- 1National Research Center for Wildlife-Borne Diseases, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 2College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3Department of Microbiology, Beijing General Station of Animal Husbandry, Beijing, China
Pigeon paramyxovirus type I (PPMV-1) causes regular outbreaks in pigeons and even poses a pandemic threat among chickens and other birds. The birds infected with PPMV-1 mainly show a pathological damage in the respiratory system, digestive system, and nervous system. However, there were few reports on the efficiency of the virus entering the host via routes of different systems. In the present study, a PPMV-1 strain was obtained from a dead wild pigeon in 2016 in Beijing, China. The mean death time (MDT) and the intracerebral pathogenicity (ICPI) of our isolate showed medium virulence. Phylogenetic analysis based on F gene sequence showed that the isolate belonged to subgenotype VIb, class II, which dominated in China in recent years. Then, we evaluated the infection efficiency of different routes. Pigeons were randomly divided into five groups of six as follows: intracephalic (IC), intranasal (IN), and intraoral (IO) infection routes, cohabitation infection (CO), and negative control (N negative). All pigeons were inoculated with 100 μl·106 EID50 PPMV-1 virus. After infection, pathological lesions, virus shedding, body weight change, survival rate, and tissue tropism were tested to compare the efficiency of the different infected routes. The mortality of groups IC, IN, IO, and CO were 100, 66.7, 50, and 33.3%, respectively. Weight loss in group IC was higher than the other groups, followed by groups IN and IO. The lesions observed in PPMV-1-infected pigeons were severe, especially in the lung and intestine in group IC. Viral shedding was observed from 2 dpi in groups IC and IN, but the shedding rate was higher in group IN than group IC. The longest period was in group CO. Tissue tropism experiment showed that our isolate has a wide range of tissue distribution, and the virus titer in the heart and intestine of group IC and in the brain of group IN was higher. Our data may help us to evaluate the risk of transmission of PPMV-1.
Introduction
Newcastle disease, caused by Newcastle disease virus (NDV), is classified as a notifiable disease by the World Organization for Animal Health (OIE), because of its high morbidity and mortality in avian species. At least 250 avian species, including chickens, ducks, and pigeons were reported to be susceptible to NDV (1–3). NDV is a single-stranded, negative-sense, and non-segment RNA virus, which belongs to the genus Orthoavulavirus, subfamily Avulavirinae, family Paramyxoviridae of the Mononegavirales order (4–7). NDV isolates have been classified into classes I and II. NDV of class I are mostly of low virulence and contain only one genotype (8), and class II virus consists of 18 genotypes (9–13) based on the phylogenetic analysis of nucleotide sequence of the F gene and genomic size (14, 15). Pigeon paramyxovirus type 1 (PPMV-1) found in birds of Columbidae family, mainly turtle dove (Streptopelia turtur) and Eurasian collared dove (Stretopelia decaocto) is a variant of Newcastle disease virus, and it is almost genotype VI, class II. PPMV-1 first reported in England during the late 1970s, affected racing pigeons with outbreaks in domestic chickens (16), which caused the third worldwide pandemics of NDV (17). In recent years, a high mortality (from 40 to 80%, even to 100% in some cases) caused by PPMV-1 has been observed in pigeons (18–22). Clinical signs of the infected birds involve nervous, respiratory, and digestive system symptoms (23), consisting of moderate to severe depression with neck twisting, ataxia, crouch, paralysis, eyelid edema, diarrhea, and green loose feces. NDV has been documented to remain infectious in feces and carcasses for at least a couple of weeks, several months in feathers, and up to 90 days in soil or water (24). Quite a few times, outbreaks in poultry have been ascribed to PPMV-1 (25–28). The virulence can be enhanced after serial passages in chickens (17, 21, 29, 30), which makes these pigeon-originated viruses a tremendous and continuous threat to the poultry industry (31–33). For these reasons, the potential transmission of the virus and the effective route where the virus will enter the host have been considered points of concern. In the present study, the pathogenicity of a PPMV-1, obtained from a dead wild pigeon in 2016 in Beijing, China, was investigated via different inoculation routes. Findings from our study showed intracephalic, intranasal, and intraoral infection routes were effective, but intracephalic was the most.
Materials and Methods
Viral Isolation, Amplification, and Full-Length Genome Sequencing
A moribund pigeon with neck twisting, diarrhea, and leg paresis or paralysis was found in Beijing, 5 September 2016. We initially diagnosed that the pigeon was infected with PPMV or avian influenza virus based on clinical symptoms, then, the avian influenza virus was excluded and PPMV-1 infection was confirmed by RT-PCR. The identification of Newcastle disease and the separation of strains are as follows: viral RNA was extracted from the tissues of the pigeon (i.e., heart, liver, spleen, lung, kidney, stomach, brain, trachea, intestine, and pancreas) using Trizol reagent (ambition by Life Technologies, Beijing, China) according to the manufacturer's instructions. Reverse transcription was performed as described (34). The detection gene (a part of fusion gene, 486 bp) was amplified from the cDNA by PCR utilizing Taq DNA Polymerase (CWBIO 2× Taq MasterMix, Cat. CW0682M), and the primers were designed according to the conserved sequence (34) (Primer sequence: F:CAGCTGCGGCCCTAATACA; R:TGGATGCCCAAGAGTTGAG). The program was as follows: 95°C for 5 min; 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 25 s; and a final extension at 72°C for 10 min. The PCR products were visualized by 1% agarose gel electrophoresis. Other pathogens (circovirus, avian influenza, and pathogenic bacteria) were negative. The viruses from different organs were plaque-purified three times on primary chicken embryo fibroblasts and inoculated into the allantoic cavity of 9-day-old specific-pathogen-free (SPF) eggs. The virus was isolated and RNA was extracted from the allantoic cavity, detected using PCR as abovementioned. The strain was designated as PPMV-1/pigeon/Beijing/China/01/2016 and abbreviated as PPMV-1/BJ-01/CH. Complete genome of our strain was amplified (primers are shown in Supplementary Table 1), sequenced, and submitted to GenBank (GenBank ID: MH807446).
