
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 22 January 2019
Sec. Veterinary Infectious Diseases
Volume 5 - 2018 | https://doi.org/10.3389/fvets.2018.00347
This article is part of the Research Topic Emerging Swine Viruses View all 14 articles
A large diversity of influenza A viruses (IAV) within the H1N1/N2 and H3N2 subtypes circulates in pigs globally, with different lineages predominating in specific regions of the globe. A common characteristic of the ecology of IAV in swine in different regions is the periodic spillover of human seasonal viruses. Such human viruses resulted in sustained transmission in swine in several countries, leading to the establishment of novel IAV lineages in the swine host and contributing to the genetic and antigenic diversity of influenza observed in pigs. In this review we discuss the frequent occurrence of reverse-zoonosis of IAV from humans to pigs that have contributed to the global viral diversity in swine in a continuous manner, describe host-range factors that may be related to the adaptation of these human-origin viruses to pigs, and how these events could affect the swine industry.
Influenza is one of the most devastating respiratory pathogens of pigs and humans and continues to threat animal and public health with the continuing possibility of outbreaks or a pandemic. The intricacies of influenza A viruses (IAV) at the human-swine interface dates back to the 1918 pandemic. For several decades, it was hypothesized that pigs played a role in the origin of the 1918 H1N1 pandemic virus (1). Although there is evidence suggesting that the pandemic virus did not originate from pigs and that the classical swine H1N1 virus was in fact derived from the 1918 human virus (2), the bias perceiving swine as the source of IAV to humans still remains.
The ecology of IAV is complex and involves a broad range of avian and mammalian host species. IAVs are enveloped, segmented RNA viruses in the family Orthomyxoviridae (3). The virus genome is composed of eight negative-sense, single-stranded viral RNA (vRNA) segments that encode between 10 and 17 viral proteins depending on the strain (4–6). Each RNA segment forms the viral ribonucleoprotein complexes (vRNPs) with the nucleoprotein (NP) and the three polymerase proteins (PB2, PB1, and PA). Two major glycoproteins are projected on the virus envelope, hemagglutinin (HA), and neuraminidase (NA) (7). Based on the antigenic properties of the HA and NA, IAV are divided into 18 HA subtypes (H1–H18) and 11 NA subtypes (N1–N11) (7–9).
Influenza viruses have high mutation rates and are constantly changing, which enables the virus to quickly adapt to changes in the host environment, as is the case during interspecies transmission. The rapid evolution results from two mechanisms: reassortment and point mutations (10). Reassortment occurs when two different strains infect the same cell of a given host, allowing for exchange of intact gene segments. When reassortment involves either the HA or NA segments, it is termed antigenic shift. Point mutations occur due to an error prone polymerase devoid of a proof-reading and correction mechanism. When point mutations are fixed in the HA or NA segments, usually a result of escape from immune pressure, it is termed antigenic drift. Both of these mechanisms play pivotal roles in the emergence of novel influenza viruses that could jump the host barrier. Once the virus jumps into a new host, it must adapt and change to be able to spread and become established in the new population.
In this review, we describe the role of pigs in the interspecies transmission of influenza and how their susceptibility to different viruses can affect the overall epidemiology of swine influenza. We discuss the factors that have been implicated in the interspecies transmission of influenza with an emphasis on the human-swine interface. We then provide an overview of human-to-swine IAV spillover events that significantly affected the epidemiology of viruses circulating in swine and how these viruses can have a negative effect on the control of influenza in pigs.
To result in a successful replicative cycle, influenza viruses must efficiently infect the host cell, replicate, and produce functional virus progeny that will be released and infect new cells. The first step for infection is the attachment of the HA protein to the cell receptor. The HA is a type I transmembrane glycoprotein, present as a homotrimer on the virus' surface, each monomer carrying a transmembrane anchor and a small cytoplasmic tail. The proteolytic cleavage of the precursor HA0 produces two subunits, HA1 (globular head) and HA2 (stem). The receptor binding site (RBS) forms a shallow pocket at the distal tip of the HA1 head and consists of a base of four highly conserved amino acid residues (Y98, W153, H183, and Y195, numbering based on the H3 subtype) that are bordered by the 130-loop, the 190-helix and the 220-loop (11–13).
Through the RBS, influenza viruses bind to terminal sialic acid (SA, N-acetylneuraminic acid) moieties in glycoprotein or glycolipid receptors on the host cell surface. The SAs are usually bound to the penultimate galactose (Gal) in two major conformations: α2,3SA or α2,6SA (13). Differences in the type of SA linkage found in receptors expressed in different host species have a major impact on the host restriction of IAVs. Sialic acids with α2,3-linkage are predominantly expressed on epithelial cells in the intestinal and respiratory tracts of birds while the epithelial cells in the upper respiratory tract of humans contains predominantly α2,6-linked SA receptors (14–17) (Figure 1). Most avian influenza viruses preferentially bind to α2,3-SA, whereas human and other mammalian influenza viruses preferentially recognize α2,6- SA receptors (21–23).
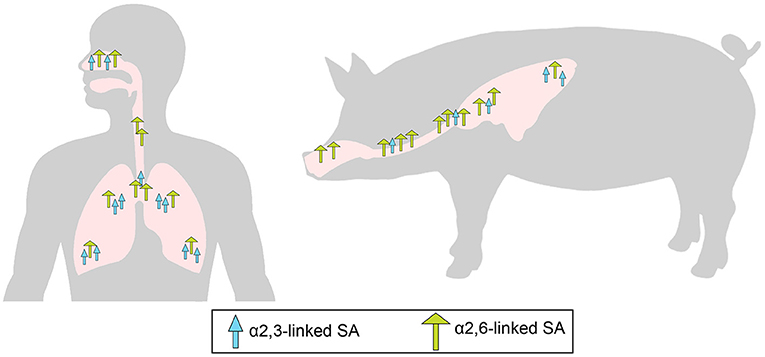
Figure 1. Overall distribution of α2,6-linked sialic acid (SA; green long arrow) and α2,3-linked SA (blue short arrow) in the epithelium of the respiratory tract of pigs (18, 19) and humans (14, 15). Adapted from de Graaf and Fouchier (20).
Pigs have been historically believed to be intermediary hosts, or “mixing vessels,” of influenza viruses due to their susceptibility to infection with both human-origin and avian-origin IAV and their propensity for the generation of reassortant viruses (24–27). Pigs have a similar distribution to humans of α2,3-SA and α2,6- SA receptors in the respiratory tract (Figure 1). As in humans, α2,6-linked SA receptors predominate in the upper respiratory tract of pigs, but α2,3-SA receptors are present in low quantities in swine tracheas, and the frequency increases toward the lower respiratory tract (18, 19) (Figure 1). The presence of both types of SA receptors in swine airways supports the potential role of pigs as “mixing vessels.” However, such distribution of α2,3- and α2,6-SA receptors is similar in swine and humans (15, 18), and it must be noted that avian viruses do not usually transmit from pig-to-pig as is also the case in humans (28, 29). Humans can also become infected with avian-origin IAVs directly from avian sources and could potentially provide the environment for the adaptation of avian viruses (30–32). Hence, generation of reassortant viruses with pandemic potential may not require swine as intermediate hosts. However, as highlighted by the 2009 pandemic (33), while swine are not required, they may serve as intermediate hosts for generation of reassortant viruses with the ability to cause human pandemics. The 2009 pandemic has led to an increased concern about the transmission of swine viruses to humans. However, improved surveillance of swine IAV after the pandemic has shown that human viruses are transmitted to pigs, and have resulted in sustained onward transmission, far more frequently than swine viruses have infected humans (34). This lower host barrier observed for human viruses in pigs can be explained in part by the similar receptor distribution in both species and the shared preference for α2,6-linked SA receptors between human and swine viruses (22, 26).
Although IAV transmission events from humans to pigs are continually detected globally and despite the similarities of receptor preference and distribution between the two species, whole human IAV rarely become established in swine. Typically, these viruses reassort and emerge with only some of the human-origin viral gene segments persisting, often with marked genetic differences from the precursor strain (34–36). This implies that adaptation factors other than the receptor linkage-type specificity are required for human-origin viruses to be transmitted and subsequently become endemic in swine populations.
