
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 21 February 2019
Sec. Veterinary Infectious Diseases
Volume 6 - 2019 | https://doi.org/10.3389/fvets.2019.00035
This article is part of the Research Topic Emerging Swine Viruses View all 14 articles
The genus Pestivirus, which belongs to the family Flaviviridae, includes ssRNA+ viruses responsible for infectious diseases in swine, cattle, sheep, goats, and other domestic and wild animals. Recently, several putative pestiviruses species have been discovered and characterized in mammalian species (giraffe pestivirus, antelope pestivirus, HoBi virus, Bungowannah virus, and Linda virus); one of these is a genetically distinct pestivirus, named atypical porcine pestivirus (APPV), discovered using the next-generation sequencing technology. APPV has been detected in piglets with congenital tremor (CT) from four different continents, including North America, South America, Europe, and Asia. There is strong evidence that experimental inoculation and in field outbreaks involving APPV induce CT in piglets. Additionally, splay leg (SL) syndrome has been observed concurrently with CT, and it was induced by APPV in experimental studies and some field cases. Animals with a persistent and/or chronic infection condition can shed the virus over time. Viral-RNA is frequently detected in different tissues from CT-piglets; however, high loads of APPV are detected most consistently in central nervous tissue. Moreover, the APPV genome has been recently detected in semen and preputial swabs from boar studs, as well as in serum and tissue samples from wild boars and domestic adult pigs, all known to be clinically healthy. Phylogenetic analysis revealed that the APPV sequence (complete or partial polyprotein) exhibits high genetic diversity between viral strains detected in different countries and formed independent clusters according to geographic location. Additional studies are needed to evaluate the molecular detection and sero-prevalence of APPV around the world. Lastly, more research is needed to understand clinical presentations associated with APPV infection, as well as the economic losses related to the virus in pig production worldwide.
Piglet pre-weaning mortality is a major problem in pig farms around the world. On average, pre-weaning mortality rates of live-born piglets can be as high as 23% and starvation and crushing are the main causes of death (1). Different etiologies may be involved in piglet pre-weaning mortality including atypical porcine pestivirus-associated congenital tremor (CT) (2–4).
Historically, CT was first reported in the literature 97 years ago, when Kinsley (5) described “dancing pigs.” Subsequently, it was characterized as tremors of intent that ceases when piglets are at rest (6). The syndrome is classified into five types according to the etiology (Type AI–AV) (Table 1); however, most contemporary CT outbreaks had been attributed to an unidentified virus, Type AII (7, 8).
Since atypical porcine pestivirus (APPV) was first identified in 2015 (9), several studies have linked this new Pestivirus with the occurrence of CT in newborn piglets. It was usually described as a temporary condition, lasting several weeks to months, and characterized by tremors of the head, limbs, and body, varying in severity and intensity. However, the clinical signs were reduced or absent during inactivity or sleep (2, 3, 10).
In general, CT is not the cause of death in affected piglets; however, their survival may be threatened because of inadequate colostrum, or milk intake, leading to severe growth retardation and death by starvation or crushing due to impairment of evasive actions (2, 7). Furthermore, APPV is capable of inducing neurological disorders, such CT, increasing piglet pre-weaning mortality and reducing reproductive performance in affected pigs (2–4, 10).
Although the impact of most exotic diseases in animal production and global economy is known (11), a more comprehensive understanding of the epidemiology, genetic variability, and economic losses associated with the role of APPV in pig production is required.
Pestiviruses are highly variable RNA viruses causing economically relevant diseases in domestic animals. The genus Pestivirus, which belongs to the family Flaviviridae (ssRNA +), includes 11 recognized species: Pestivirus A (bovine viral diarrhea virus type 1), Pestivirus B (bovine viral diarrhea virus type 2), Pestivirus C (classical swine fever virus), and Pestivirus D (border disease virus), Pestivirus E (pronghorn pestivirus), Pestivirus F (Bungowannah virus), Pestivirus G (giraffe pestivirus), Pestivirus H (Hobi-like pestivirus), Pestivirus I (Aydin-like pestivirus), Pestivirus J (rat pestivirus), and Pestivirus K (atypical porcine pestivirus) (12).
