- 1Henan Provincial Engineering Laboratory of Insects Bio-reactor, Henan Provincal Engineering and Technology Center of Health Products for Livestock and Poultry, China-UK-NYNU-RRes Joint Libratory of Insect Biology, Nanyang Normal University, Nanyang, China
- 2Department of Parasitology, School of Basic Medical Sciences, Wenzhou Medical University, Wenzhou, China
- 3Henan Provincal Engineering and Technology Center of Health Products for Livestock and Poultry, Key Laboratory of Ecological Security and Collaborative Innovation Centre of Water Security for Water Source Region of Mid-line of South-to-North Diversion Project of Henan Province, School of Agricultural Engineering, Nanyang Normal University, Nanyang, China
Bovine pestiviruses include Pestivirus A (BVDV-1), Pestivirus B (BVDV-2), and Pestivirus H, which was originally called HoBi-like pestivirus. We conducted an epidemiological investigation for pestiviruses circulating in backyard cattle farms in central China. RT-PCR assays and sequences analysis were conducted on 54 nasal swabs, 26 serum samples, and three lung samples from cattle with respiratory infections and identified 29 pestivirus strains, including 24 Pestivirus A and five Pestivirus H strains. Phylogenetic analysis based on partial 5′-UTR and Npro sequences showed that the genotypes of 24 Pestivirus A strains included Pestivirus A 1b (six isolates), Pestivirus A 1m (six isolates), Pestivirus A 1q (two isolates), Pestivirus A 1u (one isolates), and Pestivirus A 1o (nine isolates, a putative new sub-genotype). In addition, a single Pestivirus H agenotype included all five Pestivirus H strains. This study revealed extensive genetic variations within bovine pestivirus isolates derived from cattle in backyard farms in Central China, and this epidemiological information improves our understanding of the epidemics of bovine Pestiviruses, as well as will be useful in designing and evaluating diagnostic methods and developing more effective vaccines.
Introduction
Pestiviruses are single-stranded, positive-sense, enveloped RNA viruses with a genome of ~12.3 kb, which belong to the family Flaviviridae, genus Pestivirus. According to the proposed revision to its taxonomy, the Pestivirus genus includes 11 species, namely Pestivirus A (bovine viral diarrhea virus 1, BVDV-1), Pestivirus B (bovine viral diarrhea virus 2, BVDV-2), Pestivirus C (classical swine fever virus, CSFV) and Pestivirus D (border disease virus, BDV), Pestivirus E (pronghorn pestivirus), Pestivirus F (Bungowannah virus), Pestivirus G (giraffe pestivirus), Pestivirus H (Hobi-like pestivirus), Pestivirus I (Aydin-like pestivirus), Pestivirus J (rat pestivirus), and Pestivirus K (atypical porcine pestivirus) (1). Among these species, Pestivirus A, Pestivirus B, and Pestivirus H aroused great concern because these cause significant economic losses in the cattle industry worldwide (2–4). Pestivirus A and Pestivirus B are major viruses associated with a number of clinical manifestations that range from mild to severe in feedlot cattle, including respiratory disease, digestive disease, and/or reproductive system disturbances and suppression of the immune system (5–7). Natural infections in cattle involving Pestivirus H showed similar clinical signs as those of Pestivirus A or Pestivirus B infections (8–11).
To date, at least 23 genotypes of Pestivirus A (12–16) and six genotypes (17) of Pestivirus B have been classified based on sequence comparison analyses and the palindromic nucleotide substitutions (PNS) genotyping method (18, 19). Pestivirus H, first isolated from fetal calf serum (20), has spread to different continents, including North America, South America, Europe, and Asia (21–26). In China, nine genotypes of Pestivirus A (1a,1b, 1c,1d, 1m, 1o, 1p,1q, and 1u) (27–30), two genotypes (2a, 2b) of Pestivirus B (31–33), and Pestivirus H (24, 34) have been reported. Cattle production by backyard farming is a widespread cattle-keeping pattern in developing countries. In central China, which includes Henan Province, more than 3,720,000 cattle have been raised (35), and previous data showed that over 20% of cattle were kept in small farms (cattle number <10), including a large number of backyard farms in China (36), especially in the southern region of Henan Province, where free-range cattle farms are key economic sectors (35). However, the limited biosecurity measures in these farms usually lead to the introduction and spread of exotic or endemic disease (37–39). Furthermore, in backyard cattle farms in China, most of the animals graze in the wilderness, and thus come into contact with infected cattle. To our knowledge, information on the epidemiology of pestiviruses in cattle in backyard farms in China is limited. The aim of this study was to investigate the distribution of pestiviruses that are associated with respiratory disease from backyard farms in Henan Province, China.
Materials and Methods
Samples
From November 2014 to April 2019, a total of 54 nasal swabs and 26 serum samples were collected from different cattle in 41 backyard farms in Henan Province in Central China; these animals had never been vaccinated against Pestivirus A and were diagnosed with respiratory infections by rural veterinarians and treated with antibiotics for days, resulting in slow recovery. In addition, three lungs of deceased calves were collected in 2015, 2016, and 2018. All samples were stored at −80°C until analysis.
Primer Selection
The nested RT-PCR primers for genotyping bovine pestiviruses, including Pestivirus A, Pestivirus B, and Pestivirus H (40) were used to detect the pestivirus genome in the samples. For phylogenetic analysis, the BVDV-positive samples were further subjected to 5′-UTR and Npro RT-PCR using primers 324/326 (41) and BD1/BD2 (42), respectively. Because the sequences of Pestivirus H-positive samples were most closely related to the HN1507 strain (43), the positive samples further subjected to RT-PCR covering a partial 5′-UTR fragment and the entire Npro region with the primers HN-F (sense; 5′-CCTTCAGTAGGACGAGCATAA-3′) and HN-R (antisense; 5′- AGACGGGCTATACCACAATAA-3′), corresponding to nt 109–1,107 of Pestivirus H strain HN1507 (GenBank accession number: KU563155).