Virulence-Test
There are three virulence evaluation indexes on Newcastle disease virus, and virulence is usually determined by no <2 indexes. The mean death time (MDT) was determined in 9-day-old SPF chicken embryo eggs, and the intracerebral pathogenicity index (ICPI) was determined in 1-day-old chick as previously described (35). The least-fatal dose and egg-infectious dose of the virus were tested in 9-day-old SPF chicken embryo eggs by multiple proportion dilution (from 10−3 to 10−9) with five repeats per dilution and calculated using the Reed and Muench method.
Phylogenetic Analysis Based on Complete F Gene
To determine the genetic relationships of the PPMV-1/BJ-01/CH isolate to others, a phylogenetic tree was constructed using the MEGA 7.0 software with the maximum likelihood method via the Kimura two-parameter model based on the complete F gene from 21 different subgenotypes of the reference PPMV-1 isolates and 1 APMV-1 as outgroup (8, 12, 29). All complete sequences of the F gene were downloaded from GenBank and aligned utilizing ClustalW. Nucleotide sequences obtained in the present study were aligned with reference sequences available in the GenBank database to determine the subgenotypes.
Animal Experimental Infection
To further determine the pathogenicity of the virus, a total of 31-month-old pigeons, with approximately equal body weight of −5–5%, were used in our study. These pigeons were bought from a hatchery in Miyun District, Beijing, and certified by hemagglutination inhibition (HI) experiment to have no antibodies of NDV and AIV. These pigeons were randomly divided into five groups, and a marked group was placed in a separate cage in an animal room under biosafety conditions. Adequate food and drinking water were provided. Pigeons were inoculated with the virus by intranasal, intraoral, intracephalic, and cohabitation infections (signed as groups IN, IO, IC, and CO, placing groups CO and IC together), which represented through respiratory system, digestive system, nervous system, and natural infection, respectively. Additionally, the negative control group received phosphate-buffered saline (PBS) solution at pH 7.2. All infected groups were inoculated with a dose of 106 median embryo lethal dose (ELD50)/100 μl each, calculated using the Reed and Muench method. Subsequently, all pigeons were observed daily for clinical signs, and clinical symptoms, mortality, and morbidity were recorded. Oropharyngeal and cloacal swabs were taken every day, and body weight was determined every other day until day post-infection (dpi) 14 (0 day post-infection in group CO means the day the pigeons were infected), when there is only one group left to shed virus.
Virus Shedding
All of the oropharyngeal and cloacal swabs were collected, placed in tubes with phosphate-buffered saline solution and 2% fetal bovine serum and stored at −80°C until RNA extraction. RNA was extracted, and reverse transcription and RT-PCR were performed as above to test the virus shedding. In addition, cDNA of the isolate in this study and reagent-grade water were used as positive and negative control, respectively.
Gross Lesion
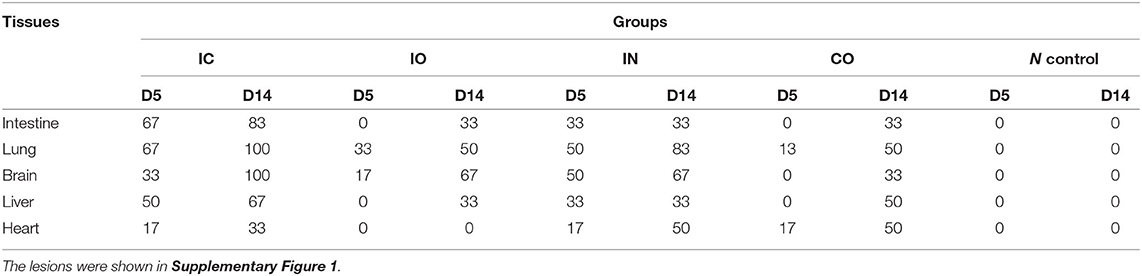
Lesion formation and lesion size are often used to quantify virulence (36). In our study, organs of all dead pigeons were collected on dpi 5 and 14. The collected tissues (including the heart, brain, lung, intestine, and liver) were pathologically lesion examined. Individuals that died ahead of time point were examined and recorded in advance. Positive rate was tallied by percentage.
Histopathology
Tissues from each group were collected and fixed with 10% neutral formalin. The sections were stained with hematoxylin and eosin (HE), and all HE stained sections were examined for the presence of microscopic lesions.
Tissue Distribution
To understand the distribution and virus load of PPMV-1 in organs from different groups at dpi 5 when clinical symptoms emerged, we randomly choose three from each group, and collected samples from the heart, brain, lung, intestine, and liver. The virus was isolated from tissues of the same weight, and TCID50 in DF-1 cells (chicken fibroblast cell line) was used to estimate viral loads of the five groups; 3 × 104 DF-1 cells were seeded in 96-well plate with five repetitions 1 day before infection. Twenty-four hours later, the cells were infected with different dilutions of the virus for 1 h at 37°C with shaking every 12 h and confirmed by the hemagglutination assay. TCID50 was calculated using the Reed-Muench method. Data were analyzed using Prism (v.5.01) software. Statistical significance was set at a P-value of <0.05.
Ethics Statement
These animal studies were performed in strict accordance with the Guidelines for the Care and Use of Animals in Research, which are issued by the Institute of Zoology, Chinese Academy of Sciences (Approval Number IOZ12017).
