- 1Equine Immunology Laboratory, Department of Clinical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, NY, United States
- 2Baker Institute for Animal Health, Cornell University, Ithaca, NY, United States
- 3Molecular Genetics Section, Laboratory of Viral Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MA, United States
Equine herpesvirus type 1 (EHV-1) is a ubiquitous and highly contagious pathogen that causes a range of disease severities with outbreaks of notable economic impact. Given the limitations in immune protection of current vaccines and the limited effectiveness of antiviral drugs on EHV-1 infections in vivo, improved treatment measures are needed to control disease. The use of drugs that alter the epigenetic state of herpes simplex virus genome has been shown to limit viral primary infection and reactivation both in vitro and in vivo. Therefore, we tested the hypothesis that maintaining a repressive epigenetic state on the EHV-1 genome in the host equine cell would decrease viral load during lytic infection. Equine fetal kidney cells (EFKCs) or isolated peripheral blood leukocytes were treated in vitro with (a) the nucleoside analog ganciclovir; (b) the histone demethylase inhibitor OG-L002; (c) both ganciclovir and OG-L002; or (d) dimethyl sulfoxide (DMSO, vehicle control); and then infected with a clinical EHV-1 isolate. Treatment of EFKCs with ganciclovir (mean 22.3 DNA copies per cell, p = 0.0005), OG-L002 (mean 25.6, p = 0.005) or both ganciclovir and OG-L002 (mean 7.1, p = 0.0001) resulted in decreased EHV-1 viral load at 24 h post-infection (hpi) in comparison with DMSO (mean 42.0), with greater impact using the combined treatment. Further, EHV-1 gene expression at 3 hpi decreased when EFKCs were infected in the presence of ganciclovir (p = 0.04) and combined treatment of ganciclovir and OG-L002 (p = 0.0003). In contrast, under similar conditions, neither ganciclovir nor OG-L002 suppressed EHV-1 infection in leukocytes. Differences between cell types, drug penetrance, or drug turnover, may have contributed to the distinct effects observed in this study.
Introduction
Equine herpesvirus 1 (EHV-1) is a serious threat to horses worldwide (1). EHV-1 outbreaks have emphasized the potential effects of this virus including: (1) severe respiratory disease, neurological disease, abortion, or death; (2) the highly contagious nature of the virus; (3) incomplete protection from vaccination and antiviral therapy; and (4) substantial economic costs for the equine industry (2–11). The prevalence of EHV-1 among 4,228 horses with fever, respiratory signs, and/or neurological deficits was 2.7% in the USA between 2008 and 2014 (12). The incidence of EHV-1-induced neurological disease was reported increased over the last decade internationally (13–15). Therefore, the global equine industry is compelled to identify more effective EHV-1 control measures and treatments.
Equine herpesvirus 1 infection is initiated by inhalation of infectious virus, whereupon the virus enters epithelial cells of the upper respiratory tract and replicates, leading to viral shedding via nasal discharge, and further spreading of the infection (16). Fever and respiratory clinical signs may ensue, although some horses experience subclinical shedding (5). EHV-1 upper respiratory tract entry facilitates infection of leukocytes, enabling the virus to circulate and infect endothelial cells of the central nervous system, leading to myeloencephalopathy; or reach the pregnant uterus, causing late-term abortion (17–19).
Although much effort has been invested in improving EHV-1 vaccines to prevent disease and curtail virus spread, current vaccines offer limited protection from infection or reactivation of the virus. Vaccines presently on the market have been shown to suppress EHV-1 disease and shedding but may not limit viral load. The level of protection against EHV-1-induced neurological disease by vaccination is unclear, and not claimed by any of the vaccine manufacturers (20–22). Further, natural exposure induces immune protection that can be as short as 3–6 months (20–23). Care of infected animals includes administration of antiviral drugs and supportive therapies (6, 24, 25). During EHV-1 infection, antiviral treatment using synthetic nucleoside analogs shows promising efficacy in cell culture models, but treatment of ponies with an experimental EHV-1 infection has been less effective (25–28). Currently used antiviral drugs are only active during lytic infection as they interfere with replication of the viral genome and, as such, they have no effect on latent EHV-1.
After EHV-1 infects the horse cell, the EHV-1 DNA genome is released into the nucleus to eventually produce more infectious EHV-1. A cost of this strategy is that the EHV-1 DNA is now subjected to the host cell’s gene regulation machinery. Recent work with human herpes simplex virus (HSV1) has demonstrated that the viral genome becomes susceptible to host-mediated epigenetic regulation, including the assembly and modulation of host-derived histones on the viral DNA genome (29–31). Subsequently, posttranslational modifications of the histone tails either permit or repress viral gene expression (29, 32, 33). Remarkably, maintaining a repressive epigenetic state of the HSV1 DNA suppressed viral gene expression during lytic infection, and suppressed reactivation from latency in vitro and in vivo (33–35). In these studies, the repressive state was preserved by preventing the removal of methyl groups from lysine 9 of histone 3 (H3K9) with the use of compounds that block the activity of the lysine-specific demethylase 1 (LSD1) protein [e.g., tranylcypromine, a monoamine oxidase inhibitor (MAOI); or a novel selective LSD1 inhibitor, OG-L002] (33–35). Because LSD1 is not the only protein that modulates histone methylation, LSD1 inhibition is not expected to have global effects. Further, the use of a histone demethylase inhibitor in conjunction with standard antiviral therapy in experimental HSV1 infection exerted a synergistic reduction of active infection, and limited reactivation from viral latency (33).