RNA Extraction, Amplification, and Sequencing
The three lung samples were first homogenized, then RNA was extracted from the lung homogenates, nasal swabs, and serum samples using an EasyPure Viral DNA/RNA Kit (Transgen Biotech, China) according to the manufacturer's instructions. The RNA was resuspended in DEPC-treated water and kept until analysis. cDNA was synthesized from RNA using Easyscript Reverse Transcriptase kit (Transgen Biotech, China) using random 9-mers as reverse transcription primer.
nRT-PCR to detect the pestivirus genome was performed as described elsewhere (40). Then, the BVDV-positive samples were further subjected to 5′-UTR and Npro RT-PCR earlier described (41, 42). The Pestivirus H-positive samples were subjected to RT-PCR in a 50-μL reaction mixture similar to the Pestivirus A reaction mixture according using the following conditions: reverse transcription at 50°C for 60 min, then denaturation at 93°C for 3 min; followed by 30 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 1 min; and a final extension at 72°C for 10 min. Then the amplified products were recovered from the agarose gel using a gel extraction kit (Omega Bio-Tek, China), and the purified amplicons were directly sequenced in both directions using an ABI automated A373 sequencer (ABI, USA). Lastly, all of the sequences were compared to the NCBI databases using a BLAST search.
Phylogenetic Analysis
The nucleotide regions of the 5′-UTR were compared and aligned using CLUSTAL W program. Molecular Evolutionary Genetics Analysis version 6 (MEGA6) (44) was used for phylogeny inference according to the neighbor-joining criterion and the Kimura 2-parameter model. The robustness of the hypothesis was tested with 1000 non-parametric bootstrap analyses.
Following strains were used for 5′-UTR Phylogenetic analysis: NADL [M31182] and Singer [L32875] are the references for the Pestivirus A 1a genotype, strains Osloss [M96687], and Draper [L32880] are the references for the Pestivirus A 1b genotype and strains Europa [AB000898], and F [AF298065] are the reference for the Pestivirus A 1.3 genotype. Strains 438/02 [AY159540], PT42-03 [AY944293], 23-15 [AF298059], so CP/75 [AB042661], Shitara-02-06 [LC089876], IS25CP/01 [AB359931], AQGN96B15 [AB300691], Bega [AF049221], Manasi [EU159702], KM [AF298068], G [AF298066], SD0803 [JN400273], isolate 6 [JX276543], 10-84 [AF298054], 3186V6 [AF298062], 11207/98 [AJ304390], 22146/81 [AJ304376], 2561 [JQ920287], 17P [AF244954], KS86-1ncp [AB042713], Deer [AB040132], TR70 [MG670547], TR75 [MG670549], ZM-95[AF526381], TJ0801 [GU120255], BJ1305 [KF925505], XZ-24 [KJ578918], TR-2007-Gu-175454-4695 [EU716150], TR16 [MG670548], TR72 [MG670546], J [AF298067], W [AF298073], BJ0702 [GU120248], BJ0703 [GU120249], A [AF298064], L [AF298069], CH-01-08 [EU180024], 71-03 [KF205294], PG/13a/07 [not deposited], GXBH-EB34 [KJ578813], GXLZ-BB4 [KJ578814], 130/15-4215 [KY085998], 130/15-5364 [KY085999], Rebe [AF299317], SuwaCp [AF117699], SuwaNcp [AF117700], CH-05-b1 [EU180030], and S153 [KF006964] are references for the Pestivirus A 1.4 to Pestivirus A 1.23.
Following strains were used for Npro Phylogenetic analysis: NADL [M31182], Oregon C24V [AF091605] and SD-1 [M96751] are the references for the Pestivirus A 1a genotype, strain Osloss [M96687] is the reference for the Pestivirus A 1b genotype. Strains F [AF287284], 10JJSKR [KC757383], 23/15 [AF287279], 58-1 [KF023454], 2541 [JQ920342], so CP/75 [AB105590] are references for the Pestivirus A 1.3, 1.5 and 1.6 genotypes. Strains IS25CP/01 [AB359931], IS26NCP/01 [AB359932], Bega [AF049221], 519 [AF144464], Deer-NZ1 [U80903], G [AF287285], CH-SM09/20 [AY895007], SD0803 [JN400273], isolate 6 [KC207072], 3186V6 [AF287282] and 26-V639 [AF287282] are references for the Pestivirus A 1.7 to Pestivirus A 1.11 genotypes. Strains Deer-GB1 [U80902] and KS86-1ncp [AB078950] are references for the genotypes Pestivirus A 1.13. Strains TR70 [KF154779], TR73 [KF154777] and TR75 [KF154778], reported as genotype R (Yesilbag et al., 2014), are references for genotype 1.14. Strains BJ1305 [KF925522], TJ0801 [GU120262] and ZM-95 [AF526381] are references for the genotype Pestivirus A 1.15. Strains TR16 [EU163964], TR27 [EU163975], TR29 [EU163977] and TR72 [KF154776] are references for the genotype Pestivirus A 1.16. Strains J [AF287286], W [AF287290], BJ0701 [GU120259], BJ0702 [GU120260], BJ0703 [GU120261], A [AF287283], L [AF287287], CH-01-08 [EU180033], 71-03 [KF205326], M31182 [JQ799141], 441/09 [KY040435], CH-Bohni [AY894997] and CH-Suwa [AY894998] are references for the Pestivirus A 1.17 to Pestivirus A 1.22 genotypes.