The adaptation of influenza viruses between humans and pigs is likely driven by selective pressures or bottlenecks imposed to the virus population during IAV host jump, as a result of the changes in the host environment (37, 38). Several factors may affect these selective pressures during interspecies transmission, either within the virus or the host. Receptor-binding specificity and affinity, balance between HA and NA content, temperature of the host, and host-specific immune factors may be some of these factors. However, the differences in the selective pressure between humans and swine and how they may differently affect virus adaptation are not entirely understood, and some of the currently know differences are discussed below.
Specific amino acid residues at the influenza HA are required for binding to either α2,3-SA or α2,6-SA receptors and specific amino acid substitutions at the RBS of the HA can alter receptor-binding specificity and facilitate host jump (Figure 2A). In H1 subtype viruses, positions 190 and 225 were shown to have an impact in receptor specificity. The combination of E190/G225, E190/D225, or D190/G225 in the RBS of the HA, found in avian viruses and late stage 2009 pandemic H1N1 strains, results in dual receptor-binding specificity, whereas D190/D225 and D190/E225, combinations found in seasonal human viruses, results in human-type receptor specificity (40–42). As for H3 and H9 viruses, positions 226 and 228 in the HA are critical for receptor specificity. Avian-adapted viruses usually present Q226/G228 and show dual-binding or α2,3-SA preference, but amino acid substitutions Q226L/G228S leads to receptor specificity switch to human-type receptor preference and is, therefore, more commonly found in human viruses (22, 43). Analysis of H1, H3, and H9 virus sequences from swine using the Influenza Research Database (44) revealed that swine viruses have mostly D190/D225 in H1 viruses, a fairly equal distribution between Q226/G228 and L226/G228 in H9 viruses, and the unique combination of amino acids in H3 viruses V226/S228 (Figure 3).
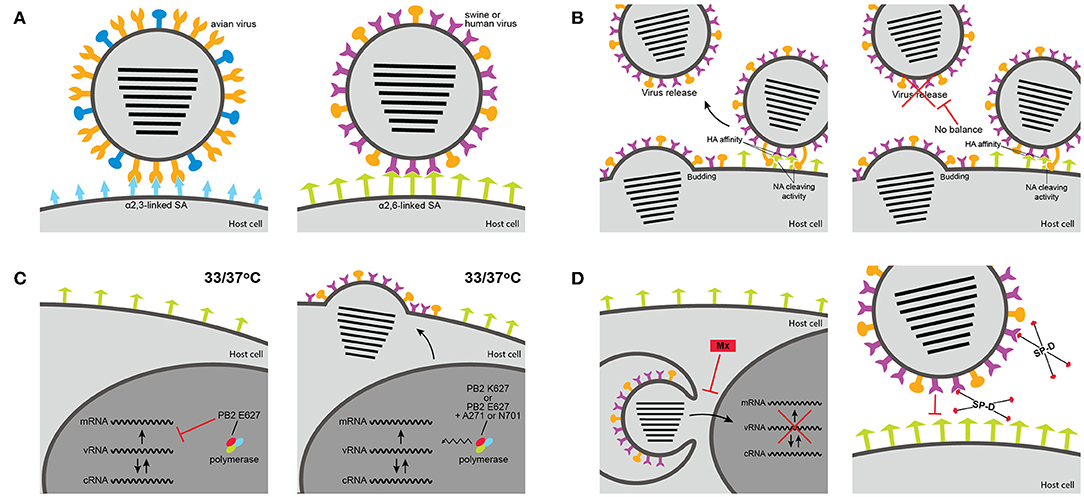
Figure 2. Host range determinants of influenza A viruses (IAV). (A) Avian influenza virus HA protein recognize short α2,3-linked sialic acid (blue), whereas HA from human and swine IAV recognize long α2,6-linked sialic acid (green). (B) The balance between the HA binding affinity and the NA activity to cleave sialic acid receptors is important for replication and adaptation to a new species. If a virus has strong biding affinity but low cleavage activity replication may be reduced. (C) The PB2 polymerase has an impact in the optimal replication temperature of IAV and can restrict host range. K627 increases replication at the low temperature of human or swine upper airway. E627 decreases replication at low temperatures, unless in combination with A271 or N701. (D) The sensitivity of a virus to host-specific innate immune factors can restrict interspecies transmission of IAV. To be able to replicate and spread in a new host, IAV must become resistant to the antiviral activity of interferon-induced Mx protein or to the neutralizing activity of surfactant protein D (SP-D) from that particular host. Adapted from Cauldwell et al. (39).
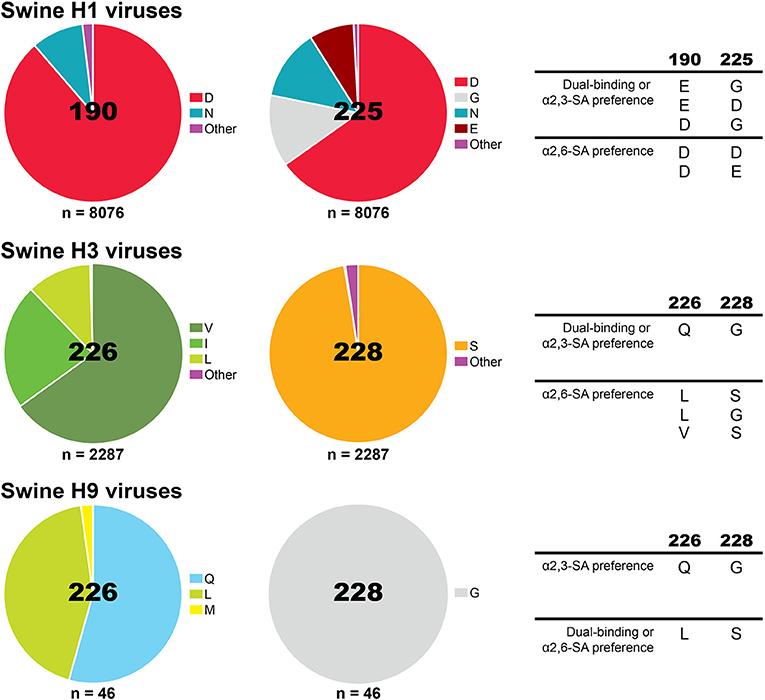
Figure 3. Proportion of amino acids found in influenza A viruses circulating in pigs globally at the HA receptor-binding site positions previously shown to impact receptor-specificity for H1, H3, and H9. Analysis was performed using the Influenza Research Database Sequence Variation (SNP) tool (44). Sequences with 100% identity were removed resulting in a set of 8076 H1 HA, 2287 H3 HA, and 46 H9 HA swine IAV sequences. The amino acids previously shown to change receptor-binding specificity are displayed on the right.
Receptor-binding specificity of influenza HA is not only mediated by changes in the sialic acid linkage, the structural length and topology of the glycans can also determine the binding specificity and affinity of IAV. Avian viruses were shown to bind to α2,3-linked SA carrying a shorter carbohydrate chain whereas human viruses bind preferentially to long α2,6-linked SA (45, 46). Moreover, avian HA binds to narrow α2,3-SA in a “cone-like” topology and human HA binds to long α2,6-SA in an “umbrella-like” topology, which are predominantly expressed in the human upper respiratory tissues (47). In general, human and swine viruses have been shown to recognize similar glycan structures on glycan microarrays, mainly branched α2,6-SA (48, 49).
While the HA is involved with binding to SA receptors, the NA cleaves α2-3 and α2-6-linked SA residues from cellular surfaces and mucus through its sialidase enzymatic activity and mediates the release of newly synthesized viruses from the host cells (7). For an optimal viral replication, balanced activities between the HA binding affinity and the NA enzymatic function are expected. The ideal HA-NA balance seems to be an important factor in host adaptation (Figure 2B). The HA-NA balance was shown to be crucial for the adaptation of the 2009 pandemic H1N1 virus to humans, since balanced HA and NA activities were seen in the human strains but not in precursor swine viruses (50) and this balance resulted in increased replication and transmissibility in ferrets (51). Additionally, adaptation of H5 and H7 viruses from wild birds to chickens led to selective changes in both HA and NA, maintaining a balance between binding and cleavage that was important for replication and transmission in the new host (52). These chicken-adapted H5 and H7 viruses possess a shorter NA due to the deletion of several residues in the stalk domain that were shown to enhance replication and virulence in chickens but block respiratory transmission in ferrets (53, 54).
