- 1Wageningen Bioveterinary Research, Department of Virology, Lelystad, Netherlands
- 2Wageningen Bioveterinary Research, Department of Bacteriology and Epidemiology, Lelystad, Netherlands
- 3Poultry Department, GD-Animal Health, Deventer, Netherlands
Wild birds are the natural reservoir of the avian influenza virus (AIV) and may transmit AIV to poultry via direct contact or indirectly through the environment. In the Netherlands, a clinically suspected free-range layer flock was reported to the veterinary authorities by the farmer. Increased mortality, a decreased feed intake, and a drop in egg production were observed. Subsequently, an infection with low pathogenic avian influenza virus was detected. This study describes the diagnostic procedures used for detection and subtyping of the virus. In addition to routine diagnostics, the potential of two different environmental diagnostic methods was investigated for detecting AIV in surface water. AIV was first detected using rRT-PCR and isolated from tracheal and cloacal swabs collected from the hens. The virus was subtyped as H10N7. Antibodies against the virus were detected in 28 of the 31 sera tested. An intravenous pathogenicity index (IVPI) experiment was performed, but no clinical signs (IVPI = 0) were observed. Post-mortem examination and histology confirmed the AIV infection. Multiple water samples were collected longitudinally from the free-range area and waterway near the farm. Both environmental diagnostic methods allowed the detection of the H10N7 virus, demonstrating the potential of these methods in detection of AIV. The described methods could be a useful additional procedure for AIV surveillance in water-rich areas with large concentrations of wild birds or in areas around poultry farms. In addition, these methods could be used as a tool to test if the environment or free-range area is virus-free again, at the end of an AIV epidemic.
Introduction
In recent decades, the avian influenza virus (AIV) has caused many infections in poultry worldwide, leading to significant economic and animal welfare implications (1, 2). Most AIVs are low pathogenic avian influenza viruses (LPAIVs), which cause no to mild clinical signs in poultry, such as respiratory symptoms, a drop in egg production and a slight increase in mortality rates (3). However, viruses of the H5 and H7 AIV subtypes can evolve to highly pathogenic avian influenza viruses (HPAIVs), which cause severe systematic infections and high mortality rates in poultry (4, 5). These severe clinical signs of these HPAIVs are the reason that prevention of AIV infections in poultry is required. If poultry is infected with H5 and H7 subtypes in spite of this, poultry must be culled, to prevent the spread of these viruses (6).
Poultry in free-range areas can come in contact with the virus through infected wild birds, especially aquatic birds from the order Anseriformes and Charadriiformes, which are the natural reservoir of AIV (7). Wild birds shed the virus via the respiratory and/or cloacal route and transmit the virus either directly by host-to-host contact, or indirectly through the environment (8). Once the virus is shed in the environment, it can remain infective for varying lengths of time depending on environmental variables such as water temperature and type (9), the number of micro-organisms present (10) and the amount of UV radiation by solar light (11). Because the virus is often shed in aquatic environments by wild birds, the detection of AIV in water is particularly relevant. Therefore, various methods have been developed for detecting AIV in water. Several studies have reported methods to detect AIV in surface water during field infections of AIV (12–15), however, most of these studies only showed the efficiency of their methods under laboratory circumstances (16–24).
In this study, we developed two methods that are able to detect AIV in natural water samples after field sampling. These methods can be useful screening tools to determine whether AIV is circulating in the environment. Such a tool has several applications. First of all, EU countries monitor the circulation of AIV in their environment using dead wild bird surveillance programs (25). In such a surveillance, dead found wild birds are examined for AIV: AIV positive dead wild birds indicate that the virus may be circulating in the area where the bird was found. Screening surface water for the presence of AIV could assist this surveillance (26) particularly when AIV surveillance fails due to HPAIVs that do not cause noticeable mortality in wild birds. The second use becomes apparent when HPAIV is already circulating: additional precautionary measures can then be taken, such as keeping free-range poultry indoors (Government of the Netherlands) (27). Screening of surface water for the absence of HPAIV can support the decision to lift the precautionary measures at the end of a HPAIV epidemic. Despite the putative advantages of screening for AIV in surface water, this is currently not performed in the Netherlands.
This manuscript describes diagnostic procedures and additional experimental research that was performed to confirm an infection with an LPAIV H10N7 virus in a free-range layer flock showing clinical signs. The infection on the farm provided an unique opportunity to test two environmental diagnostic methods that were developed, and their potential for detecting AIV in surface water. If this pilot study yields promising results, either or both of these methods could be used in the future for screening of AIV in the environment.
Materials and Methods
Diagnostics of Hens
Virus Detection
Real-time reverse transcriptase polymerase chain reaction (rRT-PCR)
Tracheal and cloacal swabs were taken from 21 hens which were lethargic and had a loss of body condition, which were the clinical signs representative for the flock. At the laboratory, the dry swabs were pooled in 10 ml 2.95% Tryptose phosphate and 1% Gentamicin. A pool consisted of five swabs of the same swab type (tracheal or cloacal). RNA was extracted from the pools using the MagNA Pure 96 System (Roche, Almere, Netherlands) and the MagNA Pure 96 DNA and Viral NA Small Volume kit (Roche) following manufacturer's instructions. Total RNA was tested by rRT-PCR targeting the influenza M gene (M-PCR), which detects all AIV subtypes, as described previously (28). In addition, the sequence of the haemagglutinin (HA) cleavage site and the neuraminidase subtype was determined by Sanger sequencing in two cloacal swab pools (29).
After sequencing, the isolated RNA was tested in an H10-specific rRT-PCR. The sequences of the primers (RF5127 5′-TGTGCATCGCATGTTTCCAT-3′ and RF5133 5′-CATCATTCTCTGGTTTAGCTTCGG-3′) and probe (RF5134 5′6-FAM-TGACAACGGCTAGAAGAACAAAACATGATGCC-BHQ-3′) were kindly provided by Ron Fouchier, Erasmus MC, Netherlands. One-step rRT-PCR was accomplished with a MX3005p cycler (Agilent, Amstelveen, Netherlands) by use of the OneStep RT-PCR Kit (Qiagen, Venlo, Netherlands). The manufacturer's instructions of the kit were followed with 1.25 mM MgCl2, 0.3 μM Rox, 0.6 μM of each primers, and 0.1 μM of the probe added to the mix. Finally, 5 μl of RNA was amplified in a volume of 25 μl by employing the following temperature profile: 50°C for 30 min and 95°C for 15 min, followed by 45 cycles of 94°C for 10 sec, 58°C for 30 sec, 72°C for 10 sec.