Additionally, three atypical pestiviruses have been characterized in pigs: Bungowannah virus (causing myocarditis), APPV and Linda virus (causing lateral shaking) (9, 13, 14). Nevertheless, several reports (experimental conditions or field cases) have demonstrated that APPV is a prominent cause of APPV-CT type II in newborn piglets around the world (2–4, 10, 15). However, no studies have provided an efficient protocol for APPV isolation in cell culture (3, 9, 16, 17), and Koch's postulate couldn't be established.
Further characterization of APPV, as well as other porcine pestiviruses linked to severe clinical diseases in pigs, is needed (18, 19). Remembering that, the continued expansion of the genus Pestivirus and its high genetic diversity constitute a worldwide concern.
APPV has been detected in four different continents, including North America, South America, Europe, and Asia, and in countries such as the United States (first report) (9, 10), the Netherlands (2), Germany (15, 16), Sweden (20), Spain (2, 21), Austria (3), China (22), South Korea (23), Brazil (4, 24, 25), Canada (26), Hungary (27), Great Britain (28, 29), Italy, the Republic of Serbia, Switzerland, and Taiwan (29). Considering all these reports, APPV has wide distribution in the world (Figure 1). Postel et al. (29) have suggested that APPV must be regarded as a pig pestivirus of likely worldwide relevance.
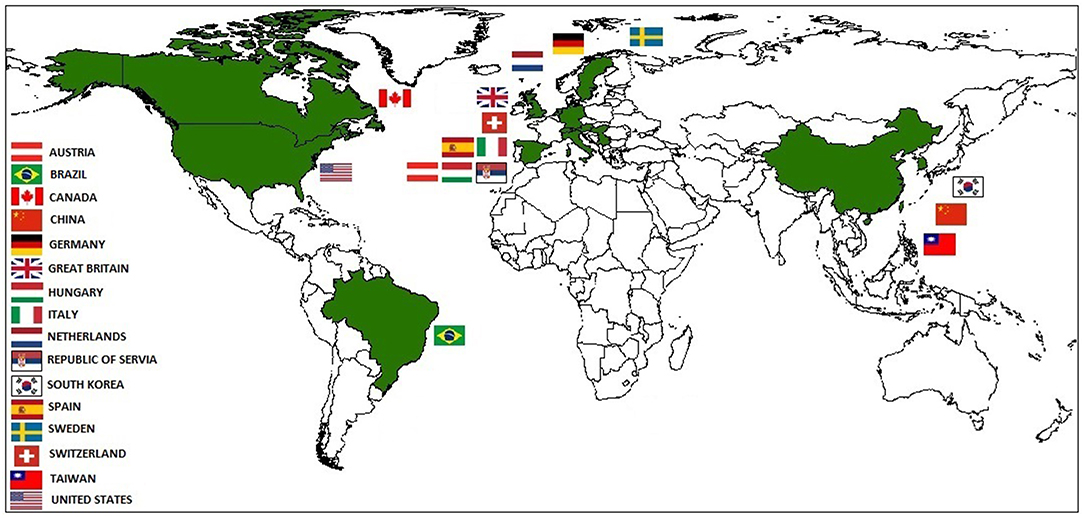
Figure 1. Geographic distribution of Atypical Porcine Pestivirus (APPV) around the world. The geographic information system ArcGIS 10.5.1 was used to generate the map.
APPV-associated CT has been reported to be more prevalent in litters of gilts than sows (2, 4), suggesting that the immune status of the dam is likely the key in disease development in piglets (4). During outbreaks, CT morbidity varies within and between litters; a few pigs in one or all pigs in several litters may be affected (2–4, 10). Overall, CT is observed in both males and females (2) and its prevalence within the litters ranges from < 10 to 100% (2–4, 10). Moreover, CT severity in piglets seems to vary within litters (10). Several recent studies have linked APPV with CT occurrence in piglets and sporadic detection in domestic and wild boars. Detailed information regarding APPV studies can be found in Table 2.
