
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 14 January 2020
Sec. Veterinary Epidemiology and Economics
Volume 6 - 2019 | https://doi.org/10.3389/fvets.2019.00501
This article is part of the Research TopicRisk-Based Evidence for Animal Health PolicyView all 18 articles
 Emily Walz1†
Emily Walz1† Jamie Middleton2†‡
Jamie Middleton2†‡ Fernando Sampedro3
Fernando Sampedro3 Kimberly VanderWaal4
Kimberly VanderWaal4 Sasidhar Malladi1
Sasidhar Malladi1 Timothy Goldsmith4*
Timothy Goldsmith4*In the event of a Food and Mouth Disease (FMD) outbreak in the United States, an infected livestock premises is likely to result in a high number of carcasses (swine and/or cattle) as a result of depopulation. If relocating infected carcasses to an off-site disposal site is allowed, the virus may have increased opportunity to spread to uninfected premises and result in exposure of susceptible livestock. A stochastic within-herd disease spread model was used to predict the time to detect the disease by observation of clinical signs within the herd, and the number of animals in different disease stages over time. Expert opinion was elicited to estimate depopulation parameters in various scenarios. Disease detection was assumed when 5% of the population showed clinical signs by direct observation. Time to detection (5 and 95th percentile values) was estimated for all swine farm sizes (500–10,000 head) ranged from 102 to 282 h, from 42 to 216 h for all dairy cattle premises sizes (100–2,000 head) and from 66 to 240 h for all beef cattle premises sizes (5,000–50,000 head). Total time from infection to beginning depopulation (including disease detection and confirmation) for the first FMD infected case was estimated between 8.5–14.3 days for swine, 6–12.8 days for dairy or beef cattle premises. Total time estimated for subsequent FMD cases was between 6.8–12.3 days for swine, 4.3–10.8 days for dairy and 4.5–10.5 days for beef cattle premises. On an average sized operation, a sizable proportion of animals in the herd (34–56% of swine, 48–60% of dairy cattle, and 47–60% of beef cattle for the first case and 49–60% of swine, 55–60% of dairy cattle, 56–59% of beef cattle for subsequent cases) would be viremic at the time of beginning depopulation. A very small fraction of body fluids from the carcasses (i.e., 1 mL) would contain virus that greatly exceeds the minimum infectious dose by oral (4–7x) or inhalation (7–13x) route for pigs and cattle.
Foot and mouth disease (FMD) is a highly contagious viral disease affecting primarily cloven-hoofed animal including key livestock production species such as cattle and swine. In the event that a case of FMD were detected in the United States (US), there would likely be serious economic impact on international trade of animals and animal products (1). The US has a preparedness and response plan for disease control and eradication in the event of a foreign animal disease event. This plan encompasses management of multiple animal species, and may include movement control, quarantine, vaccination, and depopulation measures (2).
Identification of FMD within a herd relies upon observation of clinical signs to trigger diagnostic testing of suspect individuals. Testing methods for population-level disease surveillance are lacking; this likely results in delayed detection until infection has spread at the farm level. Experimental and modeling studies of transmission in cattle (3) and swine (4) suggest that the infectious period in these species as close as under 24 h before the onset of clinical signs (fever or lesions). This underscores the important role of prompt detection by clinical signs to limit spread throughout the herd. Similarly, early detection decreased the length of epidemics in a multi-species model based on a cattle and feedlot-dense region of Texas, USA (5).
Depopulating an infected premises is performed to prevent further spread of Foot and Mouth Disease virus (FMDv) to susceptible animals and to limit additional FMDv shedding in latently or clinically infected individuals. If an outbreak were concentrated in a geographic area in which FMD can be readily contained without further spread, the response strategy of “stamping out” will likely be elected. “Stamping out,” or immediate depopulation, is the preferred control method for clinically infected and in-contact susceptible animals as a means to reduce the potential of disease spread. It is assumed that the depopulation procedures would follow the United States Department of Agriculture, Foreign Animal Disease Preparedness and Response Plan (FAD PReP) Guidance (2).
FMD was eradicated from the United States in 1929 (6); historical data specific to the modern large-scale agricultural operations most common in the US are lacking. Especially in areas where empirical data is lacking, expert opinion has been a mainstay in informing proactive planning for FMD incursions, and aspects such as disease characteristics in a naïve population and depopulation techniques have been described (7, 8). Models have been used as another means of understanding potential disease scenarios and as a way to inform planning decisions. Most models focus on between-herd spread, and incorporate aspects such as vaccination strategies, movement characteristics, and geographic proximity in areas with multiple species, such as cattle, goats, and swine (5, 9–12). In all these studies depopulation is one option to limit disease spread; however, management strategies for carcasses after depopulation was not considered.
