
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 23 June 2017
Sec. Veterinary Infectious Diseases
Volume 4 - 2017 | https://doi.org/10.3389/fvets.2017.00096
This article is part of the Research Topic Mechanisms of Persistence, Survival, and Transmission of Bacterial Foodborne Pathogens in Production Animals View all 12 articles
Salmonella remains the leading cause of foodborne illness in the United States, and the dissemination of drug-resistant Salmonellae through the food chain has important implications for treatment failure of salmonellosis. We investigated the ecology of Salmonella in integrated broiler production in order to understand the flow of antibiotic susceptible and resistant strains within this system. Data were analyzed from a retrospective study focused on antimicrobial resistant Salmonella recovered from commercial broiler chicken farms conducted during the initial years of the US FDA’s foray into retail meat surveillance by the National Antimicrobial Resistance Monitoring System (NARMS). Sixty-three percentage of Salmonella were pan-susceptible to a panel of 19 antimicrobials used by the NARMS program. Twenty-five antimicrobial resistance phenotypes were observed in Salmonella isolated from two broiler chicken farms. However, Salmonella displaying resistance to streptomycin, alone, and in combination with other antibiotics was the most prevalent (36.3%) antimicrobial resistance phenotype observed. Resistance to streptomycin and sulfadimethoxine appeared to be linked to the transposon, Tn21. Combinations of resistance against streptomycin, gentamicin, sulfadimethoxine, trimethoprim, and tetracycline were observed for a variety of Salmonella enterica serovars and genetic types as defined by pulsed-field gel electrophoresis. There were within and between farm differences in the antibiotic susceptibilities of Salmonella and some of these differences were linked to specific serovars. However, farm differences were not linked to antibiotic usage. Analysis of the temporal and spatial distribution of the endemic Salmonella serovars on these farms suggests that preventing vertical transmission of antibiotic-resistant Salmonella would reduce carcass contamination with antibiotic-resistant Salmonella and subsequently human risk exposure.
Salmonella remains the leading cause of outbreak-associated gastroenteritis in the United States, and consumption of poultry products has been implicated in several of these outbreaks (1, 2). Since implementation of the HACCP program, improvement has been made in the level of Salmonella contamination of processed chicken carcasses (3). However, a survey of retail meat from the Washington, DC, USA area revealed a surprising level of contamination of beef, pork, and poultry products with antibiotic-resistant Salmonella (4, 5). The dissemination of antibiotic-resistant Salmonella through the food chain has important public health implications considering the potential for treatment failure when cases of gastroenteritis require medical intervention, especially in children, the elderly, and the immunocompromised (6). In addition, infections with antimicrobial resistant bacteria including Salmonella have been associated with higher rates of morbidity and mortality (7–9).
The use of antibiotics in food animal production has been implicated as a contributing factor to the emergence of drug resistance in human foodborne pathogens (6, 10). The emergence and rapid worldwide spread of the multiple drug-resistant Salmonella enterica Typhimurium phage-type DT104 clone and ceftriaxone-resistant S. enterica serovars Heidelberg, Newport, and Typhimurium have underscored the threat to both animal agriculture and human health posed by multiple drug-resistant pathogens (11–15). Antimicrobial resistance genes are widely disseminated in pathogenic, commensal, and environmental bacteria (16, 17). Furthermore, it has been shown that once antimicrobial resistance has been introduced into an ecosystem, resistance can spread and persist without continuing selection pressure from antibiotics (18, 19). In addition, the reservoir of antimicrobial resistance genes is larger than previously thought (20). It is in this environment that the potential exists for Salmonella to acquire antimicrobial resistance genes from resident poultry microbiota due to selection pressure from therapeutic and non-therapeutic antibiotic usage. It follows then that the longer Salmonellae persists in the environment of an animal production facility, the chance of acquiring resistance genes increases.
We took advantage of the integrated nature of poultry production to observe the antimicrobial resistance phenotypes acquired by salmonellae during broiler chicken production in order to identify potential critical control points for Salmonella contamination and antimicrobial resistance development; ultimately in order to provide information relevant to reducing the level of carcass contamination with antibiotic-resistant Salmonella. Data were analyzed from a retrospective study focused on antimicrobial-resistant Salmonella recovered from commercial broiler chicken farms conducted during the initial years of the US FDA’s foray into National Antimicrobial Resistance Monitoring System (NARMS) retail meat surveillance (4). Despite the diversity of antimicrobial resistance profiles, poultry Salmonella recovered from these farms in 2003 were generally susceptible to the tested antimicrobials of animal and human health significance. Vertical transmission appeared to be the most important factor in chicken carcass contamination with antibiotic-resistant Salmonella.
