhengzhou, China
- 2Zhengzhou Key Laboratory of Veterinary Biological Products Technology, Zhengzhou, China
- 3College of Biology and Food, Shangqiu Normal University, Shangqiu, China
- 4College of Animal Science and Veterinary Medicine, Henan Agricultural University, Zhengzhou, China
Anaplasma capra is an emerging zoonotic pathogen that pose a risk to the health of human and veterinary animal. Numerous variants in a variety of domestic and wild animals had been reported since its discovery and confirmation in humans in 2015 and its first detection from goat blood during 2012–2013. In order to find out more A. capra variants data of A. capra in central China, 16S rRNA, gltA, groEL, and msp4 genes of this pathogen were amplified from sheep and goat samples collected during 2011–2015 and phylogenetic analysis of these sequences were conducted. The results of 16S rRNA and gltA manifested that partial sequences obtained in this study were 100% identical with A. capra isolates, while phylogenetic analysis results of groEL and msp4 showed that the obtained sequences were independent with all other Anaplasmas, formed separate branches on the evolutionary trees. What needed to be emphasized was that the 16S rRNA and gltA gene sequences of X51 (KX505302 and KX450269), a sample from Shandong in 2011, were found to be 100% identical with A. capra. Therefore, we could speculate that the occurrence of A. capra may be earlier than its first discovery and report. And the A. capra isolates in central China were novel variants which were different from known genotypes.
Introduction
Anaplasma genus members are zoonotic pathogens with remarkable importance in both human and veterinary health. Three Anaplasma species have so far been identified that infect human beings. The first discovered and the most widespread is Anaplasma phagocytophilum, which was first reported in 1994 in Wisconsin, USA, and caused a febrile illness (1). Subsequently, the disease was named human granulocytic anaplasmosis based on its main symptoms. In China, since the first HGA case was reported in Anhui province in 2006 (2), more than 90 HGA cases have been reported in Beijing, Tianjin, Shandong, Hubei, Henan, and the Inner Mongolia Autonomous Region (3). In 2007, a patient from Cyprus was diagnosed as infected with A. ovis (4); but no other human infection with A. ovis have been reported. Anaplasma capra, a novel Anaplasma member, was first identified in 28 patients from Heilongjiang province in 2015. These patients had symptoms of high fever, headache, malaise, dizziness, and muscle pain (5). Since then, numerous reports on A. capra infections in various ruminants and wild animals have been published (6–9). The widespread distribution and genetic diversity of the novel identified Anaplasma species mentioned above highlight the dangers of tick-borne disease caused by the Anaplasma genus.
In previous studies, A. capra DNA fragments were reported to be detected in domestic animals and some species of wild animals and ticks (6–10). More importantly, various genotypes of this pathogen and A. capra-like bacteria were documented, a finding similar to the case of A. phagocytophilum where its many genetic variants result in poor immunity from the vaccine based on it. In the present study, we originally planned to amplify the almost full length of Anaplasma 16S rRNA, gltA, groEL, and msp4 genes from Anaplasma-positive sheep and goats DNA samples, which was only confirmed to be positive for Anaplasma based on the short fragment of 16S rRNA gene (11) and to analyze the genetic difference of Anaplasma, especially that of A. phagocytophilum, but the results were unexpected. The sequence alignment results showed that some of the sequences we obtained had higher homology with A. capra, which may be new A. capra isolates or genotypes.
Methods
Ethics Statement
This study was conducted in accordance with the Chinese Laboratory Animal Administration Act of 1988. The research protocol was reviewed and approved by the Research Ethics Committee of Henan University of Animal Husbandry and Economy. The field studies did not involve endangered or protected species.
Specimens and DNA Samples
Altogether, 212 DNA samples obtained from sheep and goats blood and was PCR-screened to be positive for Anaplasma using primer pair EE1 and EE2 (12) was used in the present study. All DNA samples were stored at −20°C until use.
