- 1Department of Companion Animal and Wildlife Clinic, Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand
- 2Research Center of Producing and Development of Products and Innovations for Animal Health and Production, Chiang Mai University, Chiang Mai, Thailand
- 3Veterinary Diagnostic Laboratory, Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand
- 4Department of Biology, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
- 5National Primate Research Center of Thailand, Chulalongkorn University, Bangkok, Thailand
- 6Krabookkoo Wildlife Breeding Center, Chachoengsao, Thailand
- 7Department of Clinical Sciences, Colorado State University, Fort Collins, CO, United States
Intestinal parasitic infections can have an impact on health and growth of wildlife. The current study aims were to determine the prevalence of intestinal parasites and to molecular characterize Giardia duodenalis and Cryptosporidium spp. in captive gibbons at Krabokkoo Wildlife Breeding Center, Thailand. Fifty-five gibbons, 2 agile- (Hylobates agilis), 38 lar- (Hylobates lar) and 15 pileated gibbons (Hylobates pileatus) were included in this study. Fecal samples were collected individually at Krabokkoo Wildlife Breeding Center, Chachoengsao province, eastern Thailand, in November 2013. Intestinal parasitic infections were examined by zinc sulfate centrifugation flotation and by a commercially available immunofluorescent assay (IFA) for detection of G. duodenalis and Cryptosporidium spp.. Polymerase chain reaction targeting the Giardia glutamate dehydrogenase (gdh), beta- giardin (bg), triose phosphate isomerase (tpi) genes, and the Cryptosporidium small subunit-rRNA and heat-shock protein (hsp70) following by DNA sequencing were performed on the IFA positive samples. The overall prevalence of intestinal parasitic infection in gibbons at Krabokkoo Wildlife Breeding Center was 12.7% (95%CI: 5.3–24.5), Strongyloides spp. eggs or larvae were present in all positive samples. Co-infections with G. duodenalis were detected in 1.8% (95%CI: 0.1–9.7) of the samples. Based on the sequencing results of the three genes, the IFA Giardia positive isolate typed as the zoonotic genotype B. Since the data reveals the occurrence of zoonotic Giardia genotype, good hygiene management is suggested to prevent the transmission of this pathogen from gibbon to human, and vice versa.
Introduction
Intestinal parasitic infections are the most common causes of gastrointestinal diseases in captive wildlife. These infections can cause a wide range of clinical signs, from subclinical infections to malabsorption, abdominal pain, diarrhea, vomiting, anemia, severe dehydration, and death (1–3). As the living area is limited, stress and other factors such as artificial environment, poor diet or the presence of humans lead to the high risk of infection and weaken the natural resistance of the host, making the clinical illness possible (4). The weakened health condition of these captive animals can have a negative impact on their reproduction which is of major concern in the zoos and wildlife breeding facilities of captive or endangered species (3, 5).
Several studies on helminthic parasites in the free-ranging (5–10) and captive populations (4, 11–15) of non-human primates (NHP) have been conducted worldwide and they reported a high prevalence of intestinal parasites. For example, the prevalence of endoparasites in western lowland gorillas at Bai Hokou, Dzangha-Ndoki National Park, Central African Republic has been reported to be up to 100% (7). Of all intestinal parasites detected in NHP, Strongyloides spp., Oesophagostomum spp., Trichuris spp., Ascaris spp., and hookworms were the most common intestinal parasites.
Eight assemblages (A-H) of Giardia duodenalis and at least 27 Cryptosporidium spp. have been described (16, 17). Infection with G. duodenalis and Cryptosporidium ssp. in NHP are common (18–21). In wild and captive NHP, prevalence rates of these infections range from undetectable level to as high as 70% (20, 22–26). In several studies on NHP, zoonotic assemblages of G. duodenalis, assemblage A and B, were identified and the assemblage B was more prevalent in both captive and free-range animals (18, 23, 27, 28). Cryptosporidium hominis and C. parvum were commonly identified in Cryptosporidium-infected primates (29–31).
Currently, there is no information available regarding intestinal parasitic infection in captive gibbons in breeding facilities in Thailand. Knowing background prevalence of gastrointestinal parasites can be beneficial in the health management program in gibbons for the reproduction at the Krabokkoo Wildlife Breeding Center. The aims of this study were, therefore, to determine the prevalence of intestinal parasites and to molecular characterize Giardia duodenalis and Cryptosporidium spp. isolates to determine the potential of zoonotic transmissions of these pathogens from captive gibbons at Krabokkoo Wildlife Breeding Center, Thailand.