Virus isolation
A tracheal and a cloacal swab pool were each inoculated into the allantoic cavity of four 9- to 11-day-old specific pathogenic free embryonated chicken eggs (0.2 ml/egg) following the guidelines of the OIE (30). The eggs were incubated for up to 6 days at 37°C. After 6 days, or earlier when the embryo's died, the eggs were chilled at 4°C for 4 h or overnight and the allantoic fluid was harvested. The allantoic fluid was tested for the presence of haemagglutinating activity and the absence of bacterial contamination. Fluids giving a negative haemagglutination reaction were passed into another four eggs.
Serology
Two days after virus detection, blood samples were collected from 31 randomly selected hens at the farm. At the laboratory, blood was centrifuged (20 min at 1780g) to obtain sera, which were subsequently inactivated in a water bath of 56°C for 30 min. Sera were tested for the presence of antibodies against AIV using the nucleoprotein-blocking ELISA and the Haemagglutination Inhibition (HI) assay.
ELISA
The employed indirect double-antibody sandwich ELISA detects antibodies against all AIV subtypes and is a modified version of the nucleoprotein-blocking ELISA described previously (31). The antigen, an H7N1 virus treated with 1% Nonidet P40, binds to the monoclonal nucleoprotein HB65 (Wageningen Bioveterinary Research, Lelystad, Netherlands) that is used to coat the ELISA plates. Serum was diluted (1:8) with an ELISA buffer (1% Difco™ Skim Milk and 0.05% Tween 20) and was added to the antigen. If present, antibodies were bound to the antigen and were marked by the horseradish peroxidase conjugate.
HI assay
The used HI assay is a classic laboratory procedure for the subtyping of antibodies of AIV and is based on the inhibition of the agglutination reaction by haemagglutinin subtype-specific antisera (3). Auto-agglutination was avoided by incubating the sera with chicken erythrocytes as described previously (32). The HI assay was performed according to the methods described in the OIE manual using eight haemagglutination units of virus instead of four (30). Sera were tested against the antigens of subtype H10N4 and H10N9. HI assay results were reported as log2 HI titres, with titres ≥3 log2 considered positive.
Assessment of AIV Virulence: Intravenous Pathogenicity Index (IVPI)
The IVPI was determined for the virus (A/chicken/Netherlands/17013178-006010) that was obtained from the cloacal swabs. The IVPI was performed according to the method described in the “Diagnostic Manual for avian influenza” (33). Ten 6-week old specific-pathogen-free layers were inoculated intravenously with the virus, after which the layers were monitored for clinical signs and mortality for 10 days. The experiment was approved by the Central Animal Experiments Committee (license number 1631018100).
Pathological Examination
Pathology
Four hens, including one dead hen and three hens with clinical signs representative for the clinical situation in the flock, were submitted to GD Animal Health, Deventer, the Netherlands, for a general pathological examination. When arrived, the three living birds were stunned by exposure to a mixture of carbon dioxide and oxygen and then exsanguinated. Routine necropsy procedures were performed, including parasitological, and histological examination. In addition to the necropsy procedures, PCRs were executed to exclude infections with other pathogens or co-infections. Therefore, samples of the trachea were tested for the presence of Infectious bronchitis virus (34), Mycoplasma gallisepticum (35) and Mycoplasma synoviae (36). Standard PCR methods used by GD Animal Health excluded tracheal infections with Infectious laryngotracheitis virus and Avian metapneumovirus, and oviduct infections with Group I Aviadenovirus and Atadenovirus (Egg drop syndrome virus). Furthermore, immunohistochemical staining excluded infections with Ornithobacterium rhinotracheale and Chlamydia psittaci in the air sacs (data not shown).
Histology
Samples of trachea, lung, air sac, duodenum and shell gland were fixed in 4% neutral buffered formalin, embedded in paraffin, sectioned at 2 μm, and stained with hematoxylin and eosin (H&E) for light microscopic examination. In the same organs, the presence of influenza A virus antigen was investigated using immunohistochemistry (IHC). For IHC, samples were fixated for at least 24 h in buffered 10% formalin, followed by dehydration in absolute ethanol and embedding in paraffin wax, sections were cut at 4 μm and mounted on glass slides. Endogenous peroxidase activity was blocked by incubation with 1% H2O2 containing 0.1% NaN3 for 20 min at room temperature (RT) and subsequently boiled in Tris (0.01 M) EDTA (0.001 M), pH 9.0 for 10 min. The binding of Fc-receptors was blocked by incubation with 10% fetal bovine serum for 20 min at RT. The immunostaining of influenza A virus-positive-cells was performed using 1:1,000 diluted anti-influenza A virus nucleoprotein monoclonal antibody (Meridian Life Science, Memphis, USA) in Normal Antibody Diluent (Klinipath, Duiven, Netherlands) for 30 min at RT. After three subsequent wash steps with phosphate-buffered saline, the sections were treated with anti-mouse Dako EnVision+ (Dako UK Ltd, Cambridgeshire, UK) for 30 min at RT. Again, sections were washed three times with phosphate-buffered saline and then treated with DAB+ (Dako UK Ltd) for 5 min at RT. Finally, the sections were counter-stained using haematoxylin. Sections incubated in the absence of primary antibody were taken along as negative controls.
Diagnostics of Water Samples
Water Sampling
Water samples were collected from the free-range area and a waterway around the farm, 2 days after the virus detection in hens. The free-range area is a fenced grassland connected to the poultry house that allows the hens going outside during daylight. After severe and prolonged rainfall, puddles of water were formed in the free-range area (Figure 1). During the first visit, two water samples were collected from the puddles of water in the free-range area, and two water samples were collected from the waterway (Figure 2). The samples were collected with a bucket attached to a stick to avoid disturbing the sampling sites and avoid cross-contamination between the sampling sites. Samples were taken in the middle of the water puddle and at least 1 meter from the ditch side of the waterway. Within each sampling site, a 1 liter sample and a 50 liter sample of water were collected.

Figure 1. Puddles were formed in the free-range area after severe and prolonged rainfall. Water samples were collected from these puddles.