PCR Amplification
Conventional PCR was used to amplify a 1,133-bp fragment of the 16S rRNA gene from Anaplasma. The primer pair was from two previous studies (13, 14). Nested PCRs specific for three protein coding genes, citrate synthase (gltA), heat shock protein (groEL), and major surface protein 4 (msp4), were conducted using previously described primers and PCR conditions (Supplementary Table 1). PCR amplification was performed as follows: 1.0 μL of extracted DNA was added to a 25 μL PCR mixture containing 2.5 μL of 10× PCR buffer (Mg2+Plus), 2.0 μL of dNTPs, 1.0 U of Taq DNA polymerase (LA Taq for the first round, rTaq for the second round) (Takara, Dalian, China), 1.0 μL of each primer (10 pmol), and 16.5 μL of distilled water. Reactions were conducted in an automated DNA C1000 thermal cycler (Bio-Rad, Beijing, China). Each PCR reaction was conducted at least twice using nuclease-free water as the negative control in each reaction, whereas a sheep DNA sample that was preserved in the parasitology laboratory of Henan Agricultural University and previously verified as Anaplasma-positive was used as the positive control. PCR products were visualized by UV transillumination on a 1.0% agarose gel followed by electrophoresis and staining with GelRed™ (Biotium Inc., Hayward, CA, USA).
DNA Sequencing and Phylogenetic Analysis
Positive PCR products were purified using Montage PCR filters (Millipore, Bedford, MA). The products were sequenced using the BigDye Terminator v 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) on the ABI 3730 DNA analyzer (Applied Biosystems). The sequence accuracy was confirmed by two-directional sequencing and by sequencing a new PCR product when necessary. The obtained sequences were analyzed by a BLAST search of the GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequence assembly, phylogenetic analyses and evolutionary analyses were performed using Mega X software (http://www.megasoftware.net/). The number of base differences per site from averaging over all sequence pairs between groups were shown as Supplementary Tables 5–8 for each gene. These analyses involved 25 nucleotide sequences for 16S rRNA gene, 23 for gltA gene, 21 for groEL gene and 23 for msp4 gene, respectively. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 1,314, 487, 328, and 366 positions for each gene in the final dataset. Phylogenetic trees were constructed using the neighbor-joining algorithm method (15) with the Kimura two-parameter model for nucleotide sequence analysis. To examine the effect of the method of analysis on the resulting phylogeny, maximum parsimony analyses were also conducted. The stability of the trees obtained was estimated by bootstrap analysis with 1,000 replicates.
Nucleotide Sequence Accession Numbers
The representative sequences obtained in this study were deposited in the GenBank database under the following accession numbers: 16S rRNA (KX505293–KX505303), gltA (KX450266–KX450272), groEL (KX388350–KX388358), and msp4 (KX528101–KX528113).
Results
Phylogenetic Analysis of 16S rRNA and gltA Genes
Of the 212 Anaplasma–positive specimens, the 16S rRNA (~1,133 bp) gene was identified in 30. Sequence analysis showed that 11 16S rRNA sequence types, which grouped into three groups on the evolutionary tree, were obtained (Figure 1A). The sequences in group a shared 98.7–100% similarity with A. capra isolates from China (MG869594) and Korea (LC432114), but were distinct from other Anaplasma species. They represented 70% (21/30) of all the 16S rRNA genes amplified from the sheep and goat blood samples collected from several Chinses provinces between 2011 and 2015 (Supplementary Table 2). KX505300 and KX505301 were closely related to A. ovis isolates (MG869525 and KX579073, with 99.8–100% similarity) and formed a clade (group b). The sequences in group c shared 99.0% identity with the sheep and goat isolates of A. bovis from China.