Adult domestic pigs (15, 16, 21) and wild boar are susceptible to APPV infection (34, 35). Recently, APPV was detected in boar preputial fluids (2) and semen (3); both sample types came from boars that had clinical signs consistent with CT at birth. Still, APPV has been detected in semen, preputial swabs, and preputial fluids from commercial boar studs in the United States (32) and it is highly improbable that these boars had CT at the time of birth, suggesting that either transiently infected or persistently infected (PI) boars with no clinical history of CT could be shedding APPV in semen.
A retrospective analysis of cerebellum samples from Germany and another retrospective study with fresh and formaldehyde-fixed paraffin-embedded tissue samples from Hungary showed the presence of APPV in CT-affected piglets from over a decade ago (15, 27). Similarly, a study from Spain confirmed the presence of APPV nearly two decades prior to its first discovery (21).
Wild boar is also susceptible to APPV infection, although its role in the virus epidemiology is unknown (35). Limited information regarding APPV route of transmission, ecology, pathogenesis, carriage, spread, and epidemiology is available. However, piglets presenting with CT, surviving CT syndrome piglets, boars without CT (at birth), and clinically healthy adult domestic pigs, can shed moderate to high loads of virus, playing a relevant role in virus epidemiology; similar to a chronically and/or PI animal (2, 3, 32, 34).
Regarding to diagnosis, Postel et al. (17) described the presence of viral genome in serum samples with different levels of antibodies, suggesting a degree of antibody protection; and, samples with absence of antibodies and viral genome loads, indicating acutely or PI animals. Similarly, Muñoz-González et al. (21) also suggested the PI condition, which could help the APPV spread. On the other hand, the presence of systemic levels of type I Interferon in newborn piglets could lead to the activation of the immune system by APPV (21). Based on these results, more research is needed in order to better understand the role of the innate immune system response to APPV infection.
The newly discovered APPV (9) is capable of inducing neurological disorders, reducing reproductive performance, and increasing pre-weaning mortality (2–4, 10). Two independent research groups have experimentally reproduced CT using an inoculum containing APPV and observed a subset of piglets with concurrent splayleg (SL) (2, 10) and that the affected CT-litters presented as weak piglets with an abnormal posture (2). Field studies have also reported the occurrence of SL in litters with CT sporadically affecting the same piglet (3, 4). SL prevalence ranged from 6 to 55% within the affected CT litters (4).
APPV has a wide distribution in tissue samples, excretion and secretion fluids (2, 10, 15, 21, 22, 32). According to Gatto et al. (4), central nervous and lymphoid tissues appear to be suitable sites for viral replication; however, the cerebellum was the most consistently positive sample type from CT piglets and could constitute a target for APPV replication. Although a specific target of replication has not been determined (10), this may suggest that viral replication occurs systemically and has a predilection for certain types of tissues. However, the primary replication sites remain unknown. The histological findings of a number of studies are described in Table 3.
The economic relevance of an APPV-outbreak loss in pig production worldwide remains undetermined; however, it is estimated that the number of weaned piglets per sow decreases by > 10%, affecting reproductive performance. Additionally, mortality increased up to 30%, when CT-affected new-born piglets died of malnutrition in a farm in Austria (3). de Groof et al. (2) reported 26% mortality in CT-litters affected by APPV, with 60% of these deaths attributable to CT in one farm. Additionally, they showed that under experimental conditions, the affected CT-litters presented weak piglets with an abnormal posture (bent back [kyphosis] and ears on the neck) (2). The SL syndrome has been observed concomitant with some CT cases (2–4, 10), limiting the locomotion of the CT-piglets and increasing the percentage of crushing.
In China, Shen et al. (33) reported a mortality rate of 60% in CT-piglets and Dessureault et al. (26) reported an average mortality rate of 24.6% in CT-litters in Canada. According to Gatto et al. (4) the case fatality of affected CT-piglets in Brazilian pig farm production was 30%. Early data from the United States, Germany, Italy, China, and Taiwan have suggested a relatively high abundance (2.3–22%) of APPV genomes in apparently healthy pigs (9, 15, 16, 29). In addition, Gatto et al. (32) detected APPV genomes in semen (up to 34% prevalence), preputial swabs (up to 23%), and preputial fluids (up to 28%) from commercial boar studs in the United States, which could play an important epidemiological role in virus transmission route and spread. Thus, artificial insemination could constitute a potential APPV transmission route and should be considered an important factor when developing and implementing biosecurity measures to prevent APPV-infections.