Swine and cattle (beef and dairy) are the two most prevalent livestock species in the US (13, 14). If a swine or cattle premises were infected and depopulated, the option to dispose of carcasses off-site may be needed due to environmental and other limitations of disposing a large biomass on-site. It is required for trucks to be leak-proof while hauling animal carcasses according to US Code of Federal Regulations (15), however, in the event of an FMD outbreak, other means of hauling carcasses may be employed. FMDv presents a containment challenge due to its persistence in the environment, especially when it is within organic material and protected from desiccation, heat and adverse pH conditions (16). Movement of FMDv-infected carcasses represents one of the main disease spread pathways during an outbreak. Proactively evaulating the potential risk of transmission and available mitigation meaures can allow risk managers to be better prepared for these scenarios in the event of an outbreak (Figure 1).
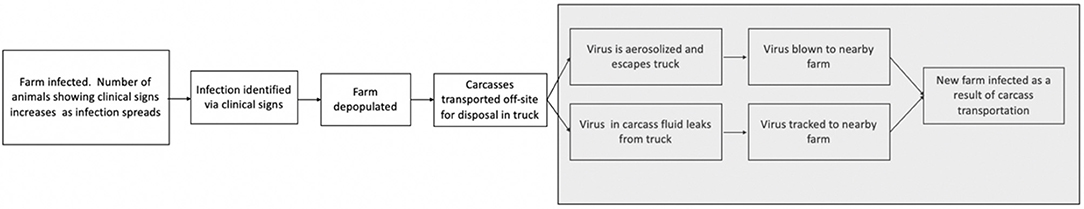
Figure 1. Disease spread pathway by which FMDv may spread during the transportation of carcasses from an infected livestock premises to an off-site disposal location. Steps in gray represent the potential transmisson risks during transportation.
The aim of this study was to evaluate the likelihood that carcasses in a truckload from a depopulated infected premises would contain an infective FMDv dose at the time of transportation to disposal. This information is an important consideration for emergency preparedness and management officials in the event of a FMD outbreak, as off-site transportation of carcasses to disposal is a potential pathway to spread virus during an outbreak.
A stochastic disease spread model was developed to simulate the transmission of FMDv within a swine, dairy or beef cattle herd and predict the proportion of viremic animals at the time of depopulation. The model was run for each of the livestock types, and it estimated the number of animals in various disease stages at each time step. Disease stages included: susceptible (S), latent (L), pre-clinical (PI), clinical (CI), and recovered (R) (17). Both pre-clinical and clinical animals were considered viremic and infectious to other susceptible animals within the herd (18, 19). The model updated the number of animals in each disease state every 6 h. The uncertainties in input variables, as well as the inherent variability associated with the course of infection in individual swine, dairy and beef cattle populations and the spread within the group were considered in the model in the form of distributions for the different parameters (transmission coefficient, duration of the latent, pre-clinical and clinical periods). Parameter distributions were obtained from previous FMD modeling studies and meta-analyses (18–20). The farm size scenarios used in the model were based on a compilation of statistics published by the National Agricultural Statistics Service (NASS) of the United States Department of Agriculture for 2014 (21). Average farm sizes were calculated for all production types within livestock category (swine, beef cattle, or dairy cattle). The model assumed that disease transmission was the same regardless of animal age. Table 1 shows the inputs used in the disease spread model.
The model assumed random mixing among the entire population. The number of susceptible animals that become infected in each time step in the model was dependent on the adequate contact rate and the proportion of infectious animals in the herd at that time step. The same contact rate was used for both index and subsequent case scenarios. The adequate contact rate (k) is defined as the mean number of other animals each infected animal comes into contact with per unit time such that the contact is adequate to transmit infection. Thus, the probability (Pt) that an animal becomes infected and the number of newly infected, latent individuals () in a given time step can be expressed as:
where N is the total population size of the farm, It is the number of infectious animals (pre-clinical or clinical) and St is the number of susceptible individuals at time t. Transitions between other disease stages (from L to PI, PI to CI and CI to R disease stages) were simulated based on the duration of each period, which was determined individually for each animal.
The disease spread model also estimated the time to detect FMD infection in the herd based on the active observation of clinical signs, which is one of the surveillance measures that may be applied in an outbreak at the herd level (23). The threshold for detection of the disease was set at 5% of the herd showing clinical signs, which was based on the percentage of naturally occurring lameness on swine and cattle farms (24, 25). A sensitivity analysis on the detection threshold (re-analyzed at 2.5 or 10%) was performed for an exemplar scenario (swine herd of 5,000 head) to ensure that time to detection distributions were not overly sensitive to changes in this threshold (Supplementary Figure 1).
Once the disease was detected at a premises, it was assumed that a depopulation protocol would be initiated by disease management officials. Total time from detection to beginning depopulation was estimated by adding each time interval by using the following equation:
where, tdet is the time elapsed to detect FMD post-infection depending on the farm size, tconf is the time interval between detection of clinical signs in a particular premises to the official laboratory confirmation of a positive sample, and tsdep is the time interval between laboratory confirmation to starting depopulation. All the time intervals were expressed in hours.