Results
Using first-round nRT-PCR to identify bovine pestivirus by amplification of a 1,013-bp fragment, 83 samples were screened and the pestivirus genome was detected in 17 out of the 54 nasal swab samples, 10 out of the 26 serum samples, and two out of three lung samples. The results of the second-round nRT-PCR showed that in 17 positive nasal swab samples, 14 were positive for Pestivirus A and the other three were positive for Pestivirus H; in the 10 positive serum samples, nine were positive for Pestivirus A and one was positive for Pestivirus H; in the two positive lung samples, one was positive for Pestivirus A and the other was positive for Pestivirus H; no Pestivirus B-positive samples were detected. The 29 pestivirus-positive samples are presented in Figure 1.
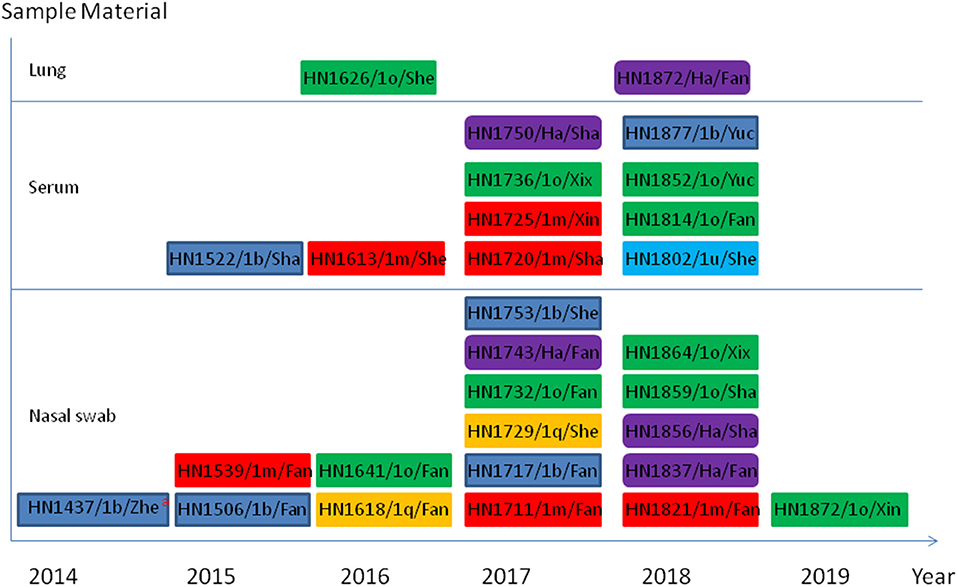
Figure 1. Schematic representation of 29 bovine Pestivirus positive in this work. aindicate the information of sample including sample number/genotype/county. The county names: Zhe, Zhenping; Fan, Fangcheng; Sha, Shangcai; She, Sheqi; Xin, Xinye; Xix, Xixia; Yuc, Yucheng. The blue rectangle represents Peativirus A 1b genotype; the red rectangle represents Peativirus A 1m genotype; the yellow rectangle represents Peativirus A 1q genotype; the green rectangle represents Peativirus A 1o genotype; the light blue rectangle represents Peativirus A 1u genotype; the purple rounded rectangle represents Peativirus H a genotype.
The sequences detected by 5′-UTR and Npro RT-PCR in 24 BVDV-positive samples were deposited in GenBank under accession numbers: MN442360–MN442383 and MN442389–MN442412. Sequence alignment of the 5′-UTR and Npro region of the 24 samples using CLUSTAL W indicated a sequence identity within the range 82.0–100% and 70.0–99.8%, respectively. BLAST analysis of the 5′-UTR and Npro sequences showed that all 24 BVDV-positive isolates belonged to Pestivirus A. The comparative analysis among Henan isolates and the reference strains of Pestivirus A (NADL, VEDEVAC) shared a 5′-UTR and Npro region sequence identity within the range 82.9–98.8%, 72.2–97.6%, respectively.
The sequences detected by 5′-UTR and Npro RT-PCR in the five Pestivirus H-positive samples were deposited in GenBank as accession numbers MN442384–MN442388 and MN442413–MN442417. Sequence alignment using CLUSTAL W program of the five samples revealed a 5′-UTR and Npro region respective sequence identity within the range 95.8–99.5% and 98.4–99.6%. BLAST analysis of the 5′-UTR and Npro sequences showed that all five Pestivirus H-positive isolates belonged to Pestivirus H. Comparative analysis of the 5′-UTR and Npro region of the Henan isolates and the reference strains of Pestivirus H (TH/04_KhonKaen, HN1507) revealed a sequence identity within the range of 87.5–99.5% and 90.3.2–99.4%, respectively. In particular, the nucleotide homologies between these isolates and the other strain (HN1507) (24, 43) isolated from goat in the same area were 97.9±1.6% and 98.6 ± 0.4% in the above two regions.
All 24 isolates from the BVDV-positive samples were classified as Pestivirus A, and on the basis of phylogenetic analysis of 5′-UTR and Npro genes (Figure 2) further classified into five genotypes: Pestivirus A 1b (six isolates), Pestivirus A 1m (six isolates), Pestivirus A 1q (two isolates), Pestivirus A 1u (one isolate), and the other nine Pestivirus A isolates cluster in the same genotype with Chinese strain XH-2 which was assigned as Pestivirus A 1o, but phylogenetic analysis showed this cluster of isolates were under different cluster from other Pestivirus A 1o strains, further analyzed by the PNS software which available at www.pns-software.com (45), these cluster should be members of a new sub-genotype (1.7.2) within the genotype 1o (1.7). In addition, five isolates from Pestivirus H-positive samples were classified as Pestivirus H, and all further were classified into genotype “Pestivirus H a” based on the results of phylogenetic analysis of the 5′-UTR and Npro genes (46) (Figure 2).