In addition to the NA, the matrix (M) gene segment has been shown to be a critical determinant of respiratory transmission efficiency of IAV in new hosts. The M segment was implicated with the increased transmissibility of the 2009 pandemic H1N1 virus in animal models (55, 56), suggesting it played an important role on the spread of the virus in humans. In pigs, the combination of the NA and M genes from the 2009 pandemic virus was essential to facilitate efficient replication and transmissibility (57). Interestingly, reassortant H1 and H3 swine-origin viruses containing the M gene of the 2009 pandemic virus have caused almost yearly zoonotic outbreaks in humans, more frequently than was observed prior to the pandemic, confirming that the M gene plays a role in adaptation and transmission of swine viruses in humans (58–60).
The virus polymerase (comprised of viral proteins PB1, PB2, and PA) was also shown to be a major determinant for host range of influenza viruses (61) (Figure 2C). This host restriction has been attributed to a single residue in the PB2 gene, amino acid 627, and is largely associated with the optimal temperature of replication of IAVs (62, 63). While the human upper respiratory tract temperature is around 33°C, in the avian intestinal tract the temperature is closer to 41°C. Therefore, enhanced replication at lower temperature should correlate with enhanced replication in the upper airway of humans and consequently improve transmission. Lysine (K) at position 627 in PB2, present in the vast majority of human viral isolates (64), was correlated with increased polymerase activity, virus replication and transmission in mammals (65–67), including enhanced replication of an avian virus in pigs (68). Replication and polymerase activity of different avian viruses, which predominantly possess glutamic acid (E) at position 627, were reduced at low temperature in mammalian cells (65, 66, 69).
The temperature of the upper respiratory tract of pigs is approximately 37°C and higher (approximately 39°C) in the lower respiratory tract. Interestingly, most swine isolates that have a PB2 of avian-origin retain the avian signature E627, including the predominant North American triple reassortant internal gene (TRIG) constellation viruses, the predominant Eurasian avian-like viruses, and even the 2009 pandemic H1N1 viruses (70, 71). The presence of the avian-like E627 in swine viruses usually does not result in the temperature sensitivity observed for avian viruses in mammalian cells (69), suggesting that these viruses can replicate at temperatures of avian intestines and human airways. Other residues, such as A271 and N701, were shown to compensate for the absence of K627 in these swine or swine-origin viruses and contribute to virus growth and transmission in swine and other mammalian species, including humans (71–74).
Following influenza infection in respiratory epithelial cells, acute inflammation leads to activation of the innate immune response through pro-inflammatory cytokines or chemokines (75). Type-I interferons (IFN-α/β) are cytokines quickly secreted after IAV infection. Type-I IFN mediated responses to IAV results in the expression of several antiviral proteins (76, 77). The Mx proteins are a family of large GTPases that are central to the antiviral activity of IFN against IAV by blocking nuclear entry of the vRNPs (78, 79). Sensitivity to interferon-induced Mx varies among different IAV strains and represents a barrier against transmission of avian influenza viruses to mammals: avian isolates are more susceptible to the antiviral action of murine Mx and human MxA proteins than human viruses (80, 81). The Mx sensitivity was shown to be determined by a cluster of surface-exposed amino acids on the viral NP (81, 82). Interestingly, serial passage in mice of a virus that is sensitive to murine/human Mx activity leads to a single amino acid adaptive NP mutation that results in escape from the Mx activity, and the same mutation is also seen in human H7N9 isolates (83). Not surprisingly, some swine IAV strains with avian-origin NP tend to have a higher sensitivity to mouse Mx1 than human isolates (84). However, the 1918 pandemic H1N1 and the 2009 pandemic H1N1 viruses acquired resistance-associated substitutions on the NP protein that allow escape from human Mx (82). The functional Mx1 protein is expressed in the lungs of pigs experimentally infected with IAV (85). It seems that the precursor of the 2009 pandemic H1N1 virus acquired Mx-resistance mutations driven by the porcine Mx1 during its circulation in pigs prior to the pandemic, being able to partially resist the human MxA (82). The Eurasian avian-like viruses are similarly resistant to human MxA, however different mutations were attributed to this phenotype (86). It remains unknown whether human and swine viruses would have different sensitivities to the porcine Mx protein.
Surfactant protein D (SP-D) is a collectin of the innate immune system that also has early strong antiviral activity against IAVs. SP-D binds to carbohydrate moieties on the surface of influenza viruses (HA and/or NA), blocking attachment to epithelial cells and inducing phagocytic responses, resulting in non-specific virus neutralization and clearance (87). The susceptibility of different IAV to SP-D activity was shown to be dependent on the glycosylation pattern of the virus, particularly on the HA (88–90). Influenza strains of the H3 subtype tend to acquire and accumulate more glycosylations on the HA head as a mechanism to evade the antibody response in humans, but this in turn may make them more susceptible to the antiviral effect of SP-D. Interestingly, porcine SP-D has a higher affinity to bind IAV glycans than human or rat SP-D, resulting in stronger neutralization activity (91, 92). Therefore, differences in susceptibility to Mx or SP-D could be an important component in host restriction of influenza viruses that needs to be overcome, usually by changes in specific viral proteins, in order for a virus to adapt to a new species (Figure 2D).
The IAV NS1 protein plays an important role as an antagonist of the host IFN response by preventing the activation of retinoic acid-inducible gene 1 (RIG-I) or inhibiting processing of mRNA (93). Differences between the NS1 amino acid sequences may affect the functional IFN-antagonistic properties of the NS1 (94, 95). Consequently, NS1 and its ability to control IFN response could play a role in host range of IAV. Indeed, although the avian NS1 protein was able to control IFN-α/β response in human cells, the human type I IFN response appeared to limit the replication of the avian viruses, suggesting that the NS1 also contributes to the host specificity of IAV (96).
The adaptive immunity of an individual or population can also have a role in host range restriction of IAV. Even when a novel virus contains an ideal combination of factors that allow replication in the new host, as discussed above, previous cross-protective immunity might block even the initial infection. In some cases, the level of cross-protective immunity of the population may still allow infection but might block virus dissemination; however, naïve individuals will be at a higher risk for infection and may serve as sources of transmission. That was the case for the zoonotic infections with swine-origin viruses in recent years, in which the majority of affected individuals were children (58, 59). For these outbreaks, infection was observed in people with close contact with pigs, and transmission from human-to-human was rare, which was attributed to low levels of cross-protective immunity in the human population due to previous exposure to seasonal viruses (97). In pigs, however, there is a continuous introduction of naïve individuals and the majority of the population does not have previous immunity to viruses circulating in humans, increasing the chances of those viruses that have ability to infect pigs to become widespread.
Human-origin viruses have been repeatedly transmitted to swine worldwide and have had a major role on the epidemiology of swine IAV (34) (Figure 4). The classical swine H1N1 virus that emerged around the 1918 pandemic remained relatively antigenically stable for eight decades without causing major problems to swine producers. A novel triple-reassortant virus with human seasonal H3N2 surface genes emerged in the late 1990's in North America (27, 98) and led to reassortment with the classical viruses and subsequently gave rise to different antigenically distinct H3N2, H1N1, and H1N2 strains (99, 100). The triple-reassortant internal gene (TRIG) constellation, containing gene segments from a complex reassortment history among swine-, human- and avian-origin IAVs, became the predominant backbone of the viruses circulating in pigs in the U.S. (101, 102). Shortly after, two additional introductions of human-origin H1N1 resulted in the establishment of two new lineages of H1N1 and H1N2 viruses after reassortment with the TRIG strains, termed δ-lineages (103). After the spread in humans, the H1N1 pandemic 2009 virus (H1N1pdm) was quickly transmitted to swine in North America (104). And recently, a novel virus derived from 2010 to 2011 human seasonal H3 IAV led to establishment of a new H3-lineage that is genetically and antigenically distinct from previously circulating strains (105). The current scenario for the epidemiology of IAV circulating in North American swine consists of a highly diverse pool of viruses, with 14 phylogenetic clades of HA co-circulating (36, 101, 105, 106). It is clear how impactful the human-to-swine transmissions were to this current epidemiology: at least 10 of these phylogenetic clades have evolved from a human virus. If considering the hypothesis that the classical swine virus originated from the human 1918 pandemic virus, all of those clades should be considered of human-origin.