Materials and Methods
Study Area
Krabokkoo Wildlife Breeding Center is located in Chachoengsao province, eastern Thailand, at the coordinates of 13°28′5.05″N, 101°35′37.30″E (Figure 1) and at 47 meters above the sea level. In November 2013, this facility accommodated four species of gibbons, 65 white-handed (Hylobates lar), 15 pileated (Hylobates pileatus), 2 agile (Hylobates agilis), and 2 crown (Nomascus spp.) gibbons. The gibbons were separated among species and were housed individually or in groups. Siblings or a family were housed together. They were fed with vegetable- and fruit-based diet and water was supplied in a bowl. Drinking water was replaced on a daily basis. All gibbons were dewormed every 3 months. The temperature are 23–27°C in winter (mid-October–mid-February), 35–40°C in summer (mid-February–mid-May), and 28–35°C in rainy season (mid-May–mid-October) (32).
Figure 1. Krabokkoo Wildlife Breeding Center is located in Chachoengsao province, eastern Thailand at 13°28′5.05″N, 101°35′37.30″E, 2.5 h from Bangkok.
Fecal Sample and Data Collection
Fifty-five fecal samples were collected from white-handed (H. lar) (n = 38), pileated (H. pileatus) (n = 15), and agile (H. agilis) (n = 2) gibbons during the November of 2013. The samples used in the study were part of the study on genetic diversity of macaques and Hylobatidae gibbons in Thailand. Each fecal sample was collected from the ground (care was taken not to have soil contamination), kept in a labeled plastic bag and stored at 4°C until examination. Sex, age, cage and identification number were recorded at the time of collection. Fecal samples were shipped on ice to the Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand within a week and the fecal consistencies were determined upon arrival.
Diagnostic Procedures
Microscopic Examination of Fecal Samples After Zinc Sulfate Centrifugal Flotation and IFA for Giardia duodenalis and Cryptosporidium spp. Detection
Fecal samples were examined for the presence of intestinal parasitic eggs, larvae, protozoal cysts and oocysts using microscopic examination after zinc sulfate centrifugal flotation (33). Giardia duodenalis and Cryptosporidium spp. infections were determined using a commercially available direct immunofluorescent assay (IFA) (MeriFluor® Cryptosporidium/Giardia Test Kit, Meridian Diagnostic Corporation, Cincinnati, OH). Prior IFA, the fecal samples (3 grams) were concentrated using sucrose gradient centrifugation technique as previously described (34). IFA was carried out according to the manufacturer's instruction.
DNA Isolation and Molecular Detection of Giardia duodenalis Infection
Three hundred microliters of each Giardia or Cryptosporidium IFA positive fecal concentrate were subjected to DNA extraction using the FastDNA® kit (MP Biomedicals, Solon, OH, USA) following an established protocol (35).
PCR assays targeting Giardia glutamate dehydrogenase (gdh), beta-giardin (bg), and triosephosphate isomerase (tpi) genes, and Cryptosporidium heat-shocked protein (hsp70), and small subunit ribosomal RNA (SSU-rRNA) were used for molecular characterization of the respective organisms in the IFA positive samples. Previously described PCR protocols were used (36–42).
DNA Sequencing and Phylogenetic Analysis
The PCR products were purified using QIAquick Gel Extraction Kit (Qiagen, OH, USA) and purified PCR product was evaluated by nucleotide sequencing using a commercially available service (1st Base Laboratory, Selangor, Malaysia). For each target gene, the obtained sequences from both directions were aligned and a consensus sequence was generated and compared with nucleotide sequences from the nucleotide database from the GenBank using BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic and molecular analyses were conducted using the MEGA 6.06 program (43). Multiple sequence alignments were performed using MUSCLE (44), and the phylogenetic analyses were performed by the Maximum Likelihood method based on the Kimura 2-parameter model. The consensus tree was obtained after bootstrap analysis with 500 replications. Reference strains of the different assemblages were retrieved from the GenBank and included for comparative phylogenetic analyses.
Statistical Analyses
A sample was considered positive for gastrointestinal parasites if parasitic eggs or larvae were detected by light microscopic examination after zinc sulfate centrifugal flotation. A sample was considered positive for Giardia and Cryptosporidium if at least one (oo)cyst was detected by either microscopic examination or IFA. Gibbons were grouped by species, age (<10 years, ≥10 years), and sex. Overall prevalence and 95% confidence interval (95%CI) were calculated. Associations of age category, sex, fecal consistency, gibbon species, and parasitic infestation results were analyzed using Fisher's Exact test. A P < 0.05 was considered statistically significant. All statistical analyses were performed using STATA statistical software release 10.1 (Stata Corp., College Station, Texas, USA).
Results
Microscopic Examination of Fecal Samples After Zinc Sulfate Centrifugal Flotation and IFA for Giardia duodenalis and Cryptosporidium spp. Infections
Characteristics of gibbons and samples and the descriptive statistics are shown in Table 1. Of 55 fecal samples, Strongyloides spp. eggs or larvae were detected in 7 samples by microscopic examination after zinc sulfate centrifugal flotation. Giardia cysts were detected in one fecal sample by IFA. Cryptosporidium oocysts were not detected by IFA in any fecal samples, therefore, PCR assays were not performed.