Expert opinion was solicited via email from five national experts in emergency management and depopulation procedures working in academia, industry and government settings to provide estimates on time intervals for laboratory confirmation after the detection of an infected premises and for starting the depopulation protocol (Supplementary Table 1). It was assumed that the time to complete indemnity or time to find disposal options were not included in the estimation of total time from infection to depopulation. Two scenarios (index case and subsequent cases) were given to the experts for estimating the time to start the depopulation procedure (tsdep). Input values of equation 3 for swine, dairy and beef cattle as the index case are shown in Tables 2–4. A Pert distribution was used to characterize the variability among experts' responses (26). The worst-case scenario was selected to populate the distribution by identifying the longest time interval estimates among all the experts for the minimum, most likely and maximum values. For subsequent cases, the time from disease detection to laboratory confirmation and the time from confirmation to beginning depopulation were each set at 24 h. A Monte Carlo simulation was carried out by using @Risk 6.2 for Excel (Palisade Corporation, NY). The analysis was performed using 1,000 iterations with Latin-hypercube method. Outputs were expressed by the mean and 90% prediction intervals as calculated by the 5th and 95th percentile values. The proportion of viremic and recovered animals at the time of starting depopulation was predicted from the disease transmission model at the time elapsed between infection and starting the depopulation.
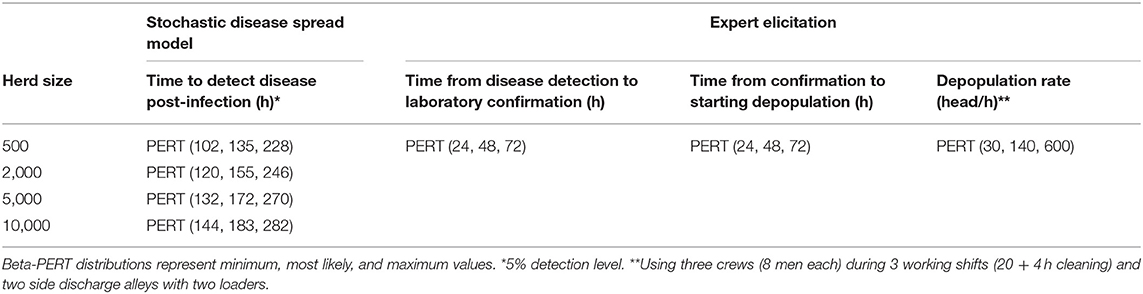
Table 2. Input values to estimate timings for depopulation procedure in case of FMD outbreak in swine premises.
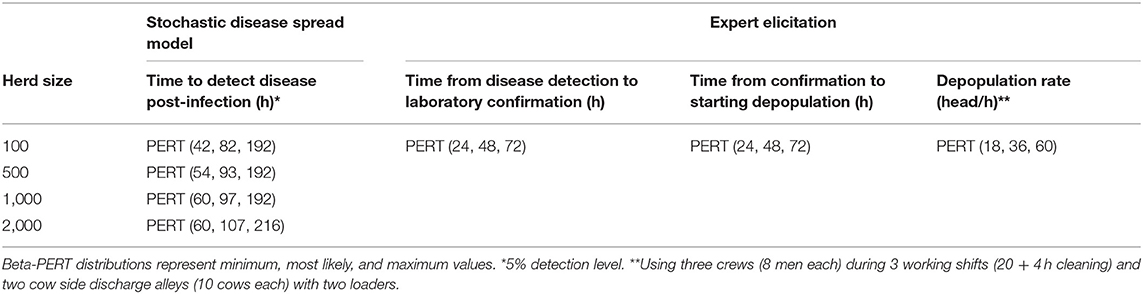
Table 3. Input values to estimate timings for depopulation procedure in case of FMD outbreak in dairy premises.
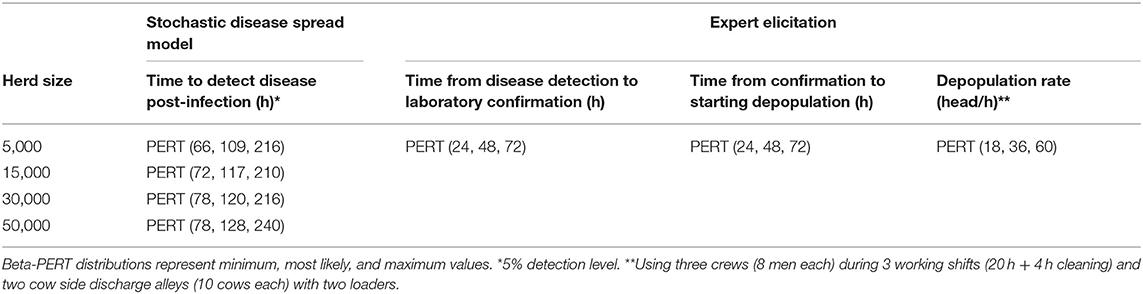
Table 4. Input values to estimate timings for depopulation procedure in case of FMD outbreak in beef cattle premises.