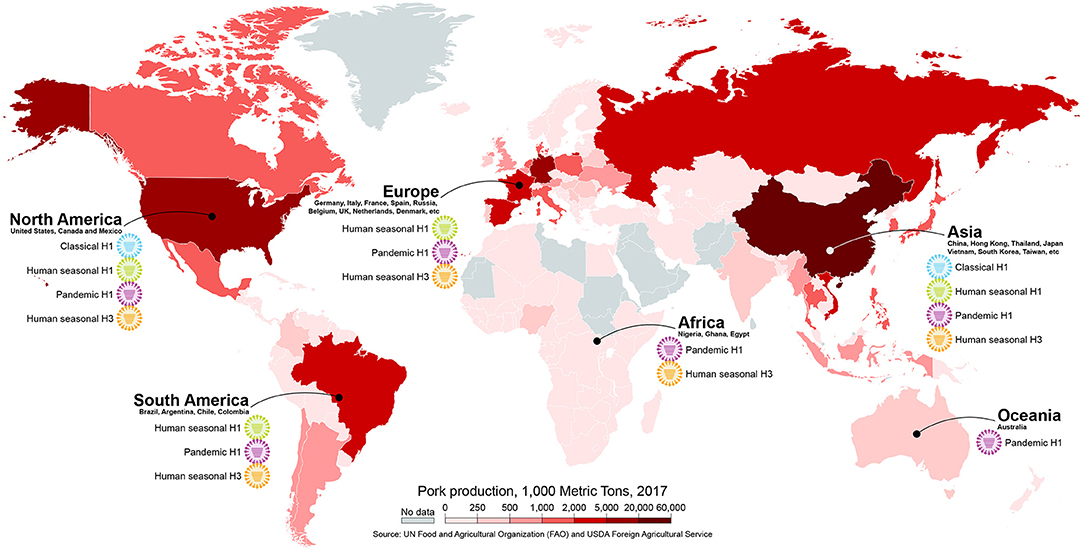
Figure 4. Different subtypes/lineages of human-origin influenza viruses circulating in swine in different continents. The map is colored according to pork production in 1,000 metric tons. Map created with mapchart.net.
In Europe, a human-origin H3N2 virus descendent from the 1968 pandemic virus was introduced in the 1980's. This virus became widespread after reassorting with an avian-origin H1N1 virus that was introduced to European swine in 1979 and remains endemic to date (107, 108). Another human-origin virus, an H1N2, was detected in 1994, containing the H1 that evolved from a 1980 human seasonal H1N1 virus and a human-origin N2 that is distinct from the previously introduced H3N2 human-like virus. This virus acquired the internal gene constellation of the 1979 avian-like virus after reassortment and is now endemic in Europe (109, 110). As in the U.S., the H1N1pdm virus has been transmitted from humans to pigs in Europe establishing a new endemic lineage (108, 111). Recently, a triple-reassortant H3N2 virus with a human-origin HA from a 2004–2005 seasonal virus, N2 from endemic swine viruses, and the internal genes from H1N1pdm has spread in Denmark swine herds (112). In China and other countries in Asia, importation of live animals has resulted in the co-circulation of both European (or Eurasian) and North American TRIG virus lineages that contain human-origin genes (113–115). Additionally, reassortant genotypes between these lineages containing HA and/or NA genes from H1N1 and H3N2 human viruses have been detected in Asia since the 1960's and have become established in pigs (34, 116, 117).
Human-origin IAVs have been reported circulating in pigs in other countries where surveillance is limited (34, 118–120), including countries with large swine populations like Brazil (121, 122), Vietnam (123), Mexico and Chile (124). But, even in some of these cases where human-origin viruses or viral genes were reported in swine, it is not possible to infer if they have become endemic or predominant. However, in several cases, such as in Latin America, the human-origin swine viruses were most closely related to human seasonal strains that circulated many years earlier and were separated by long phylogenetic branches, suggesting that these viruses have circulated undetected in pigs for years prior to their recent detection (34, 118, 121, 124). Considering the frequency of human-to-swine transmissions in highly surveilled areas, it is likely that additional human-origin viruses have gone undetected in countries with low surveillance efforts.
In addition to the recurrent seasonal virus spillover events into swine populations, the H1N1pdm has been repeatedly transmitted from humans to swine globally (125). The H1N1pdm virus originated in Mexico from the reassortment between Eurasian and North American swine viruses and this novel virus may have circulated undetected for approximately 10 years before it gained the ability to infect humans (33). Soon after the initial spread of the H1N1pdm in the human population, the H1N1pdm virus was detected in pigs and since then transmitted from human to pigs throughout the world (104, 126–131). The virus has now become endemic in humans and circulates as a seasonal strain, increasing the possibility of spillovers to swine populations during influenza season each year. Owing to its swine-origin and yearly circulation in humans, continuous and frequent detection of the H1N1pdm virus in pigs has been reported globally (125, 132). The constant circulation and re-introduction of H1N1pdm globally has led to reassortment with endemic swine viruses and changed the genotypic characteristics of swine IAV by contributing several genes, most commonly the internal genes. In the U.S., the surface genes of the H1N1pdm are not frequently maintained, however most genotypes of H1 and H3 viruses contain at least one internal gene of pandemic lineage (133, 134). In Europe, the H1N1pdm virus has reassorted with endemic European viruses and gave rise to genotypes containing the internal genes from pandemic origin and some genotypes have maintained one or both surface genes of pandemic lineage (135). Interestingly, there is recent evidence of the independent antigenic evolution of the swine H1N1pdm virus in European pigs (136). In China, although Eurasian and North American viruses circulated prior to the 2009 pandemic without substantial evidence of reassortment, the introduction of the H1N1pdm led to the establishment of reassortant genotypes containing several internal genes from pandemic lineage (117). The H1N1pdm has been reported throughout the world in swine with frequent reassortment (137–139), even in countries that were previously considered influenza free like Australia and Norway (128, 140).
The repeated transmission of human seasonal viruses to pigs has resulted in the establishment of several human-origin virus lineages globally, adding to the antigenic diversity of swine viruses. Global antigenic characterization has revealed that the antigenic diversity of H1 and H3 viruses circulating in pigs was largely a result of the frequent introductions of human-origin IAV into swine (35). These viruses then evolved antigenically, independent from human strains and often confined to their geographic areas, contributing to the overall global diversity, which consequently contributes to the challenges for effective vaccination programs in swine. Most vaccines used against influenza in swine are whole inactivated virus (WIV) vaccines combined with oil-in-water adjuvants typically given to sows to allow transfer of maternally derived antibodies to piglets (141). Recently, two novel platforms were licensed for use in pigs in the U.S. as alternatives to improve the efficacy of swine vaccines, a non-replicating alphavirus RNA vectored-vaccine and a live-attenuated influenza virus (LAIV) vaccine (142, 143).
Because most vaccines rely on the effective stimulation of the immune response against the surface HA glycoprotein, any changes that lead to antigenic drift, such as the incursion of novel human-origin viruses, can lead to vaccine mismatch. It was demonstrated that changes in only 6 amino acids in the HA account for major antigenic changes of swine H3 influenza viruses, and a single amino acid change can lead to significant antigenic drift (144, 145). Amino acids in similar positions at the HA were also associated with antigenic characteristics of H1 viruses (36). It is not surprising, therefore, that when novel human-origin viruses become established in pigs there are considerable antigenic differences from the circulating swine strains (105), and any vaccines available at the time are unlikely to provide immunity against these novel viruses. In addition to the lack of protection, vaccine mismatch can also have detrimental effects. When the vaccine stimulates a cross-reactive antibody response that fails to neutralize the virus, it can result in severe immune-mediated disease termed vaccine-associated enhanced respiratory disease (VAERD). Therefore, more effective vaccine technologies and vaccination strategies that improve the breadth of the immune response and avoid any negative effects are needed to increase protection against the antigenically diverse human-origin viruses that are continuously introduced in pigs.