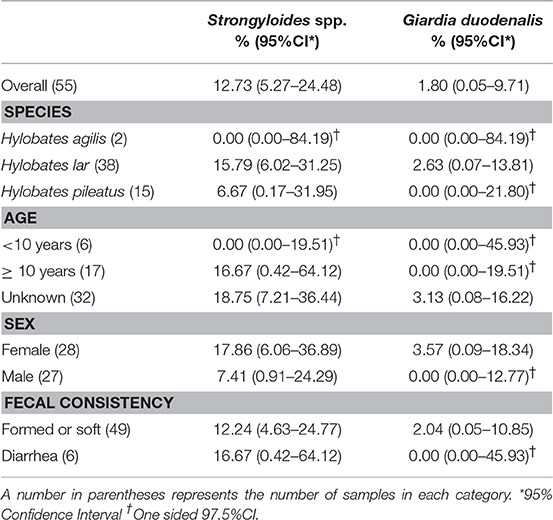
Table 1. Prevalence of intestinal parasitic infection by gibbon species, age, sex, and fecal consistency.
Giardia duodenalis Sequences and Phylogenetic Analyses
DNA fragments of the only IFA Giardia positive sample (G29) were successfully amplified and typed as assemblage B by the three genes (gdh, bg, and tpi). The gdh sequence of G29 showed 99% homology to the assemblage B gdh sequences recovered from a water sample and a beaver in Canada, an ostrich in Brazil, a human and a dog in Australia, and a human from India (Figure 2). The G29 gdh sequence has 3 SNPs (single-nucleotide polymorphism) at position 12 (A vs. T), 93 (A vs. G), and 199 (A vs. G), when compared to those sequences mentioned previously; however, neither of these SNPs resulted in amino acid change. The beta-giardin sequence of G29 showed 100% homology to the assemblage B from human from Thailand and India and 99.9% homology to assemblage B human isolates from Kenya, Egypt, Brazil, and Ethiopia (Figure 3). Sequences from tpi gene contained ambiguous nucleotides at position 108 (T or G) and 443 (A or T). When translating the G29 tpi sequence to amino acids, substitution of T with G at position 108 did not cause amino acid change, whilst substitution of A with T at position 443 resulted in an amino acid change from Valine to Aspartic acid. Variants of tpi sequences showed 99–100% homology to the tpi sequences recovered from rhesus macaque, long-tailed macaque, and gibbons from China, Sumatran Orangutan from Indonesia, beaver from Canada, cat from Japan, rabbit from Nigeria, and humans from Canada, Malaysia, and Spain (Figure 4). From the phylogenetic analyses of gdh, bg and tpi genes, the G. duodenalis isolate in this study was placed into BIV, BI, and BIV branch, respectively (Figures 2–4).
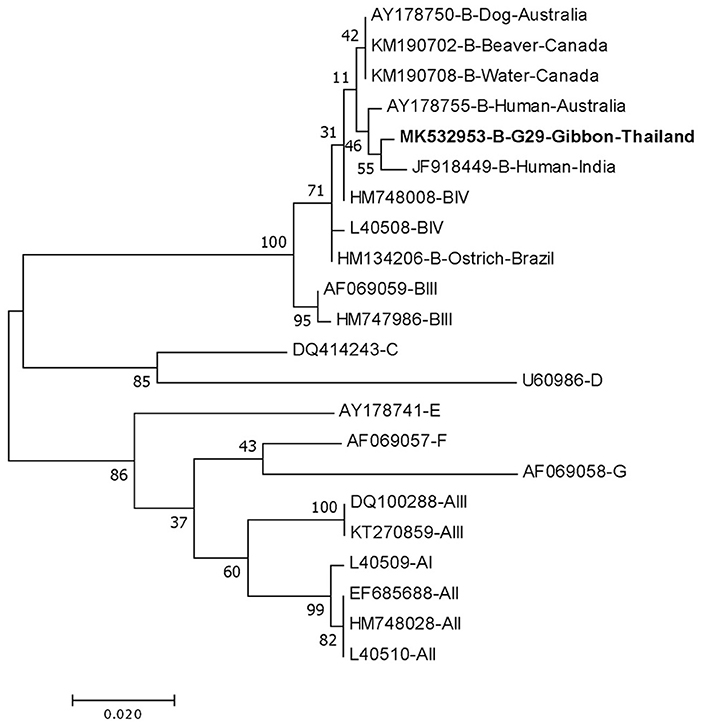
Figure 2. Phylogenetic tree of Giardia isolate based on the sequence of glutamate dehydrogenase (gdh) gene from a gibbon in this study by the Maximum Likelihood algorithm using the MEGA 6.06 program. Sequences obtained from GenBank are indicated by their accession numbers. Percentage bootstrap supports (500 replicates) are shown by numbers at the respective nodes. Bold texts represent the Giardia detected in this study.