Regulating EHV-1 by maintaining a repressive epigenetic state would offer a new strategy to combat both lytic and latent EHV-1 infections. Although the extent of epigenetic regulation of the EHV-1 genome has not been determined to date, recent work showed that enforcing a permissive epigenetic state accelerates EHV-1 protein expression and induces productive infection (36). This finding suggests that EHV-1 DNA is subject to epigenetic regulation in horse cells and EHV-1 gene expression can be modulated by altering histone modifications. Given these data, we hypothesize that maintaining a repressive epigenetic state of the EHV-1 genome in the host equine cell would decrease viral load during lytic infection. To test this hypothesis, we investigated how histone tail hypermethylation altered EHV-1 lytic infection of permissive equine cells in vitro.
Materials and Methods
This study was carried out in accordance with the recommendations of Institutional Animal Care and Use Committee for the use of vertebrates in research, and the protocol approved by the Cornell University Center for Animal Resources and Education.
EHV-1 Isolate
Our laboratory isolated the EHV-1 virus from a naturally infected, neurologically affected research horse at Cornell University in 2005 using permissive rabbit kidney 13 cell cultures [titer of 1 × 107 plaque forming units (PFU)/mL], and this isolate was confirmed to harbor the ORF30 G2254 allele by PCR performed at the Cornell University Animal Health Diagnostic Center.
Cells, Treatments, and In Vitro Cultures
Primary equine fetal kidney cells (EFKCs) were isolated in our laboratory (37). The EFKCs were frozen in cell freeze media and stored in liquid nitrogen. Upon thawing, passage 5 of EFKCs were seeded into 12-well tissue culture plates in DMEM-F12 medium containing 10% fetal bovine serum, and 1× antibiotics–antimycotics (ThermoFisher Scientific, Waltham, MA, USA) until they reached 80–90% confluence before treatment and EHV-1 infection.
Blood samples were collected by jugular venipuncture into vacutainers containing heparin sulfate from three research healthy adult horses (two Warmblood mares and one Pony gelding, age range 14–20 years) from the Cornell Equine Park, Ithaca, NY, USA. Peripheral blood mononuclear cells were isolated using a previously described protocol of Ficoll-Paque density centrifugation (38).
We selected OG-L002 for these experiments rather than tranylcypromine to avoid unintended inhibition of other MAOI targets in the equine cell. The nucleoside analog ganciclovir was used as a model antiviral drug as in vitro EHV-1 studies demonstrated that ganciclovir was the most potent of six antiviral drugs with the lowest, non-toxic effective concentration (28). A combined epigenetic and antiviral treatment was also included in our experimental design based on observed synergistic effects when dual LSD1 inhibitor and nucleoside analog treatments were studied in HSV1 infection in vivo (34).
Evaluation of Treatment Toxicity In Vitro
To determine whether the ganciclovir or histone demethylase inhibitor treatments exerted a cytotoxic effect on the cultured cells, a range of ganciclovir (0.25–4.0 µg/mL) or OG-L002 (25–400 µM) concentrations were tested on equine leukocytes (0.25 × 106 cells/well) in triplicate for 24 h of culture in RPMI medium with 10% fetal bovine serum and 1X antibiotics/antimycotics (ThermoFisher Scientific, Waltham, MA, USA). Cell metabolic activity via enzymatic reduction of the tetrazolium dye MTT to its insoluble formazan was measured in order to assess cell viability (ATCC, Manassas, VA, USA). Based on the results, the concentrations used in the subsequent infection experiments were 50 µM OG-L002 and 1 µg/ml ganciclovir (Figure 1). Comparable concentrations have been used in other studies without evidence of cytotoxicity (28, 35).
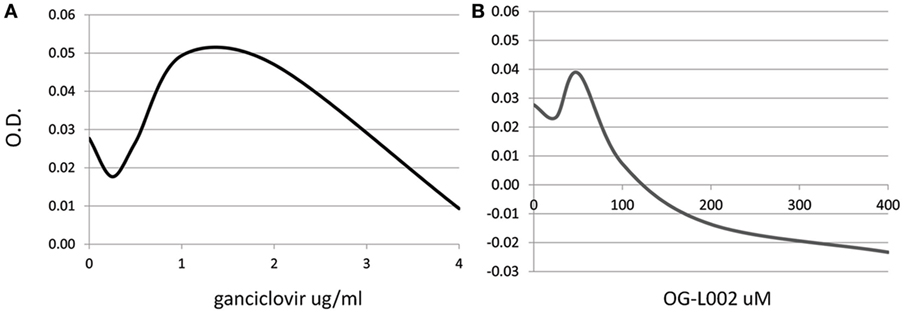
Figure 1. Dose-dependent cytotoxic effect of treatment. Cell viability of equine leukocytes was tested after 24 h of treatment with different concentrations of ganciclovir (A) or OG-L002 (B) using the MTT assay. Results indicate best-fit curve of mean values obtained from triplicate experiments at 540 nm optical density (OD).
Equine fetal kidney cell (1 × 105 cells/well) or leukocyte (5 × 105 cells/well) cultures were pretreated for 4 h with one of the following: (1) LSD1 inhibitor OG-L002 (50 µM, Selleck Chemicals, Houston, TX, USA); (2) nucleoside analog ganciclovir (1 µg/mL, Sigma-Aldrich Corporation, St. Louis, MO, USA); (3) both OG-L002 and ganciclovir at their respective concentrations; or (4) 0.005% dimethyl sulfoxide (DMSO, vehicle control, drug diluent). Cell cultures were then infected with 7.5 × 103 PFUof EHV-1 for 1 h, followed by replacement of viral inoculum with media containing the respective compounds. Replicate wells were included in each experiment and independent experiments were performed two or three times. Cells were harvested at 3 (for RNA isolation and viral gene expression analysis) or 24 h post-infection (hpi) (for DNA isolation and viral load analysis). For kinetic analysis of gene expression, samples were harvested at 1, 3, 6, 12, and 24 hpi without treatment.