Since the 2009 pandemic, renewed attention has been given to the interspecies transmission of influenza viruses between pigs and humans, bringing back the attention to the theory that pigs can serve as “mixing-vessels” of influenza viruses. However, it is not entirely clear if swine are in fact more susceptible to infection with avian viruses than humans. There is compelling evidence, though, that human viruses are frequently transmitted to pigs, and have had a significant impact on the diversity of viruses that circulate in pigs globally. Additional surveillance is necessary to understand the diversity of IAVs circulating in different regions and the participation of human-origin strains in this overall diversity. Surveillance is also critical for antigenic characterization of the strains that are circulating in a particular area to allow an accurate selection of representative vaccine strains that will provide an optimal protection. Moreover, despite the increasing evidence of the important role that human seasonal viruses have played in driving the genetic and antigenic diversity of IAV in swine, vaccine and sick leave policies for swine industry workers are not consistently employed but should be considered. Furthermore, understanding the mechanisms involved with host-range specificity and the adaptation to swine allows assessment of the risks posed by the introduction of novel viruses into the swine population, which is crucial for preparedness and to improve biosecurity measures to reduce the IAV burden to the swine industry.
DR, AV, and DP contributed to the conceptualization of the ideas, drafting and critical revision of the manuscript, and final approval. DR designed figures.
The authors were supported in part by the Center for Research on Influenza Pathogenesis (CRIP), a National Institute of Allergy and Infectious Diseases (NIAID) funded Center of Excellence for Influenza Research and Surveillance (CEIRS, HHSN272201400008C).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, et al. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci USA. (2009) 106:11709–12. doi: 10.1073/pnas.0904991106
2. Worobey M, Han GZ, Rambaut A. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci USA. (2014) 111:8107–12. doi: 10.1073/pnas.1324197111
3. Palese P, Shaw M. Orthomyxoviridae: the viruses and their replication. In: Knipe DM and Howley PM, editors. Fields Virology. Philadelphia, PA: Lippincott Williams & Wilkins (2007). p. 1647–90.
4. Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol. (2013) 87:2455–62. doi: 10.1128/JVI.02656-12
5. Shi Y, Wu Y, Zhang W, Qi J, Gao GF. Enabling the ‘host jump': structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol. (2014) 12:822–31. doi: 10.1038/nrmicro3362
6. Yamayoshi S, Watanabe M, Goto H, Kawaoka Y. Identification of a novel viral protein expressed from the PB2 segment of influenza A virus. J Virol. (2016) 90:444–56. doi: 10.1128/JVI.02175-15
7. Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. (2010) 285:28403–9. doi: 10.1074/jbc.R110.129809
8. Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci USA. (2012) 109:4269–74. doi: 10.1073/pnas.1116200109
9. Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. (2013) 9:e1003657. doi: 10.1371/journal.ppat.1003657
10. Shao W, Li X, Goraya MU, Wang S, Chen JL. Evolution of influenza A virus by mutation and re-assortment. Int J Mol Sci. (2017) 18:1650. doi: 10.3390/ijms18081650
11. Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature (1981) 289:366–73. doi: 10.1038/289366a0
12. Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Ann Rev Biochem. (1987) 56:365–94. doi: 10.1146/annurev.bi.56.070187.002053
13. Nicholls JM, Chan RW, Russell RJ, Air GM, Peiris JS. Evolving complexities of influenza virus and its receptors. Trends Microbiol. (2008) 16:149–57. doi: 10.1016/j.tim.2008.01.008
14. Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature (2006) 440:435–6. doi: 10.1038/440435a
15. Nicholls JM, Bourne AJ, Chen H, Guan Y, Peiris JS. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. (2007) 8:73. doi: 10.1186/1465-9921-8-73
16. Kimble B, Nieto GR, Perez DR. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol J. (2010) 7:365. doi: 10.1186/1743-422X-7-365
17. Pillai SP, Lee CW. Species and age related differences in the type and distribution of influenza virus receptors in different tissues of chickens, ducks and turkeys. Virol J. (2010) 7:5. doi: 10.1186/1743-422X-7-5
18. Nelli RK, Kuchipudi SV, White GA, Perez BB, Dunham SP, Chang KC. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet Res. (2010) 6:4. doi: 10.1186/1746-6148-6-4
19. Trebbien R, Larsen LE, Viuff BM. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol J. (2011) 8:434. doi: 10.1186/1743-422X-8-434
20. de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. (2014) 33:823–41. doi: 10.1002/embj.201387442
21. Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology (1983) 127:361–73. doi: 10.1016/0042-6822(83)90150-2
22. Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. (2000) 74:8502–12. doi: 10.1128/JVI.74.18.8502-8512.2000
23. Gambaryan A, Yamnikova S, Lvov D, Tuzikov A, Chinarev A, Pazynina G, et al. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology (2005) 334:276–83. doi: 10.1016/j.virol.2005.02.003
24. Scholtissek C. Pigs as ‘mixing vessels' for the creation of new pandemic influenza A viruses. Med Principles Pract. (1990) 2:65–71.
25. Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, et al. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. (1994) 75(Pt 9):2183–8. doi: 10.1099/0022-1317-75-9-2183
26. Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. (1998) 72:7367–73.
27. Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. (1999) 73:8851–6.
28. Balzli C, Lager K, Vincent A, Gauger P, Brockmeier S, Miller L, et al. Susceptibility of swine to H5 and H7 low pathogenic avian influenza viruses. Influenza Other Respir Viruses (2016) 10:346–52. doi: 10.1111/irv.12386
29. Abente EJ, Gauger PC, Walia RR, Rajao DS, Zhang J, Harmon KM, et al. Detection and characterization of an H4N6 avian-lineage influenza A virus in pigs in the Midwestern United States. Virology (2017) 511:56–65. doi: 10.1016/j.virol.2017.08.021
30. Worobey M, Han GZ, Rambaut A. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature (2014) 508:254–7. doi: 10.1038/nature13016
31. Lai S, Qin Y, Cowling BJ, Ren X, Wardrop NA, Gilbert M, et al. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997-2015: a systematic review of individual case data. Lancet Infect Dis. (2016) 16:e108–18. doi: 10.1016/S1473-3099(16)00153-5
32. Nelson M, Worobey M. Origins of the 1918 pandemic: revisiting the swine ‘mixing vessel' hypothesis. Am J Epidemiol. (2018) 187:2498–502. doi: 10.1093/aje/kwy150
33. Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernandez R, Lara-Puente JH, et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife (2016) 5:e16777. doi: 10.7554/eLife.16777
34. Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, et al. Introductions and evolution of human-origin seasonal influenza a viruses in multinational Swine populations. J Virol. (2014) 88:10110–9. doi: 10.1128/JVI.01080-14
35. Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, et al. The global antigenic diversity of swine influenza A viruses. Elife (2016) 5:e12217. doi: 10.7554/eLife.12217
36. Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS, Vincent AL. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology (2018) 518:45–54. doi: 10.1016/j.virol.2018.02.006
37. Varble A, Albrecht RA, Backes S, Crumiller M, Bouvier NM, Sachs D, et al. Influenza A virus transmission bottlenecks are defined by infection route and recipient host. Cell Host Microbe (2014) 16:691–700. doi: 10.1016/j.chom.2014.09.020
38. McCrone JT, Woods RJ, Martin ET, Malosh RE, Monto AS, Lauring AS. Stochastic processes constrain the within and between host evolution of influenza virus. Elife (2018) 7:e35962. doi: 10.7554/eLife.35962
39. Cauldwell AV, Long JS, Moncorge O, Barclay WS. Viral determinants of influenza A virus host range. J Gen Virol. (2014) 95(Pt 6):1193–210. doi: 10.1099/vir.0.062836-0
40. Lin T, Wang G, Li A, Zhang Q, Wu C, Zhang R, et al. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology (2009) 392:73–81. doi: 10.1016/j.virol.2009.06.028
41. Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol. (2012) 86:982–90. doi: 10.1128/JVI.06322-11
42. Zhang W, Shi Y, Qi J, Gao F, Li Q, Fan Z, et al. Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution. J Virol. (2013) 87:5949–58. doi: 10.1128/JVI.00545-13
43. Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology (1994) 205:17–23. doi: 10.1006/viro.1994.1615
44. Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, et al. Influenza research database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. (2016) 45:D466–74. doi: 10.1093/nar/gkw857
45. Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. (2006) 355:1143–55. doi: 10.1016/j.jmb.2005.11.002
46. Hidari KI, Shimada S, Suzuki Y, Suzuki T. Binding kinetics of influenza viruses to sialic acid-containing carbohydrates. Glycoconj J. (2007) 24:583–90. doi: 10.1007/s10719-007-9055-y
47. Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. (2008) 26:107–13. doi: 10.1038/nbt1375
48. Bradley KC, Jones CA, Tompkins SM, Tripp RA, Russell RJ, Gramer MR, et al. Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (Novel 2009 H1N1). Virology (2011) 413:169–82. doi: 10.1016/j.virol.2011.01.027
49. Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, et al. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc Natl Acad Sci USA. (2014) 111:E2241–50. doi: 10.1073/pnas.1323162111
50. Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, et al. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J Virol. (2012) 86:9221–32. doi: 10.1128/JVI.00697-12
51. Yen HL, Liang CH, Wu CY, Forrest HL, Ferguson A, Choy KT, et al. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci USA. (2011) 108:14264–9. doi: 10.1073/pnas.1111000108
52. Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. (1999) 73:1146–55.