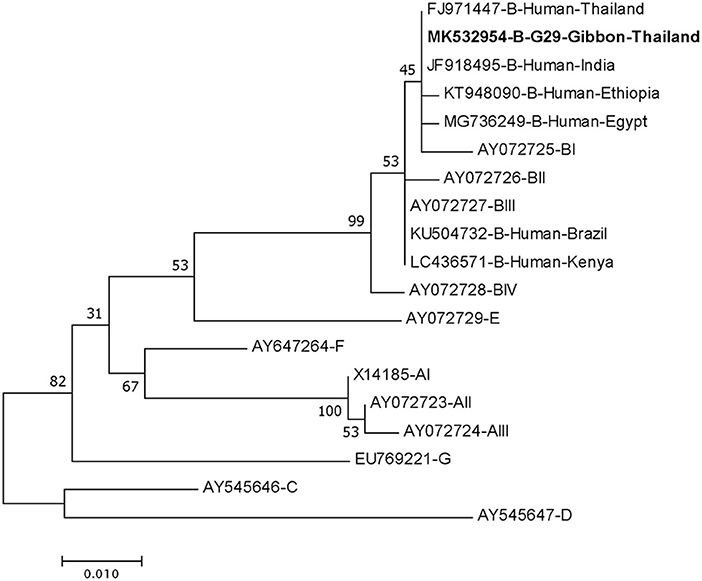
Figure 3. Phylogenetic tree of Giardia isolate based on the sequence of beta-giardin (bg) gene from a gibbon in this study by the Maximum Likelihood algorithm using the MEGA 6.06 program. Sequences obtained from GenBank are indicated by their accession numbers. Percentage bootstrap supports (500 replicates) are shown by numbers at the respective nodes. Bold texts represent the Giardia detected in this study.
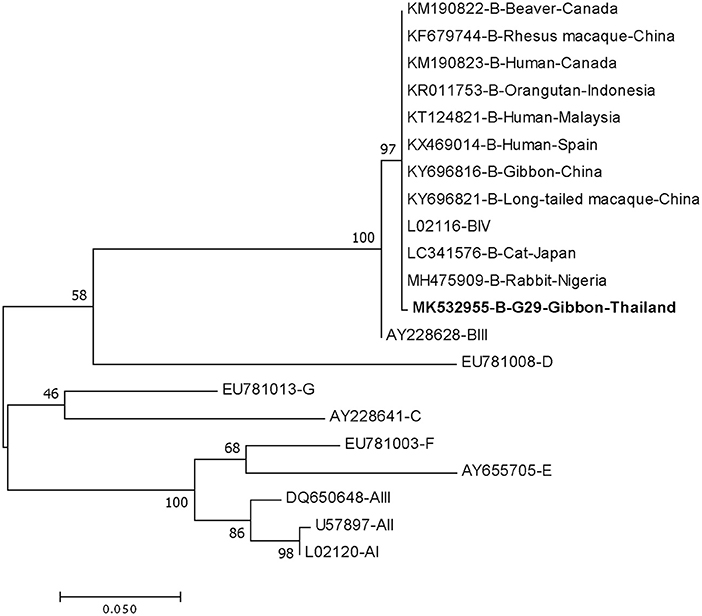
Figure 4. Phylogenetic tree of Giardia isolate based on the sequence of triose phosphate isomerase (tpi) gene from a gibbon in this study by the Maximum Likelihood algorithm using the MEGA 6.06 program. Sequences obtained from GenBank are indicated by their accession numbers. Percentage bootstrap supports (500 replicates) are shown by numbers at the respective nodes. Bold texts represent the Giardia detected in this study.
Statistical Analyses
Due to the low detection of the parasites and the small sample size, the power to detect associations between any risk factors and infections was insufficient.
Discussion
The current study represents the first report of the intestinal parasites as well as G. duodenalis and Cryptosporidium spp. prevalence rates and Giardia genotypes in captive gibbons in Thailand. The prevalence of these infections were commonly high and ranged from 25 to 100% in either free-ranging (5, 7–10, 22, 28, 31, 45, 46) or captive non-human primates (4, 7, 11, 13–15, 20, 23–26, 46, 47). In the current study, overall prevalence rates of nematodes, G. duodenalis, and Cryptosporidium spp. in gibbons were 12.7, 1.8, and 0%, respectively. These prevalence rates, however, were comparable to the previous report of 0–16.4% in gibbons in zoological parks in China (48). The low prevalence in this study may be from collecting a single fecal sample from each animal. Parasitic eggs, Giardia cysts and Cryptosporidium oocysts are intermittent shed in the feces, therefore, three or more fecal samples from animals can increase the sensitivity of intestinal parasites (33). In addition, the low detection rate of Giardia and the lack of Cryptosporidium spp. were also because of the number of cysts/oocysts were below the detection limit of the diagnostic tests used in the study (49).
