
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 03 March 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.00094
Bovine spongiform encephalopathy (BSE) is a prion disease in cattle and is classified into the classical type (C-BSE) and two atypical BSEs, designated as high type (H-BSE) and low type (L-BSE). These classifications are based on the electrophoretic migration of the proteinase K-resistant core (PrPres) of the disease-associated form of the prion protein (PrPd). In a previous study, we succeeded in transmitting the H-BSE prion from cattle to TgHaNSE mice overexpressing normal hamster cellular PrP (PrPC). Further, Western blot analysis demonstrated that PrPres banding patterns of the H-BSE prion were indistinguishable from those of the C-BSE prion in TgHaNSE mice. In addition, similar PrPres glycoprofiles were detected among H-, C-, and L-BSE prions in TgHaNSE mice. Therefore, to better understand atypical BSE prions after interspecies transmission, H-BSE prion transmission from TgHaNSE mice to hamsters was investigated, and the characteristics of classical and atypical BSE prions among hamsters, wild-type mice, and mice overexpressing bovine PrPC (TgBoPrP) were compared in this study using biochemical and neuropathological methods. Identical PrPres banding patterns were confirmed between TgHaNSE mice and hamsters in the case of all three BSE prion strains. However, these PrPres banding patterns differed from those of TgBoPrP and wild-type mice infected with the H-BSE prion. In addition, glycoprofiles of TgHaNSE mice and hamsters infected with the L-BSE prion differed from those of TgBoPrP mice infected with the L-BSE prion. These data indicate that the PrPC amino acid sequences of new host species rather than other host environmental factors may affect some molecular aspects of atypical BSE prions. Although three BSE prion strains were distinguishable based on the neuropathological features in hamsters, interspecies transmission modified some molecular properties of atypical BSE prions, and these properties were indistinguishable from those of C-BSE prions in hamsters. Taken together, PrPres banding patterns and glycoprofiles are considered to be key factors for BSE strain typing. However, this study also revealed that interspecies transmission could sometimes influence these characteristics.
Bovine spongiform encephalopathy (BSE) is a prion disease in cattle, which is characterized by spongiform changes and accumulation of a disease-associated isoform of the prion protein (PrPd) in the central nervous system (1). PrPd, a misfolded isoform of the host cellular prion protein (PrPC), is the main, if not sole, component of prions (2). Initial studies indicated that BSE is a single prion strain, named classical (C-) BSE, based on the molecular characteristics of the proteinase K (PK)-resistant core of PrPd (PrPres), neuroanatomical distribution patterns of PrPd observed in most BSE cases in cattle, and C-BSE transmitted rodent models (3). In the 2000s, atypical BSEs were detected in aged cattle worldwide and were classified into at least two forms (4, 5). These two atypical forms are commonly referred to as high-type (H-BSE) and low-type (L-BSE) BSEs based on the higher or lower molecular masses of the unglycosylated forms of PrPres, respectively (6).
It is well-known that rodent models are very useful for studying prion diseases, i.e., for prion purification and analysis of the interference between different prion strains (7–9). L-BSE prion has been shown to be directly transmitted from cattle to hamsters (10, 11). However, previous studies have demonstrated that transmission of C- and H-BSE prions from cattle to hamsters is inefficient (11, 12), whereas mouse-passaged C-BSE prion was easily transmissible to hamsters (10). Moreover, we demonstrated successful transmission of H-BSE prion from cattle to transgenic mice overexpressing hamster PrPC (TgHaNSE) (13). In a previous report, we revealed that the neuroanatomical distribution patterns of immunolabeled PrPd in the brain were different among individual BSE prion strains, even after the third passage in TgHaNSE mice. In contrast to PrPd immunopathological findings, Western blotting (WB) analyses demonstrated that the molecular mass of the unglycosylated form of PrPres and glycoform profiles of H-BSE prion were indistinguishable from those of the C-BSE prion in TgHaNSE mice. Therefore, to investigate the effect of amino acid sequences of host PrPC, host genetic factors, and host microenvironment on the molecular aspects of BSE prions after interspecies transmission, we established a transmission model of TgHaNSE-passaged H-BSE prion in hamsters in this study. Subsequently, we investigated the biological and biochemical properties of three different BSE prion strains in hamsters and compared these characteristics among hamsters, TgBoPrP, and wild-type mice.
Animal experiments were performed in strict accordance with the regulations in the Guide for the Care and Use of Laboratory Animals of the National Institute of Animal Health and in accordance with the Guidelines for Proper Conduct of Animal Experiments (2006) by the Science Council of Japan. Procedures involving animal subjects were approved by the Institutional Animal Care and Use Committee at the National Institute of Animal Health (approval ID: 15-031), with all possible effort made to minimize pain and discomfort of the animals. Six 3 week-old female hamsters were inoculated intracerebrally with 20 μl of 10% brain homogenate (BH) from TgHaNSE mice infected with H-BSE prion, which had been passaged thrice in TgHaNSE mice.
L-BSE prion was successfully transmitted from cattle to hamsters and TgHaNSE mice (10). Four female hamsters (3 weeks old) were inoculated intracerebrally with 20 μl of 10% BH from TgHaNSE mice infected with L-BSE prion. However, C-BSE prion was passaged once in CD-1 mice before transmission to TgHaNSE mice. Finally, TgHaNSE-passaged C-BSE prion was transmitted to hamsters and passaged thrice. The details of the transmission of C- and L-BSE prions to hamsters and TgHaNSE mice were described in previous studies (10, 13). To compare the characteristic changes of the three BSE prions after interspecies transmission, we intracerebrally inoculated 20 μL of 10% BH prepared from C-, H-, and L-BSE-affected cattle into TgBoPrP and C57BL/6 mice. Three BSE prions were passaged five times in TgBoPrP mice. On the other hand, C- and H-BSE prions were passaged thrice in C57BL/6 mice, and L-BSE prion was passaged twice in C57BL/6 mice, including a blind passage.