Selection and description of study farms was as previously described (21). Approximately 17,000 chicks were placed in each house on Farm One. No litter amendments were used (22). At the hatchery, gentamicin was administered in ovo (0.1 mg/egg) on day 17 of development. No antibiotics were used therapeutically on this farm to treat birds during this study. Chicks were fed starter feed containing virginiamycin (10 g/ton) (25 g/ton) for the first 2 weeks. The starter feed contained coccidiostat rotated in the following order: Flock 1; diclazuril (1 g/ton), Flock 2; narasin (72 g/ton), Flock 3; monensin (100 g/ton), Flocks 4, 5; nicarbazin (82 g/ton), and Flocks 6, 7; salinomycin (60 g/ton). Flocks were fed grower feed for the next 2 weeks containing bacitracin (25 g/ton), and other coccidiostats rotated in the following order: Flock 1; salinomycin (60 g/ton), Flocks 2, 3; narasin (72 g/ton), Flocks 4, 5; lasalocid (82 g/ton), and Flocks 6, 7; diclazuril (1 g/ton). Finisher feed containing virginiamycin (15 g/ton), without coccidiostat was fed for 1–2 weeks as birds approached market weight. Withdrawal feed containing neither antibiotics nor coccidiostats was fed for the last week of grow-out. Feed was withdrawn for 16 h prior to catch.
Approximately 20,000 chicks were placed per house on Farm Two. No litter amendments were used on Farm Two (22). At the hatchery, gentamicin (0.2 mg/chick) was injected subcutaneously into day-of-hatch chicks. Chicks were reared on starter feed containing bacitracin (25 g/ton), and salinomycin (50 g/ton) for the first 2 weeks, then grower feed containing bacitracin (25 g/ton) and salinomycin (50 g/ton) for 2 weeks, then finisher feed without growth promotant or coccidiostat for 1–2 weeks. Withdrawal feed without antibiotic or coccidiostat was fed for the last week of grow-out. Feed was withdrawn for 16 h prior to shipment. Escherichia coli airsacculitis was diagnosed in house B during week six of Flock 3 on Farm Two, and oxytetracycline was administered in drinking water at 10.4 mg/kg weight for 1 day and at 5.1 mg/kg weight for 4 days. In this work, we sampled chick box liners, the poultry environment, and chicken carcasses. The latter was provided to us by the participating poultry companies. We did not physically interact with chickens raised on these farms and, therefore, we were exempt from university guidelines and USDA/NIH regulations regarding animal use.
The 289 Salmonella strains, examined in this study, were isolated, serotyped, phage-typed, and strain-typed as previously described (21). Presence of aadA1 and merA was determined as described by Bass et al. (23).
Antibiotic susceptibility was determined for the 289 archived Salmonella isolates (21). The minimum inhibitory concentration (MIC) of the antimicrobial agents tested was determined with the Sensititre® automated antimicrobial susceptibility system (Trek Diagnostic Systems, Westlake, OH, USA) and interpreted according to the National Committee for Clinical Laboratory Standards (NCCLS) guidelines for microbroth dilution methods (24, 25). Sensititre® susceptibility testing was performed according to the manufacturer’s instructions, and susceptibility and resistance were reported as MIC (μg/ml). Three-letter abbreviations and resistance breakpoint concentration are in parentheses. The antimicrobials assayed were as follows: amikacin (AMI > 64 μg/ml), amoxicillin/clavulanic acid (AUG > 32/16 μg/ml), ampicillin (AMP > 32 μg/ml), apramycin (APR 32 µg/ml), ceftriaxone (AXO > 64 μg/ml), cefazolin (CEF 32 µg/ml), cefoxitin (FOX > 32 μg/ml), ceftiofur (TIO > 8 μg/ml), cephalothin (CEP > 32 μg/ml), chloramphenicol (CHL > 32 μg/ml), ciprofloxacin (CIP > 4 μg/ml), kanamycin (KAN 64 µg/ml), gentamicin (GEN > 16 μg/ml), imipenem (IMP > 4 μg/ml), nalidixic acid (NAL > 32 μg/ml), streptomycin (STR > 64 μg/ml), sulfadimethoxine (SMX > 512 μg/ml), tetracycline (TET > 16 μg/ml), and trimethoprim/sulfamethoxazole (TMS > 4/76 μg/ml). The antibiotics bacitracin and virginiamycin were not included with this panel as there is no breakpoint for Salmonella as their activity is specifically directed toward Gram-positive bacteria and it is used to prevent Clostridium perfringens infections in chickens.