The disease spread model estimated the time to reach 5% of clinical animals in the herd (threshold for FMD detection by active observational surveillance). Time to detection (5 and 95th percentile values) was estimated at 102–282 h for all swine farm sizes (500–10,000 head), from 42 to 216 h for all dairy cattle premises sizes (100–2,000 head), and from 66 to 240 h for all beef cattle premises sizes (5,000–50,000 head). A sensitivity analysis of the detection threshold demonstrated that the distributions for time to detection were not sensitive to the threshold.
A sensitivity analysis was carried out to identify the time interval that had the greatest influence on the total time (from detection to finalized depopulation). As it can be seen in Figure 2, the detection time was the input variable with the greatest influence for dairy and swine premises. However, given the size (i.e., large number of animals on beef premises), the time to depopulate a farm was the interval with the greatest influence on beef premises.
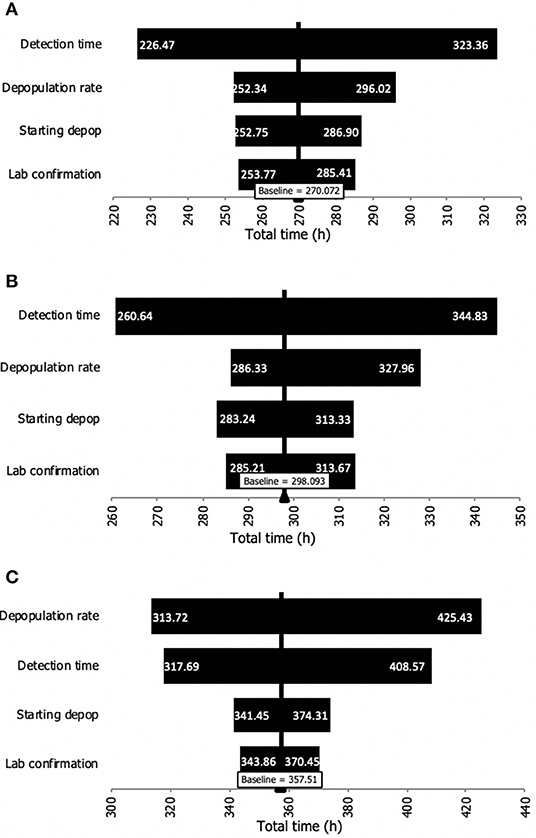
Figure 2. Sensitivity analysis of the influence of the input time intervals on the total time from detection to depopulation of premises during an FMD outbreak. (A) Dairy premises. (B) Swine premises. (C) Beef cattle premises.
Total time from infection to depopulation (90% prediction interval) for the first FMD infected case was estimated to be 8.5–14.3 days for a 3,000 head swine herd, 6.0–12.8 days for a 2,000 head dairy herd and 6.0–12.8 days for 5,000 head beef cattle premises (Tables 2–4). Total time estimated for subsequent FMD cases is reported in Table 5. A sizable proportion of animals in the herd (34–56% of swine, 48–60% of dairy cattle, and 47–60% of beef cattle for the first case, and 49–60% of swine, 55–60% of dairy cattle, 56–59% of beef cattle for subsequent cases) would be viremic at the time of depopulation (Table 6).
Model outputs suggest that if a herd is depopulated when 5% of animals show active clinical signs, a large proportion of the herd will be viremic at the time of beginning depopulation. Even in subsequent cases where it is assumed that the time to from disease detection to depopulation will be shorter (48 h), the proportion of viremic animals remains relatively unchanged. Moving infected carcasses represents a real risk for FMDv spread during an outbreak. However, in the event that “stamping out” is employed, off-site disposal is likely to be required due to the size of beef, dairy, and commercial swine premises in the US and the large amount biomass resulting from depopulation.
Virus could escape from a load of carcasses in leaked fluid, expelled fomites (e.g., dirt, feces), or jostled carcasses from the load, or via aerosolization of virus-laden particulate matter. The likelihood of a spill or aerosol event is unknown, however it is likely that even a small volume of escaped fluid may contain an infectious dose of virus. The average concentration of FMDv in a carcass in experimental inoculation studies was 103 PFU/g for a pig carcass and 106 PFU/g for a cattle carcass (27–40). Consultation with rendering industry experts revealed that for transportation of fresh, intact carcasses under normal conditions, most body fluids remain inside the carcass (personal communication, 2013). In a full load of a standard rendering truck (29–1,000 carcasses), experts estimated the amount of fluid leakage from carcasses at 20 L per load. Assuming that 1 mL of leakage contains equivalent virus to 1 g of carcass material, 1 mL of body fluids could contain 10–100,000 times higher virus quantity (103-106 PFU) than the minimum infectious dose by oral (1.4 × 104 – 1.4 × 106 PFU) and inhalation route (7–357 PFU) for pigs and cattle (41, 42). Of note, these estimates are based on literature review and experimental studies; virus loads in tissues may be different among virus strains and subtypes or in non-experimental conditions, however, this data was not available for extrapolation.