Results
Phylogenetic Analysis and Genetic Characteristics of PPMV-1/BJ-01/CH Isolate
Phylogenetic analysis showed our isolate belonged to subgenotype VIb (Figure 1). Our isolate possessed an 112R-R-Q-K-RF117 segment at the cleavage site of F gene. This is characteristic of virulent NDVs. This motif is commonly found in NDV strains that are highly or moderately virulent in chickens, especially in genotypes VII and IX viruses and some pigeon paramyxovirus strains. Amino acid residues important for receptor recognition of HN is 174R 175I 198D 236K 258E 299Y 317Y 401E 416R 498R 516R 526Y 547E (37), which is the same as most PPMV-1 strains.
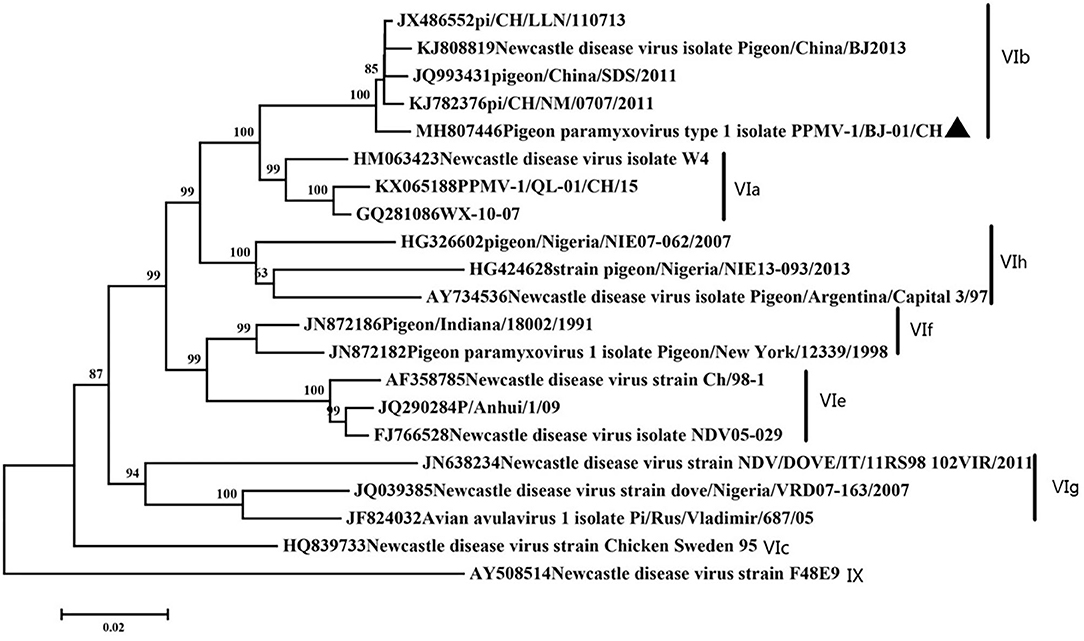
Figure 1. Classification of the virus based on genotype criteria. A maximum likelihood tree of the complete F gene nucleotide sequence. The evolutionary analyses were conducted using the Kimura two-parameter method with 1,000 bootstrap replicates in MEGA7.0. Our isolate was marked with a black triangle.
Virulence of the PPMV-1/BJ-01/CH Isolate and Sequence Analysis
The MDT and ICPI were 72 h and 0.76, respectively. The results indicated that the PPMV-1 strain was medium according to the criteria (38). Moreover, the minimum lethal dose and 50% egg infection dose of the virus in chick embryo eggs were 10−3 and 10−5.37, respectively.
Pathogenicity of the PPMV-1/BJ-01/CH in Different Routes of the Infection
Clinical signs were observed in all pigeons from 5 dpi. The pigeons lost their weight sharply once clinical symptoms emerged, then soon died. The body weight change (Figure 2A) and survival curve (Figure 2B) showed the damage of the infected pigeons from group IC were much severer than other groups. A steady increase of body weight was found in negative control group. Post-mortem examination showed the congestion or hemorrhages on the meninx and in the brain. The tracheal mucosa was congested or hemorrhagic. Extensive hemorrhages were observed in the mucosa of the small intestine. The spleens were atrophic, friable, and hemorrhagic. Congestion and hemorrhage were found in the pancreas. Our data showed the most serious tissue lesion was discovered in group IC. Groups IO and CO caused less lesions on dpi 5 (Table 1).
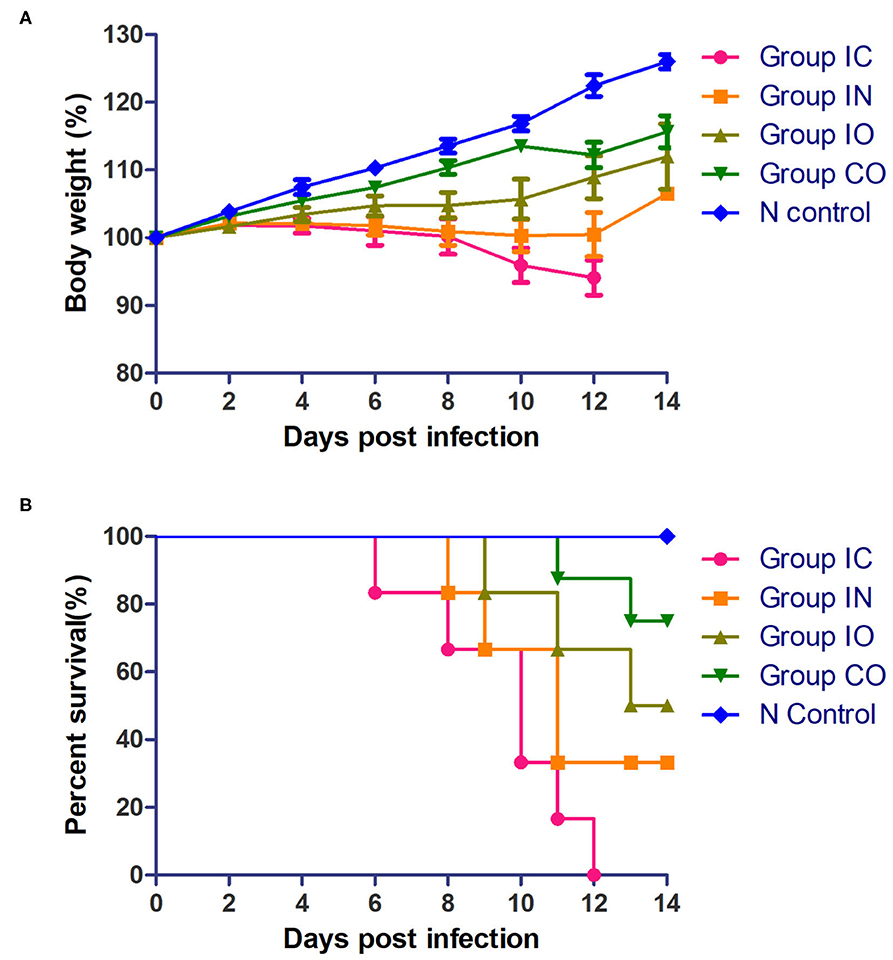
Figure 2. Pathogenicity of the PPMV-1/BJ-01/CH in different routes of infection. (A) Body weight change of different groups post-infection. (B) Survival curve of different groups post-infection.
