
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 14 February 2020
Sec. Parasitology
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.00064
 Federica Marcer1
Federica Marcer1 Federica Tosi2
Federica Tosi2 Giovanni Franzo1
Giovanni Franzo1 Alessia Vetri2
Alessia Vetri2 Silvia Ravagnan3
Silvia Ravagnan3 Mario Santoro4
Mario Santoro4 Erica Marchiori1*
Erica Marchiori1*Sulcascaris sulcata is a nematode parasite of sea turtles, widespread in neritic foraging grounds with variable prevalence, reaching 30% in loggerhead turtles Caretta caretta feeding in northern Adriatic Sea. Ulcerative gastritis associated to high intensity of infection is reported in this host species. The life cycle of S. sulcata has been elucidated in Australian and American waters, demonstrating the ability of the species of infecting a wide range of intermediate hosts, represented by bivalve and gastropod molluscs. During regular sanitary inspections, nematode larvae were found within the adductor muscle of Pecten jacobeus and Aequipecten opercularis collected from the Northern Adriatic Sea. Morphological and molecular analyses were performed for the identification of larvae, and molecular data were obtained from adult S. sulcata as well. Analysis of the sequences of ITS fragment, and mitochondrial genes cox1 and cox2, revealed high genetic similarity among all the samples, and no geographical clustering was observed between adult parasites collected in Adriatic and Tyrrhenian Seas. A common allele pool was detected also between the two developmental stages, included larvae from Pecten and Aequipecten. The comparison with other members of the family Anisakidae demonstrated that S. sulcata formed a clear monophyletic cluster. This study reports the first identification of intermediate hosts for S. sulcata within the Mediterranean Sea. Infection in edible scallops justifies the exclusion of the product from the market and zoonotic potential of larvae of this anisakid nematode are yet to be completely excluded. Fidelity of sea turtles to selected foraging grounds, such as the Northern Adriatic shelf, warrants the life cycle of S. sulcata to perpetrate in the area; at the same time, long distance migrations of individuals justify the dispersal of infecting elements over the Mediterranean basin, regardless of turtles' origin.
Sulcascaris sulcata (Rudolphi, 1819) (Nematoda: Anisakidae) is a nematode parasite of sea turtles, with a wide geographical distribution in marine ecosystems (1–8). Adult parasites have been described within the gastric lumen of loggerhead (Caretta caretta), Kemp's ridley (Lepidochelys kempii) and green turtles (Chelonia mydas), in association with variable degrees of ulcerative gastritis (3, 9, 10). Infection by S. sulcata has been extensively reported within the Mediterranean basin, in particular from neritic grounds, such as the Adriatic Sea (5, 10–13), the African shelf (1) and coastal waters off Campania region (6). Higher prevalence and abundance of S. sulcata in larger turtles, feeding in inshore waters is expected, due to the higher probability of ingestion of mollusc intermediate hosts (13). Several species of bivalves have been reported to carry infectious larvae of S. sulcata in waters off America and Australia (9, 14–18), including species of the family Pectinidae. Molluscs of this family are part of loggerhead turtle diet also in the Adriatic feeding grounds (19) but no intermediate hosts have been yet described for S. sulcata within this basin. The life cycle of S. sulcata has been experimentally studied by Berry and Cannon (3). Larvae hatched from eggs shed with turtles’ feces can infect molluscs through inhalation in the siphon; third stage larvae are generally found in the adductor muscle of the invertebrates. After molting to L4, larvae of about 5 mm length are able to infect sea turtles through ingestion and eggs deposition begins about 6 months after infection.
Even though a low risk for warm-blooded animals has been demonstrated by experimental infections (3), zoonotic potential of S. sulcata still has to be completely excluded. Besides, presence of larval stage S. sulcata in edible scallops has substantial implications in the depreciation of the product, as well as consequences for health and hygiene requirements following legislation.
Here, we report on the identification of two intermediate hosts of S. sulcata in the Adriatic Sea; samples of S. sulcata from definitive and intermediate hosts and from different geographical areas have been molecularly characterized and the genomic features have been compared between different developmental stages and marine basins (Adriatic vs. Tyrrhenian).
Intact adult specimens of S. sulcata (n = 19) were isolated from the stomach and upper part of the intestine of loggerhead turtles stranded along NW Adriatic (n = 10) and Tyrrhenian coasts (n = 9) of Italy in the period 2012–2015. Adult parasites (n = 12), with intact cephalic end and tail, were clarified in Amman's lactophenol solution and subsequently observed at light microscope (Olympus, ACH 40X-2) by NIS-Elements D software (Nikon). Before clarification with Amman's lactophenol, a fragment of all adult specimens was obtained and used for molecular study.