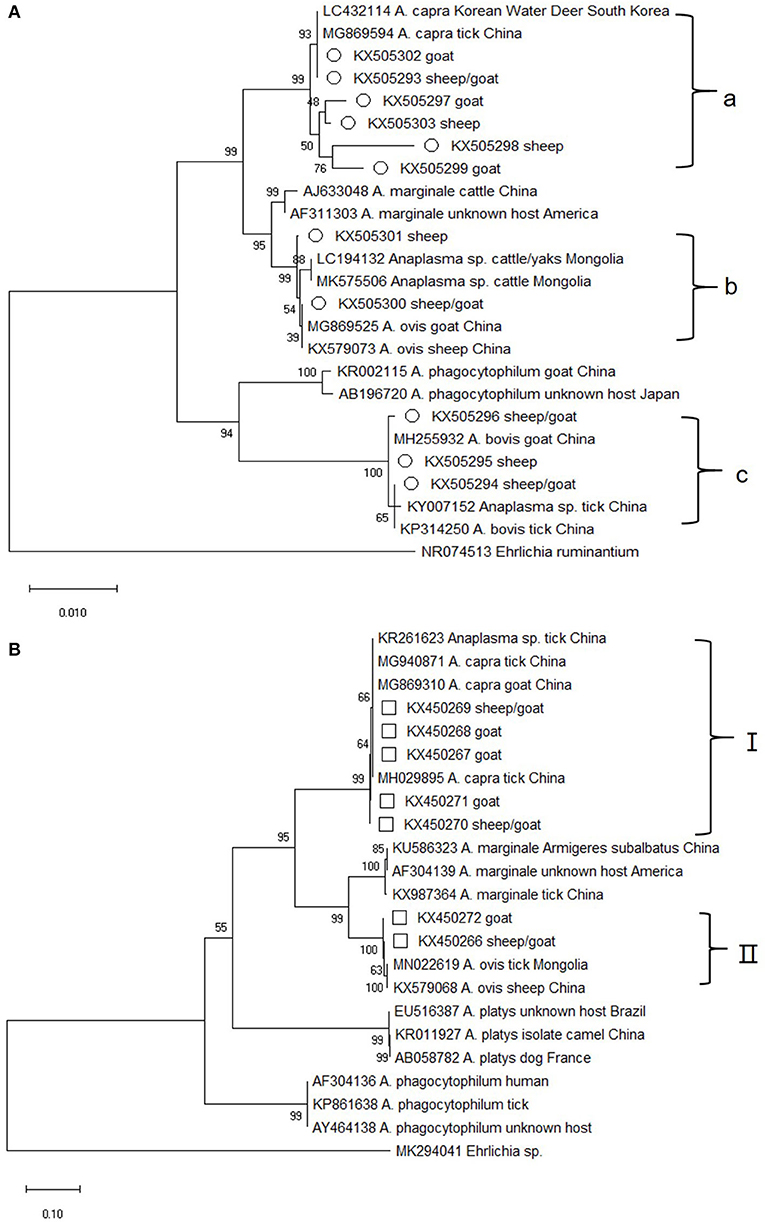
Figure 1. Phylogenetic tree of Anaplasma based on 16S rRNA and gltA gene. An alignment of 1,133 bp 16S rRNA gene sequences (A) and 759 bp gltA gene sequences (B) was used to construct the neighbor-joining phylogenetic tree using Kimura two-parameter model in Mega X. Ehrlichia ruminantium or Ehrlichia sp. was used as the outgroup. The sequences marked with hollow circle and hollow box were obtained in the present study.
Altogether, 118 of the 212 blood samples from sheep and goats were positive for the gltA gene. Sequence analysis showed that seven sequence types, which fell into two distinct groups, were obtained. The group I sequences, which represented nearly all of the gltA sequences (96%, 113/118) in this study, were highly homologous (99.6–100%) to A. capra isolates from China (MG940871, MG869310, and MH029895) and appeared on a separate evolutionary branch with them (Figure 1B). It is worth mentioning that the sequences from this group corresponded to those in group a of 16S rRNA gene (the two gene sequences could be amplified at the same time in 9 blood samples) (Supplementary Table 3). The group I specimens were collected in 2011–2015 (Supplementary Table 4), which is same as that for 16S rRNA. The remaining gltA sequences along with two A. ovis sequences from China formed group II, which was similar with the 16S rRNA gene sequences in group b. The sequences in group II shared a similarity of 99.3–99.9%.