Histopathological examination showed that the most severe lesions are found in group IC, including the desquamation of small intestine villi, lymphocyte infiltration, and severe epithelial cell loss in the intestine. The alveoli have been destroyed, and a small amount of lymphocyte infiltration and congestion were observed in the lungs. Hepatomegaly and the disappearance of hepatic cord structures were observed in the liver. A small amount of lymphocyte infiltration was also observed in the cerebrum. Milder lesions were observed in other groups (Figure 3).
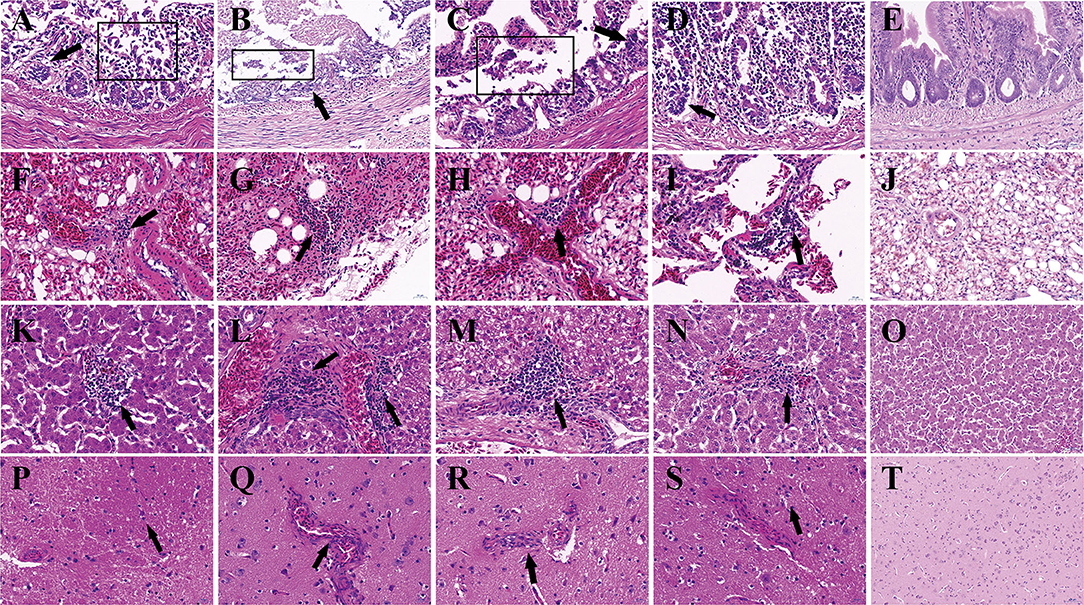
Figure 3. Histopathologic lesions in pigeons of groups. The groups and tissues were marked in the figure. Black arrow indicated the severe histopathologic. (A,F,K,P) Histopathologic lesions in group IC. (B,G,L,Q) Histopathologic lesions in Group IN. (C,H,M,R) Histopathologic lesions in group IO. (D,I,N,S) Histopathologic lesions in group CO. (E,J,O,T) Histopathologic lesions in group N. (A–E) Small intestine. (F–J) Lung. (K–O) Liver. (P–T) Cerebrum. The pictures were magnified ×40.
Virus Shedding
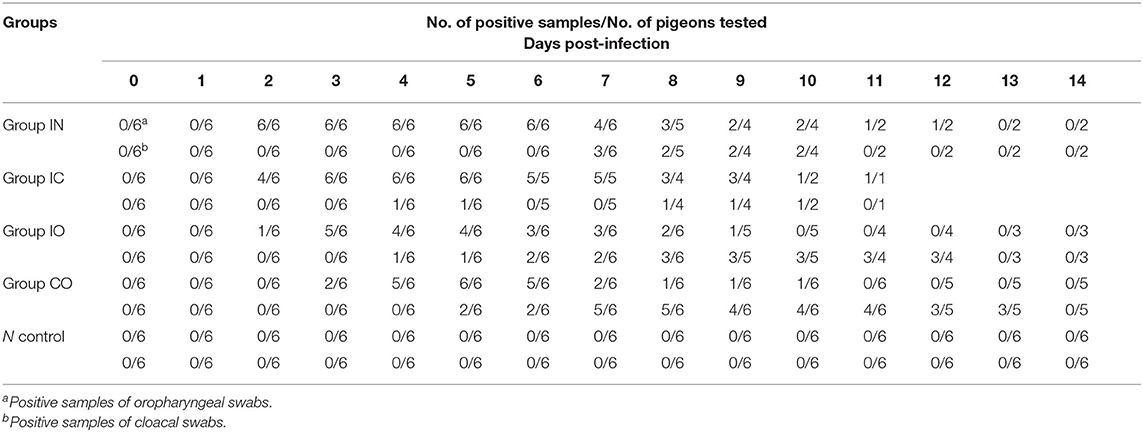
The results and mortality of each group are compiled in Table 2. Pigeons shed virus from dpi 2 to dpi 14. The virus was detected from oropharyngeal swabs earlier than cloacal swabs in all groups. The earliest start time observed was in groups IC and IN in dpi 2, but the shedding rate was higher in group IN than group IC. The highest shedding rates were observed in group IN from the throat at 2 dpi and cloaca at 7 dpi. Viral shedding ceased at 14 dpi in all groups (Table 2).