The left hemisphere and selected tissues were fixed in 10% buffered formalin containing 10% methanol. Formalin-fixed tissue specimens were immersed in 98% formic acid for 60 min to reduce the infectivity; this was followed by embedding in paraffin, sectioning, and staining with hematoxylin and eosin (HE) for histological evaluation by scoring vacuolar changes in nine different areas of the brain (14).
Dewaxed sections were placed on silanized glass slides, treated with 3% hydrogen peroxide at room temperature (R/T) for 10 min, incubated with 10 μg/ml of PK (Nacalai Tesque, Kyoto, Japan) in PBS containing 0.1% (v/v) Triton X-100 (Nacalai Tesque) at R/T for 10 min, and immersed in 150 mM sodium hydroxide solution at 60°C for 10 min for epitope retrieval (15). Immunohistochemical staining was performed using a tyramide signal amplification (TSA) system (PerkinElmer, Waltham, MA, USA). Sections were incubated with blocking reagent for 30 min at R/T, monoclonal antibody (mAb) 44B1 (Table 1) for 1 h at R/T (16, 17), and biotinylated anti-mouse IgG (PerkinElmer) for 30 min at R/T. Slides were then incubated with streptavidin–horseradish peroxidase (SA-HRP) for 30 min at R/T, treated with biotinyl tryamide as the amplification reagent for 5 min at R/T, and incubated again with SA-HRP for 30 min at R/T before visualization with 3,3′-diaminobenzidine tetrachloride containing 10 mM imidazole, as the chromogen. Finally, the sections were slightly counterstained with Mayer's hematoxylin.
In brief, 20% BH was mixed with an equal volume of detergent buffer containing 4% (w/v) Zwittergent 3–14 (Merck Japan, Tokyo, Japan), 1% (w/v) Sarkosyl (Sigma-Aldrich, St. Louis, MO, USA), 100 mM NaCl, and 50 mM Tris–HCl (pH 7.6), and incubated for 30 min with collagenase (FUJIFILM Wako Pure Chemical, Osaka, Japan; final concentration of 500 μg/ml) at 37°C. The lysate was then incubated for 30 min with PK (Roche Applied Science, Penzberg, Germany; final concentration of 40 μg/ml) at 37°C. The digestion with PK was terminated with 2 mM 4-(2-aminoethyl) benzene sulfonyl fluoride hydrochloride (Pefabloc; Roche Applied Science). Further, deglycosylation with N-glycosidase F (PNGase F; New England Biolabs, Ipswich, MA, USA) was performed per the manufacturer's instructions. The resultant solution was mixed with a 2-butanol/methanol mixture (5:1), and PrPres was precipitated by centrifugation at 20,000 × g for 10 min at R/T. The pellet was resolubilized in LDS sample loading buffer (Thermo Fisher Scientific, Waltham, MA, USA). PrPres-enriched samples were prepared from the spleen of hamsters according to the modified method described in our previous report (18). In brief, minced spleens (200 mg) were homogenized in 50 mM Tris–HCl (pH 7.6) containing 2% (v/v) Triton X-100, 0.5% (v/v) Sarkosyl, 100 mM NaCl, 5 mM MgCl2, 20 mg/ml collagenase, and 40 μg/ml DNase I (FUJIFILM Wako Pure Chemical), and then incubated at 37°C for 2 h. The homogenates were digested with 80 μg/ml PK for 1 h at 37°C and centrifuged at 68,000 × g for 30 min at 4°C. The resulting pellet was resuspended in 6.25% Sarkosyl in 10 mM Tris–HCl (pH 7.6) and centrifuged at 9,000 × g for 5 min at R/T. Sodium phosphotungstate (Na-PTA; Nacalai Tesque, final concentration of 0.3%) was added into the supernatant, and the resultant solution was incubated for 30 min at 37°C with constant rotation, followed by centrifugation at 20,000 × g for 30 min at 4°C. The pellet was resuspended in LDS sample loading buffer.
Samples were electrophoresed on NuPAGE Novex 12% Bis-Tris gels with NuPAGE MOPS-SDS running buffer in accordance with the manufacturer's instructions (Thermo Fisher Scientific). The proteins were then transferred onto Immobilon-P membranes (Merck Millipore, Burlington, MA, USA). The blotted membranes were incubated with blocking reagent for 30 min and then with the anti-PrP mAbs (Table 1) at 4°C overnight. After washing with PBS containing 0.05% (v/v) Tween 20 (PBST), the membranes were incubated with goat anti-mouse IgG-HRP (Jackson ImmunoResearch, West Grove, PA, USA) for 1 h, which was followed by washing with PBST. Signals were developed with a chemiluminescent substrate (SuperSignal; Thermo Fisher Scientific). For semi-quantification, blots were imaged using a Fluorchem system (Alpha Innotech, San Leandro, CA, USA) and analyzed using an image reader software (AlphaEaseFC; Alpha Innotech) according to the manufacturer's instructions.
Brain samples were incubated with various PK concentrations (50, 100, 500, and 1,000 μg/ml) at 37°C for 30 min. Subsequently, the samples were subjected to WB as described above. The PrPres signals were detected with mAb 6H4 and normalized by the signal intensity of PrPres digested with 50 μg/ml PK.
To determine statistical significance, Student's t-tests were applied on paired data. Differences of P < 0.05 were considered significant. Statistical analysis was performed using KaleidaGraph software (Synergy Software, Reading, PA, USA).
All hamsters inoculated with 10% BH prepared from a TgHaNSE mouse infected with the H-BSE prion showed neurological signs such as loss of appetite, wasting, inactivity, hunching, and inability to groom. They reached the terminal stage of the disease after a survival period of 445 ± 47.6 days (mean survival days ± standard deviation). Subsequent transmission to hamsters resulted in similar clinical signs, and their survival periods showed no significant differences between passage numbers (P > 0.05). The survival period of hamsters inoculated with 10% BH prepared from a TgHaNSE mouse infected with the L-BSE prion was 228 ± 11.0 days. Comparatively, hamsters inoculated with 10% hamster-BH infected with the L-BSE prion developed disease after a survival period of 208 ± 15.5 days at the second passage, which showed no statistical differences between the two groups (P > 0.05, Table 2).