Viral Load Quantification
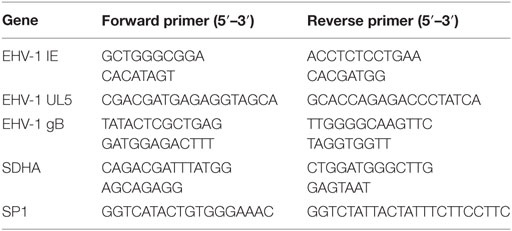
Twenty-four hours after respective treatment and EHV-1 infection, media from cultured EFKC or leukocytes were removed and cells directly lysed using a Quick-gDNA MiniPrep kit (Zymo Research, Irvine CA, USA). The DNA isolation was performed as directed by the manufacturer immediately after lysis or after homogenized samples were stored at −80°C. Quantitative PCR (qPCR) was performed to measure viral loads and equine cell genomic DNA using primers to the viral gB or the cellular SP1 genes, respectively (24). Table 1 shows the primer sequences used in this study. Reactions were performed in triplicate with iTaq Universal SYBR Green Supermix on a CFX96 Real Time System, including no template controls (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The number of EHV-1 copies per cell was calculated by dividing the number of gB copies by the number of SP1 copies, and then multiplying that value by two to account for two SP1 alleles per equine genome.
Gene Expression Quantification
Media from cultured EFKC cells were removed and cells directly lysed using Quick-RNA microprep (Zymo Research, Irvine, CA, USA). The RNA isolation was performed immediately after lysis or after homogenized samples were stored at −80°C. After isolation, RNA was treated with DNase I to degrade contaminating equine genomic and viral DNA (ThermoFisher Scientific, Waltham, MA, USA). Quantitative RT-PCR assays were designed and validated for EHV-1 detection with iTaq Universal SYBR Green One-Step kit for the EHV-1 immediate early gene (IE), an early gene (UL5), and a late gene (gB) (Table 1). Assay specificity was confirmed using uninfected cell controls and each gene was cloned to generate a standard curve. Quantitative RT-PCR was performed with these samples to identify an equine reference (housekeeping) gene. After comparing expression of six reference genes (beta-actin, c-MYB, GAPDH, SDHA, SP1, and SLC25A44), SDHA was chosen as the expression was consistent, and the primers did not amplify equine genomic DNA (data not shown; SDHA and SLC25A44 primer sequences provided by D. Miller and D. F. Antczak, Cornell University College of Veterinary Medicine, Ithaca, NY, USA). Reactions were performed in triplicate with the iTaq Universal SYBR Green One-Step kit on a CFX96 Real Time System, including no reverse-transcriptase controls and no template controls (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Relative EHV-1 gene expression was determined by calculating the difference between the EHV-1 gene, and horse SDHA cycle threshold values for each sample (normalization). For kinetic analysis, gene expression at 1 h was set to 100%, and that of later time points was determined relative to this value with the ddCt method. For experiments, expression of the IE gene in DMSO-treated EHV-1-infected cells was set to 100%, and expression in treated cells determined relative to this value.
Statistical Analysis
The Shapiro–Wilk normality test was employed to determine whether data fit a normal distribution. When data fit a normal distribution, ANOVA including Tukey’s multiple comparisons test was performed to assess effect of treatments, and mean values were provided. Alternatively, when data did not fit a normal distribution, analyses were conducted with Kruskal–Wallis tests including Dunn’s multiple comparisons test, and median values were provided. All statistical tests were performed with GraphPad Prism (GraphPad Prism v6.05 for Windows, San Diego, CA, USA). A p-value ≤0.05 was considered significant.
Results
The objective of this work was to measure the effect of inhibition of the histone demethylase LSD1 on EHV-1 lytic infection in vitro. Adherent primary EFKCs or leukocyte cultures were treated and infected, and the resulting EHV-1 viral load and gene expression quantified.
Effect of Treatment on EHV-1 Viral Load During Lytic Infection of Primary EFKCs
To quantify EHV-1 viral load in the presence of ganciclovir and/or OG-L002 during lytic infection of primary EFKC in vitro, DNA was harvested at 24 hpi and qPCR performed for EHV-1 gB and equine SP1 targets. Viral DNA loads per cell were calculated as described previously. In comparison to the DMSO vehicle control (mean 42.0, 14.6–70.2), the number EHV-1 DNA copies per cell was reduced (p = 0.0005) in the presence of ganciclovir alone (mean 22.3, range 11.4–43.2), OG-L002 alone (mean 25.6, range 13.3–53.9, p = 0.005), and when treatment included both ganciclovir and OG-L002 (mean 7.1, range 4.4–10.4, p < 0.0001) (Figure 2). Further, combining the treatments decreased the number of EHV-1 DNA copies in comparison to treatment with ganciclovir alone (p = 0.01) or OG-L002 alone (p = 0.001), whereas there was no statistical difference between ganciclovir alone and OG-L002 alone (p = 0.9).