Larvae of nematodes (n = 277) were isolated from the adductor muscle of 35 specimens of Pecten jacobaeus obtained during regular sanitary inspection by the Sanitary Services of Public Health Authority in Venice province. The specimens belonged to four different production batches, collected between May 2017 and March 2018. Additionally, two similar, non-viable larvae were recovered from two specimens of Aequipecten opercularis in May 2019. All specimens of P. jacobeus and A. opercularis were from Northern Adriatic (FAO zone 37.02.01).
An overall number of 10 larvae, selected from all batches and obtained from both mollusc species were submitted for morphological characterization. Twenty larvae, included the just aforementioned 10 individuals, were included in the molecular study. Morphometric features of adult and larvae have been compared using keys available in literature (3, 20).
Since all turtles were found to be already dead at the time of stranding, no ethical approvals were required for the development of this study.
DNA was isolated from adult nematodes (n = 19) and larvae (n = 20) using the extraction kits NucleoSpin® Tissue Kit (Macherey-Nagel, Germany) and the QIAamp DNA Mini Kit (Qiagen), respectively, according to manufacturer's instructions. The entire rDNA fragment comprising ITS1, 5.8S, and ITS2 were amplified using previously described primers NC5 (5′-GTA GGT GAA CCT GCG GAA GGA TCA TT-3′) and NC2 (5′-TTA GTT TCT TTT CCT CCG CT-3′) by polymerase chain reaction (21). The mt-cox1 and cox2 genes were amplified as well, with primers JB3 (5′-TTT TTT GGG CAT CCT GAG GTT TAT-3′) and JB4.5 (5′-TAA AGA AAG AAC ATA ATG AAA ATG-3′) (22) and primers 211F (5′-TTT TCT AGT TAT ATA GAT TGR TTY AT-3′) and 210R (5′-CAC CAA CTC TTA AAA TTA TC-3′) (23), respectively.
The PCR for ITS region was performed in a 50 μl reaction, comprising 1 μl DNA, 1.5 mM MgCl2, 0.2 mM dNTPs (Thermo Fisher Scientific), 1X PCR buffer, 0.25 μM of each forward and reverse primer, 0.025 U Platinum Taq DNA Polymerase (Invitrogen), with the remainder of the volume made of sterile water. Cycling conditions comprised an initial activation step at 95°C for 2 min., followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 75 s, with a final extension step of 72°C for 10 min.
The PCR for cox1 region was performed in a 30 μl reaction volume, comprising 1–3 μl DNA, 2.5 mM MgCl2, 0.5 mM dNTPs (MBI Fermentas, Germany), 1X PCR buffer, 1.25 μM of each forward and reverse primer, 1U Platinum Taq DNA Polymerase (Invitrogen), with the remainder of the volume made of sterile water. Cycling conditions comprised an initial activation step at 95°C for 2 min., followed by 35 cycles of 94°C for 40 s, 50°C for 30 s, 72°C for 30 s, with a final extension step of 72°C for 5 min.
The PCR for cox2 region was performed in a 50 μl reaction volume comprising 5 μl DNA, 2.5 mM MgCl2, 0.2 mM dNTPs (Thermo Fisher Scientific, USA), 1X PCR buffer, 0.5 μM of each forward and reverse primer, 2U AmpliTaq DNA Polymerase (Invitrogen™, USA), with the remainder of the volume made of sterile water. Cycling conditions comprised an initial activation step at 95°C for 10 min., followed by 50 cycles of 94°C for 30 s, 48°C for 30 s, 72°C for 45 s, with a final extension step of 72°C for 7 min. Negative controls were subjected to amplification together with experimental samples in all PCR assays, as well as a positive control, represented by DNA of Anisakis simplex sensu stricto (previously identified and provided by the National Reference Center for Anisakiasis—Istituto Zooprofilattico Sperimentale della Sicilia).
The PCR products were resolved in 1% agarose gel with GelRed® Nucleic Acid Gel Stain 10.000X (Biotium, USA) and SYBR® Safe DNA gel stain (Invitrogen™, USA). The amplicons of PCR (fragments of expected size: 900 bp for ITS, 710 bp and 610 bp for coxI and cox2, respectively) were directly sequenced by Macrogen (Macrogen Europe, the Netherland) or at the Istituto Zooprofilattico Sperimentale delle Venezie trough the ABI Prism 3130xl Genetic Analyzer (Life Technologies).
The chromatograms were corrected using the software ChromasPro version 2.4.3 (Technelysium Pty Ltd, Australia). The consensus sequences were assembled with the program SeqMan available in the DNAstar package. The consensus sequences were compared with the non-redundant data base available in the GenBank database using the software BLAST (24).