Tissue Tropism of the PPMV-1/BJ-01/CH in Different Routes of Infection
In this study, we found that the virus can effectively replicate in different tissues, no matter which route of infection, and virus titer in the brain, lung, and intestine were higher than in other tissues. The highest titer of the virus was found in the intestine in group IC and brain in group in (Figure 4).
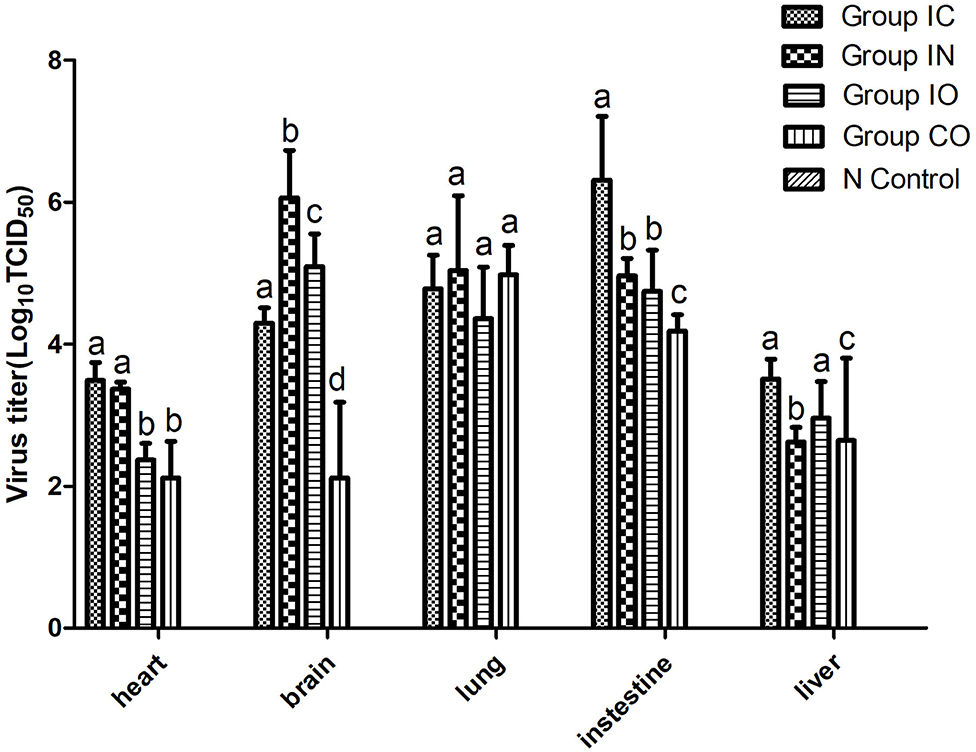
Figure 4. Viral loads in different organs of pigeons post-infection. Virus titer of different tissues from dead pigeons [log10 median tissue culture infectious dose/ml (TCID50)] were quantified by serial end-point dilution in 96-well plates using DF-1 cells. The titer was calculated by one-way analysis of variance with standard error bars. Different lowercase letters over the bars denote statistically significant differences (P < 0.05) among different tissues after infection calculated by t-test. Non-significance (P > 0.05) was marked up by the same lowercase letters on different bars. Different lowercase letters marked up on the bars have no relationship among these five organs and represent one organ.
Discussion
In the present study, a PPMV-1 was isolated from a dead pigeon. In order to better understand the genetic characteristics of the strain, the full-length genome was amplified and sequenced. Phylogenetic analysis showed that the isolate belonged to VIb, class II, which was consistent with most of the genotypes found in China (20, 21, 39–41). Amino acid sequence of F gene 112–117 is a typical virulent motif. But the MDT and ICPI showed medium virulence of our isolate. This contradiction often occurs on PPMV-1. Amino acid residues important for receptor recognition of HN are the same as most PPMV-1. All the characteristics of our strain consisted with current epidemic isolates in China (20, 42, 43).
The pathogenicity of the virus may relate to different inoculation routes. For example, the ducks infected intramuscularly with a virulent NDV strain showed the most severe clinical signs, while ducks infected intranasally and intraocularly sometimes also exhibited clinical signs but seldom died (44). To further understand the target system of the virus obtained in this study, several experiments were conducted to evaluate the pathogenicity via different system routes. Cohabitation infection was intended to mimic a natural-acquired infection. Intranasal infection was often used by labs as a substitute for aerosol infection of the virus. Oral infection mimic was used to simulate contaminated food and water. Inoculate intracranially was regularly used to evaluate NDV virulence, so it may be the most effective route of infection.