The most common helminthic species detected in NHP were Strongyloides spp., Trichuris spp., Oesophagostomum spp., Ascaris spp. and hookworms (5, 7, 22, 45, 46). A similar pattern of gastrointestinal parasites was also observed in captive gibbons in a zoo in China (13). In a study in 23 wild white-handed gibbons at Khao Yai National Park, Thailand, Trichuris spp. and Ternidens spp. were the most prevalent helminthic parasites detected (91.3%), followed by Strongyloides fuelleborni (56.5%) (9). However, in the current study, only Strongyloides spp. eggs or larvae were detected in the fecal samples. Strongyloides spp. are soil-transmitted nematodes with an estimated 370 million people infected worldwide (50). In this study, the detection of Strongyloides spp. in feces is less likely to be from contaminated soil as fecal samples were carefully collected not be contaminated with soil before the storage in a plastic bag. These nematodes can cause a chronic and persistent strongyloidiasis in the infected host because of the autoinfective life cycle (51) and cause diarrhea, hyperinfection syndrome, dissemination, and death in immunocompromised hosts. Strongyloides stercoralis is a primary species infecting human; however, the infections of primates' parasites S. fuelleborni fuelleborni and S. fuelleborni kellyi have also been reported (8). The molecular characterization of Strongyloides positive samples is suggested since microscopic identification is insufficient for species identification and determination of its zoonotic potential. In this study, the species identification was not performed but this finding has raised concerns regarding the zoonotic potential. Since the fatal strongyloidiasis cases of gibbons in Thailand has been reported (1) and Strongyloides spp. is also an important parasitic helminth of humans (8), an effective anthelminthic program is recommended.
Giardia duodenalis and Cryptosporidium spp. are important intestinal protozoans in non-human primates. These pathogens can cause a wide range of clinical signs, from subclinical to malabsorption, abdominal pain, failure to thrive, acute or chronic diarrhea especially in young, old and immune-compromised animals (3, 52, 53). The organisms are commonly found in both free-ranging and captive non-human primates with the prevalence from 0–70% to 0–48%, for Giardia and Cryptosporidium infections, respectively (19, 21, 22, 24, 31, 47, 54). In Thailand, a low prevalence rate (1/23, 4.35%) of Cryptosporidium spp. infection has been previously reported in wild white-handed gibbons at Khao Yai National Park in Thailand. IFA was used for the detection of Giardia cysts or Cryptosporidium oocysts in repeatedly collected fecal samples that ranged from 3 to 25 samples per gibbon, resulting in a total of 324 samples (9). In that study, there was no Giardia detected. In this study, in contrast, no Cryptosporidium oocysts were detected in all fecal samples and Giardia cysts were detected in only one fecal sample of 55 samples. These findings could be due to that the numbers of cysts or oocysts of these pathogens were low and were below the detection limit of the IFA tests. The Giardia positive sample, in this study, typed as assemblage BIV and BI by gdh and tpi and bg genes, respectively. This finding is in agreement with previous reports that assemblage B was predominant in NHP (18, 23, 27, 28). Although the prevalence of Giardia infection in this study is low, the identification of G. duodenalis assemblage B may suggest the potential of zoonotic or anthroponotic transmissions in this area.
The limitations of this study are the small sample size and the nature of single sample collected from each animal; selection bias may have led to an underestimation of the prevalence rates. A larger sample size or more frequent collection of gibbon's feces are needed for further studies. This study had inadequate power to detect associations between any risk factors and infections. In addition, we analyzed gibbons' species, age, sex, and diarrhea status; however, other important risk factors, e.g., season, diet, or water source could be suggested for future study to help in prevention and control of intestinal parasitic infection in this population.
Author Contributions
ST, SM, and ML designed the study. SM and RA performed fecal sample collection. DS performed the microscopic fecal examination, ST performed IFA and molecular analyses. ST analyzed sequences. ML provided laboratory supplies. ST interpreted the results and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AS declared a shared affiliation, with no collaboration, with one of the authors, ML, to the handling editor at time of review.
Acknowledgments
The authors would like to thank the administrative members of the Krabokkoo Wildlife Breeding Center for giving permission to use the animals' feces and to all animal keepers for providing assistance during fecal samples collection and Dr. Terdsak Yano for a map of Chachoengsao province. This research work was partially supported by Chiang Mai University.