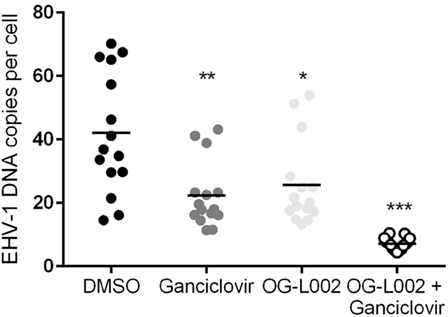
Figure 2. Effect of treatment on equine herpesvirus type 1 (EHV-1) viral load during lytic infection of primary equine fetal kidney cells. At 24 hpi, reduced EHV-1 DNA copies per cell were detected in the presence of ganciclovir (**p = 0.0005) or OG-L002 (*p = 0.005), and markedly when both treatments (***p < 0.0001) were applied simultaneously in comparison with DMSO control vehicle. Combined treatment decreased the number of EHV-1 DNA copies in comparison to treatment with ganciclovir alone (p = 0.01) or OG-L002 alone (p = 0.001). Lines represent mean values.
Kinetic Analysis of EHV-1 Gene Expression During Lytic Infection of Primary EFKCs
The reduction in EHV-1 viral load in EFKCs observed 24 h after treatment with OG-L002 suggested that this epigenetic modulator inhibited EHV-1 activity. To further study this effect, we measured how OG-L002 treatment altered EHV-1 gene expression soon after infection. First, a kinetic study without treatment of EFKC was undertaken to assess EHV-1 immediate early (IE), early (UL5), and late (gB) gene expression from 1 to 24 hpi. At 1 hpi, expression of the EHV-1 IE gene was close to the lower limit of detection, steadily increased by 2 hpi, and became robust by 3 hpi (Figure 3). Expression of UL5 gene followed a similar pattern, but consistent detection was measured at 3 hpi. The gB gene was abundantly expressed at 6 hpi and beyond. At 24 hpi, IE, UL5, and gB gene expressions were increased (p < 0.05) in comparison to all earlier time points. Since the EHV-1 immediate early and early genes were consistently detected at 3 hpi, this time point was selected for RNA harvesting and analysis of treated samples.
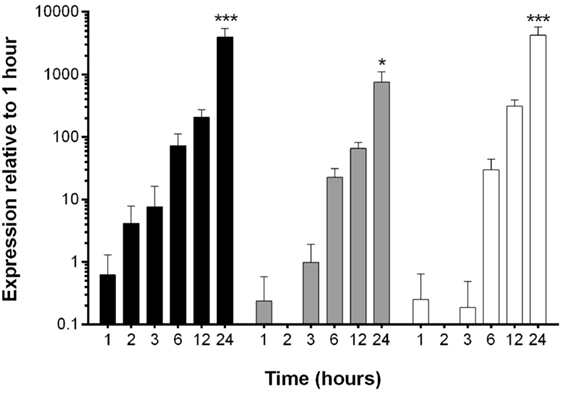
Figure 3. Kinetic analysis of equine herpesvirus type 1 gene expression during lytic infection of primary equine fetal kidney cells (EFKCs). Expression of the IE gene was detected in EFKCs starting at 1 hpi (black bars), whereas expression of UL5 (gray bars) and gB (white bars) genes was not consistently detected until 3 and 6 hpi, respectively. Data are shown as mean plus SD. Statistical significance is indicated by asterisks and denotes an increase at 24 hpi compared to all earlier time points (*p < 0.05, ***p < 0.0001).
Effect of Treatment on EHV-1 Gene Expression during Lytic Infection of Primary EFKCs
Experiments were then undertaken to test the effect of ganciclovir and/or OG-L002 treatment on EHV-1 IE gene expression during lytic infection of primary EFKC in vitro. At 3 hpi EHV-1 infection, RNA was isolated and qRT-PCR was performed for the EHV-1 IE gene and equine SDHA reference gene. Decreased (p = 0.04) EHV-1 IE gene expression was observed with ganciclovir treatment (mean 57.2, range 15.2–125.7) in comparison to DMSO vehicle control (mean 92.2, range 45.7–156.2) (Figure 4). Treatment with OG-L002 trended toward a reduction (p = 0.08) in IE gene expression (mean 60.6, range 10.9–174.4); data analysis performed when an outlier was removed in the OG-L002 resulted in significant statistical difference between OG-L002 and DMSO vehicle control (p = 0.005). Combined treatment of both ganciclovir and OG-L002 resulted in a synergistically decreased (p = 0.0003) IE gene expression (mean 35.9, range 9.8–68.7) when compared to DMSO vehicle control. Further, combining the treatments did not result in statistically significant differences in EHV-1 IE gene expression when compared to ganciclovir alone (p = 0.3) or OG-L002 alone (p = 0.2) or when ganciclovir alone was compared to OG-L002 alone (p = 1.0).
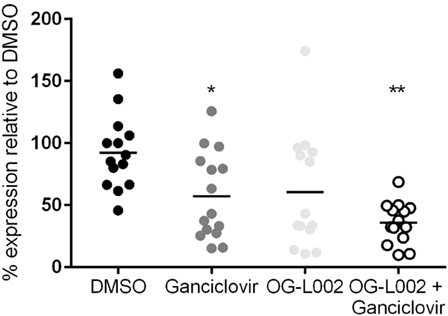
Figure 4. Effect of treatment on equine herpesvirus type 1 (EHV-1) IE gene expression during lytic infection of primary equine fetal kidney cells. EHV-1 IE gene expression was decreased in the presence of ganciclovir (*p = 0.04) and combined treatment of ganciclovir and OG-L002 (**p = 0.0003) at 3 hpi, when compared with DMSO control vehicle. Lines represent mean values.