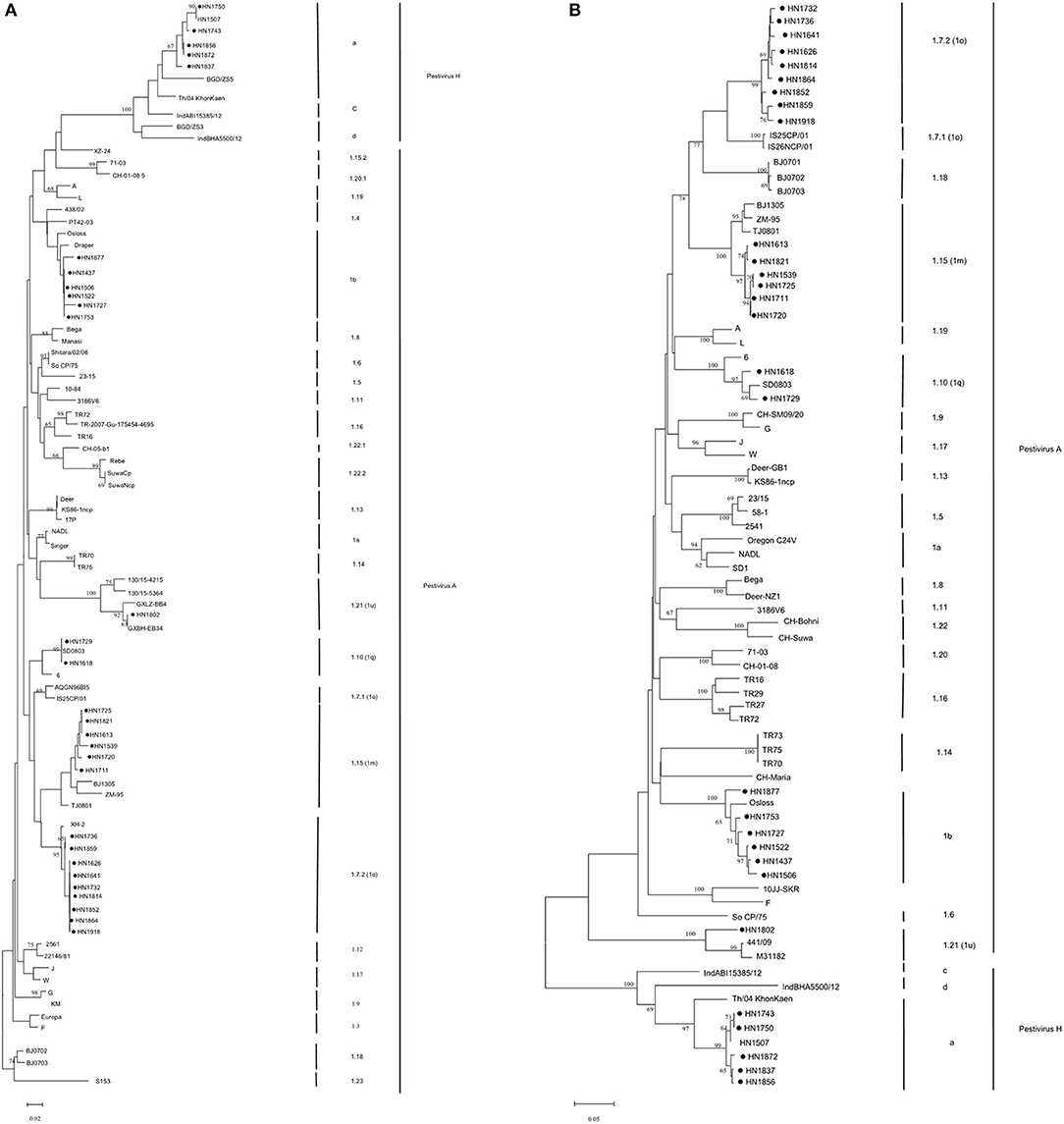
Figure 2. Phylogenetic analysis of bovine Pestivirus from cattle in backyard farms in Central China, and reference strains using the 5′-UTR (A) and Npro (B) sequences. Bootstrap analyses that were supported by >60% of 1,000 replicates are indicated in nodes. Henan pestivirus isolates in this work are highlighted with a symbol (•).
Discussion
China is one of the countries that have the largest domesticated ruminant population in the world, including a large number of backyard farms (35). In 2017, commercial Pestivirus A or Pestivirus B vaccines were released to the market. However, vaccination is not mandatory, and awareness of the importance of immunization to prevent Pestivirus infections among backyard farm keepers is rare, despite the occurrence of pestivirus epidemics in large-scale farms in China in recent years (27–29, 47). It is thus essential to investigate the genetic diversity of pestiviruses in backyard farms. Furthermore, the new genotype pestivirus might result in the immune failure of pestivirus vaccine (48). For these above reasons, the genetic diversity of 29 pestivirus-positive samples derived from infected calves in backyard farms from central China was investigated by phylogenetic analysis of the 5′-UTR and Npro partial genomic regions.
The results of phylogenetic analysis showed that Pestivirus A−1o, Pestivirus A−1b, and Pestivirus A−1m were the predominant genotypes in our samples, followed by Pestivirus A−1q and Pestivirus A−1u. Furthermore, among these three predominant genotypes, Pestivirus A−1b and Pestivirus A−1m are frequently reported in China (27–30, 49, 50) the Pestivirus A 1a found in Henan province (51) was not detected in this work.