The impact of APPV infection on pig production seems to be indirect, since the mortality of CT piglets is the only loss reported so far, and it is mainly attributed to secondary factors. So, further studies evaluating its impact on pig production should be performed in order to estimate the direct loss in affected piglets, or suggest that the indirect loss could be related to the depletion of the immune system, such as what is reported in other pestiviruses.
Since 2015, 20 complete APPV polyproteins from six countries (Table 4) have been submitted to the “GenBank” database. The virus genome of Pestivirus K species is ~10.8–11.5 kb. The APPV genome has ~25–28% pairwise identity to known pestiviruses and 68% pairwise identity to a recently partially characterized Rhinolophus affinis pestivirus, placing both viruses in a highly divergent lineage of pestiviruses (9).
Phylogenetic analysis has revealed that APPV sequences (complete or partial polyprotein) exhibit high genetic diversity between viral strains detected in different countries (4, 27, 29–31) and form independent clusters according to geographic location (Figure 2). Based on phylogenetic analysis of the Npro gene, different viral strains can be present in the same farm simultaneously and at different times (32).
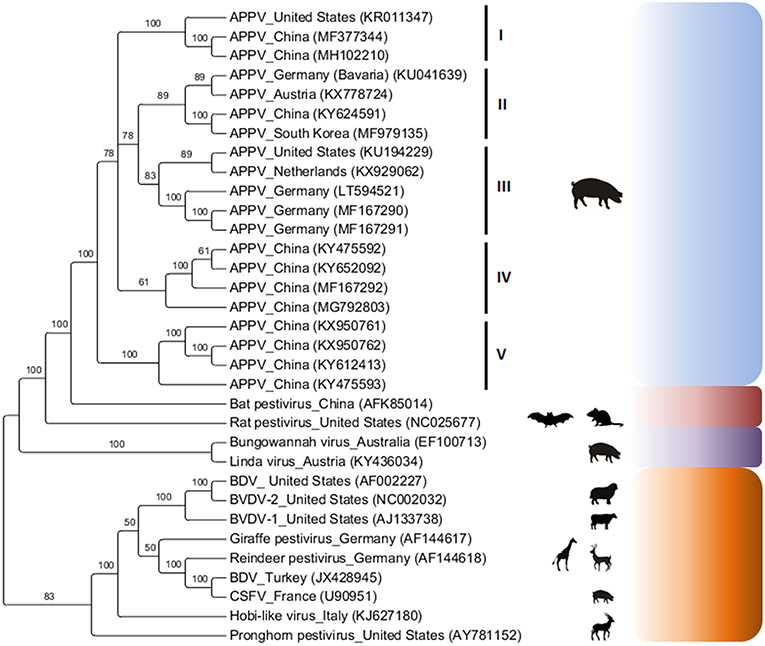
Figure 2. Phylogenetic analysis of the atypical porcine pestivirus complete polyprotein (13,695 bp after alignment). Analysis was based on the Bayesian method and the GTR + I + G4 evolutionary model using MrBayes 3·1·2 (37) via CIPRES Science Gateway (38). The numbers on the nodes correspond to clade support values accessed with 106 bootstrap replicates. Viruses of the genus Pestivirus (classical and atypical) were used as an external group.
A phylogenetic tree based on Bayesian analysis of 20 complete APPV polyprotein sequences and other pestiviruses demonstrated a monophyletic cluster topology for APPV. In addition, five distinct clusters were observed within the APPV clade (cluster I: North America and Asia, cluster II: Europe and Asia, cluster III: North America and Europe and clusters IV and V: Asia; Figure 2).
The economic impact of some pestiviruses in swine species is manifested by devastating losses worldwide. The high genetic variability of pestiviruses is the key point triggering practical consequences in epidemiology, diagnosis, control, and economic impact on livestock, especially in pigs.