Effect of Treatment on EHV-1 Viral Load During Infection of Equine Peripheral Blood Leukocytes
Determining the effect of histone demethylase inhibitor treatment on EHV-1 infection of leukocytes is relevant to the clinical disease condition because leukocytes are integral to the systemic spread of EHV-1 and harbor latent EHV-1 (17, 26, 39–41). Leukocytes were pretreated with ganciclovir, OG-L002, both ganciclovir and OG-L002, or with DMSO vehicle control, and then infected with EHV-1 as described previously. DNA was harvested at 24 hpi, and qPCR performed to quantify the EHV-1 DNA copies per cell. In contrast to the experiments performed with EFKC infection, the presence of ganciclovir (median 0.06, range 0.03–0.2) did not change (p > 0.05) the number of EHV-1 copies per leukocyte compared to DMSO vehicle control (median 0.14, range 0.01–0.24). Under these culture conditions, OG-L002 treatment (median 0.32; range 0.17–5.5) did not suppress EHV-1 viral load in leukocytes when compared with DMSO vehicle control (p = 0.02), or with ganciclovir (p = 0.002) treatment (Figure 5). Treatment of leukocytes with both ganciclovir and OG-L002 (median 0.59, range 0.05–1.3) yielded an EHV-1 viral load equivalent (p > 0.05) to DMSO vehicle control, ganciclovir alone (p = 0.2) and OG-L002 alone (p = 1.0). Viral replication in leukocytes without treatment was confirmed in independent experiments based on a 5.9-fold increase in IE expression at 24 hpi when compared to values at 3 hpi (p = 0.049).
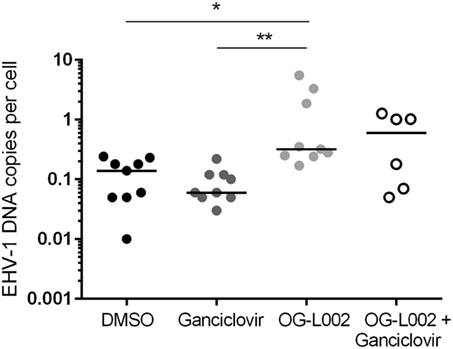
Figure 5. Effect of treatment on equine herpesvirus type 1 (EHV-1) viral load during infection of equine leukocytes. At 24 hpi, increased EHV-1 DNA copies per cell was detected in the presence of OG-L002 when compared with DMSO vehicle control (*p = 0.02) and ganciclovir (**p = 0.002). There was no difference (p > 0.05) in EHV-1 viral load between DMSO vehicle control, ganciclovir or OG-L002 plus ganciclovir treatments. Note that y-axis has a log10 scale. Lines represent median values.
Discussion
This work assessed the potential of the LSD1 inhibitor OG-L002 to limit EHV-1 viral load and gene expression during lytic infection in vitro. When used individually, OG-L002 inhibited EHV-1 activity in EFKCs to levels similar to that of ganciclovir treatment. When used in combination, a synergistic effect of ganciclovir and OG-L002 was measured. This observation reinforces the premise that these drugs inhibit viral replication through independent mechanisms, and could be an effective strategy to limit EHV-1 infection. However, under the culture conditions described herein, neither ganciclovir nor OG-L002 treatment of leukocytes suppressed EHV-1 viral load. Studies on the effect of compounds that alter epigenetic modifications on leukocytes have not been published before, and drug dosage or cell penetrance may account for the cell-specific impacts of these compounds on fetal kidney cells relative to equine leukocytes. The lack of EHV-1 inhibition by ganciclovir treatment in leukocytes was also unexpected, particularly in contrast to the results of EFKC cultures. These findings further suggest that differences between the cell types may be due to either cell bioavailability and/or length of exposure in vitro. Studies demonstrating the efficacy of ganciclovir in controlling EHV-1 in equine embryonic lung cells, equine dermal cells, pig kidney cells, or rabbit kidney cells rather than leukocytes have been reported (28, 39–41). Of note, ganciclovir treatment of human lymphoblastoid cells in vitro showed minimal antiviral effect when used for less than 7 days, even at high doses of 20 µg/mL; and, significant effects of ganciclovir treatment were observed at 14 days (42). In addition, the efficacy of antiviral drugs in vivo has not been consistent with in vitro findings, partly due to bioavailability or differential cell sensitivity (43–45). Unlikely these findings were due to lower efficiency of viral replication in leukocytes in comparison to kidney cells, since viral IE expression increased at 24 hpi.
With respect to the cell type dependence of the LSD1 inhibitor OG-L002, there is also the potential that tissue-specific expression or activities of LSD1 isoforms may have resulted in the lack of impact in leukocytes. Several LSD1 isoforms are generated by alternative splicing and these can differ in substrate specificities (H3K4 versus H3K9) and coregulatory protein interactions, resulting in differential gene regulation (46). Nevertheless, inhibition of histone demethylases, including LSD1, have been shown to inhibit HSV, human cytomegalovirus, adenovirus type 5, and varicella zoster virus (VZV) in primary fibroblasts in vitro, and to suppress HSV lytic infection and reactivation in multiple animal models (33–35).
The principal hypothesis of this study was based on previous HSV studies, in which promoting or maintaining an epigenetic repression of the virus suppressed viral infection and reactivation from latency. Therefore, it was anticipated that epigenetic suppression of EHV-1 would decrease viral load during lytic infection and prevent reactivation. Because effects were seen on the transcription of the EHV-1 IE gene in EFKC, the histone demethylase inhibitor OG-L002 was likely to be functioning as expected. In support of this observation, we identified a putative enhancer core element in the EHV-1 genome that may serve as the binding site for the protein complex needed for LSD1 activity, with the motif TAATGAGAT at positions 119,838–119,846 of the EHV-1 genome sequence NC_001491. For HSV and VZV IE gene expression, cellular transcription factors and proteins in the virion recruit host cellular transcriptional coactivator host cell factor-1, LSD1, and JMJD2 to bind the enhancer core element upstream of the IE gene (33). The presence of the enhancer core motif upstream of EHV-1 ORF64 and near other transcription factor binding motifs (SP1, CCAAT, and TATA) supports the likelihood that epigenetic mechanisms similar to those described for HSV1 also regulate EHV-1.