Six isolates of the Pestivirus A 1b genotype were detected in Zhenping, Fangcheng, Shangcai, Sheqi, and Yucheng in this study (Figures 1, 2). This widespread distribution of Pestivirus A is not surprising, as the first genotype of Pestivirus A was isolated in China in 1983. Subsequently, Pestivirus A−1b was later detected in most of the provinces, including Henan Province and the neighboring provinces numerous reports (27–29, 47, 49, 52–55). Meanwhile, a recent study has shown that 31.6% (2193:6939) of the corresponding Pestivirus A isolates around the world were Pestivirus A−1b (16). These reports indicate the strong spreading ability of Pestivirus A−1b among large-scale farms, and the high detection rate (6/24) in our study also suggests that Pestivirus A−1b is a predominant genotype among backyard farms.
Six isolates of the Pestivirus A 1m genotype were detected in Fangcheng, Sheqi, Shangcai, and Xinye in this study (Figures 1, 2). The first Pestivirus A−1m strain ZM-95 in China was isolated from pigs in 1995 (56). Subsequent reports revealed that Pestivirus A−1m is emerging in most of provinces and is considered to be a predominant Pestivirus A genotype in herds (27, 30, 50, 54, 57), and also detected in Henan province and neighbor province (29). In addition, sequences of Pestivirus A−1m strains in different regions showed high-nucleotide homology, indicating that these strains share the same origin (27, 49). Recently, other surveys on goats uncovered that the Pestivirus A−1m could infect goats naturally and cause diarrhea (58). In this study, Pestivirus A 1m strains were detected in different backyard farms that shared grassland with goats, this feeding method provided more chances for interspecies transmission of BVDV-1m, further accelerating the evolution of these viruses and more widely spreading disease.
The other nine Pestivirus A isolates shared the highest sequence identities (97–98%) in strains such as XH-6, XH-5, XH-1, XH-2, and BJ09 that were isolated from other provinces in China which was assigned as Pestivirus A 1o. Furthermore, the Npro sequences of these strains were not found in GenBank, and thus, confirmation could not be done based on the Npro phylogenetic tree analysis. The Npro sequences of the 21 isolates were also analyzed by BLAST, and the highest identities (86–91%) were observed in strain IS26/01ncp from Japan, and BJ0703, BJ0702, BJ0701, and JS12/02 that were isolated from other provinces including the Jiangsu province near to the Henan province in China and were classified as BVDV-1o or BVDV-1p (27, 58, 59). Then the PNS method was used to analyze the existing strain such as XH-2, then the genotype Pestivirus A 1.7 (1o) was verified, but the cluster in phylogenetic tree this cluster was in different cluster from other Pestivirus A 1.7 (1o) strains, this result indicated these strains form a new sub-genotype (1.7.2). (Figures 2A,B). Pestivirus A 1o was first isolated from a calf that developed a mucosal disease and from PI calves in Japan (60), and has been detected in camels, goats, and pigs in China (30, 50, 58). In this study the new sub-genotype Pestivirus A 1o were detected in different backyard farms in a few of counties, and the Pestivirus A 1o could infect goats and sheep, it is in need that necessary measures should be taken to avoid this new sub-genotype Pestivirus A 1o spread in other hosts.
Five isolates of the Pestivirus H “a” genotype were detected in Fangcheng and Shangcai in this study (Figures 1, 2). Before this research, Pestivirus H has been previously detected in goats and sheep in Fangcheng (24), and phylogenetic analysis showed that these five isolates were closely related to the HN1507 strain isolated from goat (43). These results indicated that these isolates shared the same origin. Furthermore, considering that these strains were all isolated from animals raised in backyard farms where livestock commonly grazed in the mix, this specific feeding method provides a convenient route for interspecies transmission of Pestivirus H. To date, in China, Pestivirus H has been reported in contaminated cells, commercial FCS, goats, and sheep (24, 34, 61). This study showed that Pestivirus H could be detected in cattle immediately after being detected in goats and sheep in Central China (24).
Bovine pestivirus isolates in backyard farms exhibited a high level of genetic diversity, as indicated in the novel epidemic genotypes of Pestivirus A and Pestivirus H first emerging in cattle in China. These results indicate that backyard cattle farms could be a special reservoir for the evolution of bovine Pestivirus and provide an important complement to understand the epidemics of bovine Pestivirus. Furthermore, this study will be useful in designing and evaluating diagnostic methods and in developing more effective vaccines.
Conclusion
Several genotypes of Pestivirus A and Pestivirus H infections were identified in cattle with respiratory diseases and kept in backyard farms by RT-PCR, sequencing, and phylogenetic analysis. This is the first report on the molecular evidences on natural infections of Pestivirus H in cattle in China.
Data Availability Statement
Datasets are in a publicly accessible repository: The datasets generated for this study can be found in GenBank: https://www.ncbi.nlm.nih.gov/genbank/. The Genbank accession numbers are mentioned in the Results of the article.
Ethics Statement
The processes of nasal swabs and blood from cattle were approved by their hosts, and all lungs were from animals found dead in the study. The study was approved by the Animal Welfare and Ethics Committee of Nanyang Normal University (No 14027).
Author Contributions
HS participated in the design of the study, and drafted the main parts of the manuscript. HL, YZ, and LYan participated in the sample collection and PCR detection. YH, ZW, LD, and CL participated in the data analyzing. BY and LYao participated in revised the manuscript and supervised the project.