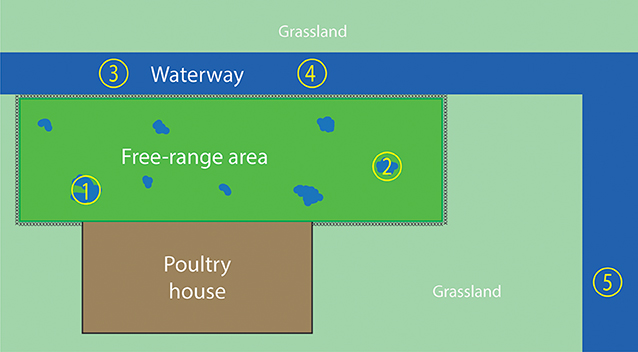
Figure 2. Overview of the water sampling sites. The fenced free-range area is enclosed by the poultry house and waterway. Water samples 1 and 2 were collected from the puddles of water in the free-range area, which were caused by severe rainfall. Water samples 3, 4, and 5 were collected from the waterway. Sampling sites 1–4 were sampled during both farm visits; sampling site 5 was sampled only during the second visit.
After the virus was detected in hens, they were prohibited to enter the free-range area any longer. Because the hens could not excrete new virus in the free-range area, there was a possibility to examine the survival of the virus in the environment. Therefore, the farm was visited 2 weeks after the first water sampling moment. During the second visit, 14 days after the first visit, the sampling sites of the first visit were sampled again plus an additional site of the waterway (sample site number 5 in Figure 2).
Detection of AIV in 1 Liter Water Samples: “The 1L Method”
Water samples were stored at 4°C before being filtered within 12 h of sampling. One liter of water was divided into two conical centrifuge tubes (Corning® 500 mL PP Centrifuge Tubes, Thermofisher, Breda, Netherlands) and centrifuged in a Mistral 6000 (MSE, Heathfield, UK) for 15 min at 1781 g at 4°C. After centrifuging, the supernatant was filtrated using a Sterifil™ 47 mm Aseptic Vacuum Filter System and Holder (Millipore, Merck, Darmstadt, Germany) and two different types of filters. The first filter, a hydrophilic Glass Fiber Filter with a pore size of 1.0 μm without binder resins (Millipore, Merck), was used for liquid clarification and was replaced when the filter was constipated. The second filter, a hydrophilic mixed cellulose esters membrane (MF-Millipore™ Membrane Filter, Merck) with a pore size of 5.0 μm, is able to catch the virus if present. After processing the sample, the membrane was collected from the filter system into a bead tube and frozen (−80°C) until the start of the RNA isolation. In addition, the filter system was cleaned by soaking the system in a chlorine solution (Suma Tb D4, Diversey, Utrecht, Netherlands)for 10 min, followed by rinsing the system with demi water.
RNA was isolated from the membrane with the PowerWater® RNA Isolation kit (Qiagen). Manufacturer's instructions were followed, with several modifications: (1) the prescribed vortex adapter was not used; instead the bead tubes were horizontally vortexed manually on the vortex (IKA® MS1 minishaker, Merck), (2) the fluid was centrifuged for 3 min at 3000 g instead of 1 min at 4000 g at point 9, and 3) the RNA was eluted in 50 μl RNase-free water to obtain a higher concentration of RNA. The eluate was frozen (-80°C) until it was processed in the M-PCR.
Detection of AIV in 50 Liter Water Samples: “The 50L Method”
Water samples were stored at 4°C before filtering for a maximum of 24 h. First, large substances, such as water plants, were filtered out of the water samples by flowing the water through a clean towel. Secondly, 200 μl of Porcine Epidemic Diarrhea virus (PEDV) with a titer of 6.3 × 104 plaque-forming units/ml was added as an intern control. After these preparations, the water was filtered through a Rexeed™-25A hemodialyzer dead-end hollow fiber ultrafilter (Asahi Kasei Medical, Brussel, Belgium) using a peristaltic pump (Geopump™ series II, Geotech, Haarlem, Netherlands) and sterilized silicon hoses (drive 1, speed 2.5). After processing the sample, the filter system was rinsed with 0.5L of tap water to flush the last part of the sample from the silicon hoses into the filter. The filter was stored overnight at 4°C, when necessary. The virus was flushed from the filter during a 20-min back flush with 500 ml TGBE buffer [500 ml RNase-free water, 6.05 gram Tromethamine, 1.9 gram glycine, and 10 gram beef extract (Bacto™ Beef Extract Dessicated, Life Technologies, Thermofisher)]. After back flushing, the TGBE buffer containing virus was divided over two conical centrifuge tubes (Corning® 500 mL PP Centrifuge Tubes, ThermoFisher) and centrifuged for 2 h at 4500 g at 4°C. After centrifuging, the pH was set to a pH of 7 by adding 5M HCL and 125 ml 5x PEG/NaCl (500 gram PEG 8000 plus 87 gram NaCl and filled with distilled water up to 500 ml). After an incubation period of 1 h (or overnight) at 4°C, the centrifuge step was repeated and the supernatant was removed. To make the pellets more compact, the tubes were centrifuged for another 10 min at 4500 g at 4°C and again the supernatant was removed. Each pellet was resuspended in 500 μl Dulbecco's Modified Eagle Medium (Gibco, ThermoFisher). Finally, 500 μl of the fluid was added to 2 ml of NucliSens lysis buffer (bioMérieux, Boxtel, The Netherlands) and stored for maximum 24 h at 4°C.
RNA was isolated using the Nuclisens® Magnetic Extraction Reagents and the Nuclisens® MiniMAG® (bioMérieux) according to the manufacturer's instructions. The eluate was frozen (−80°C) until it was processed in the M-PCR.
Full Genome Sequencing and Phylogenetic Analysis
Next-generation sequencing (NGS) was performed to determine the full genome sequences of the AIV isolated from the hen (EPI_ISL_394155) and from the 1L water sample that was collected at sampling site 1 during the first visit (EPI_ISL_394156). Illumina sequencing was performed as described previously (37).
For phylogenetic analysis, a dataset was composed of HA nucleotide sequences of the top 100 blast hits (on March 10, 2020) from the GISAID EpiFLu Database (https://www.gisaid.org), using the HA segment of A/Chicken/Netherlands/17013178-006-010/2017 as query. The collected sequences were aligned using Multiple Alignment with Fast Fourier Transformation [MAFFT v7.427; Katoh and Standley (38)]. Subsequently, a maximum likelihood tree was constructed by using RaxML version 8.2.12 (39) based on the general time-reversible (GTR) model of nucleotide substitutions with a gamma-distributed variaten of rates and 1,000 bootstrap replicates. The tree with the highest log likelihood was visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/~figtree).
Results
Outbreak Description
In September 2017, the veterinary authorities received a report of clinical signs in hens of a commercial free-range layer farm. At the time of reporting, the farm housed 35,388 brown Novogen laying hens of 82 weeks of age in one house. The flock showed increased mortality, decreased feed intake and a drop in egg production as shown in Figure 3. The increased daily mortality, which started at 81 weeks of age, peaked with 0.24% between 82 and 83 weeks of age. During the mortality peak, mean daily feed intake decreased from approximately 120 gram per day to 100 gram per day per hen. The mean egg production per week decreased from about 78% at 80 and 81 weeks of age to 68% at 82 weeks of age. It is not likely that the virus was present in the flock for more than 8 weeks, because the flock had tested negative for antibodies against AIV during the national serological screening for AIV in poultry at 74 weeks of age (40).