53. Hoffmann TW, Munier S, Larcher T, Soubieux D, Ledevin M, Esnault E, et al. Length variations in the NA stalk of an H7N1 influenza virus have opposite effects on viral excretion in chickens and ducks. J Virol. (2012) 86:584–8. doi: 10.1128/JVI.05474-11
54. Blumenkrantz D, Roberts KL, Shelton H, Lycett S, Barclay WS. The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J Virol. (2013) 87:10539–51. doi: 10.1128/JVI.00967-13
55. Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, Garcia-Sastre A, et al. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol. (2011) 85:11235–41. doi: 10.1128/JVI.05794-11
56. Campbell PJ, Danzy S, Kyriakis CS, Deymier MJ, Lowen AC, Steel J. The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. J Virol. (2014) 88:3802–14. doi: 10.1128/JVI.03607-13
57. Ma W, Liu Q, Bawa B, Qiao C, Qi W, Shen H, et al. The neuraminidase and matrix genes of the 2009 pandemic influenza H1N1 virus cooperate functionally to facilitate efficient replication and transmissibility in pigs. J Gen Virol. (2012) 93(Pt 6):1261–8. doi: 10.1099/vir.0.040535-0
58. Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, et al. Human infections with influenza A(H3N2) variant virus in the United States, 2011-2012. Clin Infect Dis. (2013) 57(Suppl. 1):S4–11. doi: 10.1093/cid/cit272
59. Jhung MA, Epperson S, Biggerstaff M, Allen D, Balish A, Barnes N, et al. Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis. (2013) 57:1703–12. doi: 10.1093/cid/cit649
60. Centers for Disease Control Prevention. Reported Infections with Variant Influenza Viruses in the United States since 2005 [Online]. (2017) Available online at: https://www.cdc.gov/flu/swineflu/variant-cases-us.htm (Accessed August 28, 2018).
61. Neumann G, Kawaoka Y. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg Infect Dis. (2006) 12:881–6. doi: 10.3201/eid1206.051336
62. Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. (1993) 67:1761–4.
63. Massin P, van der Werf S, Naffakh N. Residue 627 of PB2 is a determinant of cold sensitivity in RNA replication of avian influenza viruses. J Virol. (2001) 75:5398–404. doi: 10.1128/JVI.75.11.5398-5404.2001
64. Chen GW, Chang SC, Mok CK, Lo YL, Kung YN, Huang JH, et al. Genomic signatures of human versus avian influenza A viruses. Emerg Infect Dis. (2006) 12:1353–60. doi: 10.3201/eid1209.060276
65. Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, et al. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. (2007) 3:1374–9. doi: 10.1371/journal.ppat.0030133
66. Labadie K, Dos Santos Afonso E, Rameix-Welti MA, van der Werf S, Naffakh N. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology (2007) 362:271–82. doi: 10.1016/j.virol.2006.12.027
67. Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. (2009) 5:e1000252. doi: 10.1371/journal.ppat.1000252
68. Manzoor R, Sakoda Y, Nomura N, Tsuda Y, Ozaki H, Okamatsu M, et al. PB2 protein of a highly pathogenic avian influenza virus strain A/chicken/Yamaguchi/7/2004 (H5N1) determines its replication potential in pigs. J Virol. (2009) 83:1572–8. doi: 10.1128/JVI.01879-08
69. Massin P, Kuntz-Simon G, Barbezange C, Deblanc C, Oger A, Marquet-Blouin E, et al. Temperature sensitivity on growth and/or replication of H1N1, H1N2 and H3N2 influenza A viruses isolated from pigs and birds in mammalian cells. Vet Microbiol. (2010) 142:232–41. doi: 10.1016/j.vetmic.2009.10.012
70. Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science (2009) 325:197–201. doi: 10.1126/science.1176225
71. Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J Virol. (2010) 84:4395–406. doi: 10.1128/JVI.02642-09
72. Moncorge O, Long JS, Cauldwell AV, Zhou H, Lycett SJ, Barclay WS. Investigation of influenza virus polymerase activity in pig cells. J Virol. (2013) 87:384–94. doi: 10.1128/JVI.01633-12
73. Ma J, Shen H, Liu Q, Bawa B, Qi W, Duff M, et al. Pathogenicity and transmissibility of novel reassortant H3N2 influenza viruses with 2009 pandemic H1N1 genes in pigs. J Virol. (2015) 89:2831–41. doi: 10.1128/JVI.03355-14
74. Liu S, Zhu W, Feng Z, Gao R, Guo J, Li X, et al. Substitution of D701N in the PB2 protein could enhance the viral replication and pathogenicity of Eurasian avian-like H1N1 swine influenza viruses. Emerg Microbes Infect. (2018) 7:75. doi: 10.1038/s41426-018-0073-6
75. La Gruta NL, Kedzierska K, Stambas J, Doherty PC. A question of self-preservation: immunopathology in influenza virus infection. Immunol Cell Biol. (2007) 85:85–92. doi: 10.1038/sj.icb.7100026
76. Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. (2008) 89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0
77. Killip MJ, Fodor E, Randall RE. Influenza virus activation of the interferon system. Virus Res. (2015) 209:11–22. doi: 10.1016/j.virusres.2015.02.003
78. Xiao H, Killip MJ, Staeheli P, Randall RE, Jackson D. The human interferon-induced MxA protein inhibits early stages of influenza A virus infection by retaining the incoming viral genome in the cytoplasm. J Virol. (2013) 87:13053–8. doi: 10.1128/JVI.02220-13
79. Ciancanelli MJ, Abel L, Zhang SY, Casanova JL. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr Opin Immunol. (2016) 38:109–20. doi: 10.1016/j.coi.2015.12.002
80. Dittmann J, Stertz S, Grimm D, Steel J, Garcia-Sastre A, Haller O, et al. Influenza A virus strains differ in sensitivity to the antiviral action of Mx-GTPase. J Virol. (2008) 82:3624–31. doi: 10.1128/JVI.01753-07
81. Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J Virol. (2011) 85:8133–40. doi: 10.1128/JVI.00712-11
82. Manz B, Dornfeld D, Gotz V, Zell R, Zimmermann P, Haller O, et al. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. (2013) 9:e1003279. doi: 10.1371/journal.ppat.1003279
83. Riegger D, Hai R, Dornfeld D, Manz B, Leyva-Grado V, Sanchez-Aparicio MT, et al. The nucleoprotein of newly emerged H7N9 influenza A virus harbors a unique motif conferring resistance to antiviral human MxA. J Virol. (2015) 89:2241–52. doi: 10.1128/JVI.02406-14
84. Verhelst J, Parthoens E, Schepens B, Fiers W, Saelens X. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J Virol. (2012) 86:13445–55. doi: 10.1128/JVI.01682-12
85. Jung K, Chae C. Expression of Mx protein and interferon-alpha in pigs experimentally infected with swine influenza virus. Vet Pathol. (2006) 43:161–7. doi: 10.1354/vp.43-2-161
86. Dornfeld D, Petric PP, Hassan E, Zell R, Schwemmle M. Eurasian avian-like swine influenza A viruses escape human MxA restriction by distinct mutations in their nucleoprotein. J Virol. (2018). doi: 10.1128/JVI.00997-18. [Epub ahead of print].