Phylogenetic Analysis of the groEL Gene
Altogether, 162 groEL gene sequences were amplified from the Anaplasma–positive DNA samples. Although the sequence alignment analysis showed that nine different sequence types contained 1–7 nucleotides difference, the sequences were almost identical at amino acid level (data not shown). Furthermore, the online sequence alignment showed that the obtained sequences were distinct with those of other known Anaplasma species and strains, with the highest similarity (90.0%) with an Anaplasma sp. isolate from deer in Japan (JN055360). Phylogenetic analysis based on the groEL gene sequences also revealed that the isolates from this study were evolutionarily distant from other Anaplasma species and strains and formed an independent branch on the evolutionary tree, which was supported by high bootstrap values (100%) (Figure 2).
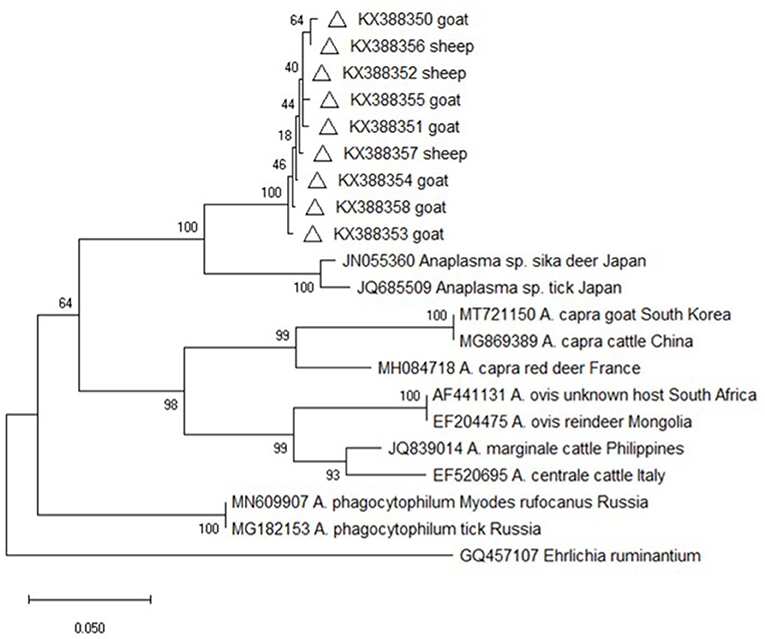
Figure 2. Phylogenetic tree of Anaplasma based on groEL gene. An alignment of 341 bp groEL gene sequences was used to construct the neighbor-joining phylogenetic tree using Kimura two-parameter model in Mega X. Ehrlichia ruminantium was used as the outgroup. The sequences marked with hollow triangle were obtained in the present study.
Phylogenetic Analysis of the msp4 Gene
Altogether, 54 msp4 gene sequences were amplified from the DNA samples, and the sequence alignment analysis conducted on them showed that 13 sequence types were present. The pairwise sequence alignments revealed that the sequences fell into two groups (group i and group ii) with a similarity score of 98.4–99.7% within each group, and a score of 71.8–76.0% between the groups. The online sequence alignment results showed that the sequences obtained in this study differed greatly from the known sequences, and the highest similarity score (75.7% for group i and 75.0% for group ii) was seen with two A. phagocytopuilum isolates (CP015376 and KM205444). Phylogenetic analysis based on the msp4 gene sequences also revealed that the isolates from the present study were distinct from other well-defined Anaplasma species and fell into two separate clades on the evolutionary tree (Figure 3).