Obtained sequences were aligned using MAFFTv7.450 (25) and the alignment was visually inspected to evaluate the quality and identify potential misalignments, suggestive of poor quality sequences. Sequences ends were trimmed to obtain a final alignment where a full coverage was present in the whole considered region. An additional alignment obtained concatenating cox1, cox2 and ITS genes was also created. Raw genetic distances were calculated using the ape package in R.
The sequence suitability for phylogenetic analyses was assessed by likelihood mapping analysis, performed using IQ-TREE (26) and phylogenetic tree were reconstructed with the same software, selecting as substitution model the one with the lowest Akaike Information Criterion (AIC), calculated using jModelTest (27). To evaluate if considered genes could be suitable for species identification, a collection of reference sequences of the same genes from individuals belonging to the family Anisakidae was downloaded from Genbank. Obtained sequences were aligned with those obtained in the present study and trimmed to obtain a final alignment where a full coverage was present. To reduce the number of sequences and increase the results interpretability, only one individual was selected as representative of all unique sequences (i.e., 100% percentage of identity). Phylogenetic trees were reconstructed using the previously described approach.
Adult and immature adult specimens were found free within gastric lumen or fixed with the cephalic end in the stomach wall. Morphological features were in accordance with the description of S. sulcata by Berry and Cannon (3) and are here briefly reported, together with measurements (in μm).
Cephalic end with three conspicuous lips and interlabia. Excretory pore observable at the base of the lips. Nerve ring at 828 ± 171 from the apical end. Esophagus, 4,705 ± 1,234 long, terminates in the ventriculum (length 995 ± 542, width 512 ± 215), without any diverticula. An intestinal caecum is present as a small sac-like structure parallel to the ventriculum. Spicules well-visible, cuticularized (1,735 ± 884 long); several pre-cloacal and post-cloacal papillae are visible.
Larvae collected from the adductor muscle of P. jacobeus and A. opercularis showed morphological features compatible with stage L4 of S. sulcata (Figure 1). More in detail, the larvae, from 9,737 to 21,752 μm long, showed rudimental lips, well-separated from the body, with no ventral tooth; excretory pore was visible at 92.4 ± 33.2 from the extremity, slightly caudal to the lips. A ventriculum (304.4 ± 72 μm long) without lateral diverticula was visible, ending with a rounded fund; intestinal caecum was visible in some specimens as a rudimental sac lateral to the ventriculum, developing cranially. Esophagus 1,522 ± 378 long. Nerve ring at 92 ± 33 from the apical end. A mucron was visible on the tip of the tail.
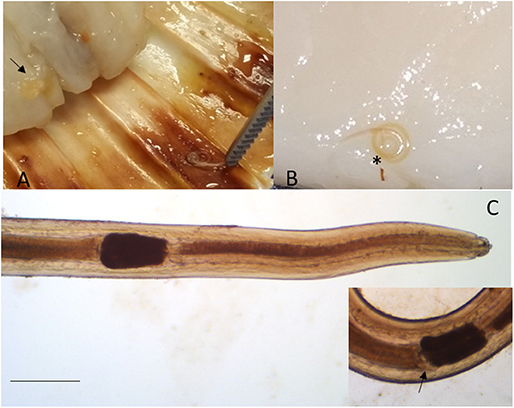
Figure 1. Sulcascaris sulcata larvae L4. In (A), larvae (black arrow) are visible within the adductor muscle of the scallop P. jacobeus. In (B), enlargement of the dissected muscle with one coiled larva (*). In (C), anterior part of L4 showing the ventricle (*) and caecum (inset, black arrow) (scale bar: 500 μm).
Amplification and sequencing of ITS, cox1 and cox2 regions resulted in 39 good quality consensus sequences for each marker. The BLAST research in GenBank revealed Contracaecum spp. to provide the most significant alignment with ITS and cox1 sequences from our specimens (86% of alignment with Contracaecum osculatum [accession number: MN428821] for ITS, 87–88% of alignment for cox1 with Contracaecum ogmorhini [AJ616898]), being the sequences of these two DNA fragments absent in Genbank for the species S. sulcata. Conversely, cox2 sequences blasted with 100% of identity with sequences of S. sulcata already present in the database (HQ328505). Newly produced sequences were deposited in GenBank under accession numbers MN712341-MN712379 (ITS), MN713844-MN713882 (cox1), and MN713883-MN713921 (cox2).
Independently of the considered gene, the collected samples displayed a high genetic similarity (Table 1). No geographic clustering could be identified between sequences obtained from samples collected in the Adriatic or Tyrrhenian Sea (Figure 2), and a clear overlapping in genetic distances within and between basins could be observed (Supplementary Figure 1). Especially, some parasites sampled in the two considered basins showed identical or extremely related sequences (p-distance lower than 0.005), at least in the considered genes.