87. Hartshorn KL, White MR, Voelker DR, Coburn J, Zaner K, Crouch EC. Mechanism of binding of surfactant protein D to influenza A viruses: importance of binding to haemagglutinin to antiviral activity. Biochem J. (2000) 351(Pt 2):449–58. doi: 10.1042/bj3510449
88. Vigerust DJ, Ulett KB, Boyd KL, Madsen J, Hawgood S, McCullers JA. N-linked glycosylation attenuates H3N2 influenza viruses. J Virol. (2007) 81:8593–600. doi: 10.1128/JVI.00769-07
89. Hartshorn KL, Webby R, White MR, Tecle T, Pan C, Boucher S, et al. Role of viral hemagglutinin glycosylation in anti-influenza activities of recombinant surfactant protein D. Respir Res. (2008) 9:65. doi: 10.1186/1465-9921-9-65
90. Hillaire ML, van Eijk M, Nieuwkoop NJ, Vogelzang-van Trierum SE, Fouchier RA, Osterhaus AD, et al. The number and position of N-linked glycosylation sites in the hemagglutinin determine differential recognition of seasonal and 2009 pandemic H1N1 influenza virus by porcine surfactant protein D. Virus Res. (2012) 169:301–5. doi: 10.1016/j.virusres.2012.08.003
91. van Eijk M, White MR, Crouch EC, Batenburg JJ, Vaandrager AB, Van Golde LM, et al. Porcine pulmonary collectins show distinct interactions with influenza A viruses: role of the N-linked oligosaccharides in the carbohydrate recognition domain. J Immunol. (2003) 171:1431–40. doi: 10.4049/jimmunol.171.3.1431
92. Hillaire ML, van Eijk M, van Trierum SE, van Riel D, Saelens X, Romijn RA, et al. Assessment of the antiviral properties of recombinant porcine SP-D against various influenza A viruses in vitro. PLoS ONE (2011) 6:e25005. doi: 10.1371/journal.pone.0025005
93. Hale BG, Albrecht RA, Garcia-Sastre A. Innate immune evasion strategies of influenza viruses. Future Microbiol. (2010) 5:23–41. doi: 10.2217/fmb.09.108
94. Noronha JM, Liu M, Squires RB, Pickett BE, Hale BG, Air GM, et al. Influenza virus sequence feature variant type analysis: evidence of a role for NS1 in influenza virus host range restriction. J Virol. (2012) 86:5857–66. doi: 10.1128/JVI.06901-11
95. Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, et al. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. (2012) 8:e1003059. doi: 10.1371/journal.ppat.1003059
96. Hayman A, Comely S, Lackenby A, Hartgroves LC, Goodbourn S, McCauley JW, et al. NS1 proteins of avian influenza A viruses can act as antagonists of the human alpha/beta interferon response. J Virol. (2007) 81:2318–27. doi: 10.1128/JVI.01856-06
97. Houser KV, Pearce MB, Katz JM, Tumpey TM. Impact of prior seasonal H3N2 influenza vaccination or infection on protection and transmission of emerging variants of influenza A(H3N2)v virus in ferrets. J Virol. (2013) 87:13480–9. doi: 10.1128/JVI.02434-13
98. Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res. (2002) 85:199–210. doi: 10.1016/S0168-1702(02)00027-8
99. Karasin AI, Landgraf J, Swenson S, Erickson G, Goyal S, Woodruff M, et al. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J Clin Microbiol. (2002) 40:1073–9. doi: 10.1128/JCM.40.3.1073-1079.2002
100. Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res. (2004) 103:67–73. doi: 10.1016/j.virusres.2004.02.015
101. Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses (2013) 7(Suppl. 4):42–51. doi: 10.1111/irv.12193
102. Lorusso A, Vincent AL, Gramer ME, Lager KM, Ciacci-Zanella JR. Contemporary epidemiology of North American lineage triple reassortant influenza A viruses in pigs. In: Richt JA and Webby RJ, editors. Swine Influenza. Berlin; Heidelberg: Springer (2013). p. 113–32.
103. Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes (2009) 39:176–85. doi: 10.1007/s11262-009-0386-6
104. Howden KJ, Brockhoff EJ, Caya FD, McLeod LJ, Lavoie M, Ing JD, et al. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J. (2009) 50:1153–61.
105. Rajao DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, et al. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol. (2015) 89:11213–22. doi: 10.1128/JVI.01675-15
106. Walia RR, Anderson TK, Vincent AL. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses (2018) doi: 10.1111/irv.12559. [Epub ahead of print].
107. Castrucci MR, Campitelli L, Ruggieri A, Barigazzi G, Sidoli L, Daniels R, et al. Antigenic and sequence analysis of H3 influenza virus haemagglutinins from pigs in Italy. J Gen Virol. (1994) 75(Pt 2):371–9. doi: 10.1099/0022-1317-75-2-371
108. Simon G, Larsen LE, Durrwald R, Foni E, Harder T, Van Reeth K, et al. European surveillance network for influenza in pigs: surveillance programs, diagnostic tools and swine influenza virus subtypes identified in 14 European countries from 2010 to 2013. PLoS ONE (2014) 9:e115815. doi: 10.1371/journal.pone.0115815
109. Brown IH, Harris PA, McCauley JW, Alexander DJ. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J Gen Virol. (1998) 79(Pt 12):2947–55. doi: 10.1099/0022-1317-79-12-2947
110. Marozin S, Gregory V, Cameron K, Bennett M, Valette M, Aymard M, et al. Antigenic and genetic diversity among swine influenza A H1N1 and H1N2 viruses in Europe. J Gen Virol. (2002) 83(Pt 4):735–45. doi: 10.1099/0022-1317-83-4-735
111. Welsh MD, Baird PM, Guelbenzu-Gonzalo MP, Hanna A, Reid SM, Essen S, et al. Initial incursion of pandemic (H1N1) 2009 influenza A virus into European pigs. Vet Rec. (2010) 166:642–5. doi: 10.1136/vr.4851
112. Krog JS, Hjulsager CK, Larsen MA, Larsen LE. Triple-reassortant influenza A virus with H3 of human seasonal origin, NA of swine origin, and internal A(H1N1) pandemic 2009 genes is established in Danish pigs. Influenza Other Respir Viruses (2017) 11:298–303. doi: 10.1111/irv.12451
113. Poonsuk S, Sangthong P, Petcharat N, Lekcharoensuk P. Genesis and genetic constellations of swine influenza viruses in Thailand. Vet Microbiol. (2013) 167:314–26. doi: 10.1016/j.vetmic.2013.09.007
114. Zhu H, Webby R, Lam TT, Smith DK, Peiris JS, Guan Y. History of swine influenza viruses in Asia. Curr Top Microbiol Immunol. (2013) 370:57–68. doi: 10.1007/82_2011_179
115. Nelson MI, Viboud C, Vincent AL, Culhane MR, Detmer SE, Wentworth DE, et al. Global migration of influenza A viruses in swine. Nat Commun. (2015) 6:6696. doi: 10.1038/ncomms7696
116. Vijaykrishna D, Smith GJ, Pybus OG, Zhu H, Bhatt S, Poon LL, et al. Long-term evolution and transmission dynamics of swine influenza A virus. Nature (2011) 473:519–22. doi: 10.1038/nature10004
117. Liang H, Lam TT, Fan X, Chen X, Zeng Y, Zhou J, et al. Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J Virol. (2014) 88:10864–74. doi: 10.1128/JVI.01327-14