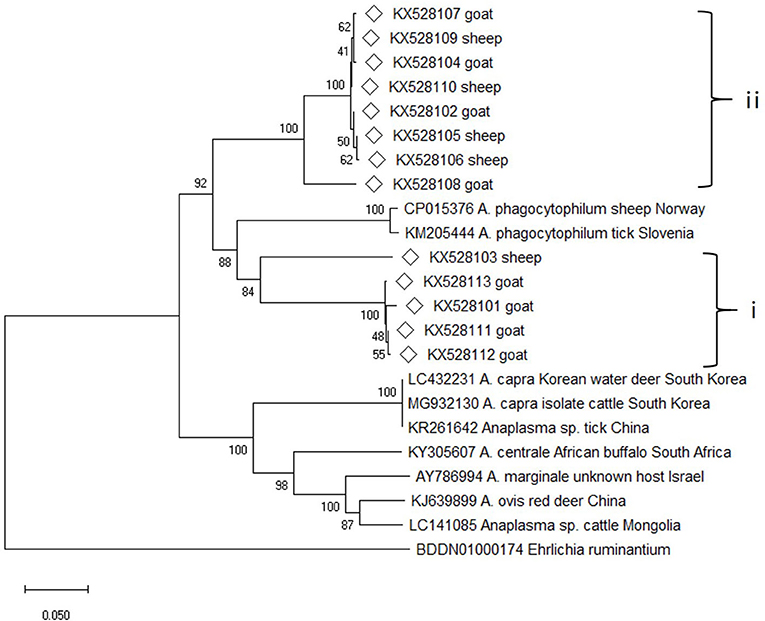
Figure 3. Phylogenetic tree of Anaplasma based on msp4 gene. An alignment of 350 bp msp4 gene sequences was used to construct the neighbor-joining phylogenetic tree using Kimura two-parameter model in Mega X. Ehrlichia ruminantium was used as the outgroup. The sequences marked with hollow diamond were obtained in the present study.
Discussion
In 2015, Li et al. identified a novel tick-borne Anaplasma species they called “A. capra” (5). Furthermore, the unknown bacterium was able to be propagated in HL-60 cells, which mainly allow for the growth of human isolates. These results verified their former findings that the agent was a new species of Anaplasma genus with zoonotic potential, just like A. phagocytophilum. They also mentioned that 37 blood samples from asymptomatic goats were positive for an unknown Anapalsma species during 2012–2013. Following its first report as a human pathogen in 2015, A. capra has been detected in several tick species, including Ixodes persulcatus, Haemaphysalis longicornis, H. qinghaiensis, and Rhipicephalus microplus (5, 10, 16, 17), in sheep and goats (9), in cattle (18), in dogs (19), in Korean water deer (20). These findings together with the results obtained herein suggest that additional domestic and wild animals and/or tick species might be the hosts and tick vectors of A. capra and are involved in its transmission and maintenance, a possibility the needs further investigation.
In the present study, 16S rRNA and gltA gene fragments were also PCR-amplified from the samples from sheep and goats collected in 2012–2015, and even from the blood samples collected in 2011, which is earlier than the first literature to report the origin of this bacterium. Therefore, we can speculate that the novel species may have existed in animals/ticks for a long time and had either not been detected or was mistaken for other related species. Our results showed that the gltA sequences in group I were 100% identical to another Anaplasma sp. isolate (KJ700628) and share 99% sequence identity with an A. centrale isolate (AF304141). In 2018, Khumalo et al. reported a similar finding and confirmed that the A. centrale 16S rRNA gene sequence (Aomori strain, AF283007) from cattle in Japan that was acquired in 2001 was actually A. capra (21).
As shown in Supplementary Table 3, the 16S rRNA sequences in group a and the gltA sequences in group I could correspond to each other. The two genes from nine of the samples were both closely related to the human A. capra pathogen and group in a separate clade with other A. capra isolates, but were distinct from other Anaplasma species, indicating the novelty of this pathogen (Figure 1). In addition, sequences in 16S rRNA group b and gltA group II were both closely related to A. ovis, although these two sets of sequences could not correspond to a same sample. This result along with the fact that KX505294–KX505296 obtained in this study were grouped with A. bovis isolates from China suggested that there may be new A. bovis and A. ovis isolates in the samples collected.