This study was performed in 2003, early in the US Food and Drug Administration’s survey of antimicrobial-resistant foodborne bacteria recovered from retail meats, using the same methods and antimicrobial resistance break points recommended by NCCLS (Clinical and Laboratory Standards Institute) at that time.
The Fisher’s exact test with α = 0.05 and Mantel–Haenszel chi-squared test were used to test for non-random associations between specific data values. Salmonella Typhimurium PFGE types 1.1, 1.2, and 1.3 were ranked with regard to multiple drug resistance as determined by fitting linear model: log (μi) = βo = β1*PFGE typei, μi = mean number antimicrobial resistances or resistance type, with assumption that data conformed to Poisson distribution.
There is ample opportunity for antibiotic-resistant Salmonella to emerge on poultry farms due to the combination of on farm antibiotic usage and the significant reservoir of antimicrobial resistance genes present in poultry litter. We examined the antibiotic susceptibility of Salmonella collected from two commercial broiler farms in northeast Georgia in relation to on-farm antibiotic usage. The majority of Salmonella isolates (62.6%; n = 172) were susceptible to all 19 antimicrobials tested, with the remainder displaying resistance to streptomycin (30.9%), gentamicin (12.6%), sulfadimethoxine (20.9%), tetracycline (13.9%), and trimethoprim/sulfamethoxazole (8.6%) (Table 1). Salmonella resistance to streptomycin alone was the most prevalent antimicrobial resistance phenotype (30.9%) (Tables 1 and 2).
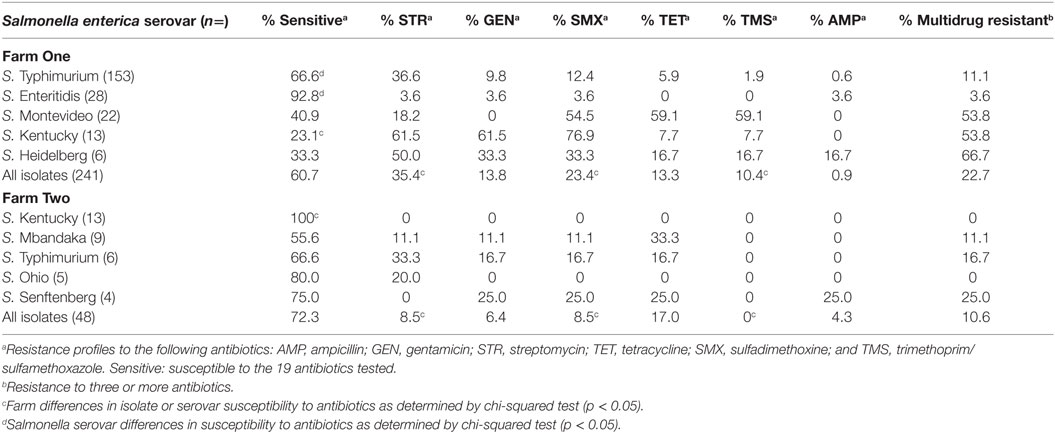
Table 1. Most prevalent antimicrobial resistance phenotypes observed in the Salmonella serovars isolated from production and processing of seven consecutive commercial broiler flocks.