The environmental conditions which favor airborne FMDv spread are high humidity, low precipitation, low to moderate wind speed, and flat terrain (43). Suitable conditions of relative humidity (RH) above 60% and temperatures below 33°C (91°F) are needed for long-range airborne transmission to be possible. FMDv bioaerosols degrade quickly in RH below 55% due to desiccation (44). Precipitation generally reduces atmospheric bioaerosol concentrations, while high levels of turbulence temporarily increase aerosolized concentrations when dust is raised (45). In longer-range airflows, turbulence eventually causes dilution of FMD bioaerosol concentrations and higher gravitational sedimentation, especially in particles smaller than 10 micrometers (46). Sunlight has minimal effect on the aerosol spread of FMDv, and instead mostly affects survival on surfaces (46). While beyond the scope of this study, further work on the risks of aerosol spread may be warranted if off-site carcass transportation is considered.
A standard rendering truck is outfitted with sealed tailgate and tarp cover to prevent spills or aerosolization, however, it is unlikely that this will completely mitigate risk of virus escape from a load. In the event of an outbreak, other truck types may be employed due to increased demand for timely carcass disposal. A standard rendering truck, roll-off, or dump truck without tarp covering would have an increased likelihood of spillage, due to the proximity of carcasses and other contaminated debris to the top of the trailer in a full load. The use of a sealed plastic bag suitable for the disposal of biological residues is an option provide full protection against spillage and aerosolization. In the event that new or different types of equipment are employed, or that new personnel lacking adequate training are used during an outbreak, the potential that standard mitigation measures may be misused due to human error cannot be underestimated.
Due to the proactive nature of this assessment, some assumptions in calculations were made which may limit this model's applicability in the event of an outbreak. For example, in estimating the time until FMD detection on a farm, only direct animal-animal contact was considered in disease spread, however, in some geographies or production systems, aerosol or fomite (contaminated person/equipment) may also contribute to spread. In addition, the presence of segregated or sub-herds within a population would change the contact rate and the number of animals with viremia at different time points. However, an analysis by Kinsley showed that adding within-farm population structure did not substantially influence time to detection or time to the peak of the epidemic (47). Additionally, although a change in time to detection (either shorter or longer) could influence our results, we did not find time to detection to be influenced by the detection threshold (percent of animals clinical). Part of the reason for this is the high transmission rate of the virus. By the time 5% of the animal are clinical, transmission is in its exponential growth phase (48), and the difference between time until 5 vs. 10% are clinically infected is very small. In addition, our results for time to detection, derived from the stochastic model, were consistent with an analysis of real-world data from the UK epidemic, where the probability of a farm escaping detection fell sharply at around 7 days and was negligible by 12–13 days (49).
In the event of an especially large infected premises, such as a feedlot operation or an integrated farrow to finish swine operation, depopulation (even at efficient speeds) may last weeks to months. The proportion of viremic animals near the end of a depopulation effort and after significant time has elapsed in disease progression is likely markedly different than that which was calculated at the start of depopulation. Further modeling of this disease progression is an area for further work which may be instrumental in planning for management of large infected premises.
In calculating length of time to depopulation, it was assumed that the disposal site was identified and secured before the outbreak, and no additional delays in depopulation or transportation of carcasses occurred as a result of having to locate an acceptable disposal site. It was assumed that the time from depopulation to movement of carcasses to the disposal site would be very short (a matter of hours), so the potential for body fluids to escape from carcasses (leakage) will be minimized. In the event that depopulation or movement of carcasses from euthanasia location into transport vehicle is delayed, it is likely that larger amounts of body fluids may be present, and risks associated with leakage from carcasses may become more significant. Additional delays in transportation or increased duration of transportation to distant disposal sites can be expected to have similar effects on increased leakage as additional body fluids and products of autolysis escape from a carcass.
Finally, this study did not consider issues related to capacity, resource availability, and resource depletion. A large number of infected premises over an extended time period would have the potential to deplete available resources as well as capacity. This would likely result in longer delays in identification, depopulation and disposal. In this event, the herds continue to progress toward a recovered stage, and the proportion of the herd which is viremic will continue to decrease, while the potential for viral contamination of the premises will increase. Issues of logistics and animal welfare must be balanced with the potential for depopulation to decrease the number of potential animal hosts in a local area.