References
1. DePaoli A, Johnsen DO. Fatal strongyloidiasis in Gibbons (Hylobates lar). Vet Pathol. (1978) 15:31–9. doi: 10.1177/030098587801500104
2. Penner LR. Concerning Threadworm (Strongyloides stercoralis) in Great Apes: Lowland Gorillas (Gorilla gorilla) and Chimpanzees (Pan troglodytes). J Zoo Anim Med. (1981) 12:128–31. doi: 10.2307/20094543
3. Panayotova-Pencheva MS. Parasites in captive animals: a review of studies in some European zoos. Der Zoologische Garten. (2013) 82:60–71. doi: 10.1016/j.zoolgart.2013.04.005
4. Darabus G, Afrenie M, Hotea I, Imre M, Morariu S. Endoparasites in mammals from seven zoological gardens in Romania. J Zoo Wildl Med. (2014) 45:239–46. doi: 10.1638/2012-0170.1
5. Martin-Solano S, Carrillo-Bilbao GA, Ramirez W, Celi-Erazo M, Huynen MC, Levecke B, et al. Gastrointestinal parasites in captive and free-ranging Cebus albifrons in the Western Amazon, Ecuador. Int J Parasitol Parasites Wildl. (2017) 6:209–18. doi: 10.1016/j.ijppaw.2017.06.004
6. Landsoud-Soukate J, Tutin CE, Fernandez M. Intestinal parasites of sympatric gorillas and chimpanzees in the Lope Reserve, Gabon. Ann Trop Med Parasitol. (1995) 89:73–9.
7. Freeman AS, Kinsella JM, Cipolletta C, Deem SL, Karesh WB. Endoparasites of Western Lowland Gorillas (Gorilla gorilla gorilla) at Bai Hokou, Central African Republic. J Wildl Dis. (2004) 40:775–81. doi: 10.7589/0090-3558-40.4.775
8. Anderson J, Upadhayay R, Sudimack D, Nair S, Leland M, Williams JT, et al. Trichuris sp. and Strongyloides sp. infections in a free-ranging baboon colony. J Parasitol. (2012) 98:205–8. doi: 10.1645/GE-2493.1
9. Gillespie TR, Barelli C, Heistermann M. Effects of social status and stress on patterns of gastrointestinal parasitism in wild white-handed gibbons (Hylobates lar). Am J Phys Anthropol. (2013) 150:602–8. doi: 10.1002/ajpa.22232
10. Klaus A, Zimmermann E, Roper KM, Radespiel U, Nathan S, Goossens B, et al. Co-infection patterns of intestinal parasites in arboreal primates (proboscis monkeys, Nasalis larvatus) in Borneo. Int J Parasitol Parasites Wildl. (2017) 6:320–9. doi: 10.1016/j.ijppaw.2017.09.005
11. Sanchez VVV, Patino AS, Sandoval JAC, Esquivel CVC, Sanchez TAC. Prevalence of gastrointestinal parasites among captive primates in Panama. J Anim Vet Adv. (2009) 8:2644–9.
12. Fagiolini M, Lia RP, Laricchiuta P, Cavicchio P, Mannella R, Cafarchia C, et al. Gastrointestinal parasites in mammals of two Italian zoological gardens. J Zoo Wildl Med. (2010) 41:662–70. doi: 10.1638/2010-0049.1
13. Li M, Zhao B, Li B, Wang Q, Niu L, Deng J, et al. Prevalence of gastrointestinal parasites in captive non-human primates of twenty-four zoological gardens in China. J Med Primatol. (2015) 44:168–73. doi: 10.1111/jmp.12170
14. Zanzani SA, Gazzonis AL, Epis S, Manfredi MT. Study of the gastrointestinal parasitic fauna of captive non-human primates (Macaca fascicularis). Parasitol Res. (2016) 115:1432–955. doi: 10.1007/s00436-015-4748-9
15. Kvapil P, Kastelic M, Dovc A, Bartova E, Cizek P, Lima N, et al. An eight-year survey of the intestinal parasites of carnivores, hoofed mammals, primates, ratites and reptiles in the Ljubljana zoo in Slovenia. Folia Parasitol. (2017) 64:2017.013. doi: 10.14411/fp.2017.013
16. Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. (2011) 24:110–40. doi: 10.1128/CMR.00033-10
17. Slapeta J. Cryptosporidiosis and Cryptosporidium species in animals and humans: a thirty colour rainbow? Int J Parasitol. (2013) 43:957–70. doi: 10.1016/j.ijpara.2013.07.005
18. Beck R, Sprong H, Pozio E, Caccio SM. Genotyping Giardia duodenalis isolates from dogs: lessons from a multilocus sequence typing study. Vector Borne Zoonotic Dis. (2012) 12:206–13. doi: 10.1089/vbz.2011.0751
19. Karim MR, Zhang S, Jian F, Li J, Zhou C, Zhang L, et al. Multilocus typing of Cryptosporidium spp. and Giardia duodenalis from non-human primates in China. Int J Parasitol. (2014) 44:1039–47. doi: 10.1016/j.ijpara.2014.07.006