All hamsters infected with the H-BSE prion accumulated PrPres, and the major triplet PrPres banding pattern detected with mAb 6H4 was identical to that in TgHaNSE mice infected with the H-BSE prion (Figure 1A). However, in WB analysis using mAb SAF84, the multiple PrPres bands were not detected in TgHaNSE mice and hamsters infected with the H-BSE prion, including an additional ~12-kDa small PrPres fragment, which was generally identified in cattle, TgBoPrP, and wild-type mice infected with the H-BSE prion (arrow in Figure 1B). Further, transmission of the L-BSE prion from cattle to wild-type mice did not occur, as previously reported (19). Deglycosylation of PrPres with PNGase F demonstrated that the molecular mass of the unglycosylated PrPres of H-BSE-infected hamsters was indistinguishable from that of C-BSE-infected hamsters and slightly higher than that of L-BSE-infected hamsters (Figure 1C). Furthermore, deglycosylation experiments confirmed the lack of the ~12-kDa small PrPres fragment in hamsters infected with the H-BSE prion (arrow in Figure 1C). Three BSE prion strains showed individual glycoform profiles in cattle (Figure 2A). Conversely, glycoprofiles were identical among BSE prion strains in TgHaNSE mice and hamsters, in which the diglycosylated form of PrPres was dominant (Figures 2B,C). Because the molecular mass of the unglycosylated PrPres and the glycoprofiles were identical between C-BSE- and H-BSE-infected hamsters, the relative PK resistance of their PrPd was compared. In contrast to C-and H-BSE prions in cattle, PrPd of hamster H-BSE showed higher resistance to PK digestion than that of hamster C-BSE (Figure 3).
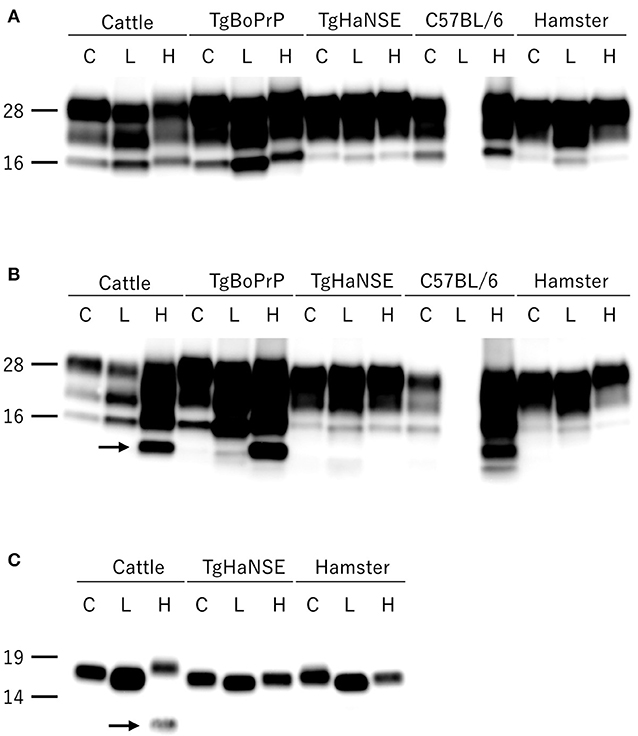
Figure 1. Comparison of PrPres profiles among BSE strains in different host species. PrPres was detected with mAb 6H4 (A) and SAF84 (B). PrPres was also detected with mAb SAF84 after PNGase deglycosylation (C). PrPres banding patterns, which were specific to individual BSE strains in cattle, were conserved in TgBoPrP and wild-type (C57BL/6) mice, but not in TgHaNSE mice and Syrian hamsters. Further, the L-BSE prion was not transmissible to wild-type mice. All BSE strains showed similar molecular features between TgHaNSE mice and hamsters. Arrows indicate the ~12-kDa small C-terminus PrPres fragment that is specific for the H-BSE strain, but was not detected in TgHaNSE mice and hamsters infected with the H-BSE prion. C, L, and H on the top of each panel indicate C-BSE, L-BSE, and H-BSE, respectively. Molecular markers are shown to the left (kDa).
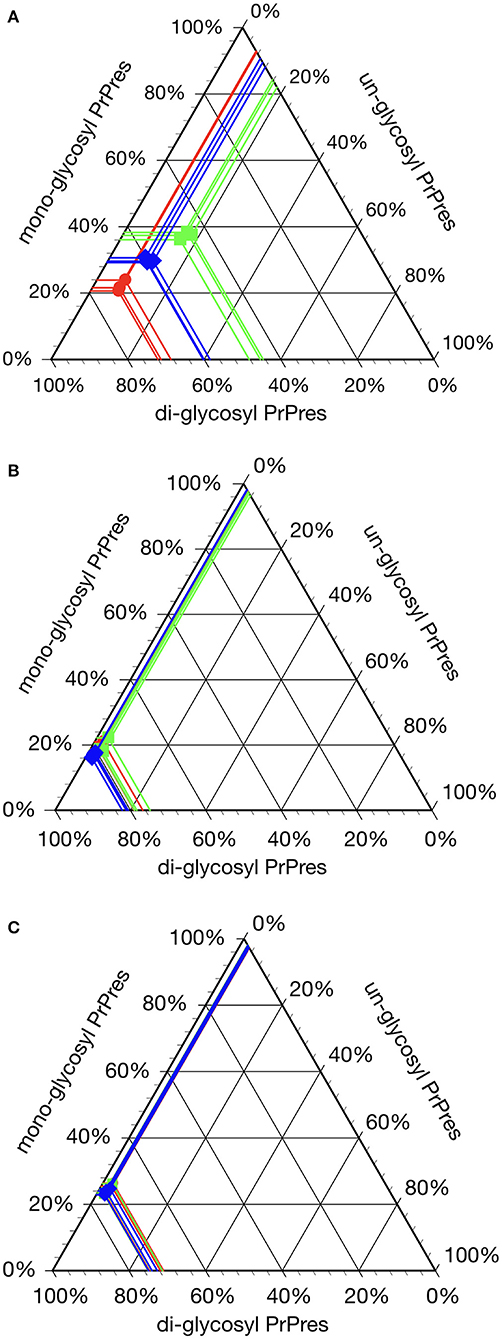
Figure 2. Glycoprofile analysis of C-, L-, and H-BSE strains in different host species. The relative quantity of the three major PrPres bands was determined using an image reader software (AlphaEaseFC; Alpha Innotech) for glycosylation analysis (percentage of the di-, mono-, and unglycosylated PrPres band). Glycoprofiles of BSE strains in cattle (A) were distinctly different from those in TgHaNSE mice (B) and hamsters (C). Di-glycosylated form of PrPres was dominant in all the three BSE strains in TgHaNSE mice and hamsters. Symbols: red circles, C-BSE; blue triangles, L-BSE; green squares. PrPres was detected with mAb 6H4.