In the event of an FMD outbreak in the US, significant time will lapse between infection of a livestock premises and beginning depopulation. During this time, disease continues to spread throughout a herd, and it is likely that a large proportion of animals will be viremic at the time of depopulation, even if disease confirmation and beginning depopulation occurs in a timely manner. Given that even a small amount of leakage from viremic carcasses is likely to contain FMDv concentrations that will exceed the minimum FMD infective dose for pigs and cattle by several degrees of magnitude, it appears that leakage from vehicles transporting viremic carcasses to off-site disposal locations represent a real risk for virus spread during an outbreak. Delays in identification, depopulation and disposal will likely result in greater number of animals that are in the recovered stage.
This study can inform the risk assessment of FMD transmission during the movement of infected carcasses, and should be valuable for risk managers when considering emergency response options. In addition, this can help federal and state agencies to adopt additional risk mitigation measures to reduce the likelihood of infection of susceptible livestock during an FMD outbreak in the US.
All datasets generated for this study are included in the article/Supplementary Material.
TG and FS led project development and provided oversight for this work. FS, SM, and KV developed the modeling components for this study. JM participated in drafting initial reports and literature review. EW updated and synthesized previous work and wrote the manuscript. All authors reviewed and provided critical feedback on the manuscript before submission.
This project was developed and funded through a sub-award 000000042653 with West Texas A&M University through primary award # 12-9100-1366-CA from USDA-APHIS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We sincerely thank all those who contributed to expert opinion Dyan L. Pratt (University of Saskatchewan), Tom Beseman (Central Bi-Products), Thomas Kuehn and Peter Raynor (University of Minnesota), Shawn M. Guetter (Redwood Metal Works), Lori P. Miller (Department of Homeland Security), Robert E. De Otte Jr. and Donald Topliff (West Texas A&M University), David Finch (Texas Animal Health Commission), Curtis Morgan (Texas A&M Transportation Institute), Michael Mays and Jimmy Tickel (North Carolina Department of Agriculture and Consumer Services), Darrel Styles (USDA APHIS), Peter Davies (University of Minnesota), Vanessa Spradlin (West Texas A&M University), and Michael Mays (North Carolina Department of Agriculture and Consumer Services).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2019.00501/full#supplementary-material
1. OIE. Disease Card: Foot and Mouth Disease. World Organization for Animal Health (2013). Available online at: http://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/FOOT_AND_MOUTH_DISEASE.pdf
3. Charleston B, Bankowski BM, Gubbins S, Chase-Topping ME, Schley D, Howey R, et al. Relationship between clinical signs and transmission of an infectious disease and the implications for control. Science. (2011) 332:726–9. doi: 10.1126/science.1199884
4. Arzt J, Branan MA, Delgado AH, Yadav S, Moreno-Torres KI, Tildesley MJ, et al. Quantitative impacts of incubation phase transmission of foot-and-mouth disease virus. Sci Rep. (2019) 9:2707. doi: 10.1038/s41598-019-39029-0
5. Ward MP, Highfield LD, Vongseng P, Graeme Garner M. Simulation of foot-and-mouth disease spread within an integrated livestock system in Texas, USA. Prev Vet Med. (2009) 88:286–97. doi: 10.1016/j.prevetmed.2008.12.006
6. APHIS. Foot and Mouth Disease (2018). Available online at: https://www.aphis.usda.gov/publications/animal_health/fs-fmd-general.pdf
7. McReynolds SW, Sanderson MW. Feasibility of depopulation of a large feedlot during a foot-and-mouth disease outbreak. J Am Vet Med Assoc. (2014) 244:291–8. doi: 10.2460/javma.244.3.291
8. Cabezas AH, Sanderson MW, Jaberi-Douraki M, Volkova V V. Clinical and infection dynamics of foot-and-mouth disease in beef feedlot cattle: an expert survey. Prev Vet Med. (2018) 158:160–8. doi: 10.1016/j.prevetmed.2018.08.007
9. Sanson RL, Dubé C, Cork SC, Frederickson R, Morley C. Simulation modelling of a hypothetical introduction of foot-and-mouth disease into Alberta. Prev Vet Med. (2014) 114:151–63. doi: 10.1016/j.prevetmed.2014.03.005
10. McReynolds SW, Sanderson MW, Reeves A, Hill AE. Modeling the impact of vaccination control strategies on a foot and mouth disease outbreak in the Central United States. Prev Vet Med. (2014) 117:487–504. doi: 10.1016/j.prevetmed.2014.10.005
11. Buhnerkempe MG, Tildesley MJ, Lindström T, Grear DA, Portacci K, Miller RS, et al. The impact of movements and animal density on continental scale cattle disease outbreaks in the United States. PLoS ONE. (2014) 9:e91724. doi: 10.1371/journal.pone.0091724
12. Lindström T, Grear DA, Buhnerkempe M, Webb CT, Miller RS, Portacci K, et al. A bayesian approach for modeling cattle movements in the United States: scaling up a partially observed network. PLoS ONE. (2013) 8:e53432. doi: 10.1371/journal.pone.0053432
15. 21 9 CFR §. Means of Conveyance in Which Dead, Dying, Disabled, or Diseased Livestock and Parts of Carcasses Thereof Shall be Transported. (2010). Available online at: www.ecfr.gov (accessed December 27, 2019).