Given the wide distribution of APPV and its genetic variability in different countries, studies regarding the epidemiology, ecology, pathogenesis, pathophysiology, transmission routes, and the impact of this virus on swine farms are necessary. Recent detection of APPV in semen from commercial boar studs has sparked further investigations to clarify whether APPV can be transmitted through artificial insemination or reproductive biotechnologies, commonly used in pig breeding, which might play a significant role in the dissemination of pathogens.
Another interesting epidemiological issue is the recent detection of APPV in wild boars, demonstrating the ability of the virus to infect domestic and wild pigs. To date, the role of this wild species in the epidemiology of APPV remains unknown and further research should be conducted with this species, as the world's population of wild boars is increasing. Moreover, the impact of this ecological imbalance on the maintenance and spread of pathogens could be devastating, as in the case of classical swine fever virus spread in Europe.
Based on the available data, we highlighted some key points related to APPV infection, such as: (1) adaptive immunity of gilts/and sows; (2) possible transmission of the virus by semen; (3) absence of a commercial vaccine. For this reason, we hypothesize and suggest a few insights that may have a positive impact on the control of the infection. Since the highest prevalence of CT-piglets occurs in gilts, we suggest that the immune status of the dam and the time of infection are the key factors related to disease development. So, we recommend the use of an acclimatization strategy for replacement gilts, similarly to the protocol used for Mycoplasma hyopneumoniae control. In addition, to reduce the risk of APPV transmission by semen, it is advised that batches used in artificial insemination protocols are previously tested for the presence of APPV genomes. Likewise, based on the high genetic variability of APPV and recent research about the development of a subunit vaccine against APPV based on the E2 protein (39), we strongly recommend the implementation of a feedback management in farms with CT-cases, strategy that could be used until the development of an efficient commercial vaccine. Therefore, additional epidemiological information is required in order to develop strategies of control and eradication of APPV in pig production.
Finally, even though the potential for intercontinental spread of some viruses and the impact of exotic and emerging diseases on worldwide pig production is known, we strongly recommend additional epidemiological studies that will provide current essential information regarding APPV and elucidate possible routes of entry, dissemination, and genetic evolution of APPV, as well as other viral agents. This data will aid in the active surveillance of pathogens considered exotic and/or emerging around the world (i.e., porcine epidemic diarrhea virus, transmissible gastroenteritis virus, porcine reproductive and respiratory syndrome virus, and African swine fever virus).
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Kielland C, Wisløff H, Valheim M, Fauske A, Reksen O, Framstad T. Preweaning mortality in piglets in loose-housed herds: etiology and prevalence. Animal (2018) 12:1950–7. doi: 10.1017/S1751731117003536
2. de Groof A, Deijs M, Guelen L, van Grinsven L, van Os-Galdos L, Vogels W, et al. Atypical porcine pestivirus: a possible cause of congenital tremor type A-II in newborn piglets. Viruses (2016) 8:271. doi: 10.3390/v8100271
3. Schwarz L, Riedel C, Högler S, Sinn LJ, Voglmayr T, Wöchtl B, et al. Congenital infection with atypical porcine pestivirus (APPV) is associated with disease and viral persistence. Vet Res. (2017) 48:51. doi: 10.1186/s13567-016-0406-1
4. Gatto IRH, Harmon K, Bradner L, Silva P, Linhares DCL, Arruda PH, et al. Detection of atypical porcine pestivirus in Brazil in the central nervous system of suckling piglets with congenital tremor. Transbound Emerg Dis. (2018) 65:375–80. doi: 10.1111/tbed.12824
6. Stromberg MW, Kitchell RL. Studies on myoclonia congenita. I Review of literature and field investigations. Am J Vet Res. (1958) 19:377–82.
7. Bolin SR. Congenital tremors virus. In: Leman AD, Straw BE, Mengeling WL, D'Allaire S, Taylor DJ, editors. Diseases of Swine. London: Wolfe Publishing Ltd. (1992). p. 247–9.
8. Done S, Williamson SM, Strugnell BW. Nervous and locomotor systems. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Diseases of Swine. Ames, IA: Wiley-Blackwell (2012). p. 294–328.