Additional experiments are required to confirm the inhibition of histone demethylation in the equine system, and optimization of the type of drug and dosage for different cell types. Inhibitors of other proteins in the same pathway also exert antiviral activity, and it would be of interest to investigate their impact on EHV-1 infection (47). If testing other compounds would show an effect on viral load in equine leukocytes, the use of respective drug inhibitor controls and additional experiments testing mechanisms of action should be pursued. The use of compounds that alter epigenetic modifications in the host cell rather than targeting viral protein(s) is advantageous in that it is not likely to elicit resistant viral strains. Identifying treatments that simultaneously impair EHV-1 lytic infection and reactivation from latency through a mechanism complementary to current antiviral drugs would be of great value in gaining control of EHV-1 disease severity and spread.
In conclusion, this study showed a favorable effect of the histone demethylase inhibitor OG-L002 in controlling EHV-1 load and gene expression in equine kidney cells, and these results support our hypothesis that maintaining a repressive epigenetic state on the EHV-1 genome in the host equine cell decreases viral load during lytic infection. However, this same effect was not observed in EHV-1 infected equine leukocytes, and additional compounds that alter epigenetic status should be tested.
Ethics Statement
This study was carried out in accordance with the recommendations of Institutional Animal Care and Use Committee for the use of vertebrates in research, and the protocol approved by the Cornell University Center for Animal Resources and Education.
Author Contributions
RT: designed and prepared grant proposal that funded study, including hypothesis, specific aims, and methods; performed molecular experiments; compiled data; performed statistical analyses; and prepared manuscript draft. EŽ: performed part of the molecular experiments; compiled data; performed statistical analyses; participated in discussions; and edited manuscript draft. GW and TK: provided expertise, input and editing of grant proposal preparation (coinvestigator), including methods and experimental design; discussed results and conclusions; and edited manuscript draft. MF: designed and prepared grant proposal that funded study, including hypothesis, specific aims, and methods; performed cellular experiments; compiled data; and edited manuscript draft.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Donald Miller and Dr. D. F. Antczak at Cornell University College of Veterinary Medicine, Ithaca, NY, USA, for kindly sharing qPCR primer sequences.
Funding
This work was funded by the Harry M. Zweig Memorial Fund for Equine Research (MF) and, in part, by the Intramural Research Program of the NIH, NIAID.
References
1. Lunn DP, Davis-Poynter N, Flaminio MJ, Horohov DW, Osterrieder K, Pusterla N, et al. Equine herpesvirus-1 consensus statement. J Vet Intern Med (2009) 23:450–61. doi:10.1111/j.1939-1676.2009.0304.x
2. van Maanen C, Willink DL, Smeenk LA, Brinkhof J, Terpstra C. An equine herpesvirus 1 (EHV1) abortion storm at a riding school. Vet Q (2000) 22:83–7. doi:10.1080/01652176.2000.9695030
3. Studdert MJ, Hartley CA, Dynon K, Sandy JR, Slocombe RF, Charles JA, et al. Outbreak of equine herpesvirus type 1 myeloencephalitis: new insights from virus identification by PCR and the application of an EHV-1-specific antibody detection ELISA. Vet Rec (2003) 153:417–23. doi:10.1136/vr.153.14.417
4. van Maanen C, Oldruitenborgh-Oosterbaan MMS, van, Damen EA, Derksen AG. Neurological disease associated with EHV-1-infection in a riding school: clinical and virological characteristics. Equine Vet J (2001) 33:191–6. doi:10.1111/j.2042-3306.2001.tb00600.x
5. Burgess BA, Tokateloff N, Manning S, Lohmann K, Lunn DP, Hussey SB, et al. Nasal shedding of equine herpesvirus-1 from horses in an outbreak of equine herpes myeloencephalopathy in Western Canada. J Vet Intern Med (2012) 26:384–92. doi:10.1111/j.1939-1676.2012.00885.x
6. Henninger RW, Reed SM, Saville WJ, Allen GP, Hass GF, Kohn CW, et al. Outbreak of neurologic disease caused by equine herpesvirus-1 at a university equestrian center. J Vet Intern Med (2007) 21:157–65. doi:10.1111/j.1939-1676.2007.tb02942.x
7. Damiani AM, de Vries M, Reimers G, Winkler S, Osterrieder N. A severe equine herpesvirus type 1 (EHV-1) abortion outbreak caused by a neuropathogenic strain at a breeding farm in northern Germany. Vet Microbiol (2014) 172:555–62. doi:10.1016/j.vetmic.2014.06.023
8. Walter J, Seeh C, Fey K, Bleul U, Osterrieder N. Clinical observations and management of a severe equine herpesvirus type 1 outbreak with abortion and encephalomyelitis. Acta Vet Scand (2013) 55:19. doi:10.1186/1751-0147-55-19