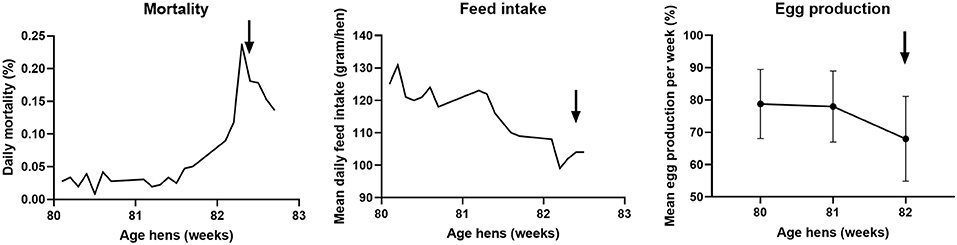
Figure 3. Course of daily mortality, mean feed intake and mean egg production of the flock between 80 and 83 weeks of age at the free-range layer farm. Daily mortality is expressed as percentage of the total amount of hens present, the mean daily feed intake in gram per day per hen, and the mean egg production per week as a percentage of the total amount of hens present ± standard deviation. The arrow indicates the day of reporting the clinically suspect situation.
Before reporting, the commercial free-range layer farm had experienced already four LPAIV introductions since the start of production in 2008. An H6N1 virus was detected in 2010 (41), followed by an H6N2 virus in 2012, an H9N2 virus in 2013, and an H6N2 virus in 2014. The farm is located in the northern part of the Netherlands, <2 km from the North Sea coast, and this area has plenty of waterways and lakes. This water-rich area provides a good habitat for wild (water) birds, which live in large quantities near the farm. Elbers and Gonzales (42) monitored the free-range area of this particularly farm with video-cameras for 1 year and reported various visits of wild birds to the free-range area (42). No direct contact between hens and wild birds was observed, and it is hypothesized that wild birds infected with AIV probably contaminate the environment—soil and water present in the free-range area—with AIV. Consequently, poultry may be infected through contact with contaminated soil or water in the free-range area. The type of farm, a free-range farm, together with the location of the farm, and therefore the presence of wild aquatic birds, may contribute to the multiple LPAIV introductions in the poultry farm. The presence of wild birds in the free-range area was also observed during the ifirst visit for water sampling: two mallards (Anas plathyrhynchos) were observed swimming in the puddle of water at sampling site 1 (Figure 2).
Diagnosis of Hens
AIV was detected in all five tracheal and five cloacal swab pools using the M-PCR. The virus was subtyped as LPAIV H10N7 using Sanger Sequencing, and all swab pools tested positive in the H10-specific PCR. The virus was isolated by inoculation into specific pathogenic free embryonated chicken eggs and also subtyped as H10N7. The serum samples which were collected at the farm, tested positive for antibodies against AIV. The ELISA detected antibodies against AIV in 28 of the 31 sera. The HI assay detected H10 antibodies in 17 sera when tested against the H10N4 antigen, while 12 sera tested positive against the H10N9 antigen. The positive log2 HI titres ranged from 3 to 8. The pathogenicity of the isolated H10N7 virus was tested using an IVPI experiment. An IVPI score of 0.0 was measured. This score indicates that no clinical signs were observed during the experiment, which is in contrast to field observations.
Pathological Examination
Pathology
The hen that was submitted dead showed severe loss of body condition with loss of productivity. The ovary contained an ovarian adenocarcinoma with extensive peritoneal metastases. The euthanized hens presented a loss of body condition, characterized by muscular atrophy predominantly of the pectoral muscles. The head and appendices showed no abnormalities, the same for the lungs, but the tracheas were hyperemic. One hen had an extensive fibrinous airsacculitis, mainly of the thoracic air sacs. All hens had productive ovaries. Gastrointestinal contents were normal and despite a mild infection of small numbers of Ascaridia galli and Heterakis gallinarum no lesions were found. The liver, spleen and kidneys were normal. The abdominal fat pads were relatively normal in size. No other pathogens were detected by PCR in samples of the trachea and oviduct.
Histology
All four hens showed a mild lymphoplasmacytic tracheitis, with intact epithelium and erythrocyte engorged capillaries (hyperemia). However, no viral antigen was demonstrated with IHC staining for influenza A virus in the tracheas. One of the four hens had a mild pneumonia, consisting of collapse of multiple atria and capillaries by moderate infiltrates of mainly macrophages, lymphocytes, less heterophils and fibrinous and necrotic debris with organization (Figure 4A). In addition, the epithelium of the parabronchi of this hen was hypertrophic and hyperplastic, while the lumen contained protein rich fibrillar debris (fibrin) with heterophils and macrophages. Although no viral antigen was present in epithelial cells or endothelial cells of this hen, the mononuclear infiltrate scattered cells contained viral antigen by IHC staining for influenza A virus (Figure 4B). Furthermore, this affected hen had a lymphohistiocytic airsaculitis with protein rich edema and accumulation with intraluminal protein rich debris and many large macrophages with foamy cytoplasm (active macrophages with signs of phagocytosis of debris). No viral antigen could be demonstrated by IHC staining for influenza A virus in the airsac.
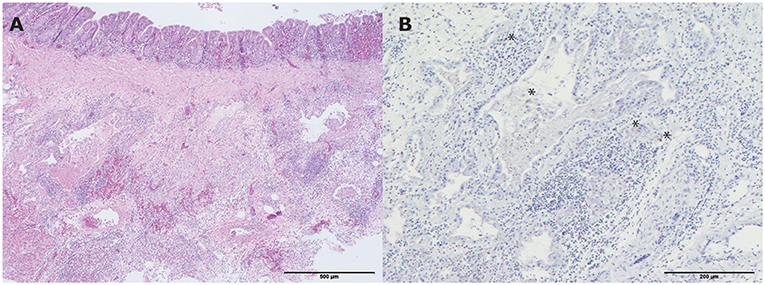
Figure 4. Histopathologic and immunohistochemical (IHC) testing results of a lung from one of the living hens that were submitted for pathological examination. (A) Mild pneumonia consisting of collapse of multiple atria and capillaries by moderate infiltrates of mainly macrophages, lymphocytes, less heterophils and fibrinous and necrotic debris with organization (hematoxylin and eosin (H&E) stain). Bar < 500 μm; (B) Influenza A virus antigen within the mononuclear infiltrate scattered cells. Bar < 200 μm. *indicates the colored cells.