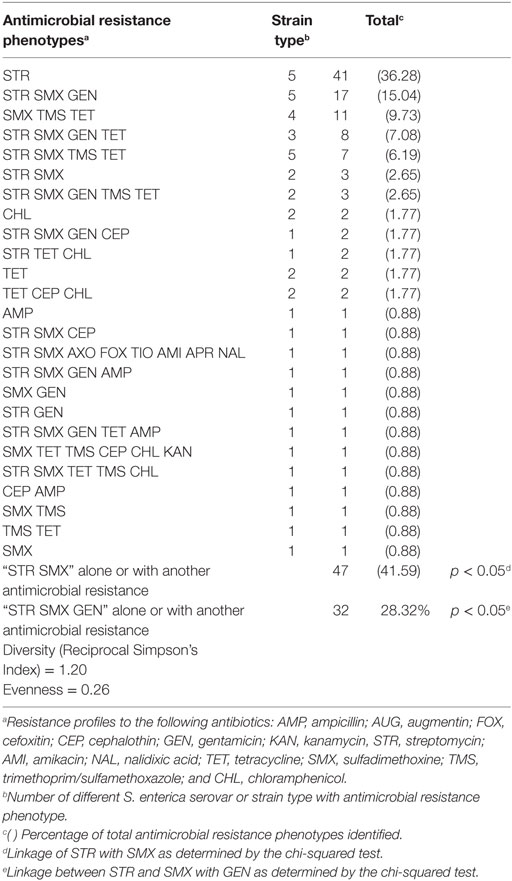
Table 2. Diversity of antimicrobial resistance phenotypes in Salmonella isolated from two commercial poultry farms.
A diversity of antimicrobial resistance phenotypes (n = 25) was observed among the Salmonella isolated from commercial broiler chicken farms (Table 2). Twenty percentage of our poultry Salmonella isolates were resistant to three or more antibiotics (Table 1). The most common antimicrobial resistance phenotypes identified were to streptomycin (36.28%); streptomycin and sulfadimethoxine, alone or in combination with other antibiotics (41.59%); and streptomycin, sulfadimethoxine, and gentamicin, alone or in combination with other antibiotics (28.32%) (Table 2). There was a statistically significant association between Salmonella isolates displaying resistance to streptomycin and sulfadimethoxine; and streptomycin, sulfadimethoxine, and gentamicin (chi-squared test: p < 0.05). While antimicrobial resistance phenotype diversity was high (Reciprocal Simpson’s Index: 1.20), evenness in distribution of these phenotypes among Salmonella was low (0.26). The low evenness score may be a reflection of the broad distribution of certain antimicrobial resistance phenotypes compared to others [streptomycin resistance, alone (41 strain types); streptomycin, sulfadimethoxine, and gentamicin resistance (17 strain types); sulfadimethoxine, trimethoprim/sulfamethoxazole, and tetracycline resistance (11 strain types); streptomycin, sulfadimethoxine, gentamicin, and tetracycline resistance (8 strain types); streptomycin, sulfadimethoxine trimethoprim/sulfamethoxazole, and tetracycline resistance (7 strain types)].
Two of the common antimicrobial resistances identified, streptomycin and sulfadimethoxine resistance, are commonly associated with the transposon, Tn21. The resistance genes merA and aadA1 are resident on this mobile genetic element and the distribution of these loci was 17.86 and 10.56%, respectively, in the recovered poultry isolates. There was a significant association between these resistance genes and resistance to streptomycin or sulfadimethoxine (chi-squared test; p < 0.05).
Differences were observed within and between poultry farms in antibiotic susceptibilities of Salmonella isolates. Antimicrobial susceptibility patterns differed between farms as Salmonella isolates from Farm One were more likely to be resistant to streptomycin, sulfadimethoxine, and trimethoprim/sulfamethoxazole compared to those recovered from Farm Two (chi-squared test: p < 0.05) (Table 1). There were also differences in antibiotic susceptibilities among certain Salmonella serovars within farms as well as between farms. S. Typhimurium isolated from Farm One were less susceptible to antibiotics, tested in this study, than S. Enteritidis isolated from the same farm. Salmonella Kentucky isolated from Farm One exhibited significantly more antimicrobial resistance than other Salmonella isolated from the same farm as well as S. Kentucky isolated from Farm Two (Table 1). Following tetracycline treatment on Farm Two, Salmonella isolates were less likely to be resistant to tetracycline, as determined using one-sided, Fisher’s exact test at α = 0.05 (p = 0.0046), or to other antibiotics (Cochran–Mantel–Haenszel method, p = 0.0046). The therapeutic treatment of E. coli airsacculitis with tetracycline did not seem to selectively enrich for antimicrobial resistance in Salmonella isolated from subsequent flocks. In addition, there was no statistically significant difference in Salmonella isolates displaying resistance to tetracycline between the two poultry farms (chi-squared test; p = 0.34).