Funding
The National Natural Science Foundation of China (Grant no. 31902263 and 31870917), the program for Innovative Research Team of Science and Technology in University of Henan Province (No. 20IRTSTHN024), the Henan Provincial Scientific and technological research project (Grant no. 182102110084) and Nanyang Normal University (CN) (Grant no. 15081) supported this study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
References
1. Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Scott Muerhoff A, et al. Proposed revision to the taxonomy of the genus pestivirus, family flaviviridae. J Gen Virol. (2017) 98:2106–12. doi: 10.1099/jgv.0.000873
2. Decaro N, Lucente MS, Losurdo M, Larocca V, Elia G, Occhiogrosso L, et al. HoBi-like pestivirus and its impact on cattle productivity. Transbound Emerg Dis. (2016) 63:469–73. doi: 10.1111/tbed.12529
3. Gates MC, Humphry RW, Gunn GJ, Woolhouse ME. Not all cows are epidemiologically equal: quantifying the risks of bovine viral diarrhoea virus (BVDV) transmission through cattle movements. Vet Res. (2014) 45:110. doi: 10.1186/s13567-014-0110-y
4. Pinior B, Firth CL, Richter V, Lebl K, Trauffler M, Dzieciol M, et al. A systematic review of financial and economic assessments of bovine viral diarrhea virus (BVDV) prevention and mitigation activities worldwide. Prev Vet Med. (2017) 137(Pt. A):77–92. doi: 10.1016/j.prevetmed.2016.12.014
5. Scharnbock B, Roch FF, Richter V, Funke C, Firth CL. A meta-analysis of Bovine Viral Diarrhoea Virus (BVDV) prevalences in the global cattle population. Sci Rep. (2018) 8:14420. doi: 10.1038/s41598-018-32831-2
6. Obritzhauser W, Baumgartner W, Kasbohrer A, Pinior B, Hancock AS, Younis PJ, et al. Infectious reproductive disease pathogens in dairy herd bulls. Sci Rep. (2015) 93:349–53. doi: 10.1038/s41598-018-32831-210.1111/avj.12369
7. Silveira S, Weber MN, Mosena AC, da Silva MS, Streck AF, Pescador CA, et al. Genetic diversity of brazilian bovine pestiviruses detected between 1995 and 2014. Transbound Emerg Dis. (2017) 64:613–23. doi: 10.1111/tbed.12427
8. Decaro N, Lanave G, Lucente MS, Mari V, Varello K, Losurdo M, et al. Mucosal disease-like syndrome in a calf persistently infected by Hobi-like pestivirus. J Clin Microbiol. (2014) 52:2946–54. doi: 10.1128/JCM.00986-14
9. Decaro N, Mari V, Pinto P, Lucente MS, Sciarretta R, Cirone F, et al. Hobi-like pestivirus: both biotypes isolated from a diseased animal. J Gen Virol. (2012) 93:1976–83. doi: 10.1099/vir.0.044552-0
10. Decaro N, Lucente MS, Mari V, Sciarretta R, Pinto P, Buonavoglia D, et al. Hobi-like pestivirus in aborted bovine fetuses. J Clin Microbiol. (2012) 50:509–12. doi: 10.1128/JCM.05887-11
11. Decaro N, Lucente MS, Mari V, Cirone F, Cordioli P, Camero M, et al. A typical pestivirus and severe respiratory disease in calves, Europe. Emerg Infect Dis. (2011) 17:1549–52. doi: 10.3201/eid1708.101447
12. Booth RE, Thomas CJ, El-Attar LM, Gunn G, Brownlie J. A phylogenetic analysis of Bovine Viral Diarrhoea Virus (BVDV) isolates from six different regions of the UK and links to animal movement data. Vet Res. (2013) 44:43. doi: 10.1186/1297-9716-44-43
13. Giangaspero M, Yesilbag K, Apicella C. Who's who in the Bovine viral diarrhea virus type 1 species: genotypes L and R. Virus Res. (2018) 256:50–75. doi: 10.1016/j.virusres.2018.07.009
14. Vilcek S, Durkovic B, Kolesarova M, Paton DJ. Genetic diversity of BVDV: consequences for classification and molecular epidemiology. Prev Vet Med. (2005) 72:31–5. doi: 10.1016/j.prevetmed.2005.08.004
15. Vilcek S, Paton DJ, Durkovic B, Strojny L, Ibata G, Moussa A, et al. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol. (2001) 146:99–115. doi: 10.1007/s007050170194
16. Yesilbag K, Alpay G, Becher P. Variability and global distribution of subgenotypes of bovine viral diarrhea virus. Viruses. (2017) 9:128. doi: 10.3390/v9060128
17. Giangaspero M, Decaro N, Turno P, Apicella C, Gargano P, Buonavoglia C. Pathogen spread and globalization: the case of pestivirus heterogeneity in southern Italy. Res Vet Sci. (2019) 125:100–12. doi: 10.1016/j.rvsc.2019.05.017
18. Harasawa R, Giangaspero M. A novel method for pestivirus genotyping based on palindromic nucleotide substitutions in the 5'-untranslated region. J Virol Methods. (1998) 70:225–30. doi: 10.1016/s0166-0934(97)00180-8
19. Giangaspero M, Harasawa R. Numerical taxonomy of the genus pestivirus based on palindromic nucleotide substitutions in the 5' untranslated region. J Virol Methods. (2007) 146:375–88. doi: 10.1016/j.jviromet.2007.07.009
20. Schirrmeier H, Strebelow G, Depner K, Hoffmann B, Beer M. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J Gen Virol. (2004) 85:3647–52. doi: 10.1099/vir.0.80238-0