Nevertheless, the groEL and msp4 gene sequences from this study were distinct from all other known Anaplasma species and strains with each having low sequence similarity values (90% for groEL and 74.6% for msp4), indicating the possibility of novel Anaplasma genotypes or species in sheep and goats in China and suggesting a high degree of genetic diversity and host tropisms in Anapalsma, as has already been documented for A. phagocytophilum (22). Because it's unclear whether these new genotypes or species vary in their pathogenicity profiles toward non-human and human animals, these profiles warrant further investigation.
In summary, novel Anaplasma genotypes or species closely related to human A. capra pathogen were identified with divergent groEL and msp4 genes in sheep and goats in central China. Further studies should be conducted to fully elucidate the host range, vector ticks, pathogenicity characteristics and geographic distribution of this bacterium.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Research Ethics Committee of Henan University of Animal Husbandry and Economy. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
CN conceived the study. CN and YZ designed the experiments. YZ and YC performed the experiments. YZ, YS, and HJ performed data analysis. HJ and CN wrote the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by the Earmarked Fund for China Modern Agro-industry Technology Research System (nycytx-38) and Key Research Projects of Institutions of higher learning in Henan (20A230004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sandra Cheesman, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.580007/full#supplementary-material
References
1. Chen MS, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. (1994) 32:589–95. doi: 10.1128/JCM.32.3.589-595.1994
2. Zhang LJ, Liu Y, Ni D, Li Q, Yu Y, Yu XJ, et al. Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA. (2008) 300:2263–70. doi: 10.1001/jama.2008.626
3. Fang LQ, Liu K, Li XL, Liang S, Yang Y, Yao HW, et al. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis. (2015) 15:1467–79. doi: 10.1016/S1473-3099(15)00177-2
4. Chochlakis D, Ioannou I, Tselentis Y, Psaroulaki A. Human anaplasmosis and Anaplasma ovis variant. Emerg Infect Dis. (2010) 16:1031–2. doi: 10.3201/eid1606.090175
5. Li H, Zheng YC, Ma L, Jia N, Jiang BG, Jiang RR, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. (2015) 15:663–70. doi: 10.1016/S1473-3099(15)70051-4
6. Yang J, Liu Z, Niu Q, Liu J, Han R, Guan G, et al. A novel zoonotic Anaplasma species is prevalent in small ruminants: potential public health implications. Parasit Vectors. (2017) 10:264. doi: 10.1186/s13071-017-2182-9
7. Seo MG, Ouh IO, Lee H, Geraldino PJL, Rhee MH, Kwon OD, et al. Differential identification of Anaplasma in cattle and potential of cattle to serve as reservoirs of Anaplasma capra, an emerging tick-borne zoonotic pathogen. Vet Microbiol. (2018) 226:15–22. doi: 10.1016/j.vetmic.2018.10.008
8. Yang J, Liu Z, Niu Q, Mukhtar MU, Guan G, Liu G, et al. A novel genotype of “Anaplasma capra” in wildlife and its phylogenetic relationship with the human genotypes. Emerg Microbes Infect. (2018) 7:210. doi: 10.1038/s41426-018-0212-0
9. Peng Y, Wang K, Zhao S, Yan Y, Wang H, Jing J, et al. Detection and phylogenetic characterization of Anaplasma capra: an emerging pathogen in sheep and goats in China. Front Cell Infect Microbiol. (2018) 8:283. doi: 10.3389/fcimb.2018.00283