20. Du SZ, Zhao GH, Shao JF, Fang YQ, Tian GR, Zhang LX, et al. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-human primates in Qinling mountains. Korean J Parasitol. (2015) 53:395–402. doi: 10.3347/kjp.2015.53.4.395
21. Li J, Qi M, Chang Y, Wang R, Li T, Dong H, et al. Molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Captive Wildlife at Zhengzhou Zoo, China. J Eukaryot Microbiol. (2015) 62:833–9. doi: 10.1111/jeu.12269
22. Ekanayake DK, Arulkanthan A, Horadagoda NU, Sanjeevani GK, Kieft R, Gunatilake S, et al. Prevalence of Cryptosporidium and other enteric parasites among wild non-human primates in Polonnaruwa, Sri Lanka. Am J Trop Med Hyg. (2006) 74, 322–9. doi: 10.4269/ajtmh.2006.74.322
23. Berrilli F, Prisco C, Friedrich KG, Di Cerbo P, Di Cave D, De Liberato C. Giardia duodenalis assemblages and Entamoeba species infecting non-human primates in an Italian zoological garden: zoonotic potential and management traits. Parasites Vectors. (2011) 4:199. doi: 10.1186/1756-3305-4-199
24. Martinez-Diaz RA, Sansano-Maestre J, Martinez-Herrero Mdel C, Ponce-Gordo F, Gomez-Munoz MT. Occurrence and genetic characterization of Giardia duodenalis from captive nonhuman primates by multi-locus sequence analysis. Parasitol Res. (2011) 109:539–44. doi: 10.1007/s00436-011-2281-z
25. David EB, Patti M, Coradi ST, Oliveira-Sequeira TC, Ribolla PE, Guimaraes S. Molecular typing of Giardia duodenalis isolates from nonhuman primates housed in a Brazilian zoo. Rev Inst Med Trop Sao Paulo. (2014) 56:49–54. doi: 10.1590/S0036-46652014000100007
26. Debenham JJ, Atencia R, Midtgaard F, Robertson LJ. Occurrence of Giardia and Cryptosporidium in captive chimpanzees (Pan troglodytes), mandrills (Mandrillus sphinx) and wild Zanzibar red colobus monkeys (Procolobus kirkii). J Med Primatol. (2015) 44:60–5. doi: 10.1111/jmp.12158
27. Cacciò SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. (2008) 38:1523–31. doi: 10.1016/j.ijpara.2008.04.008
28. Sricharern W, Inpankaew T, Keawmongkol S, Supanam J, Stich RW, Jittapalapong S. Molecular detection and prevalence of Giardia duodenalis and Cryptosporidium spp. among long-tailed macaques (Macaca fascicularis) in Thailand. Infect Genet Evol. (2016) 40:310–4. doi: 10.1016/j.meegid.2016.02.004
29. Phillips KA, Haas ME, Grafton BW, Yrivarren M. Survey of the gastrointestinal parasites of the primate community at Tambopata National Reserve, Peru. J Zool. (2004) 264:149–51. doi: 10.1017/S0952836904005680
30. Ye J, Xiao L, Li J, Huang W, Amer SE, Guo Y, et al. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int. (2014) 63:132–7. doi: 10.1016/j.parint.2013.10.007
31. Parsons MB, Travis D, Lonsdorf EV, Lipende I, Roellig DM, Collins A, et al. Epidemiology and molecular characterization of Cryptosporidium spp. in humans, wild primates, and domesticated animals in the Greater Gombe Ecosystem, Tanzania. PLoS Negl Trop Dis. (2015) 9:e0003529. doi: 10.1371/journal.pntd.0003529
32. Thai Meteorological Department (2018). Chachoengsao Climate [Online]. Available online at: http://climate.tmd.go.th/data/province .pdf (accessed November 19, 2018).
.pdf (accessed November 19, 2018).
34. O'Handley RM, Olson ME, Fraser D, Adams P, Thompson RC. Prevalence and genotypic characterisation of Giardia in dairy calves from Western Australia and Western Canada. Vet Parasitol. (2000) 90:193–200. doi: 10.1016/S0304-4017(00)00235-1
35. da Silva AJ, Caccio S, Williams C, Won KY, Nace EK, Whittier C, et al. Molecular and morphologic characterization of a Cryptosporidium genotype identified in lemurs. Vet Parasitol. (2003) 111:297–307. doi: 10.1016/S0304-4017(02)00384-9
36. Morgan UM, Constantine CC, Forbes DA, Thompson RC. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. (1997) 83:825–30.
37. Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, et al. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. (1999) 65:3386–91.
38. Morgan UM, Monis PT, Xiao L, Limor J, Sulaiman I, Raidal S, et al. Molecular and phylogenetic characterisation of Cryptosporidium from birds. Int J Parasitol. (2001) 31:289–96.
39. Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. (2003) 9:1444–52. doi: 10.3201/eid0911.030084
40. Read CM, Monis PT, Thompson RC. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. (2004) 4:125–30. doi: 10.1016/j.meegid.2004.02.001
41. Lalle M, Pozio E, Capelli G, Bruschi F, Crotti D, Caccio SM. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int J Parasitol. (2005) 35:207–13. doi: 10.1016/j.ijpara.2004.10.022
42. Lebbad M, Mattsson JG, Christensson B, Ljungstrom B, Backhans A, Andersson JO, et al. From mouse to moose: multilocus genotyping of Giardia isolates from various animal species. Vet Parasitol. (2010) 168:231–9. doi: 10.1016/j.vetpar.2009.11.003
43. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30:2725–9. doi: 10.1093/molbev/mst197
44. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7. doi: 10.1093/nar/gkh340
45. Murray S, Stem C, Boudreau B, Goodall J. Intestinal parasites of baboons (Papio cynocephalus anubis) and chimpanzees (Pan troglodytes) in Gombe National Park. J Zoo Wildl Med. (2000) 31:176–8. doi: 10.1638/1042-7260(2000)031[0176:IPOBPC]2.0.CO;2
46. Adrus M, Zainudin R, Ahamad M, Jayasilan MA, Abdullah MT. Gastrointestinal parasites of zoonotic importance observed in the wild, urban, and captive populations of non-human primates in Malaysia. J Med Primatol. (2018) 48:22–31. doi: 10.1111/jmp.12389
47. Zhong Z, Tian Y, Li W, Huang X, Deng L, Cao S, et al. Multilocus genotyping of Giardia duodenalis in captive non-human primates in Sichuan and Guizhou provinces, Southwestern China. PLoS ONE. (2017) 12:e0184913. doi: 10.1371/journal.pone.0184913
48. Li J, Dong H, Wang R, Yu F, Wu Y, Chang Y, et al. An investigation of parasitic infections and review of molecular characterization of the intestinal protozoa in nonhuman primates in China from 2009 to 2015. Int J Parasitol Parasites Wildl. (2017) 6:8–15. doi: 10.1016/j.ijppaw.2016.12.003
49. Rimhanen-Finne R, Ronkainen P, Hanninen ML. Simultaneous detection of Cryptosporidium parvum and Giardia in sewage sludge by IC-PCR. J Appl Microbiol. (2001) 91:1030–5. doi: 10.1046/j.1365-2672.2001.01468.x
50. Bisoffi Z, Buonfrate D, Montresor A, Requena-Méndez A, Muñoz J, Krolewiecki AJ, et al. Strongyloides stercoralis: a plea for action. PLoS Negl Trop Dis. (2013) 7:e2214. doi: 10.1371/journal.pntd.0002214
51. Page W, Judd JA, Bradbury RS. The unique life cycle of Strongyloides stercoralis and implications for public health action. Trop Med Infect Dis. (2018) 3:53. doi: 10.3390/tropicalmed3020053
52. Levecke B, Geldhof P, Claerebout E, Dorny P, Vercammen F, Cacciò SM, et al. Molecular characterisation of Giardia duodenalis in captive non-human primates reveals mixed assemblage A and B infections and novel polymorphisms. Int J Parasitol. (2009) 39:1595–601. doi: 10.1016/j.ijpara.2009.05.013
53. Thompson RC, Lymbery AJ, Smith A. Parasites, emerging disease and wildlife conservation. Int J Parasitol. (2010) 40:1163–70. doi: 10.1016/j.ijpara.2010.04.009
Keywords: intestinal parasites, Giardia duodenalis, captive, gibbons, Thailand
Citation: Tangtrongsup S, Sripakdee D, Malaivijitnond S, Angkuratipakorn R and Lappin M (2019) Intestinal Parasites and the Occurrence of Zoonotic Giardia duodenalis Genotype in Captive Gibbons at Krabokkoo Wildlife Breeding Center, Thailand. Front. Vet. Sci. 6:110. doi: 10.3389/fvets.2019.00110
Received: 15 December 2018; Accepted: 25 March 2019;
Published: 17 April 2019.
Edited by:
Michael Kosoy, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Antonio Frangipane di Regalbono, University of Padova, ItalyAndrea Valeria Scorza, Colorado State University, United States
Copyright © 2019 Tangtrongsup, Sripakdee, Malaivijitnond, Angkuratipakorn and Lappin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sahatchai Tangtrongsup, c2FoYXRjaGFpQGdtYWlsLmNvbQ==;c2FoYXRjaGFpLnRAY211LmFjLnRo
 Sahatchai Tangtrongsup
Sahatchai Tangtrongsup Duanghatai Sripakdee3
Duanghatai Sripakdee3