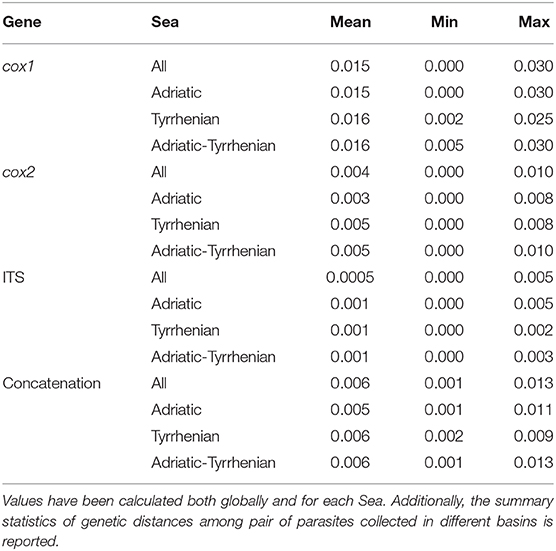
Table 1. Summary statistics of the pairwise genetic distance (p-distance) calculated for cox1, cox2, ITS and their concatenation.
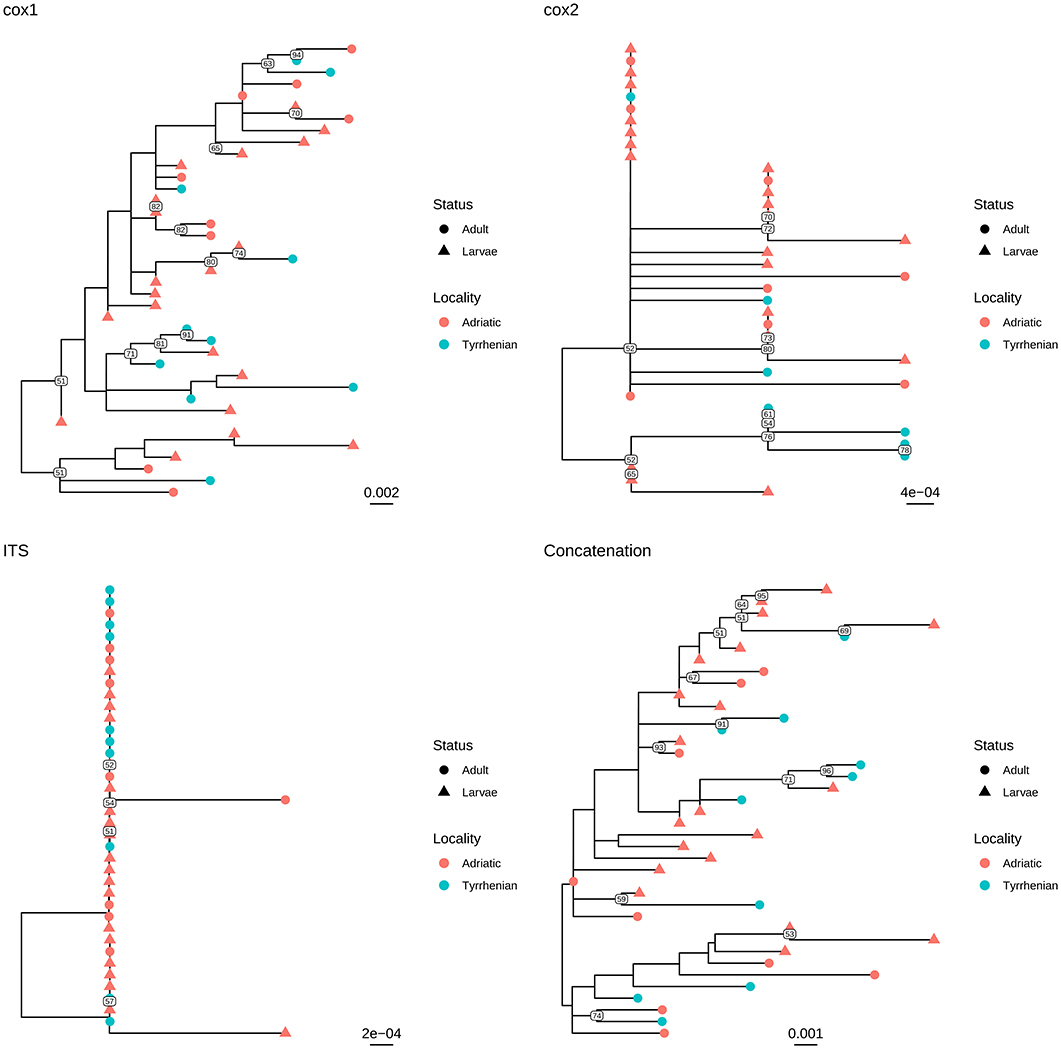
Figure 2. Maximum likelihood phylogenetic tree reconstructed based on the partial sequences of cox1, cox2, and ITS genes obtained in the present study. A phylogenetic tree was reconstructed also based on the concatenation of the above mentioned genes. The different collection localities have been color coded, while developmental stages have been marked with different tip symbols.