Viral shedding evaluates the effective replication and transmission of the virus in different groups. The results of our study showed infected pigeons of different groups except group CO which shed virus through the larynx from 2 dpi and through cloaca from 4 dpi. Pigeons shed virus through cloaca from 2 dpi after isolating NDV in previous studies (45, 46). The earliest start time observed was in groups IC and IN, and the shedding rate was higher in group IN than group IC; the highest shedding rates were observed in group IN from the throat at 2 dpi and cloaca at 7 dpi. The result indicated that the respiratory tract is the fastest way for PPMV-1 to spread. The shedding rate in group IC remained high, which may confirm that virus can efficiently enter into the hosts through the nervous system. All infection groups shed virus, and the longest persisted until 14 days. Continuous virus shedding may contribute to circulatory infection. Additionally, we used body weight and survival rate to test the pathogenicity. Death occurred in dpi 6, but previous study of a PPMV-1 was inoculated in pigeons through the intranasal route did not result in mortality up to 31 dpi. (47), it may be caused by the differences in virulence of the isolates used (48). The rate of body weight decline of group IC was higher than in the other groups, followed by groups IN and IO. According to the result of TCID50 in DF-1 cells, PPMV-1 was able to cause systemic infection in a relatively short time, and virus titer in groups IC and IN was higher than in other groups, which was broadly consistent with gross lesions at dpi 5, resulting in an effective infection process. Groups IO and CO caused less lesions which might be due to lower viral load in the organs at these days. By comparison of the viral load in different tissues, the virus had a wide range of tissue distribution, especially in the lung, brain, and intestine. It can be inferred that the virus could replicate well in these three organs during early infection. At 14 dpi, the gross lesions were more severe in these groups, but the rate of virus shedding was reduced which may be due to higher mortality rates. The results indicated that the virus can effectively infect pigeon via different routes, and the most pathogenic was infection through the nervous system and respiratory system, but infection through the nervous showed stronger pathogenicity. Previous studies described neurological lesions in NDV-infected birds, and virulent virus was capable of replication in the brain, but not the avirulent virus (49–51), thus successful replication in the nervous system determines its pathogenicity.
Overall, pigeons play an important role in the epidemiology of PPMV-1. The routes of inoculation greatly influenced pathogenicity of the pathogenic strain isolated in the present study. Considering the growing number of PPMV-1 cases in recent years, it is necessary to develop effective vaccines or other prevention and control methods.
Data Availability Statement
Complete genome of our strain was amplified (primers were showed in Supplementary Table 1), sequenced and submitted to GenBank (GenBank ID: MH807446).
Ethics Statement
The animal study was reviewed and approved by Institute of Zoology, Chinese Academy of Sciences (Approval Number IOZ12017).
Author Contributions
HH and HC designed the experiments. HC and SF conducted the experiments and analyzed the data. YW and FL provided the animals. BW performed the RNA extraction and PCR. HC wrote the manuscript. SF, QS, and JD revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by Beijing Innovation Consortium of Agriculture Research System (BAIC04-2020), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA19050204) and Chinese Academy of Sciences (CZBZX-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.569901/full#supplementary-material
References
1. Alexander DJ. Newcastle disease and other avian paramyxoviruses. Rev Sci Tech Oie. (2000) 19:443–62. doi: 10.20506/rst.19.2.1231
2. Kaleta EF, Baldauf C. Newcastle disease in free-living and pet birds. Dev Vet Virol. (1988) 8:197–246. doi: 10.1007/978-1-4613-1759-3_12
3. Wan HQ, Chen LG, Wu LL, Liu XF. Newcastle disease in geese: natural occurrence and experimental infection. Avian Pathol. (2004) 33:216–21. doi: 10.1080/0307945042000195803
4. Amarasinghe GK, Ceballos, NG, Kuhn JH. Taxonomy of the order mononegavirales: update 2018. Arch Virol. (2018) 163:2283–94. doi: 10.1007/s00705-018-3814-x
5. Maes P, Gaya KA, Jens HK. Taxonomy of the order mononegavirales: second update 2018. Arch Virol. (2019) 164:1233–44. doi: 10.1007/s00705-018-04126-4
6. Dimitrov KM, Abolnik C, Afonso CL, Albina E. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect Genet Evol. (2019) 74:103917. doi: 10.1016/j.meegid.2019.103917
7. Kuhn JH, Wolf YI, Krupovic M, Zhang YZ, Maes P, Dolja VV, et al. Classify viruses—the gain is worth the pain. Nature. (2019) 566:318–20. doi: 10.1038/d41586-019-00599-8
8. Diel DG, Silva LH, Liu HL, Miller PJ, Afonso CL. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol. (2012) 12:1770–9. doi: 10.1016/j.meegid.2012.07.012