21. Silveira S, Falkenberg SM, Elderbrook MJ, Sondgeroth KS, Dassanayake RP, Neill JD, et al. Serological survey for antibodies against pestiviruses in Wyoming domestic sheep. Vet Microbiol. (2018) 219:96–9. doi: 10.1016/j.vetmic.2018.04.019
22. Flores EF, Cargnelutti JF, Monteiro FL, Bauermann FV, Ridpath JF, Weiblen R. A genetic profile of bovine pestiviruses circulating in Brazil (1998–2018). Anim Health Res Rev. (2018) 19:134–41. doi: 10.1017/s1466252318000130
23. Lanave G, Decaro N, Lucente MS, Guercio A, Cavaliere N, Purpari G, et al. Circulation of multiple subtypes of bovine viral diarrhoea virus type 1 with no evidence for HoBi-like pestivirus in cattle herds of southern Italy. Infect Genet Evol. (2017) 50:1–6. doi: 10.1016/j.meegid.2017.02.009
24. Shi H, Kan Y, Yao L, Leng C, Tang Q, Ji J, et al. Identification of natural infections in sheep/Goats with HoBi-like pestiviruses in China. Transbound Emerg Dis. (2016) 63:480–4. doi: 10.1111/tbed.12551
25. Mishra N, Rajukumar K, Pateriya A, Kumar M, Dubey P, Behera SP, et al. Identification and molecular characterization of novel and divergent HoBi-like pestiviruses from naturally infected cattle in India. Vet Microbiol. (2014) 174:239–46. doi: 10.1016/j.vetmic.2014.09.017
26. Decaro N, Mari V, Lucente MS, Sciarretta R, Elia G, Ridpath JF, et al. Detection of a Hobi-like virus in archival samples suggests circulation of this emerging pestivirus species in Europe prior to (2007). Vet Microbiol. (2013) 167:307–13. doi: 10.1016/j.vetmic.2013.09.006
27. Xue F, Zhu YM, Li J, Zhu LC, Ren XG, Feng JK, et al. Genotyping of bovine viral diarrhea viruses from cattle in China between 2005 and (2008). Vet Microbiol. (2010) 143:379–83. doi: 10.1016/j.vetmic.2009.11.010
28. Gong X, Liu L, Zheng F, Chen Q, Li Z, Cao X, et al. Molecular investigation of bovine viral diarrhea virus infection in yaks (Bos gruniens) from Qinghai, China. Virol J. (2014) 11:29. doi: 10.1186/1743-422x-11-67
29. Deng M, Ji S, Fei W, Raza S, He C, Chen Y, et al. Prevalence study and genetic typing of bovine viral diarrhea virus (BVDV) in four bovine species in China. PLoS ONE. (2015) 10:e0121718. doi: 10.1371/journal.pone.0121718
30. Gao S, Luo J, Du J, Lang Y, Cong G, Shao J, et al. Serological and molecular evidence for natural infection of Bactrian camels with multiple subgenotypes of bovine viral diarrhea virus in Western China. Vet Microbiol. (2013) 163:172–6. doi: 10.1016/j.vetmic.2012.12.015
31. Zhu LQ, Lin YQ, Ding XY, Ren M, Tao J, Wang JY, et al. Genomic sequencing and characterization of a Chinese isolate of Bovine viral diarrhea virus 2. Acta Virol. (2009) 53:197–202. doi: 10.4149/av_2009_03_197
32. Tao J, Wang Y, Wang J, Wang JY, Zhu GQ. Identification and genetic characterization of new bovine viral diarrhea virus genotype 2 strains in pigs isolated in China. Virus Genes. (2013) 46:81–7. doi: 10.1007/s11262-012-0837-3
33. Zhu LQ, Ren M, Lin YQ, Ding XY, Zhang GP, Zhao X, et al. Identification of a bovine viral diarrhea virus 2 isolated from cattle in China. Acta Virol. (2009) 53:131–4. doi: 10.4149/av_2009_02_131
34. Mao L, Li W, Zhang W, Yang L, Jiang J. Genome sequence of a novel Hobi-like pestivirus in China. Virol J. (2012) 86:12444. doi: 10.1128/JVI.02159-12
35. N. National Bureau of statistics of China. No. of Livestock and Poultry. (2017). Available online at: http://data.stats.gov.cn/easyquery.htm?cn=E0103
36. A. Agricultural Commodity Production Cost and Return Data. Agricultural Commodity Production Cost and Return Data. Beijing: China Statistical Publisher (2009).
37. Pires FA, Peterson A, Baron JN, Adams R, Martinez-Lopez B, Moore D. Small-scale and backyard livestock owners needs assessment in the western United States. PLoS ONE. (2019) 14:e0212372. doi: 10.1371/journal.pone.0212372
38. Hernandez-Jover M, Schembri N, Holyoake PK, Toribio JL, Martin PA. A comparative assessment of the risks of introduction and spread of foot-and-mouth disease among different pig sectors in Australia. Front Vet Sci. (2016) 3:85. doi: 10.3389/fvets.2016.00085
39. Kuster K, Cousin ME, Jemmi T, Schupbach-Regula G, Magouras I. Expert opinion on the perceived effectiveness and importance of on-farm biosecurity measures for cattle and swine farms in Switzerland. PLoS ONE. (2015) 10:e0144533. doi: 10.1371/journal.pone.0144533
40. Decaro N, Sciarretta R, Lucente MS, Mari V, Amorisco F, Colaianni ML, et al. A nested PCR approach for unambiguous typing of pestiviruses infecting cattle. Mol Cell Probes. (2012) 26:42–6. doi: 10.1016/j.mcp.2011.11.003
41. Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol. (1994) 136:309–23.