9. Traub-Dargatz JL, Pelzel-McCluskey AM, Creekmore LH, Geiser-Novotny S, Kasari TR, Wiedenheft AM, et al. Case-control study of a multistate equine herpesvirus myeloencephalopathy outbreak. J Vet Intern Med Am Coll Vet Intern Med (2013) 27:339–46. doi:10.1111/jvim.12051
10. Pronost S, Legrand L, Pitel PH, Wegge B, Lissens J, Freymuth F, et al. Outbreak of equine herpesvirus myeloencephalopathy in France: a clinical and molecular investigation. Transbound Emerg Dis (2012) 59:256–63. doi:10.1111/j.1865-1682.2011.01263.x
11. Goehring LS, Landolt GA, Morley PS. Detection and management of an outbreak of equine herpesvirus type 1 infection and associated neurological disease in a veterinary teaching hospital. J Vet Intern Med (2010) 24:1176–83. doi:10.1111/j.1939-1676.2010.0558.x
12. Pusterla N, Mapes S, Akana N, Barnett C, MacKenzie C, Gaughan E, et al. Prevalence factors associated with equine herpesvirus type 1 infection in equids with upper respiratory tract infection and/or acute onset of neurological signs from 2008 to 2014. Vet Rec (2016) 178:70. doi:10.1136/vr.103424
13. Allen GP. Risk factors for development of neurologic disease after experimental exposure to equine herpesvirus-1 in horses. Am J Vet Res (2008) 69:1595–600. doi:10.2460/ajvr.69.12.1595
14. Perkins GA, Goodman LB, Tsujimura K, de Walle GRV, Kim SG, Dubovi EJ, et al. Investigation of the prevalence of neurologic equine herpes virus type 1 (EHV-1) in a 23-year retrospective analysis (1984-2007). Vet Microbiol (2009) 139:375–8. doi:10.1016/j.vetmic.2009.06.033
15. Pusterla N, Wilson WD, Madigan JE, Ferraro GL. Equine herpesvirus-1 myeloencephalopathy: a review of recent developments. Vet J (2009) 180:279–89. doi:10.1016/j.tvjl.2008.08.004
16. Kydd JH, Smith KC, Hannant D, Livesay GJ, Mumford JA. Distribution of equid herpesvirus-1 (EHV-1) in the respiratory tract of ponies: implications for vaccination strategies. Equine Vet J (1994) 26:466–9. doi:10.1111/j.2042-3306.1994.tb04051.x
17. Kydd JH, Smith KC, Hannant D, Livesay GJ, Mumford JA. Distribution of equid herpesvirus-1 (EHV-1) in respiratory tract associated lymphoid tissue: implications for cellular immunity. Equine Vet J (1994) 26:470–3. doi:10.1111/j.2042-3306.1994.tb04052.x
18. Smith KC, Whitwell KE, Binns MM, Dolby CA, Hannant D, Mumford JA. Abortion of virologically negative foetuses following experimental challenge of pregnant pony mares with equid herpesvirus 1. Equine Vet J (1992) 24:256–9. doi:10.1111/j.2042-3306.1992.tb02830.x
19. Edington N, Bridges CG, Patel JR. Endothelial cell infection and thrombosis in paralysis caused by equid herpesvirus-1: equine stroke. Arch Virol (1986) 90:111–24. doi:10.1007/BF01314149
20. Goehring LS, Wagner B, Bigbie R, Hussey SB, Rao S, Morley PS, et al. Control of EHV-1 viremia and nasal shedding by commercial vaccines. Vaccine (2010) 28:5203–11. doi:10.1016/j.vaccine.2010.05.065
21. Goodman LB, Wagner B, Flaminio MJ, Sussman KH, Metzger SM, Holland R, et al. Comparison of the efficacy of inactivated combination and modified-live virus vaccines against challenge infection with neuropathogenic equine herpesvirus type 1 (EHV-1). Vaccine (2006) 24:3636–45. doi:10.1016/j.vaccine.2006.01.062
22. Bresgen C, Lammer M, Wagner B, Osterrieder N, Damiani AM. Serological responses and clinical outcome after vaccination of mares and foals with equine herpesvirus type 1 and 4 (EHV-1 and EHV-4) vaccines. Vet Microbiol (2012) 160:9–16. doi:10.1016/j.vetmic.2012.04.042
23. Kydd JH, Townsend HG, Hannant D. The equine immune response to equine herpesvirus-1: the virus and its vaccines. Vet Immunol Immunopathol (2006) 111:15–30. doi:10.1016/j.vetimm.2006.01.005
24. Pusterla N, Wilson WD, Mapes S, Finno C, Isbell D, Arthur RM, et al. Characterization of viral loads, strain and state of equine herpesvirus-1 using real-time PCR in horses following natural exposure at a racetrack in California. Vet J (2009) 179:230–9. doi:10.1016/j.tvjl.2007.09.018
25. Maxwell LK. Antiherpetic drugs in equine medicine. Vet Clin North Am Equine Pract (2017) 33:99–125. doi:10.1016/j.cveq.2016.12.002
26. Garre B, Gryspeerdt A, Croubels S, Backer PD, Nauwynck H. Evaluation of orally administered valacyclovir in experimentally EHV1-infected ponies. Vet Microbiol (2009) 135:214–21. doi:10.1016/j.vetmic.2008.09.062
27. Glorieux S, Vandekerckhove AP, Goris N, Yang XY, Steukers L, de Walle GRV, et al. Evaluation of the antiviral activity of (1’S,2’R)-9-[[1’,2’-bis(hydroxymethyl)cycloprop-1’-yl]methyl]guanine (A-5021) against equine herpesvirus type 1 in cell monolayers and equine nasal mucosal explants. Antiviral Res (2012) 93:234–8. doi:10.1016/j.antiviral.2011.11.016
28. Garre B, van der Meulen K, Nugent J, Neyts J, Croubels S, Backer PD, et al. In vitro susceptibility of six isolates of equine herpesvirus 1 to acyclovir, ganciclovir, cidofovir, adefovir, PMEDAP and foscarnet. Vet Microbiol (2007) 122:43–51. doi:10.1016/j.vetmic.2007.01.004