Similar results were observed when adults and larvae were compared. Identical sequences could be observed also between larvae collected from Aequipecten and Pecten scallops (Figure 3 and Supplementary Figure 2). Therefore, a common allele pool could be detected in the two basins (with the exception of cox1 gene) and in the different developmental stages.
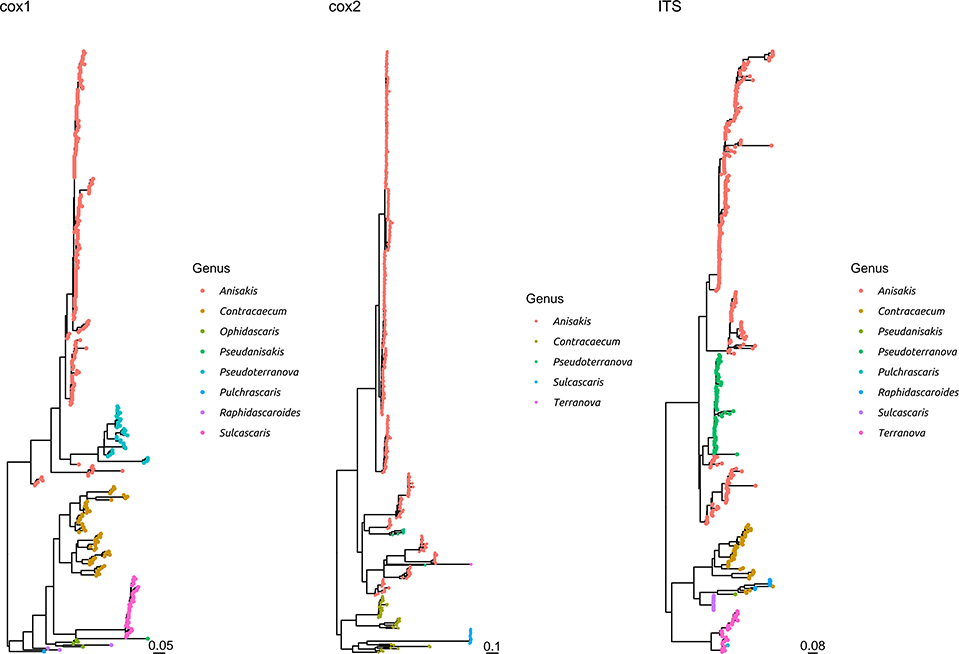
Figure 3. Maximum likelihood phylogenetic tree reconstructed based on the partial sequences of cox1, cox2, and ITS genes. The sequences obtained in the present study plus a representative dataset of other Anisakidae members have been included in the analysis. The different genera have been color coded.
The comparison with other members of the same family demonstrated that S. sulcata formed a clear monophyletic cluster, distantly related to other genera, independently from the considered gene (Figure 3 and Supplementary Figure 3).
This study reports the first identification of two intermediate hosts for S. sulcata within the Mediterranean basin, both belonging to the family Pectinidae. Larvae of S. sulcata have been reported in Australian and Western Atlantic waters from several bivalve species of Pectinidae including Pecten spp. (28), Argopecten gibbus (18), and Amusium balloti (29), and Mactridae including Spisula solidissima (2, 30). Additionally, experimental infection was successful in Isognomon ephippium (Isognomonidae) and Pinctada spp. (Pteriidae) (3) and species of Gastropoda Cypraea tigris and Fasciolaria sp. are as well included in the list of intermediate hosts of S. sulcata (15, 31). In this scenario, we can easily hypothesize that several other intermediate hosts probably exist within the Mediterranean basin for S. sulcata. Even though P. jacobeus and A. opercularis are common commercial species for human consumption, little likelihood of danger for human health exists after ingestion of S. sulcata larvae, being the elective definitive host of this nematode represented by cold-blooded vertebrates. Nevertheless, the anisakid Hysterothylacium aduncum, whose definitive hosts are cold-blooded species, is reported in humans as a rare agent of disease (32, 33) and, similarly, larvae of the nematode Echinocephalus sinensis, a parasite of elasmobranchs, have been demonstrated to be able to infect primates (34). The habits of cooking scallops before consuming them is protective toward accidental infections, nevertheless, presence of larvae or discolouration of the adductor muscle, due to the presence of the symbiont protozoan Urosporidium spisuli (35), justifies the depreciation when not the exclusion of the product from the market when infected by S. sulcata larvae.