42. Vilcek S, Nettleton PF, Paton DJ, Belak S. Molecular characterization of ovine pestiviruses. J Gen Virol. (1997) 78:725–35. doi: 10.1099/0022-1317-78-4-725
43. Shi H, Leng C, Xu Q, Shi H, Sun S, Kan Y, et al. Characterization of a pestivirus H isolate originating from goats. Virus Genes. (2018) 54:603–7. doi: 10.1007/s11262-018-1579-7
44. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. (2013) 30:2725–9. doi: 10.1093/molbev/mst197
45. Giangaspero M, Apicella C, Harasawa R. Numerical taxonomy of the genus pestivirus: new software for genotyping based on the palindromic nucleotide substitutions method. J Virol Methods. (2013) 192:59–67. doi: 10.1016/j.jviromet.2013.04.023
46. Silveira S, Baumbach LF, Weber MN, Mosena ACS, da Silva MS, Cibulski SP, et al. HoBi-like is the most prevalent ruminant pestivirus in Northeastern Brazil. Transbound Emerg Dis. (2018) 65:e113–20. doi: 10.1111/tbed.12689
47. Gong X, Cao X, Zheng F, Chen Q, Zhou J, Yin H, et al. Identification and characterization of a novel subgenotype of bovine viral diarrhea virus isolated from dairy cattle in Northwestern China. Virus Genes. (2013) 46:375–6. doi: 10.1007/s11262-012-0861-3
48. Ridpath JF. Bovine viral diarrhea virus: global status. Vet Clin North Am Food Anim Pract. (2010) 26:105–21. doi: 10.1016/j.cvfa.2009.10.007
49. Weng XG, Song QJ, Wu Q, Liu MC, Wang ML, Wang JF. Genetic characterization of bovine viral diarrhea virus strains in Beijing, China and innate immune responses of peripheral blood mononuclear cells in persistently infected dairy cattle. J Vet Sci. (2015) 16:491–500. doi: 10.4142/jvs.2015.16.4.491
50. Deng Y, Sun CQ, Cao SJ, Lin T, Yuan SS, Zhang HB, et al. High prevalence of bovine viral diarrhea virus 1 in Chinese swine herds. Vet Microbiol. (2012) 159:490–3. doi: 10.1016/j.vetmic.2012.04.023
51. Wang W, Shi X, Tong Q, Wu Y, Xia MQ, Ji Y, et al. A bovine viral diarrhea virus type 1a strain in China: isolation, identification, and experimental infection in calves. Virol J. (2014) 11:8. doi: 10.1186/1743-422x-11-8
52. Zhang S, Tan B, Ding Y, Wang F, Guo L, Wen Y, et al. Complete genome sequence and pathogenesis of bovine viral diarrhea virus JL-1 isolate from cattle in China. BioMed Res Int. (2014) 11:67. doi: 10.1155/2014/147145/10.1186/1743-422x-11-67
53. Xie Z, Fan Q, Xie Z, Liu J, Pang Y, Deng X, et al. Complete genome sequence of a bovine viral diarrhea virus strain isolated in southern china. Genome Announc. (2014) 2:e00512–14. doi: 10.1128/genomeA.00512-14
54. Zhong F, Li N, Huang X, Guo Y, Chen H, Wang X, et al. Genetic typing and epidemiologic observation of bovine viral diarrhea virus in Western China. Virus Genes. (2011) 42:204–7. doi: 10.1007/s11262-010-0558-4
55. Wang L, Wu X, Wang C, Song C, Bao J, Du J. Origin and transmission of bovine viral diarrhea virus type 1 in China revealed by phylodynamic analysis. Res Vet Sci. (2020) 128:162–9. doi: 10.1016/j.rvsc.2019.11.015
56. Wang X, Tu C, Li H, Jin K, Xuan H, Chang G, et al. Detection and isolation of bovine viral diarrhea virus from classical swine fever suspected pigs. Chinese J Vet Sci. (1996) 16:341–5.
57. Zhu L, Lu H, Cao Y, Gai X, Guo C, Liu Y, et al. Molecular characterization of a novel Bovine viral diarrhea virus isolate SD-15. PLoS ONE. (2016) 11:e0165044. doi: 10.1371/journal.pone.0165044
58. Mao L, Li W, Yang L, Wang J, Cheng S, Wei Y, et al. Primary surveys on molecular epidemiology of bovine viral diarrhea virus 1 infecting goats in Jiangsu province, China. BMC Vet Res. (2016) 12:181. doi: 10.1186/s12917-016-0820-7
59. Sato A, Tateishi K, Shinohara M, Naoi Y, Shiokawa M, Aoki H, et al. Complete genome sequencing of bovine viral diarrhea virus 1, subgenotypes 1n and 1o. Genome Announc. (2016) 4:e01744–15. doi: 10.1128/genomeA.01744-15
60. Nagai M, Hayashi M, Itou M, Fukutomi T, Akashi H, Kida H, et al. Identification of new genetic subtypes of bovine viral diarrhea virus genotype 1 isolated in Japan. Virus Genes. (2008) 36:135–9. doi: 10.1007/s11262-007-0190-0
Keywords: Pestivirus A, Pestivirus H, cattle, genotype, China
Citation: Shi H, Li H, Zhang Y, Yang L, Hu Y, Wang Z, Duan L, Leng C, Yan B and Yao L (2020) Genetic Diversity of Bovine Pestiviruses Detected in Backyard Cattle Farms Between 2014 and 2019 in Henan Province, China. Front. Vet. Sci. 7:197. doi: 10.3389/fvets.2020.00197
Received: 03 January 2020; Accepted: 25 March 2020;
Published: 17 April 2020.
Edited by:
Satoshi Sekiguchi, University of Miyazaki, JapanReviewed by:
Norikazu Isoda, Hokkaido University, JapanMassimo Giangaspero, Independent Researcher, Teramo, Italy
Copyright © 2020 Shi, Li, Zhang, Yang, Hu, Wang, Duan, Leng, Yan and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongfei Shi, a2NuMUAxNjMuY29t; NTA0MjM5MDMxQHFxLmNvbQ==; Baolong Yan, MTk4MnlibGxvZ0AxNjMuY29t; Lunguang Yao, bHVuZ3Vhbmd5YW9AMTYzLmNvbQ==
 Hongfei Shi
Hongfei Shi Huan Li1
Huan Li1 Baolong Yan
Baolong Yan