9. Hause BM, Collin EA, Peddireddi L, Yuan F, Chen Z, Hesse RA, et al. Discovery of a novel putative atypical porcine pestivirus in pigs in the USA. J Gen Virol. (2015) 96:2994–8. doi: 10.1099/jgv.0.000251
10. Arruda BL, Arruda PH, Magstadt DR, Schwartz KJ, Dohlman T, Schleining JA, et al. Identification of a divergent lineage porcine pestivirus in nursing piglets with congenital tremors and reproduction of disease following experimental inoculation. PLoS ONE. 11:e0150104. doi: 10.1371/journal.pone.0150104
11. Waage JK, Mumford JD. Agricultural biosecurity. Philos. Trans. R. Soc. Lond. B. Biol. Sci. (2008) 363:863–76. doi: 10.1098/rstb.2007.2188
12. Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Scott Muerhoff A, et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J Gen Virol. (2017) 98:2106–12. doi: 10.1099/jgv.0.000873
13. Kirkland PD, Frost MJ, Finlaison DS, King KR, Ridpath JF, Gu X. Identification of a novel virus in pigs - Bungowannah virus: a possible new species of pestivirus. Virus Res. (2007) 129:26–34. doi: 10.1016/j.virusres.2007.05.002
14. Lamp B, Schwarz L, Högler S, Riedel C, Sinn L, Rebel-Bauder B, et al. Novel Pestivirus species in pigs, Austria, 2015. Emerg Infect Dis. (2017) 23:1176–9. doi: 10.3201/eid2307.170163
15. Postel A, Hansmann F, Baechlein C, Fischer N, Alawi M, Grundhoff A, et al. Presence of atypical porcine pestivirus (APPV) genomes in newborn piglets correlates with congenital tremor. Sci Rep. 6:27735. doi: 10.1038/srep27735
16. Beer M, Wernike K, Drager C, Hoper D, Pohlmann A, Bergermann C, et al. High prevalence of highly variable atypical porcine pestiviruses found in Germany. Transbound Emerg Dis. (2017) 64:e22–e26. doi: 10.1111/tbed.12532
17. Postel A, Meyer D, Petrov A, Becher P. Recent emergence of a novel porcine pestivirus: interference with classical swine fever diagnosis? Emerg Microbes Infect. 6:e19. doi: 10.1038/emi.2017.5
18. Moennig V, Becher P. Pestivirus control programs: how far have we come and where are we going? Anim Health Res Rev. (2015) 16:83–7. doi: 10.1017/S1466252315000092
19. Kirkland PD, Read AJ, Frost MJ, Finlaison DS. Bungowannah virus–a probable new species of pestivirus–what have we found in the last 10 years? Anim Health Res Rev. (2015) 16:60–3. doi: 10.1017/S1466252315000031
20. Blomström AL, Fossum C, Wallgren P, Berg M. Viral metagenomic analysis displays the co-infection situation in healthy and PMWS Affected pigs. PLoS ONE (2016) 11:e0166863. doi: 10.1371/journal.pone.0166863
21. Muñoz-González S, Canturri A, Pérez-Simó M, Bohórquez JA, Rosell R, Cabezón O, et al. First report of the novel atypical porcine pestivirus in Spain and a retrospective study. Transbound Emerg Dis. (2017) 64:1645–9. doi: 10.1111/tbed.12699
22. Yuan J, Han Z, Li J, Huang Y, Yang J, Ding H, et al. Atypical porcine pestivirus as a novel type of pestivirus in pigs in China. Front Microbiol. 8:862. doi: 10.3389/fmicb.2017.00862
23. Kim S, Jeong C, Yoon S, Lee K, Yang M, Kim B, et al. Detection of atypical porcine pestivirus (APPV) from a case of congenital tremor in Korea. Korean J Vet Serv. (2017) 40:209–13. doi: 10.7853/kjvs.2017.40.3.209
24. Mósena ACS, Weber MN, da Cruz RAS, Cibulski SP, da Silva MS, Puhl DE, et al. Presence of atypical porcine pestivirus (APPV) in Brazilian pigs. Transbound Emerg Dis. (2018) 1:22–6. doi: 10.1111/tbed.12753
25. Possatti F, Headley SA, Leme RA, Dall Agnol AM, Zotti E, de Oliveira TES, et al. Viruses associated with congenital tremor and high lethality in piglets. Transbound Emerg Dis. (2018) 65:331–7. doi: 10.1111/tbed.12807
26. Dessureault FG, Choinière M, Provost C, Gagnon CA. First report of atypical porcine pestivirus in piglets with congenital tremor in Canada. Can Vet J. (2018) 59:429–32.