The duodenum of all investigated hens showed mild villus atrophy and a moderate lymphoplasmacytic infiltrate with scattered heterophils within the lamina propria. The epithelium was intact, but numbers of intraepithelial lymphocytes were multifocally increased. Crypts appeared normal. No Influenza A viral antigen was demonstrated by IHC. The shell gland of two hens did not show any abnormalities, while in one hen that arrived alive scattered perivascular lymphocytic infiltrates were present in het deep stromal region of the secondary folds. The glandular and surface epithelium were not altered and Influenza A viral antigen could not be demonstrated by IHC staining.
Diagnosis of Water Samples
Water samples collected on both sampling days from the free-range area and waterway were examined for the presence of AIV. The results of the M-PCR and H10-specific rRT-PCR for both methods are shown in Table 1. From the samples collected during the first sampling day, sampling site 1 resulted in positive H10-specific rRT-PCR results for both methods. It was sampling site 2—the other puddle in the free-range area—that was positive for both PCRs in both methods. No AIV was detected in the waterway samples. On day 14, the second sampling day, AIV was detected at sampling site 1 by both methods and PCRs. At sampling site 2, AIV was detected by the 1L method in both PCRs and by the 50L method only in the H10-specific rRT-PCR. The positive results of the second sampling day indicated that the virus is still detectable for at least 2 weeks. Again, no virus was detected in the samples collected from the waterway.
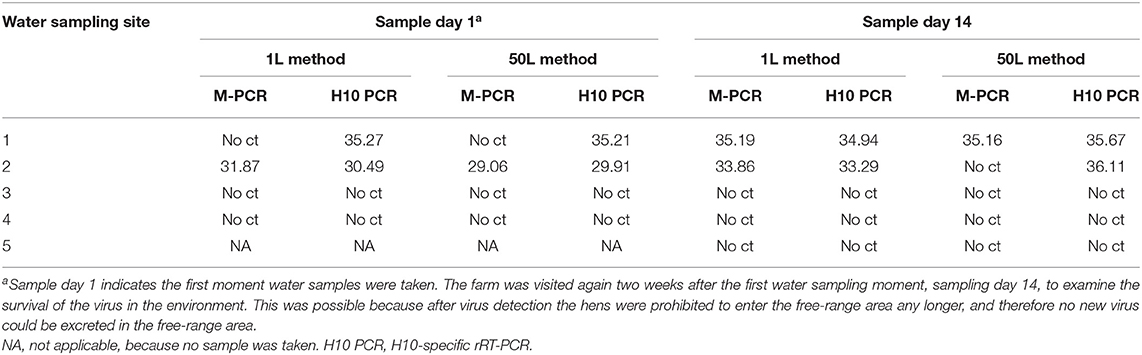
Table 1. Detection of AIV by the M-PCR and H10-specific rRT-PCR in water samples using the 1L and 50L water sample method.
Full Genome Sequencing and Phylogenetic Analysis
The full genome sequences of the viruses detected in the hens and in the water samples collected in the free-range area were determined by NGS. The genome sequences of these viruses were similar, and no specific mutations or adaptations which lead to any changes in amino acids sequences were identified. Therefore, it remains unclear whether the water in the free-range area was contaminated by infected wild birds, resulting in introduction of the virus into the poultry farm. Or, whether the free-range area was contaminated later, by the already infected chickens. Phylogenetic analysis of the HA gene segment was performed to study the relationship between the H10N7 virus detected at the farm and other poultry and wild bird viruses. The most closely related virus based on the HA gene segment was a H10N6 virus detected in a mallard in Denmark in 2011. No closely related H10N7 viruses were detected in poultry or wild birds in 2017 (Figure S1).
Discussion
Post-mortem examination, diagnostics and additional experimental research confirmed an LPAIV H10N7 infection in a commercial free-range layer flock in the Netherlands. The circumstances surrounding this particular outbreak enabled us to investigate the potential of two methods for detecting AIV in the environmental water samples during a natural AIV infection. The described methods can be a useful additional procedure for AIV surveillance, or can be used as a tool to decide when precautionary measures, like keeping free-range chickens indoors, could be lifted at the end of an AIV epidemic. In contrast to the waterways near the farm, in which no AIV was detected, H10N7 virus was detected in the puddles of water in the free-range area. Both methods used for detecting AIV in water allowed the detection of H10N7 virus, demonstrating the potential of these diagnostic methods. Unfortunately, full genome sequencing did not provide conclusive insight into the epidemiology of the infection. Phylogenetic analysis did not reveal circulation of similar H10 viruses in wild birds or poultry in Europe in 2017.
The clinically suspect situation, as a result of the infection with the LPAIV H10N7 was observed and officially reported by the farmer. He observed an increase in mortality, a decrease in feed intake and a drop in egg production although none of these three parameters reached the official reporting thresholds described in the acting Statutory Regulation (43). These thresholds, originally implemented for the early detection of HPAIV, are set for laying flocks at ≥0.5% mortality per day for two consecutive days and ≥5% decrease in egg production or feed/water intake per day for two consecutive days. New effective thresholds for an early detection for HPAIV and LPAIV infections in poultry have recently been suggested (44). If those newly suggested thresholds would have been operational, the observed clinical signs would have led to mandatory reporting.
In contrast to the clinical signs observed on the farm, no clinical signs were observed during the IVPI experiment. There are several possible explanations for the observed difference between the field situation and the experiment. Firstly, the AIV infection in the field may have been accompanied by other viral or bacterial respiratory pathogens leading to more severe clinical signs. These kind of co-infections for AIV have previously been described for infectious bronchitis disease, Staphylococcus sp., Ornitholobacterium rhinotracheale, Mycoplasma gallisepticum, and Escherichia coli (45). However, no evidence was found for such co-infections during diagnostics. Secondly, non-pathogenic stress factors, such as dietary calcium stress, could have caused more severe clinical signs of the AIV infection in the field (46). Thirdly, the difference in clinical signs may be caused by the different ages of the infected hens: the hens in the field were 82 weeks old, while the hens in the experiment were only 6 weeks old. It has previously been demonstrated that non-reproductive hens shed less LPAIV than reproductive hens (47). Moreover, it has been shown that non-reproductive hens infected with an H3N1 virus infection, which circulated in poultry in Belgium in 2019, showed less severe clinical signs than hens that were laying eggs (48).