16. Geering WA, Lubroth J. Preparation of Foot-and-Mouth Disease Contingency Plans FAO Animal Health Manual. Rome: Food and Agriculture Organization of the United Nations; Emergency Prevention System for Transboundary Animal and Plant Pests and Diseases (2002).
17. Carpenter TE, Thurmond MC, Bates TW. A simulation model of intraherd transmission of foot and mouth disease with reference to disease spread before and after clinical diagnosis. J Vet Diagnostic Investig. (2004) 16:11–6. doi: 10.1177/104063870401600103
18. Mardones F, Perez A, Sanchez J, Alkhamis M, Carpenter T. Parameterization of the duration of infection stages of serotype O foot-and-mouth disease virus: an analytical review and meta-analysis with application to simulation models. Vet Res. (2010) 41:45. doi: 10.1051/vetres/2010017
19. Kinsley AC, Patterson G, VanderWaal KL, Craft ME, Perez AM. Parameter values for epidemiological models of foot-and-mouth disease in swine. Front Vet Sci. (2016) 3:44. doi: 10.3389/fvets.2016.00044
20. USDA. Parameters Used to Simulate the Spread of Foot and Mouth Disease in Texas Using the North American Animal Disease Spread Model (NAADSM). Fort Collins, CO: USDA (2012).
21. NAHMS. Overview of U.S. Livestock, Poultry, and Aquaculture Production in 2014 (2014). Available online at: https://www.aphis.usda.gov/animal_health/nahms/downloads/Demographics2014.pdf
22. Eblé P, de Koeijer A, Bouma A, Stegeman A, Dekker A. Quantification of within- and between-pen transmission of Foot-and-Mouth disease virus in pigs. Vet Res. (2006) 37:647–54. doi: 10.1051/vetres:2006026
23. Iowa State University. Secure Pork Supply (SPS) Plan for Continuity of Business. (2017). Available online at: http://www.securepork.org/Resources/Secure-Pork-Supply_Plan_for_COB.pdf (accessed December 27, 2019).
24. USDA. Dairy 2007, Part I: Reference of Dairy Cattle Health and Management Practices in the United States, 2007. Fort Collins, CO: USDA-APHIS (2007). Available online at: https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy07/Dairy07_dr_PartI_1.pdf
25. USDA. Swine 2006, Part 1: Baseline Reference of Swine Health and Management in the United States, 2006. Fort Collins, CO (2006). Available online at: https://www.aphis.usda.gov/animal_health/nahms/swine/downloads/swine2006/Swine2006_dr_PartI_1.pdf
26. Vose D. Risk Analysis: A Quantitative Guide, 3rd Edn. Chichester; Hoboken, NJ: John Wiley & Sons (2008).
27. Savi P, Baldelli B, Morozzi A. Presence and persistence of foot-and-mouth disease virus in meat and meat products from cattle and pigs. Bull Off Inter Epizoot. (1962) 57:853–90.
28. Scott F, Cottral G, Gailiunas P. Presence of foot-and-mouth disease virus in the pituitary and central nervous system of experimentally infected cattle. In: Proceedings of the Annual Meeting of the United States Livestock Sanitary Association. Lansing, MI (1965). p. 87–93.
29. Mebus CA, House C, Gonzalvo FR, Pineda JM, Tapiador J, Pire JJ, et al. Survival of foot-and-mouth disease, African swine fever, and hog cholera viruses in Spanish serrano cured hams and Iberian cured hams, shoulders and loins. Food Microbiol. (1993) 10:133–43. doi: 10.1006/fmic.1993.1014
30. Mebus C, Arias M, Pineda JM, Tapiador J, House C, Sanchez-Vizcaino JM. Survival of several porcine viruses in different Spanish dry-cured meat products. Food Chem. (1997) 59:555–9. doi: 10.1016/S0308-8146(97)00006-X
31. Alexandersen S, Oleksiewicz MB, Donaldson AI. The early pathogenesis of foot-and-mouth disease in pigs infected by contact: a quantitative time-course study using TaqMan RT–PCR. J Gen Virol. (2001) 82:747–55. doi: 10.1099/0022-1317-82-4-747
32. Chou C., Yang S. Inactivation and degradation of O Taiwan97 foot-and-mouth disease virus in pork sausage processing. Food Microbiol. (2004) 21:737–42. doi: 10.1016/j.fm.2004.02.002
33. Burrows R. Studies on the carrier state of cattle exposed to foot-and-mouth disease virus. J Hyg. (1966) 64:81–90. doi: 10.1017/S0022172400040365
34. Gailiunas P, Cottral GE. Presence and persistence of foot-and-mouth disease virus in bovine skin. J Bacteriol. (1966) 91:2333–8.