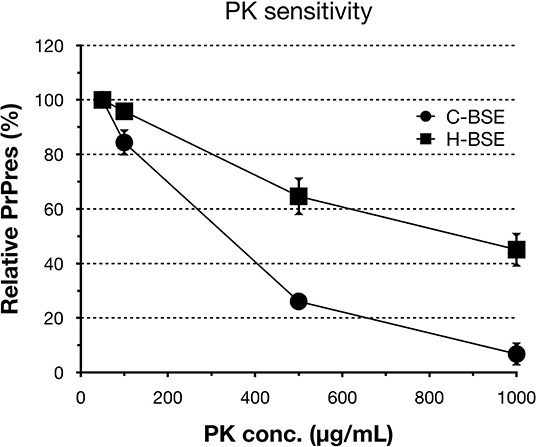
Figure 3. PrPd sensitivity to different PK concentrations of hamster-adapted H-BSE. PK susceptibility was compared between H- and C-BSE strains in hamsters. The signal intensities were quantified by an image reader software (AlphaEaseFC; Alpha Innotech), whereby the intensity of 50 μg/ml PK concentration was set as the reference point (100%). PrPres was detected with mAb 6H4. Symbols: black circles, C-BSE; black squares, H-BSE.
Spongiform changes, represented by vacuolar lesion profiles in the brain, at both first and second passage in H-BSE-infected hamsters were very similar, and the lesion profiles were clearly different from those of hamsters infected with C- and L-BSE prions (Figure 4). The extent of vacuolation was mild to moderate throughout the brains of H-BSE-infected hamsters. Furthermore, PrPd distribution patterns in the brains of H-BSE-infected hamsters were distinguishable from those in the brains of hamsters infected with C- and L-BSE prions (Figures 5A–C). No specific background staining was detected in the brain of hamsters intracranially inoculated with PBS (Figure 5D). In H-BSE prion infection, the most frequent and prominent deposits of PrPd were coalescing or plaque-like deposits in the periventricular, subependymal, and subpial regions of the brain, which were not detected in C- and L-BSE-infected hamsters (Figures 5A,E,F). However, fine granular PrPd was also present in the neuropil of the gray matter of the brain (Figure 5F).
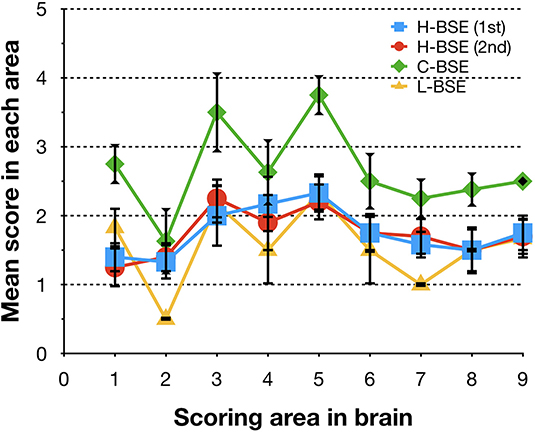
Figure 4. Comparison of lesion profiles of hamsters infected with three different BSE strains. Lesion profiles of hamsters inoculated with H-BSE prion passaged in TgHaNSE mice (first passage, blue square; second passage, red circle), Mouse-adapted C-BSE prion passaged in TgHaNSE mice (green diamond), and L-BSE prion from cattle (yellow triangle). Vacuolation was scored on a scale of 0–5 in the following brain areas: 1, dorsal medulla; 2, cerebellum; 3, midbrain; 4, hypothalamus; 5, thalamus; 6, hippocampus; 7, septal nuclei of the paraterminal body; 8 caudal cerebral cortex; and 9, rostral cerebral cortex. Data represent mean ± standard deviation (n = 4–6). The vacuolar lesion scores for C-BSE and L-BSE were taken from a previous study (10).