Both environmental diagnostic methods showed their potential for detection of AIV in water. The H10-specific rRT-PCR proved more sensitive than the M-PCR, which is likely due to differences in the match between this virus strain and the primers and probe used in these PCRs. The biggest advantage of the 1L method is the relatively small sampling volume, which makes the sampling and filtering process manageable. Another advantage of the 1L method is that the procedure takes less time than the procedure of the 50L method. However, the 50L method makes it possible to filter the surface water directly in the field, using portable equipment. After filtering, only the filter needs to be transported to the laboratory. Moreover, it has been shown that the 50L method can also be used for filtering other viruses, such as PEDV and Arboviruses (data not shown). PEDV, which was also used as an intern control, was detectable with CTs of around 20, when 200 μl of virus with a titer of 6.3 × 104 plaque-forming units/ml was added to a surface water sample of 50L. No detection limit of the 50L method was determined for AIV, but for the 1L method the detection limit was determined using environmental water samples that were spiked with LPAIV H5N7 virus. The detection limit was determined at 0.33 log10 egg infectious dose (EID50)/ml, which means that the 1L method is able to detect a minimum of 2140 virus particles in 1L of water. Looking at the mean cloacal HPAIV shedding of ducks—which is 3.8 log10 EID50/ml and takes on average 6.6 days (47)—this environmental diagnostic method may still be able to detect AIV when 9 infected ducks shed virus in a pond or waterway of 10,000L (assuming that a duck produce on average 60 ml feces per day). As discussed, AIV can remain infective for varying lengths of time in water. This was shown for several AIVs which were added to different types—fresh, brackish and salt—of natural water: for three LPAIVs the viral titer decreased with 1 log10 TCID50/ml within five to seven days (49). When the survival of two other LPAIVs was investigated, a maximum survival of 2 weeks was defined (10). Our study showed that AIV was still detectable with the environmental diagnostic methods after at least 2 weeks. During these 2 weeks, the mean ambient temperature, measured by the nearest weather station, was 12.6°C (50). Due to the relatively low detection limit and long persistence of the virus, it appears likely that in areas with several infected wild birds, the presence of AIV in water can be measured using these environmental diagnostic methods. Therefore, these methods could be useful for screening purposes of AIV in wetlands with high bird numbers, important wintering sites for migratory water birds or in free-range areas of farms. The use of environmental diagnostic methods was also shown in other studies. AIV was detected in 12 out of 597 water samples collected from the wetlands in California (14) and in 4 out of 9 water samples collected from the Dombes ponds in France during fall migration of wild birds in 2009 (13) using similar methods. Furthermore, AIV was isolated from 12 out of 102 water samples collected from different lakes in Alaska which were important breeding sites for migratory water birds (12), and from 1 out of 265 water samples collected from the Izumi plain, an overwintering site of endangered cranes and of many other migratory birds (15).
To conclude, this study showed the potential of two methods to detect AIV in water during field infections. Detecting AIV in the environment using water samples may be a good tool to improve AIV surveillance and may assist in deciding to lift precautionary measures in outbreak situations.
Data Availability Statement
All datasets analyzed for this study are included in the article.
Ethics Statement
The animal study was reviewed and approved by the Centrale Commissie Dierproeven (CCD), Den Haag, Netherlands.
Author Contributions
EG, AE, WP, and NB: conceptualization. EG, NDB, RH, RHH, and WV: methodology. RH, RHH, WP, and WV: validation. EG: data curation. AE, WP, and NB: supervision. SB and ME: phylogenetic analysis. EG and NDB: writing—original draft preparation. EG, AE, NDB, RH, WV, SB, ME, RHH, WP, and NB: writing—review and editing.
Funding
This work was funded by the Dutch Ministry of Agriculture, Nature and Food Quality (project WOT-1600014-01 and WOT-160068-01).
Conflict of Interest
NDB was employed by the company GD-Animal Health, Deventer, Netherlands.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kimberly Verboon (student of Inholland University of Applied Sciences, Delft) for her help with taking water samples at the poultry farm. We thank the poultry farmer for his hospitality and collaboration. We thank Ron Fouchier, Erasmus Medical Center, Rotterdam, for sharing the sequences of the primers and probe for the specific H10 rRT-PCR. We thank Eline Verheij (WBVR) for her practical help with the water samples. We acknowledge the authors and submitting laboratories of the sequences from the GISAID EpiFlu Database.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00237/full#supplementary-material
Figure S1. Maximum likelihood phylogenetic tree based on the nucleotide sequence of the HA segments from influenza strains related to A/Chicken/Netherlands/17013178-006-010/2017. The evolutionary history was inferred by using RaxML (39) and utilizing the maximum likelihood method based on the General Time Reversible (GTR) model with a gamma-distributed variation of rates and 1000 bootstrap replicates. Bootstrap support values are indicated at the corresponding branch. The optimal phylogenetic tree is shown, and is drawn to scale. The GISAID accession numbers of the viruses are shown in the trees. The H10N7 viruses isolated from the hens and from the water sample are marked in red.
Abbreviations
AIV, avian influenza virus; EID50, egg infectious dose; HA, haemagglutinin; H&E, hematoxylin and eosin; HI, Haemagglutination Inhibition; HPAIV, highly pathogenic avian influenza virus; IHC, immunohistochemistry; IVPI, intravenous pathogenicity index; LPAIV, low pathogenic avian influenza virus; M-PCR, rRT-PCR targeting the Influenza M gene; NA, not applicable; NGS, next-generation-sequencing; PEDV, Porcine Epidemic Diarrhea virus; rRT-PCR, Real-time Reverse Transcriptase Polymerase Chain Reaction; RT, room temperature.
References
1. Elbers AR, Fabri TH, De Vries TS, De Wit JJ, Pijpers A, Koch G. The highly pathogenic avian influenza A (H7N7) virus epidemic in The Netherlands in 2003: lessons learned from the first five outbreaks. Avian Dis. (2004) 48:691–705. doi: 10.1637/7149
2. Van Asseldonk MA, Meuwissen MP, Mourits MC, Huirne RB. Economics of controlling avian influenza epidemics. In: Schrijver RK, editors. Avian Influenza Prevention and Control. Dordrecht: Springer (2005). p. 139–148. doi: 10.1007/1-4020-3441-5_15
3. Spackman E. Avian Influenza Virus. Totowa, NJ: Human Press (2008). doi: 10.1007/978-1-59745-279-3
4. Webster RG, Rott R. Influenza virus a pathogenicity: the pivotal role of hemagglutinin. Cell. (1987) 50:665–6. doi: 10.1016/0092-8674(87)90321-7
5. Erasmus Medical Centre (NL) Istituto Zooprofilattico Sperimentale delle Venezie (IT). Mechanisms and Risk Factors for Mutation From Low to Highly Pathogenic Avian Influenza Virus. EFSA supporting publication 2017:EN-1287 (2017). p. 26. doi: 10.2903/sp.efsa.2017.EN-1287
6. EU. Council Directive 2005/94/EC of 20 December 2005 on Community measures for the control of avian influenza and repealing Directive 92/40/EEC. J Eur Union. (2005) 10:16–64.