Infections by S. sulcata are described in the Mediterranean population of loggerhead turtles with prevalence ranging around 20% (6, 10, 13), with significantly higher values among loggerhead turtles feeding in Adriatic compared to Tyrrhenian, up to 30% (10). The particular ecological features of the Adriatic basin together with its variety and abundance of suitable preys, make it one of most populated neritic foraging ground for loggerhead turtles within the Mediterranean Sea. Several studies have emphasized the importance of benthic molluscs in loggerhead turtle diet, with large sized species being highly represented and constituting an energetically valuable food item (36, 37). A study by Lazar et al. (19) upon loggerhead turtles feeding in the Northern Adriatic grounds also reported molluscs to be an essential part of sea turtles' diet. Crushed shells of the large-sized bivalves A. opercularis and P. jacobaeus were identified among the preys in that study, thus we can assume that larvae of S. sulcata within the adductor muscle of specimens of the aforementioned bivalve species reasonably can reach the definitive hosts during the turtles' foraging in Northern Adriatic waters. The fidelity of sea turtles to selected foraging grounds indeed promotes completion of the life cycle of the nematode in specific areas, with turtles acting as perpetrators of the parasites life cycle by shedding negatively-buoyant eggs with their feces. Nevertheless, long distance migration of turtles could justify also the expansion of the geographical range of the parasites. This is supported by molecular evidence in this study, which highlights the genetic similarity between parasites isolated in Adriatic and Tyrrhenian waters. In fact, the analysis of three different genes demonstrated the presence of an essentially common allele pool in the two basins, as well as the absence of any geographical clustering among the sequences collected in the Adriatic and Tyrrhenian Seas (Figure 2). Although the limited phylogenetic signal warrants caution in phylogenetic analysis evaluation, the results of individual and concatenated genes analysis suggest a wide circulation of those parasites in the Mediterranean, likely mediated by final host migration. While studies on loggerhead turtles movements within Mediterranean region suggest a tendency to foraging grounds fidelity and shorter range movements, exceptions have been reported, especially with juveniles showing a propensity to wander over quite large areas. Moreover, a clear overlapping is present between typical movements areas of northern and southern Adriatic, Ionian, south-central Mediterranean and Tyrrhenian Seas, which could facilitate parasite transmission and dispersal (38). Unfortunately, the absence of S. sulcata sequences collected in other regions hinders a more detailed analysis and further, collaborative, efforts will be necessary to understand the molecular epidemiology and phylogeography of this parasite.
The high genetic homogeneity, especially in the ITS and, to a lesser extent, cox2 genes (lower than 1% maximum genetic distance) demonstrates that the investigated genes could be considered an adequate target for species identification. This is confirmed by the comparison with other Anisakidae members, highlighting that S. sulcata formed a clearly independent cluster. Nevertheless, a broader collection of S. sulcata sequences from individuals sampled all around the world would be of benefit to establish clear species boundaries.
Based on these evidences, the high percentage of identity of the analyzed genes between adults and larvae confirmed the morphological classification of the latter as S. sulcata (Figure 2). Molecular methods could thus safely replace morphological methods for larvae identification, coupling a greater easiness of execution and reliability. Additionally, the clear overlapping in genetic distance distribution (Supplementary Figure 2) between strain collected in Aequipecten or Pecten scallops demonstrates for the first time that this parasite is able to infect also Aequipecten species and confirms the broad intermediate host tropism.
The datasets generated for this study can be found in the GenBank MNMN712341–MN712379, GenBank MN713844–MN713882, GenBank MN713883–MN713921.
FM contributed to the conceptualization of the study, coordinated sample collection, performed morphological studies on the parasites and provided funding for the study. FT provided samples, performed molecular analyses and morphological observations on larvae, and revised the paper. GF performed sequences and phylogenetic analyses and drafted the paper. SR and AV provided samples, performed molecular analyses and morphological observations, and revised the paper. MS provided samples and revised the paper. EM performed morphological and molecular analyses and drafted the paper.
This work was financially supported by a Research Project of Padova University (Prot. DOR1901948/19).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge Dr. Giorgia Dotto and Dr. Cinzia Tessarin (Department of Animal Medicine, Production and Health, University of Padova, Italy) for their kind support in the production and elaboration of molecular data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00064/full#supplementary-material
1. Sey O. Examination of helminth parasites of marine turtles caught along the Egyptian coast. Acta Zool Hung. (1977) 23:387–94.