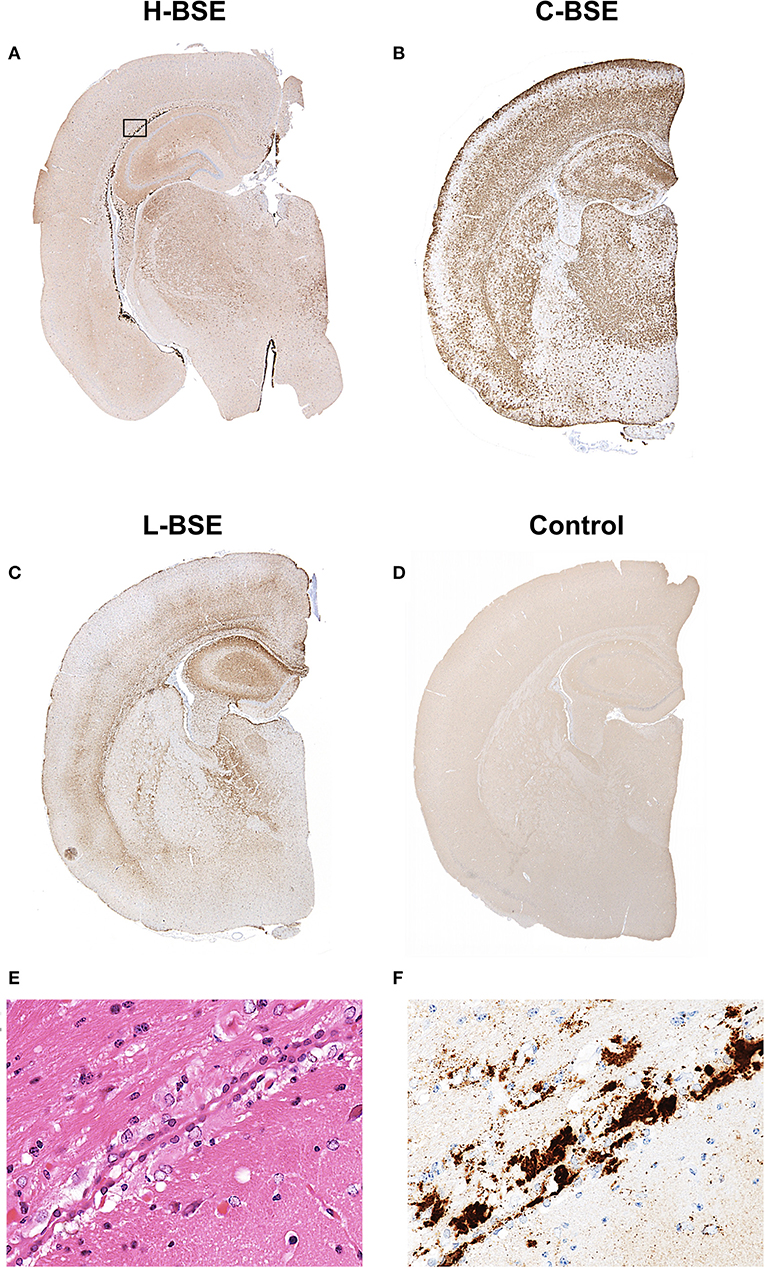
Figure 5. Comparison of PrPd distribution in the brains of hamsters infected with three different BSE strains. Representative PrPd distribution in a coronal brain slice (section) at the thalamic level of a hamster infected with H-, C-, and L-BSE is shown (A–C). A brain section from a hamster inoculated with PBS is shown as a negative control for PrPd immunohistochemistry (D). Dense plaque-like deposits and coalescing PrPd aggregates were mainly distributed in the periventricular and subcallosal regions of the H-BSE-infected hamster, but granular PrPd was deposited throughout the brain of the C-BSE- and L-BSE-infected hamsters. The area within the box is shown enlarged (E,F). Immunohistochemistry was performed with mAb 44B1.
Granular PrPd deposits were detected in the brain and extracerebral tissues, including the spinal cord (Figure 6A), retina (Figure 6B), trigeminal ganglia (Figure 6C), adrenal medulla (Figure 6D), muscle bundles or skeletal and lingual muscle fibers (Figures 6E,F), enteric plexuses (Figures 6G,H), olfactory sensory epithelium (Figure 6I), and Peyer's patches, presumably corresponding to follicular dendritic cells (Figure 6J). In addition, WB analysis demonstrated the accumulation of PrPres in the spleens of H-BSE-infected hamsters (Figure 7). The results for immunohistochemical detection of PrPd in negative tissue controls are shown in Figure S1. Weak diffuse background labeling was present in the gray matter of the spinal cord (Figure S1A) and adrenal medulla (Figure S1D). Non-specific background immunolabeling was also detected in the olfactory glomeruli (Figure S1I; asterisks).
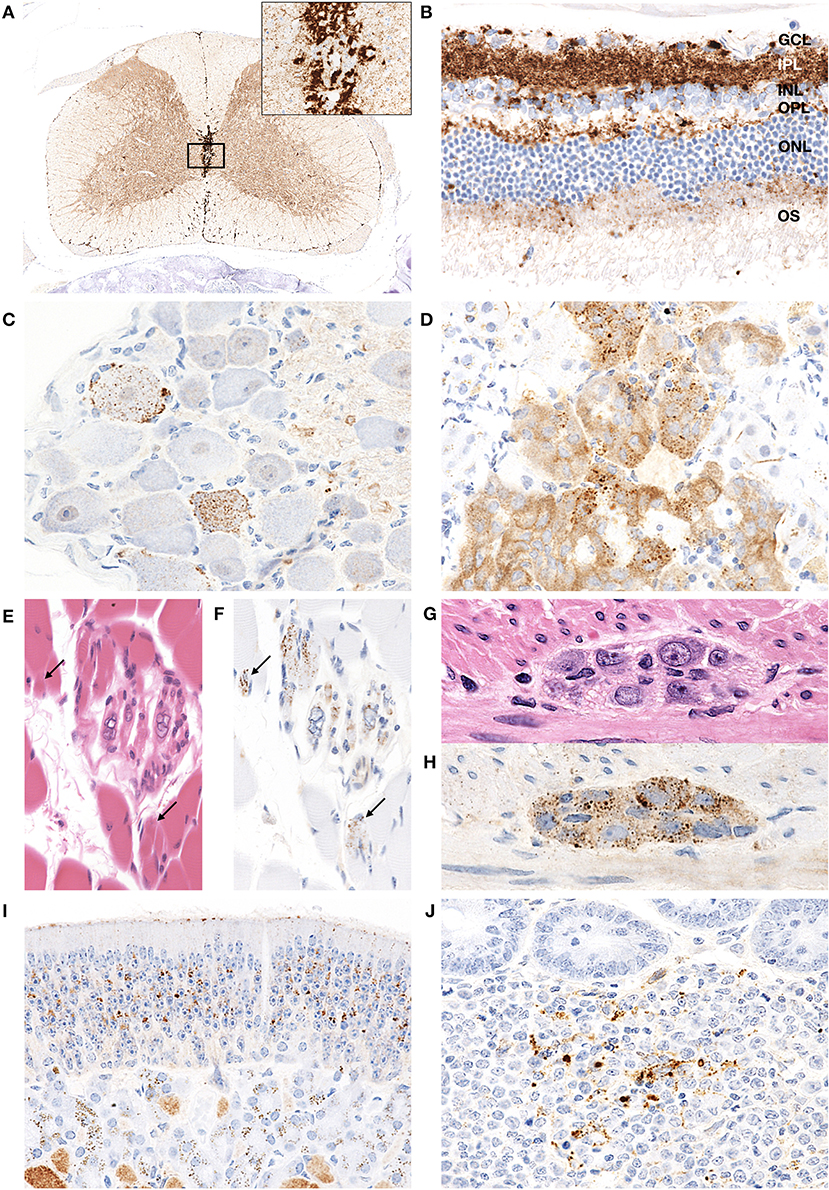
Figure 6. PrPd distribution in extracerebral tissues of H-BSE-infected hamsters. Dense PrPd aggregates are observed in the peri-central canal and subpial regions, and granular PrPd deposits are conspicuous in the gray matter (A). The area within the box is shown enlarged in the inset in the upper right corner. Diffuse granular deposition of PrPd is observed in all layers of the retina (B). GCL, ganglion cell layer; INL, inner nucleus layer; IPL, inner plexiform layer; NFL, nerve fiber layer; ONL, outer nucleus layer; OPL, outer plexiform layer; OS, outer segments. Granular PrPd deposits are observed in the ganglionic and satellite cells of the trigeminal ganglion (C), chromaffin cells of the adrenal medulla (D), intrafusal myofiber of the muscle spindle (E,F), skeletal muscle fibers (E,F; arrows), and neuronal cells of the myenteric plexus in the colon (G,H). PrPd deposits are observed in the olfactory sensory epithelium of the nasal mucosa (I) and the Peyer's patch, presumably corresponding to follicular dendritic cells (J). Immunohistochemistry was performed with mAb 44B1.
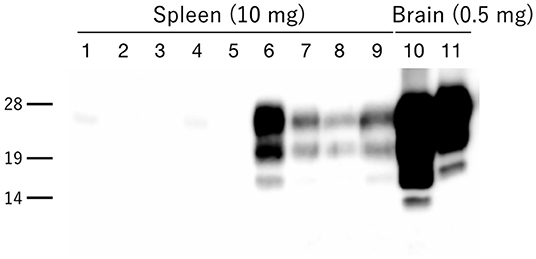
Figure 7. PrPres accumulation in the spleens of H-BSE-infected hamsters. PrPres was detected using mAb 6H4. PrPres signals were detected in the spleens of four individual hamsters infected with the H-BSE prion (lanes 6–9), but not in five individual hamsters infected with the L-BSE prion (lanes 1–5). Brain samples from hamsters infected with L- and H-BSE prions were loaded as positive controls, respectively (lanes 10 and 11). The equivalent tissue quantities loaded per lane are indicated at the top, and the molecular markers are shown to the left of the panel (kDa).