10. Guo WP, Zhang B, Wang YH, Xu G, Wang X, Ni X, et al. Molecular identification and characterization of Anaplasma capra and Anaplasma platys-like in Rhipicephalus microplus in Ankang, Northwest China. BMC Infect Dis. (2019) 19:434. doi: 10.1186/s12879-019-4075-3
11. Zhang Y, Lv Y, Zhang F, Zhang W, Wang J, Cui Y, et al. Molecular and phylogenetic analysis of Anaplasma spp. in sheep and goats from six provinces of China. J Vet Sci. (2016) 17:523–9. doi: 10.4142/jvs.2016.17.4.523
12. Barlough JE, Madigan JE, DeRock E, Bigornia L. Nested polymerase chain reaction for detection of Ehrlichia equi 4 genomic DNA in horses and ticks (Ixodes pacificus). Vet Parasitol. (1996) 63:319–29. doi: 10.1016/0304-4017(95)00904-3
13. Wen B, Jian R, Zhang Y, Chen R. Simultaneous detection of Anaplasma marginale and a new Ehrlichia species closely related to Ehrlichia chaffeensis by sequence analyses of 16S ribosomal DNA in Boophilus microplus ticks from Tibet. J Clin Microbiol. (2002) 40:3286–90. doi: 10.1128/JCM.40.9.3286-3290.2002
14. Li H, Jiang JF, Liu W, Zheng YC, Huo QB, Tang K, et al. Human infection with Candidatus Neoehrlichia mikurensis, China. Emerg Infect Dis. (2012) 18:1636–9. doi: 10.3201/eid1810.120594
15. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. (1987) 4:406–25.
16. Sun XF, Zhao L, Wen HL, Luo LM, Yu XJ. Anaplasma species in China. Lancet Infect Dis. (2015) 15:1263–4. doi: 10.1016/S1473-3099(15)00377-1
17. Yang J, Liu Z, Niu Q, Liu J, Han R, Liu G, et al. Molecular survey and characterization of a novel Anaplasma species closely related to Anaplasma capra in ticks, northwestern China. Parasit Vectors. (2016) 9:603. doi: 10.1186/s13071-016-1886-6
18. Xian KF, Chandrawathani P, Thomas SF, Tee TS. Molecular investigation of Anaplasma spp. in domestic and wildlife animals in Peninsular Malaysia. Inter J Syst Evol Microbiol. (2018) 68:2682–91. doi: 10.1016/j.vprsr.2018.05.006
19. Shi K, Li JQ, Yan YQ, Chen Q, Wang KL, Zhou YC, et al. Dogs as new hosts for the emerging zoonotic pathogen Anaplasma capra in China. Front Cell Infect Microbiol. (2019) 9:394. doi: 10.3389/fcimb.2019.00394
20. Amer S, Kim S, Yun Y, Na KJ. Novel variants of the newly emerged Anaplasma capra from Korean water deer (Hydropotes inermis argyropus) in South Korea. Parasit Vectors. (2019) 12:365. doi: 10.1186/s13071-019-3622-5
21. Khumalo ZTH, Brayton KA, Collins NE, Chaisi ME, Oosthuizen MC. Evidence confirming the phylogenetic position of Anaplasma centrale (ex Theiler 1911) Ristic and Kreier 1984. Inter J Syst Evol Microbiol. (2018) 68:2682–91. doi: 10.1099/ijsem.0.002832
Keywords: phylogenetic analysis, 16S rRNA, variants, Anaplasma capra, gltA
Citation: Zhang Y, Cui Y, Sun Y, Jing H and Ning C (2020) Novel Anaplasma Variants in Small Ruminants From Central China. Front. Vet. Sci. 7:580007. doi: 10.3389/fvets.2020.580007
Received: 04 July 2020; Accepted: 06 November 2020;
Published: 26 November 2020.
Edited by:
Camilla Luzzago, University of Milan, ItalyReviewed by:
Marcos Rogério André, São Paulo State University, BrazilKevin Bown, University of Salford, United Kingdom
Copyright © 2020 Zhang, Cui, Sun, Jing and Ning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changshen Ning, bm5sMTk4NkAxNjMuY29t
 Yan Zhang
Yan Zhang Yanyan Cui
Yanyan Cui Yanting Sun1
Yanting Sun1 Changshen Ning
Changshen Ning