2. Lichtenfels JR, Bier JW, Madden PA. Larval anisakid (Sulcascaris) nematode from atlantic molluscs with marine turtles as definitive host. Trans Am Microsc Soc. (1978) 97:199–207. doi: 10.2307/3225593
3. Berry GN, Cannon LRG. The life history of Sulcascaris sulcata (Nematoda: Ascaridoidea), a parasite of marine molluscs and turtles. Int J Parasitol. (1981) 11:43–54. doi: 10.1016/0020-7519(81)90024-2
4. Vicente JJ, Rodrigues HO, Gomes DC, Pinto RM. Nematóides do Brasil. III Nematóides de répteis. Rev Bras Zool. (1993) 10:19–168. doi: 10.1590/S0101-81751993000100003
5. Manfredi MT, Piccolo G, Meotti C. Parasites of sea turtles. II. Loggerhead turtles (Caretta caretta [Linnaeus, 1758]). Parassitologia. (1998) 40:305–8.
6. Santoro M, Badillo FJ, Mattiucci S, Nascetti G, Bentivegna F, Insacco G, et al. Helminth communities of loggerhead turtles (Caretta caretta) from Central and Western Mediterranean Sea: the importance of host's ontogeny. Parasitol Int. (2010) 59:367–75. doi: 10.1016/j.parint.2010.04.009
7. Werneck MR, Martos Thomazini C, Shigueru Mori E, Thereza Gonçalves V, Gomes Gallo BM, Da Silva RJ. Gastrointestinal helminth parasites of loggerhead turtle Caretta caretta Linnaeus 1758 (Testudines, Cheloniidae) in Brazil. Pan-Ame J Aquat Sci. (2008) 3:351–4.
8. Greiner EC. “Parasites of marine turtles.” In: Wyneken J, Lohmann KJ, Musick JA, editors. The Biology of Sea Turtles. Boca Raton, FL: CRC Press (2013). p. 427–46.
9. Lester RJG, Blair D, Heald D. Nematodes from scallops and turtles from Shark Bay, Western Australia. Aust J Mar Freshwater Res. (1980) 3:713–7. doi: 10.1071/MF9800713
10. Santoro M, Marchiori E, Iaccarino D, Uberti BD, Cassini R, Di Nocera F, et al. Epidemiology of Sulcascaris sulcata (Nematoda: Anisakidae) ulcerous gastritis in the Mediterranean loggerhead sea turtle (Caretta caretta). Parasitol Res. (2019) 118:1457–63. doi: 10.1007/s00436-019-06283-0
11. Piccolo G, Manfredi MT. New reports on parasites of marine turtles stranded along the Italian coasts (2001). In: Proceedings of the First Mediterranean Conference on Marine Turtles. Rome. p. 271.
12. Scaravelli D, Gustinelli A, Nardini G, Cucinotta G, Affronte M, Trentini M, et al. “A parasitological survey of loggerhead turtles (Caretta caretta) from the northern Adriatic Sea.” In: Demetropoulos A, Türkozan O, editors. Proceedings of 2nd Mediterranean Conference on Marine Turtles. Antalya: Barcelona Convention — Bern Convention – Bonn Convention (CMS). p. 45.
13. Gračan R, Buršić M, Mladineo I, Kučinić M, Lazar B, Lacković G. Gastrointestinal helminth community of loggerhead sea turtles Caretta caretta in the Adriatic Sea. Dis Aquat Org. (2012) 99:227–36. doi: 10.3354/dao02490
14. Lichtenfels JR, Kern FG, Zwerner DE, Bier JW, Madden PA. Anisakid nematodes in shellfish of Atlantic continental shelf of North America. Trans Am Microsc Soc. (1976) 95:265–6.
15. Lichtenfels JR, Sawyer TK, Miller GC. New hosts for Larval Sulcascaris sp. (Nematoda, Anisakidae) and prevalence in the Calico Scallop (Argopecten gibbus). Trans Am Microsc Soc. (1980) 99:448–51. doi: 10.2307/3225656
16. Amato JFR, Amato SB. Sulcascaris sulcata (Nematoda, Anisakinae) infecting sea scallops being exported from southeastern Brazil. Arquivos da Universidade Federal Rural do Rio de Janeiro. (1982) 5:61–7.
17. Barber BJ, Blake NJ, Moyer MA, Rodrick GE. Larval Sulcascaris sulcata from calico scallops, Argopecten gibbus, along the Southeast coast of the United States. J Parasitol. (1987) 73:476–80. doi: 10.2307/3282124
18. Deardorff TL. Occurrence of larval Sulcascaris sulcata (Nematoda: Anisakidae) in the calico scallop, Argopecten gibbus, collected along the eastern coast of Florida, with comments on histopatology. Proc Helminthol Soc Wash. (1989) 56:82–5.