27. Dénes L, Biksi I, Albert M, Szeredi L, Knapp DG, Szilasi A, et al. Detection and phylogenetic characterization of atypical porcine pestivirus strains in Hungary. Transbound Emerg Dis. (2018) 65:2039–42. doi: 10.1111/tbed.12981
28. Williamson S, Group PE. Congenital tremor associated with atypical porcine pestivirus. Vet Rec. (2017) 180:42–3. doi: 10.1136/vr.j121
29. Postel A, Meyer D, Cagatay GN, Feliziani F, De Mia GM, Fischer N, et al. High abundance and genetic variability of atypical porcine pestivirus in pigs from Europe and Asia. Emerg Infect Dis. (2017) 23:2104–7. doi: 10.3201/eid2312.170951
30. Zhang K, Wu K, Liu J, Ge S, Xiao Y, Shang Y, et al. Identification of atypical porcine pestivirus infection in swine herds in China. Transbound Emerg Dis. (2017) 64:1020–3. doi: 10.1111/tbed.12659
31. Zhang H, Wen W, Hao G, Hu Y, Chen H, Qian P, et al. Phylogenetic and genomic characterization of a novel atypical porcine pestivirus in China. Transbound Emerg Dis. (2018) 65:e202–e204. doi: 10.1111/tbed.12675
32. Gatto IRH, Arruda PH, Visek CA, Victoria JG, Patterson AR, Krull AC, et al. Detection of atypical porcine pestivirus in semen from commercial boar studs in the United States. Transbound Emerg Dis. (2018) 65:339–43. doi: 10.1111/tbed.12759
33. Shen H, Liu X, Zhang P, Wang L, Liu Y, Zhang L, et al. Identification and characterization of atypical porcine pestivirus genomes in newborn piglets with congenital tremor in China. J Vet Sci. (2018) 19:468–71. doi: 10.4142/jvs.2018.19.3.468
34. Cagatay GN, Antos A, Meyer D, Maistrelli C, Keuling O, Becher P, et al. Frequent infection of wild boar with atypical porcine pestivirus (APPV). Transbound Emerg Dis. (2018) 65:1087–93. doi: 10.1111/tbed.12854
35. Colom-Cadena A, Ganges L, Muñoz-González S, Castillo-Contreras R, Bohórquez JÁ, Rosell R, et al. Atypical porcine pestivirus in wild boar (Sus scrofa). Spain Vet Rec. (2018) 183:569. doi: 10.1136/vr.104824
36. Possatti F, Oliveira TES, Leme RA, Zotti E, Dall Agnol AM, Alfieri AF. Pathologic and molecular findings associated with atypical porcine pestivirus infection in newborn piglets. Vet Microbiol. (2018) 227:41–4. doi: 10.1016/j.vetmic.2018.10.026
37. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (2003) 19:1572–4. doi: 10.1093/bioinformatics/btg180
38. Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE). New Orleans, LA (2010). p. 1–8.
Keywords: atypical porcine pestivirus, congenital tremor, pestiviruses, pig production, pre-weaning mortality
Citation: Gatto IRH, Sonálio K and de Oliveira LG (2019) Atypical Porcine Pestivirus (APPV) as a New Species of Pestivirus in Pig Production. Front. Vet. Sci. 6:35. doi: 10.3389/fvets.2019.00035
Received: 10 October 2018; Accepted: 28 January 2019;
Published: 21 February 2019.
Edited by:
Jingyun Ma, South China Agricultural University, ChinaReviewed by:
Benjamin Lamp, University of Veterinary Medicine Vienna, AustriaCopyright © 2019 Gatto, Sonálio and de Oliveira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luís Guilherme de Oliveira, bHVpcy5ndWlsaGVybWVAdW5lc3AuYnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.