35. Sellers R, Burrows R, Mann J, Dawe P. Recovery of virus from bulls affected with foot-and-mouth disease. Vet Rec. (1968) 83:303. doi: 10.1136/vr.83.12.303
36. Cottral GE. Persistence of foot-and-mouth disease virus in animals, their products and the environment. Off Int Epizoot Bull. (1969) 71:549–68.
37. Sellers R. Quantitative aspects of the spread of foot and mouth disease. Vet Bull. (1971) 41:431–9.
38. Dhennin L, Frouin A, Gicquel B, Bidard J, Labie J. Risques de dissémination de la fièvre aphteuse par des produits de charcuterie. Survie du virus aphteux dans le saucisson sec. Bull Acad Vet. (1980) 53:349–55.
39. Burrows R, Mann JA, Garland AJM, Greig A, Goodridge D. The pathogenesis of natural and simulated natural foot-and-mouth disease infection in cattle. J Comp Pathol. (1981) 91:599–609. doi: 10.1016/0021-9975(81)90089-X
40. Panina G, Civardi A, Massirio I, Scatozza F, Baldini P, Palmia F. Survival of foot-and-mouth disease virus in sausage meat products (Italian salami). Int J Food Microbiol. (1989) 8:141–148. doi: 10.1016/0168-1605(89)90068-8
41. Donaldson AI, Gibson CF, Oliver R, Hamblin C, Kitching RP. Infection of cattle by airborne foot-and-mouth disease virus: minimal doses with O1 and sat 2 strains. Res Vet Sci. (1987) 43:339–46. doi: 10.1016/S0034-5288(18)30804-X
42. Alexandersen S, Quan M, Murphy C, Knight J, Zhang Z. Studies of quantitative parameters of virus excretion and transmission in pigs and cattle experimentally infected with foot-and-mouth disease virus. J Comp Pathol. (2003) 129:268–82. doi: 10.1016/S0021-9975(03)00045-8
43. Donaldson AI, Alexandersen S. Predicting the spread of foot and mouth disease by airborne virus. Rev Sci Tech. (2002) 21:569–75. doi: 10.20506/rst.21.3.1362
44. Donaldson A. Aerobiology of foot-and-mouth disease (FMD): an outline and recent advances [airborne infection, aphthovirus]. Rev Sci Tech. (1986) 5:315–21. doi: 10.20506/rst.5.2.234
45. Jones AM, Harrison RM. The effects of meteorological factors on atmospheric bioaerosol concentrations—a review. Sci Total Environ. (2004) 326:151–80. doi: 10.1016/j.scitotenv.2003.11.021
46. Casal J, Planas-Cuchi E, Moreso JM, Casal J. Forecasting virus atmospherical dispersion. Studies with foot-and-mouth disease. J Hazard Mater. (1995) 43:229–44. doi: 10.1016/0304-3894(95)00040-2
47. Kinsley AC, VanderWaal K, Craft ME, Morrison RB, Perez AM. Managing complexity: Simplifying assumptions of foot-and-mouth disease models for swine. Transbound Emerg Dis. (2018) 65:1307–17. doi: 10.1111/tbed.12880
48. Hayer SS, VanderWaal K, Ranjan R, Biswal JK, Subramaniam S, Mohapatra JK, et al. Foot-and-mouth disease virus transmission dynamics and persistence in a herd of vaccinated dairy cattle in India. Transbound Emerg Dis. (2018) 65:e404–15. doi: 10.1111/tbed.12774
Keywords: foot and mouth disease, FMDv, carcass, cattle, swine
Citation: Walz E, Middleton J, Sampedro F, VanderWaal K, Malladi S and Goldsmith T (2020) Modeling the Transmission of Foot and Mouth Disease to Inform Transportation of Infected Carcasses to a Disposal Site During an Outbreak Event. Front. Vet. Sci. 6:501. doi: 10.3389/fvets.2019.00501
Received: 06 August 2019; Accepted: 18 December 2019;
Published: 14 January 2020.
Edited by:
Lisa Boden, University of Edinburgh, United KingdomReviewed by:
Michael Tildesley, University of Warwick, United KingdomCopyright © 2020 Walz, Middleton, Sampedro, VanderWaal, Malladi and Goldsmith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Timothy Goldsmith, Z29sZDAxODhAdW1uLmVkdQ==
†These authors have contributed equally to this work
‡Present address: Jamie Middleton, Los Angeles County Department of Public Health, Los Angeles, CA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.