In this study, we demonstrate that H-BSE prion could transmit to wild-type hamsters by passaging in TgHaNSE mice prior to hamster transmission. Because no statistically significant difference was found in the survival periods between the primary and secondary passage in hamsters infected with the H-BSE prion, there is probably no strain or species barrier for H-BSE transmission between TgHaNSE mice and hamsters. Furthermore, no species barrier effect was observed in L-BSE infection from TgHaNSE mice to hamsters, indicating that the PrPC amino acid sequence might primarily contribute to the adaptation of atypical BSE prions to hamsters. In contrast, survival periods have been reported to be shortened in cases of atypical BSE prion transmission from cattle to TgBoPrP mice (20, 21), suggesting that important factors involved in the adaptation of atypical BSE prions to recipients after interspecies transmission depend on the host species, although the PrPC amino acid sequence would be a key factor for adaptation of atypical BSE prions.
The biological properties, such as survival periods, lesion profiles, and neuroanatomical distribution patterns, of PrPd in hamsters infected with the H-BSE prion were distinct from those of hamsters infected with C- and L-BSE prions. The most characteristic finding in H-BSE-infected hamsters was the presence of plaque-like deposits, which were absent in TgHaNSE mice infected with the H-BSE prion (13). By contrast, plaque-like deposits were present in TgHaNSE mice infected with the L-BSE prion, but not in L-BSE-infected hamsters (10, 13). Therefore, differences in their PrPC expression level and brain distributions may underlie these differences between transgenic mice and wild-type hamsters.
Interestingly, the biochemical properties of PrPd in the brains of H-BSE-infected hamsters, including PrPres banding patterns and the molecular mass of unglycosylated PrPres, were distinct from those of cattle and TgBoPrP mice infected with the H-BSE prion (20, 22). In addition, our data demonstrate that PK susceptibility of the three BSE prions was remarkably different between cattle and hamsters (10, 23). These results indicate that the PrPd structure of each BSE prion could undergo modification from the original structure in cattle to that in hamsters through interspecies transmission. Furthermore, this change in the structure of PrPd in hamsters may explain the change of the glycoprofiles in L-BSE infection between cattle and hamsters. PrPC is post-translationally modified with two terminally sialylated N-linked glycans (24). Katorcha et al. proposed that one prion strain with minor constraints between glycans belonging to neighboring PrPd monomers could recruit diglycosylated PrPC for replication; however, another prion strain with substantial spatial constraints between glycans from neighboring PrPd monomers would limit the percentage of diglycosylated PrPC due to high electrostatic repulsion between sialic acids from neighboring N-glycans (25). L-BSE prion showed the strongest exclusion of the diglycosylated form of PrPC in cattle (6, 26, 27), but L-BSE prion in hamsters favored the diglycosylated form for replication (Figures 1, 2), indicating that the L-BSE prion-specific alignment of glycan moieties of PrPd might change after interspecies transmission to hamsters, which may allow the L-BSE prion in hamsters to efficiently recruit diglycosylated PrPC molecules for its replication. Although glycoprofiles have sometimes been used for prion strain typing (28), our results demonstrated that the glycoform ratio of atypical BSE prions, like other properties (19), changed after interspecies transmission. Therefore, it does not necessarily reflect the characteristics of the original BSE prion strains.