29. Kristie TM. Dynamic modulation of HSV chromatin drives initiation of infection and provides targets for epigenetic therapies. Virology (2015) 47(9–480):555–61. doi:10.1016/j.virol.2015.01.026
30. Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol (2008) 6:211–21. doi:10.1038/nrmicro1794
31. Lieberman PM. Epigenetics and genetics of viral latency. Cell Host Microbe (2016) 19:619–28. doi:10.1016/j.chom.2016.04.008
32. Lee JS, Raja P, Knipe DM. Herpesviral ICP0 protein promotes two waves of heterochromatin removal on an early viral promoter during lytic infection. MBio (2016) 7:e2007–15. doi:10.1128/mBio.02007-15
33. Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med (2009) 15:1312–7. doi:10.1038/nm.2051
34. Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, et al. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med (2014) 6:265ra169. doi:10.1126/scitranslmed.3010643
35. Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. MBio (2013) 4:e558–512. doi:10.1128/mBio.00558-12
36. Laval K, Favoreel HW, Nauwynck HJ. Equine herpesvirus type 1 replication is delayed in CD172a+ monocytic cells and controlled by histone deacetylases. J Gen Virol (2015) 96:118–30. doi:10.1099/vir.0.067363-0
37. Battista JM, Tallmadge RL, Stokol T, Felippe MJ. Hematopoiesis in the equine fetal liver suggests immune preparedness. Immunogenetics (2014) 66:635–49. doi:10.1007/s00251-014-0799-9
38. Flaminio MJ, Rush BR, Davis EG, Hennessy K, Shuman W, Wilkerson MJ. Characterization of peripheral blood and pulmonary leukocyte function in healthy foals. Vet Immunol Immunopathol (2000) 73:267–85. doi:10.1016/S0165-2427(00)00149-5
39. Carvalho R, Oliveira AM, Souza AM, Passos LM, Martins AS. Prevalence of equine herpesvirus type 1 latency detected by polymerase chain reaction. Arch Virol (2000) 145:1773–87. doi:10.1007/s007050070055
40. Smith DJ, Iqbal J, Purewal A, Hamblin AS, Edington N. In vitro reactivation of latent equid herpesvirus-1 from CD5+/CD8+ leukocytes indirectly by IL-2 or chorionic gonadotrophin. J Gen Virol (1998) 79(Pt 12):2997–3004. doi:10.1099/0022-1317-79-12-2997
41. Welch HM, Bridges CG, Lyon AM, Griffiths L, Edington N. Latent equid herpesviruses 1 and 4: detection and distinction using the polymerase chain reaction and co-cultivation from lymphoid tissues. J Gen Virol (1992) 73(Pt 2):261–8. doi:10.1099/0022-1317-73-2-261
42. Janoly-Dumenil A, Rouvet I, Bleyzac N, Morfin F, Zabot M-T, Tod M. A pharmacodynamic model of ganciclovir antiviral effect and toxicity for lymphoblastoid cells suggests a new dosing regimen to treat cytomegalovirus infection. Antimicrob Agents Chemother (2012) 56:3732–8. doi:10.1128/AAC.06423-11
43. Rollinson EA, White G. Relative activities of acyclovir and BW759 against Aujeszky’s disease and equine rhinopneumonitis viruses. Antimicrob Agents Chemother (1983) 24:221–6. doi:10.1128/AAC.24.2.221
44. Rollinson EA. Comparative efficacy of three 2’-fluoropyrimidine nucleosides and 9-(1,3-dihydroxy-2-propoxymethyl) guanine (BW B759U) against pseudorabies and equine rhinopneumonitis virus infection in vitro and in laboratory animals. Antiviral Res (1987) 7:25–33. doi:10.1016/0166-3542(87)90036-2
45. Smith KO, Galloway KS, Hodges SL, Ogilvie KK, Radatus BK, Kalter SS, et al. Sensitivity of equine herpesviruses 1 and 3 in vitro to a new nucleoside analogue, 9-[[2-hydroxy-1-(hydroxymethyl) ethoxy] methyl] guanine. Am J Vet Res (1983) 44:1032–5.
46. Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, et al. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell (2015) 57:957–70. doi:10.1016/j.molcel.2015.01.010
47. Rai G, Kawamura A, Tumber A, Liang Y, Vogel JL, Arbuckle JH, et al. Discovery of ML324, a JMJD2 demethylase inhibitor with demonstrated antiviral activity. Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda, MD: National Center for Biotechnology Information (US) (2013). Available from: https://www.ncbi.nlm.nih.gov/books/NBK169450/
Keywords: equine, horse diseases, Equine herpesvirus type 1, ganciclovir, histone demethylase inhibitor, epigenetic, viral load
Citation: Tallmadge RL, Žygelytė E, Van de Walle GR, Kristie TM and Felippe MJB (2018) Effect of a Histone Demethylase Inhibitor on Equine Herpesvirus-1 Activity In Vitro. Front. Vet. Sci. 5:34. doi: 10.3389/fvets.2018.00034
Received: 24 September 2017; Accepted: 16 February 2018;
Published: 12 March 2018
Edited by:
Gisela Soboll Hussey, Michigan State University, United StatesReviewed by:
Junjie Zhang, University of Southern California, United StatesAkhalesh Kumar Shakya, Louisiana State University Health Sciences Center Shreveport, United States
Copyright: © 2018 Tallmadge, Žygelytė, Van de Walle, Kristie and Felippe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Julia B. Felippe, bWJmNkBjb3JuZWxsLmVkdQ==
 Rebecca L. Tallmadge
Rebecca L. Tallmadge Emilija Žygelytė1
Emilija Žygelytė1 Gerlinde R. Van de Walle
Gerlinde R. Van de Walle Thomas M. Kristie
Thomas M. Kristie M. Julia B. Felippe
M. Julia B. Felippe