9. Diel DG, Susta L, Stivalis CGA, Killian ML, Brown CC, Miller PJ, et al. Complete genome and clinicopathological characterization of a virulent Newcastle disease virus isolate from South America. J Clin Microbiol. (2012) 50:378–87. doi: 10.1128/JCM.06018-11
10. Kim LM, King DJ, Curry PE, Suarez DL, Swayne DE, Stallknecht DE, et al. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J Virol. (2007) 81:12641–53. doi: 10.1128/JVI.00843-07
11. Liu XF, Wan HQ, Ni XX, Wu YT, Liu WB. Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985–2001. Arch Virol. (2003) 148:1387–403. doi: 10.1007/s00705-003-0014-z
12. Snoeck CJ, Owoade AA, Couacy-Hymann E, Alkali BR, Okwen MP, Adeyanju AT, et al. High genetic diversity of Newcastle disease virus in Poultry in West and Central Africa: cocirculation of genotype XIV and newly defined genotypes XVII and XVIII. J Clin Microbiol. (2013) 51:2250–60. doi: 10.1128/JCM.00684-13
13. Dimitrov KM, Ramey MA, Qiu XT, Bahl J, Afonso CL. Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infect Genet Evol. (2016) 39:22–34. doi: 10.1016/j.meegid.2016.01.008
14. Czegledi A, Ujvari D, Somogyi E, Wehmann E, Werner O, Lomniczi B. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. (2006) 120:36–48. doi: 10.1016/j.virusres.2005.11.009
15. Dortmans JCFM, Rottier PJM, Koch G, Peeters BPH. The viral replication complex is associated with the virulence of Newcastle disease virus. J Virol. (2010) 84:10113–20. doi: 10.1128/JVI.00097-10
16. Meulemans G, Gonze M, Carlier MC, Petit P, Burny A, Long L. Antigenic and biological characterization of avian paramyxovirus type-1 isolates from pigeons. Arch Virol. (1986) 87:151–61. doi: 10.1007/BF01315295
17. Alexander DJ, Parsons G. Avian paramyxovirus type-1 infections of racing pigeons pathogenicity experiments in pigeons and chickens. Vet Rec. (1984) 114:466–9. doi: 10.1136/vr.114.19.466
18. Guo HB, Liu XL, Xu Y, Han ZX, Shao YH, Kong XG, et al. A comparative study of pigeons and chickens experimentally infected with PPMV-1 to determine antigenic relationships between PPMV-1 and NDV strains. Vet Microbiol. (2014) 168:88–97. doi: 10.1016/j.vetmic.2013.11.002
19. Perec-Matysiak A, Wesolowska M, Lesnianska K, Bunkowska-Gawlik K, Hildebrand J, Kicia M. Survey for zoonotic microsporidian pathogens in wild living urban rooks (Corvus frugilegus). J Eukaryot Microbiol. (2017) 64:721–4. doi: 10.1111/jeu.12402
20. Qiu XS, Meng CC, Zhan Y, Yu SQ, Li SC, Ren TT, et al. Phylogenetic, antigenic and biological characterization of pigeon paramyxovirus type 1 circulating in China. Virol J. (2017) 14:186. doi: 10.1186/s12985-017-0857-7
21. Wang JJ, Liu HL, Liu W, Zheng DX, Zhao YL, Li Y, et al. Genomic characterizations of six pigeon paramyxovirus type 1 viruses isolated from live bird markets in China during 2011 to 2013. PLoS ONE. (2015) 10:e0124261. doi: 10.1371/journal.pone.0124261
22. Zhao BH, Yuan-Hua FU, Fan JH, Bu XU, Zhu BU, Gao MY, et al. Development of bivalent inactivated oil-emulsion vaccine against Pigeon Paramyxovirus disease. Jiangsu J Agric Sci. (2010) 26:1293–7.
23. Vindevogel H, Marlier D. Viral infections in pigeons. Vet J. (2006) 172:40–51. doi: 10.1016/j.tvjl.2005.02.026
24. Leighton FA and Heckert RA. Newcastle disease and related avian paramyxoviruses. Infect Dis Wild Birds. (2008) 1–16. doi: 10.1002/9780470344668.ch1
25. Kim LM, King DJ, Guzman H, Tesh RB, Travassos da Rosa AP, Bueno R, Jr., et al. Biological and phylogenetic characterization of pigeon paramyxovirus serotype 1 circulating in wild North American pigeons and doves. J Clin Microbiol. (2008) 46:3303–10. doi: 10.1128/JCM.00644-08
26. Abolnik C, Gerdes GH, Kitching J, Swanepoel S, Romito M, Bisschop SP. Characterization of pigeon paramyxoviruses (Newcastle disease virus) isolated in South Africa from 2001 to 2006. Onderstepoort J Vet Res. (2008) 75:147–52. doi: 10.4102/ojvr.v75i2.13
27. Hassan W, Khair SAM, Mochotlhoane B, Abolnik C. Newcastle disease outbreaks in the Sudan from 2003 to 2006 were caused by viruses of genotype 5d. Virus Genes. (2010) 40:106–10. doi: 10.1007/s11262-009-0424-4
28. Pedersen JC, Senne DA, Woolcock PR, Kinde H, King DJ, Wise MG, et al. Phylogenetic relationships among virulent Newcastle disease virus isolates from the 2002–2003 outbreak in California and other recent outbreaks in North America. J Clin Microbiol. (2004) 42:2329–34. doi: 10.1128/JCM.42.5.2329-2334.2004
29. Dortmans JCFM, Rottier PJM, Koch G, Peeters BPH. Passaging of a Newcastle disease virus pigeon variant in chickens results in selection of viruses with mutations in the polymerase complex enhancing virus replication and virulence. J Gen Virol. (2011) 92:336–45. doi: 10.1099/vir.0.026344-0
30. Kommers GD, King DJ, Seal BS, Brown CC. Virulence of pigeon-origin 404 Newcastle disease virus isolates for domestic chickens. Avian Dis. (2001) 45:906–21. doi: 10.2307/1592870
31. Collins MS, Strong I, Alexander DJ. Evaluation of the molecular-basis of pathogenicity of the variant Newcastle-disease viruses termed “Pigeon Pmv-1 Viruses”. Arch Virol. (1994) 134:403–11. doi: 10.1007/BF01310577
32. Collins MS, Strong I, Alexander DJ. Pathogenicity and phylogenetic evaluation of the variant Newcastle disease viruses termed “pigeon PMV-1 viruses” based on the nucleotide sequence of the fusion protein gene. Arch Virol. (1996) 141:635–47. doi: 10.1007/BF01718322