Despite intracerebral inoculation, PrPd was present in the lymphoid tissues of H-BSE-infected hamsters, in contrast to the absence of PrPd lymphotropism in cattle (29, 30) and wild-type mice (31) infected with the H-BSE prion. However, PrPres was not detected in the lymphoid tissues of intracranially inoculated L-BSE hamsters, as previously reported (32). Although the mechanism for lymphotropism in hamsters is unclear, there is a possibility that not only the prion strain-specific properties but also host-species genetic factors and the host microenvironment might contribute to the lymphotropism of atypical BSE prions. In future studies, PrPd identified in the lymphoid tissues should be biologically characterized for a better understanding of the pathogenesis of H-BSE prion infection after interspecies transmission to hamsters.
In summary, we successfully established an H-BSE hamster model and compared the properties of PrPd among H-, C-, and L-BSE prions in hamsters. Each BSE prion strain could be distinguished by its individual pathological features, including the patterns of spongiform changes and neuroanatomical distribution of PrPd in the brain of affected hamsters. Furthermore, our datasets suggest that some molecular characteristics of atypical BSE prions are modified because of the influence of interspecies transmission from cattle to hamsters, resulting in some indistinguishable molecular characteristics of PrPd compared to C-BSE prion in hamsters. These hamster models can be useful for further research on the pathology of BSE, a disease that poses significant challenges to the cattle industry.
All datasets generated for this study are included in the article/Supplementary Material.
The animal study was reviewed and approved by The Institutional Animal Care and Use Committee at the National Institute of Animal Health. Written informed consent was obtained from the owners for the participation of their animals in this study.
KMi, KMa, and HO conceived and designed the experiments. KMi, KMa, HO, and YM performed the experiments. All the authors analyzed the data. KMi and HO wrote the manuscript. All the authors have read and approved the manuscript.
This study was supported by grants-in-aid from the research project for improving food safety and animal health from the Ministry of Agriculture, Forestry and Fisheries of Japan and was partially supported by AMED under Grant Number JP160109044.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Naomi Furuya, Naoko Tabeta, Junko Yamada, and the animal caretakers for their expert technical assistance. We would also like to thank Editage (www.editage.com) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00094/full#supplementary-material
1. Balkema-Buschmann A, Fast C, Kaatz M, Eiden M, Ziegler U, McIntyre L, et al. Pathogenesis of classical and atypical BSE in cattle. Prev Vet Med. (2011) 102:112–17. doi: 10.1016/j.prevetmed.2011.04.006
2. Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. (2011) 3:a006833. doi: 10.1101/cshperspect.a006833
3. Bruce ME, Boyle A, Cousens S, McConnell I, Foster J, Goldmann W, et al. Strain characterization of natural sheep scrapie and comparison with BSE. J Gen Virol. (2002) 83:695–704. doi: 10.1099/0022-1317-83-3-695
4. Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, et al. Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. (2004) 101:3065–70. doi: 10.1073/pnas.0305777101
5. Biacabe AG, Laplanche JL, Ryder S, Baron T. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep. (2004) 5:110–15. doi: 10.1038/sj.embor.7400054
6. Jacobs JG, Langeveld JP, Biacabe AG, Acutis PL, Polak MP, Gavier-Widen D, et al. Molecular discrimination of atypical bovine spongiform encephalopathy strains from a geographical region spanning a wide area in Europe. J Clin Microbiol. (2007) 45:1821–29. doi: 10.1128/JCM.00160-07
7. Prusiner SB, Cochran SP, Alpers MP. Transmission of scrapie in hamsters. J Infect Dis. (1985) 152:971–8. doi: 10.1093/infdis/152.5.971
8. Bessen RA, Marsh RF. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol. (1992) 73:329–34. doi: 10.1099/0022-1317-73-2-329
9. Bartz JC, Aiken JM, Bessen RA. Delay in onset of prion disease for the HY strain of transmissible mink encephalopathy as a result of prior peripheral inoculation with the replication-deficient DY strain. J Gen Virol. (2004) 85:265–73. doi: 10.1099/vir.0.19394-0
10. Shu Y, Masujin K, Okada H, Iwamaru Y, Imamura M, Matsuura Y, et al. Characterization of Syrian hamster adapted prions derived from L-type and C-type bovine spongiform encephalopathies. Prion. (2011) 5:103–8. doi: 10.4161/pri.5.2.15847
11. Nicot S, Baron T. Strain-specific barriers against bovine prions in hamsters. J Virol. (2011) 85:1906–8. doi: 10.1128/JVI.01872-10
12. Yokoyama T, Masujin K, Iwamaru Y, Imamura M, Mohri S. Alteration of the biological and biochemical characteristics of bovine spongiform encephalopathy prions during interspecies transmission in transgenic mice models. J Gen Virol. (2009) 90:261–8. doi: 10.1099/vir.0.004754-0