7. Stallknecht DE, Shane SM. Host range of avian influenza virus in free-living birds. Vet Res Commun. (1988) 12:125–41. doi: 10.1007/BF00362792
8. Breban R, Drake JM, Stallknecht DE, Rohani P. The role of environmental transmission in recurrent avian influenza epidemics. PLoS Comput Biol. (2009) 5:e1000346. doi: 10.1371/journal.pcbi.1000346
9. Dalziel AE, Delean S, Heinrich S, Cassey P. Persistence of low pathogenic influenza a virus in water: a systematic review and quantitative meta-analysis. PLoS ONE. (2016) 11:e0161929. doi: 10.1371/journal.pone.0161929
10. Zarkov IS. Survival of avian influenza viruses in filtered and natural surface, waters of different physical and chemical parameters. Rev Med Vet. (2006) 157:471–6.
11. Pinon A, Vialette M. Survival of viruses in water. Intervirology. (2018) 61:214–22. doi: 10.1159/000484899
12. Ito T, Okazaki K, Kawaoka Y, Takada A, Webster RG, Kida H. Perpetuation of influenza a viruses in alaskan waterfowl reservoirs. Arch Virol. (1995) 140:1163–72. doi: 10.1007/BF01322743
13. Deboosere N, Horm SV, Pinon A, Gachet J, Coldefy C, Buchy P, et al. Development and validation of a concentration method for the detection of influenza a viruses from large volumes of surface water. Appl Environ Microbiol. (2011) 77:3802–8. doi: 10.1128/AEM.02484-10
14. Henaux V, Samuel MD, Dusek RJ, Fleskes JP, Ip HS. Presence of avian influenza viruses in waterfowl and wetlands during summer 2010 in California: are resident birds a potential reservoir? PLoS ONE. (2012) 7:e31471. doi: 10.1371/journal.pone.0031471
15. Nakagawa H, Okuya K, Kawabata T, Matsuu A, Takase K, Kuwahara M, et al. Genetic characterization of low-pathogenic avian influenza viruses isolated on the Izumi plain in Japan: possible association of dynamic movements of wild birds with AIV evolution. Arch Virol. (2018) 163:911–23. doi: 10.1007/s00705-017-3698-1
16. Khalenkov A, Laver WG, Webster RG. Detection and isolation of H5N1 influenza virus from large volumes of natural water. J Virol Methods. (2008) 149:180–3. doi: 10.1016/j.jviromet.2008.01.001
17. Dovas CI, Papanastassopoulou M, Georgiadis MP, Chatzinasiou E, Maliogka VI, Georgiades GK. Detection and quantification of infectious avian influenza A (H5N1) virus in environmental water by using real-time reverse transcription-PCR. Appl Environ Microbiol. (2010) 76:2165–74. doi: 10.1128/AEM.01929-09
18. Horm SV, Gutierrez RA, Sorn S, Buchy P. Environment: a potential source of animal and human infection with influenza A (H5N1) virus. Influenza Other Respir Viruses. (2012) 6:442–8. doi: 10.1111/j.1750-2659.2012.00338.x
19. Keeler SP, Berghaus RD, Stallknecht DE. Persistence of low pathogenic avian influenza viruses in filtered surface water from waterfowl habitats in Georgia, USA. J Wildl Dis. (2012) 48:999–1009. doi: 10.7589/2011-11-314
20. Ronnqvist M, Ziegler T, Von Bonsdorff CH, Maunula L. Detection method for avian influenza viruses in water. Food Environ Virol. (2012) 4:26–33. doi: 10.1007/s12560-011-9075-4
21. Francy DS, Stelzer EA, Brady AM, Huitger C, Bushon RN, Ip HS, et al. Comparison of filters for concentrating microbial indicators and pathogens in lake water samples. Appl Environ Microbiol. (2013) 79:1342–52. doi: 10.1128/AEM.03117-12
22. Nielsen AA, Jensen TH, Stockmarr A, Jorgensen PH. Persistence of low-pathogenic H5N7 and H7N1 avian influenza subtypes in filtered natural waters. Vet Microbiol. (2013) 166:419–28. doi: 10.1016/j.vetmic.2013.06.024
23. Zhang H, Chen Q, Chen Z. A simple and efficient method for detecting avian influenza virus in water samples. J Virol Methods. (2014) 199:124–8. doi: 10.1016/j.jviromet.2014.01.013
24. Numberger D, Dreier C, Vullioud C, Gabriel G, Greenwood AD, Grossart HP. Recovery of influenza A viruses from lake water and sediments by experimental inoculation. PLoS ONE. (2019) 14:e0216880. doi: 10.1371/journal.pone.0216880
25. European Food Safety Authority, Brouwer A, Gonzales J, Huneau A, Mulatti P, Kuiken T, et al. Annual Report on surveillance for avian influenza in poultry and wild birds in Member States of the European Union in 2018. EFSA J. (2019) 17:e05945. doi: 10.2903/j.efsa.2019.5945
26. Stallknecht DE, Goekjian VH, Wilcox BR, Poulson RL, Brown JD. Avian influenza virus in aquatic habitats: what do we need to learn? Avian Dis. (2010) 54:461–5. doi: 10.1637/8760-033109-Reg.1
27. Government of the Netherlands Measures to combat bird flu (2020). Available online at: https://www.government.nl/topics/animal-diseases/measures-to-combat-bird-flu (accessed March 1, 2020).