19. Lazar B, Gračan R, Katić J, Zavodnik D, Jaklin A, Tvrtković N. Loggerhead sea turtles (Caretta caretta) as bioturbators in neritic habitats: an insight through the analysis of benthic molluscs in the diet. Mar Ecol. (2010) 32:65–74. doi: 10.1111/j.1439-0485.2010.00402.x
20. Anderson RC, Chabaud AG, Willmott S. Keys to the Nematode Parasites of Vertebrates. Wallingford: CABI International (2009).
21. Zhu X, D'Amelio S, Paggi L, Gasser RB. Assessing sequence variation in the internal transcribed spacers of ribosomal DNA within and among members of the Contracaecum osculatum complex (Nematoda: Ascaridoidea: Anisakidae). Parasitol Res. (2000) 86:677–83. doi: 10.1007/PL00008551
22. Bowles J, Blair D, McManus DF. Genetic variants of the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. (1992) 54:165–73. doi: 10.1016/0166-6851(92)90109-W
23. Garbin L, Mattiucci S, Paoletti M, Gonzàlez-Acuña D, Nascetti G. Genetic and morphological evidences for the existence of a new species of Contracaecum (Nematoda: Anisakidae) parasite of Phalacrocorax brasilianus (Gmelin) from Chile and its genetic relationships with congeners from fish-eating birds. J Parasitol. (2011) 97:476–92. doi: 10.1645/GE-2450.1
24. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
25. Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. (2019) 20:1160–6. doi: 10.1093/bib/bbx108
26. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. (2015) 32:268–74. doi: 10.1093/molbev/msu300
27. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. (2012) 9:772. doi: 10.1038/nmeth.2109
29. Sprent JFA. Ascaridoid nematodes of amphibians and reptiles: Sulcascaris. J Helminthol. (1977) 51:379–87. doi: 10.1017/S0022149X00007744
30. Payne WL, Gerding TA, Dent RG, Bier JW, Jackson GJ. Survey of the U.S. Atlantic coast surf clam, Spisula solidissima, and clam products for Anisakine nematodes and hyperparasitic protozoa. J. Parasitol. (1980) 66:150–3. doi: 10.2307/3280608
31. Cannon LRG. A larval ascaridoid nematode from Queensland scallops. Int J Parasitol. (1978) 8:75–80. doi: 10.1016/0020-7519(78)90055-3
32. Yagi K, Nagasawa K, Ishikura H, Nakagawa A, Sato N, Kikuchi K, et al. Female worm Hysterothylacium aduncum excreted from human: a case report. Japan J Parasitol. (1996) 45:12–23.
33. González-Amores Y, Clavijo-Frutos E, Salas-Casanova C, Alcain-Martínez G. Direct parasitological diagnosis of infection with Hysterothylacium aduncum in a patient with epigastralgia. Rev Esp Enferm Dig. (2017) 107:699–700.
34. Ko RC. Experimental infection of mammals with larval Echinocephalus sinensis (Nematoda: Gnathostomatidae) from oysters (Crassostrea gigas). Can J Zool. (2011) 54:597–609. doi: 10.1139/z76-070
35. Gosling E. Reproduction, settlement and recruitment. In: Gosling E, editor. Bivalve Molluscs: Biology, Ecology and Culture. Oxford, UK: Blackwell Science: Fishing News Books (2003). p. 390–1.
36. Laurent L, Lescure J. L'hivernage des tortues marines caouannes Caretta caretta (L.) dans le sud de Tunisien. Rev. Ecol.- Terre Vie. (1994) 49:63–86.
37. Casale P, Abbate G, Freggi D, Conte N, Oliverio M, Argano R. Foraging ecology of loggerhead sea turtles Caretta caretta in the central Mediterranean Sea: evidence for a relaxed life history model. Mar Ecol Prog Ser. (2008) 372:265–76. doi: 10.3354/meps07702
Keywords: Sulcascaris sulcata, life cycle, Pectinidae, molluscs, Anisakidae, Mediterranean Sea, Caretta caretta
Citation: Marcer F, Tosi F, Franzo G, Vetri A, Ravagnan S, Santoro M and Marchiori E (2020) Updates on Ecology and Life Cycle of Sulcascaris sulcata (Nematoda: Anisakidae) in Mediterranean Grounds: Molecular Identification of Larvae Infecting Edible Scallops. Front. Vet. Sci. 7:64. doi: 10.3389/fvets.2020.00064
Received: 11 December 2019; Accepted: 27 January 2020;
Published: 14 February 2020.
Edited by:
Donato Traversa, University of Teramo, ItalyReviewed by:
Jan Slapeta, University of Sydney, AustraliaCopyright © 2020 Marcer, Tosi, Franzo, Vetri, Ravagnan, Santoro and Marchiori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erica Marchiori, ZXJpY2EubWFyY2hpb3JpQHVuaXBkLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.