13. Okada H, Masujin K, Miyazawa K, Yokoyama T. Transmissibility of H-type bovine spongiform encephalopathy to hamster PrP transgenic mice. PLoS ONE. (2015) 10:e0138977. doi: 10.1371/journal.pone.0138977
14. Fraser H, Dickinson AG. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol. (1968) 78:301–11. doi: 10.1016/0021-9975(68)90006-6
15. Okada H, Sato Y, Sata T, Sakurai M, Endo J, Yokoyama T, et al. Antigen retrieval using sodium hydroxide for prion immunohistochemistry in bovine spongiform encephalopathy and scrapie. J Comp Pathol. (2011) 144:251–6. doi: 10.1016/j.jcpa.2010.10.001
16. Furuoka H, Yabuzoe A, Horiuchi M, Tagawa Y, Yokoyama T, Yamakawa Y, et al. Effective antigen-retrieval method for immunohistochemical detection of abnormal isoform of prion proteins in animals. Acta Neuropathol. (2005) 109:263–71. doi: 10.1007/s00401-004-0944-x
17. Kim CL, Umetani A, Matsui T, Ishiguro N, Shinagawa M, Horiuchi M. Antigenic characterization of an abnormal isoform of prion protein using a new diverse panel of monoclonal antibodies. Virology. (2004) 320:40–51. doi: 10.1016/j.virol.2003.10.026
18. Shimada K, Hayashi HK, Ookubo Y, Iwamaru Y, Imamura M, Takata M, et al. Rapid PrP(Sc) detection in lymphoid tissue and application to scrapie surveillance of fallen stock in Japan: variable PrP(Sc) accumulation in palatal tonsil in natural scrapie. Microbiol Immunol. (2005) 49:801–4. doi: 10.1111/j.1348-0421.2005.tb03660.x
19. Okada H, Masujin K, Miyazawa K, Yokoyama T. Acquired transmissibility of sheep-passaged L-type bovine spongiform encephalopathy prion to wild-type mice. Vet Res. (2015) 46:81. doi: 10.1186/s13567-015-0211-2
20. Okada H, Masujin K, Iwamaru Y, Imamura M, Matsuura Y, Mohri S, et al. Experimental transmission of H-type bovine spongiform encephalopathy to bovinized transgenic mice. Vet Pathol. (2011) 48:942–7. doi: 10.1177/0300985810382672
21. Masujin K, Miwa R, Okada H, Mohri S, Yokoyama T. Comparative analysis of Japanese and foreign L-type BSE prions. Prion. (2012) 6:89–93. doi: 10.4161/pri.6.1.18429
22. Biacabe AG, Jacobs JG, Bencsik A, Langeveld JP, Baron TG. H-type bovine spongiform encephalopathy: complex molecular features and similarities with human prion diseases. Prion. (2007) 1:61–8. doi: 10.4161/pri.1.1.3828
23. Priemer G, Balkema-Buschmann A, Hills B, Groschup MH. Biochemical characteristics and PrP(Sc) distribution pattern in the brains of cattle experimentally challenged with H-type and L-type atypical BSE. PLoS ONE. (2013) 8:e67599. doi: 10.1371/journal.pone.0067599
24. Wulf MA, Senatore A, Aguzzi A. The biological function of the cellular prion protein: an update. BMC Biol. (2017) 15:34. doi: 10.1186/s12915-017-0375-5
25. Katorcha E, Makarava N, Savtchenko R, Baskakov IV. Sialylation of the prion protein glycans controls prion replication rate and glycoform ratio. Sci Rep. (2015) 5:16912. doi: 10.1038/srep16912
26. Fukuda S, Iwamaru Y, Imamura M, Masujin K, Shimizu Y, Matsuura Y, et al. Intraspecies transmission of L-type-like bovine spongiform encephalopathy detected in Japan. Microbiol Immunol. (2009) 53:704–7. doi: 10.1111/j.1348-0421.2009.00169.x
27. Baron T, Biacabe AG, Arsac JN, Benestad S, Groschup MH. Atypical transmissible spongiform encephalopathies (TSEs) in ruminants. Vaccine. (2007) 25:5625–30. doi: 10.1016/j.vaccine.2006.10.058
28. Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. (1996) 383:685–90. doi: 10.1038/383685a0
29. Okada H, Iwamaru Y, Imamura M, Masujin K, Matsuura Y, Shimizu Y, et al. Experimental H-type bovine spongiform encephalopathy characterized by plaques and glial- and stellate-type prion protein deposits. Vet Res. (2011) 42:79. doi: 10.1186/1297-9716-42-79
30. Konold T, Bone GE, Clifford D, Chaplin MJ, Cawthraw S, Stack MJ, et al. Experimental H-type and L-type bovine spongiform encephalopathy in cattle: observation of two clinical syndromes and diagnostic challenges. BMC Vet Res. (2012) 8:22. doi: 10.1186/1746-6148-8-22
31. Baron T, Vulin J, Biacabe AG, Lakhdar L, Verchere J, Torres JM, et al. Emergence of classical BSE strain properties during serial passages of H-BSE in wild-type mice. PLoS ONE. (2011) 6:e15839. doi: 10.1371/journal.pone.0015839
Keywords: atypical bovine spongiform encephalopathy, H-BSE, L-BSE, interspecies transmission, prion strain, hamsters
Citation: Miyazawa K, Masujin K, Matsuura Y, Iwamaru Y and Okada H (2020) Influence of Interspecies Transmission of Atypical Bovine Spongiform Encephalopathy Prions to Hamsters on Prion Characteristics. Front. Vet. Sci. 7:94. doi: 10.3389/fvets.2020.00094
Received: 06 December 2019; Accepted: 07 February 2020;
Published: 03 March 2020.
Edited by:
Rohana P. Dassanayake, National Animal Disease Center (USDA ARS), United StatesReviewed by:
Byron Caughey, Rocky Mountain Laboratories (NIAID), United StatesCopyright © 2020 Miyazawa, Masujin, Matsuura, Iwamaru and Okada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kohtaro Miyazawa, bWl5YXphd2FrQGFmZnJjLmdvLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.