118. Cappuccio JA, Pena L, Dibarbora M, Rimondi A, Pineyro P, Insarralde L, et al. Outbreak of swine influenza in argentina reveals a non-contemporary human H3N2 virus highly transmissible among pigs. J Gen Virol. (2011) 92(Pt 12):2871–8. doi: 10.1099/vir.0.036590-0
119. Adeola OA, Olugasa BO, Emikpe BO. Antigenic detection of human strain of influenza virus A (H3N2) in swine populations at three locations in nigeria and ghana during the dry early months of 2014. Zoonoses Public Health (2016) 63:106–11. doi: 10.1111/zph.12210
120. Gomaa MR, Kandeil A, El-Shesheny R, Shehata MM, McKenzie PP, Webby RJ, et al. Evidence of infection with avian, human, and swine influenza viruses in pigs in Cairo, Egypt. Arch Virol. (2018) 163:359–64. doi: 10.1007/s00705-017-3619-3
121. Nelson MI, Schaefer R, Gava D, Cantao ME, Ciacci-Zanella JR. Influenza A viruses of human origin in swine, Brazil. Emerg Infect Dis. (2015) 21:1339–47. doi: 10.3201/eid2108.141891
122. Schaefer R, Rech RR, Gava D, Cantao ME, da Silva MC, Silveira S, et al. A human-like H1N2 influenza virus detected during an outbreak of acute respiratory disease in swine in Brazil. Arch Virol. (2015) 160:29–38. doi: 10.1007/s00705-014-2223-z
123. Baudon E, Chu DKW, Tung DD, Thi Nga P, Vu Mai Phuong H, Le Khanh Hang N, et al. Swine influenza viruses in Northern Vietnam in 2013-2014. Emerg Microbes Infect. (2018) 7:123. doi: 10.1038/s41426-018-0109-y
124. Nelson M, Culhane MR, Rovira A, Torremorell M, Guerrero P, Norambuena J. Novel human-like influenza A viruses circulate in swine in Mexico and Chile. PLoS Curr. (2015) 7:33ca6. doi: 10.1371/currents.outbreaks.c8b3207c9bad98474eca3013fa933ca6
125. Nelson MI, Gramer MR, Vincent AL, Holmes EC. Global transmission of influenza viruses from humans to swine. J Gen Virol. (2012) 93(Pt 10):2195–203. doi: 10.1099/vir.0.044974-0
126. Pereda A, Cappuccio J, Quiroga MA, Baumeister E, Insarralde L, Ibar M, et al. Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg Infect Dis. (2010) 16:304–7. doi: 10.3201/eid1602.091230
127. Sreta D, Tantawet S, Na Ayudhya SN, Thontiravong A, Wongphatcharachai M, Lapkuntod J, et al. Pandemic (H1N1) 2009 virus on commercial swine farm, Thailand. Emerg Infect Dis. (2010) 16:1587–90. doi: 10.3201/eid1610.100665
128. Holyoake PK, Kirkland PD, Davis RJ, Arzey KE, Watson J, Lunt RA, et al. The first identified case of pandemic H1N1 influenza in pigs in Australia. Aust Vet J. (2011) 89:427–31. doi: 10.1111/j.1751-0813.2011.00844.x
129. Njabo KY, Fuller TL, Chasar A, Pollinger JP, Cattoli G, Terregino C, et al. Pandemic A/H1N1/2009 influenza virus in swine, Cameroon, 2010. Vet Microbiol. (2012) 156:189–92. doi: 10.1016/j.vetmic.2011.09.003
130. Rajao DS, Costa AT, Brasil BS, Del Puerto HL, Oliveira FG, Alves F, et al. Genetic characterization of influenza virus circulating in Brazilian pigs during 2009 and 2010 reveals a high prevalence of the pandemic H1N1 subtype. Influenza Other Respir Viruses (2013) 7:783–90. doi: 10.1111/irv.12072
131. Adeola OA, Olugasa BO, Emikpe BO. Molecular detection of influenza A(H1N1)pdm09 viruses with M genes from human pandemic strains among Nigerian pigs, 2013-2015: implications and associated risk factors. Epidemiol Infect. (2017) 145:3345–60. doi: 10.1017/S0950268817002503
132. Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL. Continual reintroduction of human pandemic H1N1 influenza A viruses into swine in the United States, 2009 to 2014. J Virol. (2015) 89:6218–26. doi: 10.1128/JVI.00459-15
133. Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A, Vincent AL. The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol. (2017) 98:2001–10. doi: 10.1099/jgv.0.000885
134. Rajao DS, Walia RR, Campbell B, Gauger PC, Janas-Martindale A, Killian ML, et al. Reassortment between Swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol. (2017) 91:e01763–16. doi: 10.1128/JVI.01763-16
135. Watson SJ, Langat P, Reid SM, Lam TTY, Cotten M, Kelly M, et al. Molecular epidemiology and evolution of influenza viruses circulating within european swine between 2009 and 2013. J Virol. (2015) 89:9920–31. doi: 10.1128/JVI.00840-15
136. Chastagner A, Herve S, Bonin E, Queguiner S, Hirchaud E, Henritzi D, et al. Spatio-temporal distribution and evolution of the A/H1N1 2009 pandemic virus in pigs in France from 2009 to 2017: identification of a potential swine-specific lineage. J Virol. (2018) 92:e00988–18. doi: 10.1128/JVI.00988-18
137. Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science (2010) 328:1529. doi: 10.1126/science.1189132
138. Pereda A, Rimondi A, Cappuccio J, Sanguinetti R, Angel M, Ye J, et al. Evidence of reassortment of pandemic H1N1 influenza virus in swine in Argentina: are we facing the expansion of potential epicenters of influenza emergence? Influenza Other Respir Viruses (2011) 5:409–12. doi: 10.1111/j.1750-2659.2011.00246.x
139. Zhu H, Zhou B, Fan X, Lam TT, Wang J, Chen A, et al. Novel reassortment of Eurasian avian-like and pandemic/2009 influenza viruses in swine: infectious potential for humans. J Virol. (2011) 85:10432–9. doi: 10.1128/JVI.05352-11
140. Hofshagen M, Gjerset B, Er C, Tarpai A, Brun E, Dannevig B, et al. Pandemic influenza A(H1N1)v: human to pig transmission in Norway? Euro Surveill. (2009) 14:19406. doi: 10.2807/ese.14.45.19406-en
141. Sandbulte MR, Spickler AR, Zaabel PK, Roth JA. Optimal use of vaccines for control of influenza A virus in swine. Vaccines (2015) 3:22–73. doi: 10.3390/vaccines3010022
142. Vander Veen RL, Loynachan AT, Mogler MA, Russell BJ, Harris DL, Kamrud KI. Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine (2012) 30:1944–50. doi: 10.1016/j.vaccine.2012.01.030
143. Genzow M, Goodell C, Kaiser TJ, Johnson W, Eichmeyer M. Live attenuated influenza virus vaccine reduces virus shedding of newborn piglets in the presence of maternal antibody. Influenza Other Respir Viruses (2018) 12:353–9. doi: 10.1111/irv.12531
144. Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. Swine. J Virol. (2014) 88:4752–63. doi: 10.1128/JVI.03805-13
Keywords: influenza A virus, swine, human, interspecies, adaptation, host range
Citation: Rajao DS, Vincent AL and Perez DR (2019) Adaptation of Human Influenza Viruses to Swine. Front. Vet. Sci. 5:347. doi: 10.3389/fvets.2018.00347
Received: 08 November 2018; Accepted: 31 December 2018;
Published: 22 January 2019.
Edited by:
Lester J. Perez, Dalhousie University, CanadaReviewed by:
Takashi Irie, Hiroshima University, JapanCopyright © 2019 Rajao, Vincent and Perez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela S. Rajao, ZGFuaWVsYS5yYWphb0B1Z2EuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.