28. Bouwstra R, Heutink R, Bossers A, Harders F, Koch G, Elbers A. Full-Genome Sequence of Influenza A(H5N8) Virus in Poultry Linked to Sequences of Strains from Asia, the Netherlands, 2014. Emerg Infect Dis. (2015) 21:872–4. doi: 10.3201/eid2105.141839
29. Beerens N, Koch G, Heutink R, Harders F, Vries DPE, Ho C, et al. Novel Highly Pathogenic Avian Influenza A(H5N6) Virus in the Netherlands, December 2017. Emerg Infect Dis. (2018) 24:770–3. doi: 10.3201/eid2404.172124
30. Oie (2017). “Chapter 2.3.4. Avian Influenza (infection with avian influenza viruses),” in Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (2017). Available online at: https://www.oie.int/standard-setting/terrestrial-manual/access-online/
31. De Boer GF, Back W, Osterhaus AD. An ELISA for detection of antibodies against influenza a nucleoprotein in humans and various animal species. Arch Virol. (1990) 115:47–61. doi: 10.1007/BF01310622
32. Germeraad EA, Achterberg R, Venema S, Post J, De Leeuw O, Koch G, et al. The development of a multiplex serological assay for avian influenza based on Luminex technology. Methods. (2019) 158:54–60. doi: 10.1016/j.ymeth.2019.01.012
33. Eu. 2006/437/EC: Commission Decision of 4 August 2006 approving a Diagnostic Manual for avian influenza as provided for in Council Directive 2005/94/EC. (2006) 49:L237–1
34. De Wit JJ. Detection of infectious bronchitis virus. Avian Pathology. (2000) 29:71–93. doi: 10.1080/03079450094108
35. Feberwee A, Mekkes DR, De Wit JJ, Hartman EG, Pijpers A. Comparison of culture, PCR, and different serologic tests for detection of Mycoplasma gallisepticum and Mycoplasma synoviae infections. Avian Dis. (2005) 49:260–8. doi: 10.1637/7274-090804R
36. Dijkman R, Feberwee A, Landman WJM. Development, validation and field evaluation of a quantitative real-time PCR able to differentiate between field Mycoplasma synoviae and the MS-H-live vaccine strain. Avian Pathol. (2017) 46:403–15. doi: 10.1080/03079457.2017.1296105
37. Beerens N, Heutink R, Bergervoet SA, Harders F, Bossers A, Koch G. Multiple Reassorted Viruses as Cause of Highly Pathogenic Avian Influenza A(H5N8) Virus Epidemic, the Netherlands, 2016. Emerg Infect Dis J. (2017) 23:1974–81. doi: 10.3201/eid2312.171062
38. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/molbev/mst010
39. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
40. Staatscourant. Regeling van de Minister van Economische Zaken van 5 Juni 2014, nr. WJZ/14045056, Houdende Wijziging Van Diverse Regelingen in Verband Met de Overname Van Taken Van de Bedrijfslichamen. No. 11860. The Hague: Dutch Government (2014).
41. Landman WJM, Germeraad EA, Kense MJ. An avian influenza virus H6N1 outbreak in commercial layers: case report and reproduction of the disease. Avian Pathol. (2018) 1–35. doi: 10.1080/03079457.2018.1551612
42. Elbers ARW, Gonzales JL. Quantification of visits of wild fauna to a commercial free-range layer farm in the Netherlands located in an avian influenza hot-spot area assessed by video-camera monitoring. Transbound Emerg Dis. (2019) 67:661–77. doi: 10.1111/tbed.13382
43. De Minister Van Landbouw Natuur En Voedselkwaliteit. Regeling van 7 juni 2005, nr. TRCJZ/2005/1411 Regeling Preventie, Bestrijding en Monitoring van Besmettelijke Dierziekten en Zoönosen en TSE's. (2005). Available online at: http://wetten.overheid.nl/BWBR0018397/2018-01-01#Hoofdstuk3 (accessed September 1, 2019).
44. Gonzales JL, Elbers ARW. Effective thresholds for reporting suspicions and improve early detection of avian influenza outbreaks in layer chickens. Sci Rep. (2018) 8:8533. doi: 10.1038/s41598-018-26954-9
45. Samy A, Naguib MM. Avian respiratory coinfection and impact on avian influenza pathogenicity in domestic poultry: field and experimental findings. Vet Sci. (2018) 5:23. doi: 10.3390/vetsci5010023
46. Brugh M, Beard CW. Influence of dietary calcium stress on lethality of avian influenza viruses for laying chickens. Avian Dis. (1986) 30:672–8. doi: 10.2307/1590566
47. Germeraad EA, Sanders P, Hagenaars TJ, Jong MCM, Beerens N, Gonzales JL. Virus shedding of avian influenza in poultry: a systematic review and meta-analysis. Viruses. (2019) 11:812. doi: 10.3390/v11090812
48. Boerderij. Laagpathogene H3N1 zorgt voor klinische verschijnselen. (2019). Available online at: https://www.boerderij.nl/Pluimveehouderij/Nieuws/2019/6/Laagpathogene-H3N1-zorgt-voor-klinische-verschijnselen-438764E/ (accessed December 1, 2019).
49. Keeler SP, Dalton MS, Cressler AM, Berghaus RD, Stallknecht DE. Abiotic factors affecting the persistence of avian influenza virus in surface waters of waterfowl habitats. Appl Environ Microbiol. (2014) 80:2910–7. doi: 10.1128/AEM.03790-13
50. Knmi. Maandoverzicht van Het Weer in Nederland. (2020). Available online at: https://www.knmi.nl/nederland-nu/klimatologie/gegevens/mow (accessed March 1, 2020).
Keywords: LPAIV, water, environmental sampling, poultry, outbreak
Citation: Germeraad EA, Elbers ARW, de Bruijn ND, Heutink R, van Voorst W, Hakze-van der Honing R, Bergervoet SA, Engelsma MY, van der Poel WHM and Beerens N (2020) Detection of Low Pathogenic Avian Influenza Virus Subtype H10N7 in Poultry and Environmental Water Samples During a Clinical Outbreak in Commercial Free-Range Layers, Netherlands 2017. Front. Vet. Sci. 7:237. doi: 10.3389/fvets.2020.00237
Received: 24 January 2020; Accepted: 07 April 2020;
Published: 05 May 2020.
Edited by:
Jesus Hernandez, Centro de Investigación en Alimentación y Desarrollo (CIAD), MexicoReviewed by:
El-Sayed M. Abdelwhab, Friedrich Loeffler Institute, GermanyTimm Harder, Friedrich Loeffler Institute, Germany
Copyright © 2020 Germeraad, Elbers, de Bruijn, Heutink, van Voorst, Hakze-van der Honing, Bergervoet, Engelsma, van der Poel and Beerens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nancy Beerens, bmFuY3kuYmVlcmVuc0B3dXIubmw=
†These authors have contributed equally to this work
 Evelien A. Germeraad
Evelien A. Germeraad Armin R. W. Elbers
Armin R. W. Elbers Naomi D. de Bruijn
Naomi D. de Bruijn Rene Heutink1
Rene Heutink1 Marc Y. Engelsma
Marc Y. Engelsma Wim H. M. van der Poel
Wim H. M. van der Poel Nancy Beerens
Nancy Beerens