33. Irvine RM, Aldous EW, Marvell RJ, Cox WJ, Ceeraz V, Fuller CM, et al. Outbreak of Newcastle disease due to pigeon paramyxovirus type I in grey partridges (Perdix perdix) in Scotland in October 2006. Vet Rec. (2009) 165:531–5. doi: 10.1136/vr.165.18.531
34. Qiu XS, Sun Q, Wu SA, Dong L, Hu SL, Meng CC, et al. Entire genome sequence analysis of genotype IX Newcastle disease viruses reveals their early-genotype phylogenetic position and recent-genotype genome size. Virol J. (2011) 8:117. doi: 10.1186/1743-422X-8-117
35. OIE (World Organisation for Animal Health). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 5th ed. Paris: OIE (2008). p. 957. Available online at: http://www.oie.int/eng/en_index.htm
36. Tscharke DC, Reading PC, Smith GL. Dermal infection with vaccinia virus reveals roles for virus proteins not seen using other inoculation routes. J Gen Virol. (2002) 83:1977–86. doi: 10.1099/0022-1317-83-8-1977
37. Munir M, Abbas M, Khan MT, Zohari S, Berg M. Genomic and biological characterization of a velogenic Newcastle disease virus isolated from a healthy backyard poultry flock in 2010. Virol J. (2012) 9:46. doi: 10.1186/1743-422X-9-46
38. Zhang S, Wang X, Zhao C, Liu D, Hu Y, Zhao J, et al. Phylogenetic and pathotypical analysis of two virulent Newcastle disease viruses isolated from domestic ducks in China. PLoS ONE. (2011) 6:e25000. doi: 10.1371/journal.pone.0025000
39. Hongbo G, Xiaoli L, Zongxi H, Yuhao S, Jinding C, Shasha Z, et al. Phylogenetic analysis and comparison of eight strains of pigeon paramyxovirus type 1 (PPMV-1) isolated in China between 2010 and 2012. Arch Virol. (2013) 158:1121–31. doi: 10.1007/s00705-012-1572-8
40. Chen L, Gorman JJ, McKimm-Breschkin J, Lawrence LJ, Tulloch PA, Smith BJ, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. (2001) 9:255–66. doi: 10.1016/S0969-2126(01)00581-0
41. Hualei L, Zhiliang W, Cuiping S, Yongkun W, Bin Y, Dongxia Z, et al. Characterization of pigeon-origin Newcastle disease virus isolated in China. Avian Dis. (2006) 50:636–40. doi: 10.1637/7618-042606R1.1
42. Wang X, Ren S, Wang X, Wang CY, Fan M, Jia Y, et al. Genomic characterization of a wild-bird-origin pigeon paramyxovirus type 1 (PPMV-1) first isolated in the northwest region of China. Arch Virol. (2017) 162:749–61. doi: 10.1007/s00705-016-3156-5
43. Ren S, Wang C, Zhang X, Zhao L, Wang X, Yao W, et al. Phylogenetic and pathogenic characterization of a pigeon paramyxovirus type 1 isolate reveals cross-species transmission and potential outbreak risks in the northwest region of China. Arch Virol. (2017) 162:2755–67. doi: 10.1007/s00705-017-3422-1
44. Dai YB, Cheng X, Liu M, Shen XY, Li JM, Yu SQ, et al. Experimental infection of duck origin virulent Newcastle disease virus strain in ducks. BMC Vet Res. (2014) 10:164. doi: 10.1186/1746-6148-10-164
45. King D. Susceptibility and protection of naive and vaccinated racing pigeons (Columba liva) against exotic Newcastle disease virus from the California 2002–2003 outbreak. Avian Dis. (2006) 50:336–41. doi: 10.1637/7479-112905R.1
46. Wakamatsu N, King DJ, Kapczynski DR, Seal BS, Brown CC. Experimental pathogenesis for chickens, turkeys, and pigeons of exotic Newcastle disease virus from an outbreak in California during 2002–2003. Vet Pathol. (2006) 43:925–33. doi: 10.1354/vp.43-6-925
47. Liu M, Cheng X, ShenXY, Zhou S, Wei YY, Liu JS, et al. Pathogenicity of duck origin virulent Newcastle disease virus strain in ducks. Chin J Vet Sci. (2014) 10:164.
48. Awu A, Shao MY, Liu MM, Hu YX, Qin ZM, Tian FL, et al. Characterization of two pigeon paramyxovirus type 1 isolates in China. Avian Pathol. (2015) 44:204–11. doi: 10.1080/03079457.2015.1025255
49. Ecco R, Susta L, Afonso CL, Miller PJ, Brown C. Neurological lesions in chickens experimentally infected with virulent Newcastle disease virus isolates. Avian Pathol. (2011) 40:145–52. doi: 10.1080/03079457.2010.544289
50. Wang W, Chang X, Yao W, Wei N, Huo N, Wang Y, et al. Host CARD11 inhibits Newcastle disease virus replication by suppressing viral polymerase activity in neurons. J Virol. (2019) 93:e01499–19. doi: 10.1128/JVI.01499-19
Keywords: pigeon, paramyxovirus, virus, Newcastle disease, pathogenicity, route
Citation: Chang H, Feng S, Wang Y, Li F, Su Q, Wang B, Du J and He H (2021) Isolation and Pathogenic Characterization of Pigeon Paramyxovirus Type 1 via Different Inoculation Routes in Pigeons. Front. Vet. Sci. 7:569901. doi: 10.3389/fvets.2020.569901
Received: 05 June 2020; Accepted: 28 October 2020;
Published: 17 February 2021.
Edited by:
Van Giap Nguyen, Vietnam National University of Agriculture, VietnamReviewed by:
Francisco Rivera-Benítez, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP), MexicoHyuk-Joon Kwon, Seoul National University, South Korea
Copyright © 2021 Chang, Feng, Wang, Li, Su, Wang, Du and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongxuan He, aGVoeEBpb3ouYWMuY24=
†These authors have contributed equally to this work
 Han Chang
Han Chang Shengyong Feng
Shengyong Feng Yutian Wang3
Yutian Wang3 Hongxuan He
Hongxuan He