S. Typhimurium (n = 159) was the most prevalent serovar isolated in this study, and this serovar was frequently isolated from Farm One. Serovar Typhimurium isolates were largely pan-susceptible (66.6%); however, the most prevalent antimicrobial resistance phenotypes were to streptomycin (6.6%), sulfadimethoxine (12.4%), gentamicin (9.4%), and tetracycline (6.4%) (Table 3). Resistance to the other 14 antimicrobials tested was not observed that often (≤5%). Eleven percentage of S. Typhimurium isolates were resistant to three or more antibiotics. The most prevalent S. Typhimurium resistance phenotypes observed were as follows: streptomycin alone (23.7%) and the multi-drug resistant phenotype to streptomycin, gentamicin, sulfadimethoxine, and tetracycline (5.3%). A diversity of antimicrobial resistance phenotypes (n = 9) was observed for the three related S. Typhimurium strain types identified by PFGE (Table 3). Combinations of resistance against streptomycin, gentamicin, sulfadimethoxine, and tetracycline, accounted for 85.3% of the resistance phenotypes (Table 3). There was no significant difference in resistance phenotypes between the three S. Typhimurium genetic types isolated from Farm One with the exception that PFGE type T1.3 was significantly more likely to be ampicillin resistant (α = 0.05).
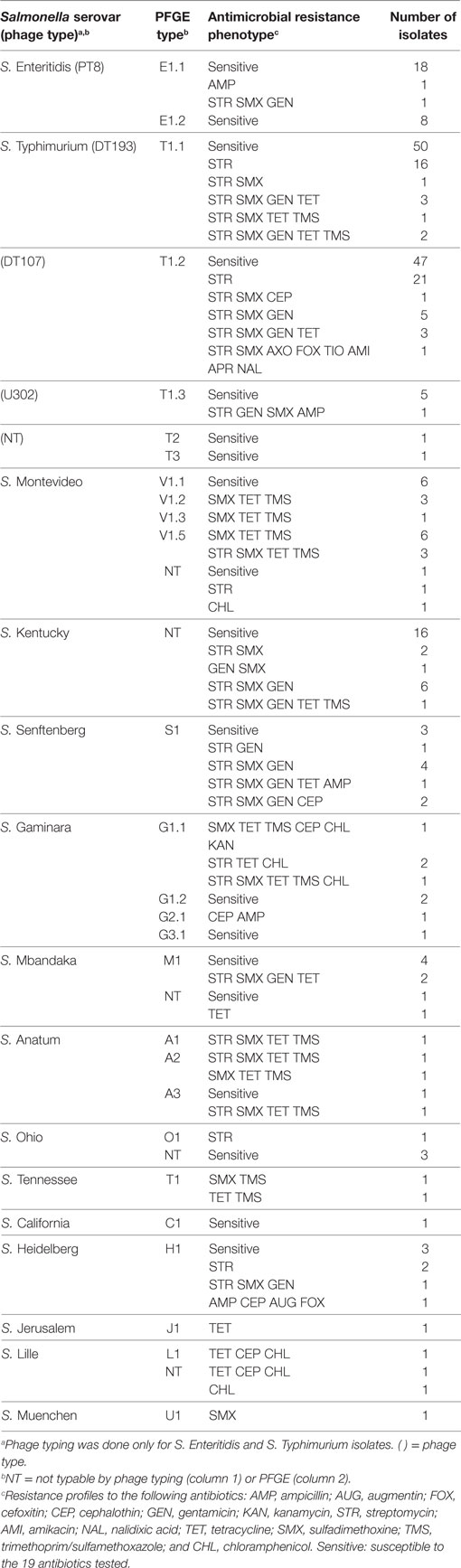
Table 3. Antimicrobial resistance phenotypes of Salmonella enterica serovars and strain types isolated from commercial broiler chicken farms.
Of the three S. Typhimurium strain types (T1.1, T1.2, and T1.3) present on Farm One, there were three instances where two of these strain types were present with chicks on the broiler chicken farm (T1.1 and T1.2) and chicken carcasses derived from these flocks (Table 4). There were also three other situations where these same S. Typhimurium strain types were only isolated from the farm environment and then chicken carcasses at processing. The only antibiotic resistant S. Typhimurium strain types found on chicken carcasses matched with those isolated from chicks at farm placement indicating that resistant S. Typhimurium strains were likely vertically transferred from the breeder flock.
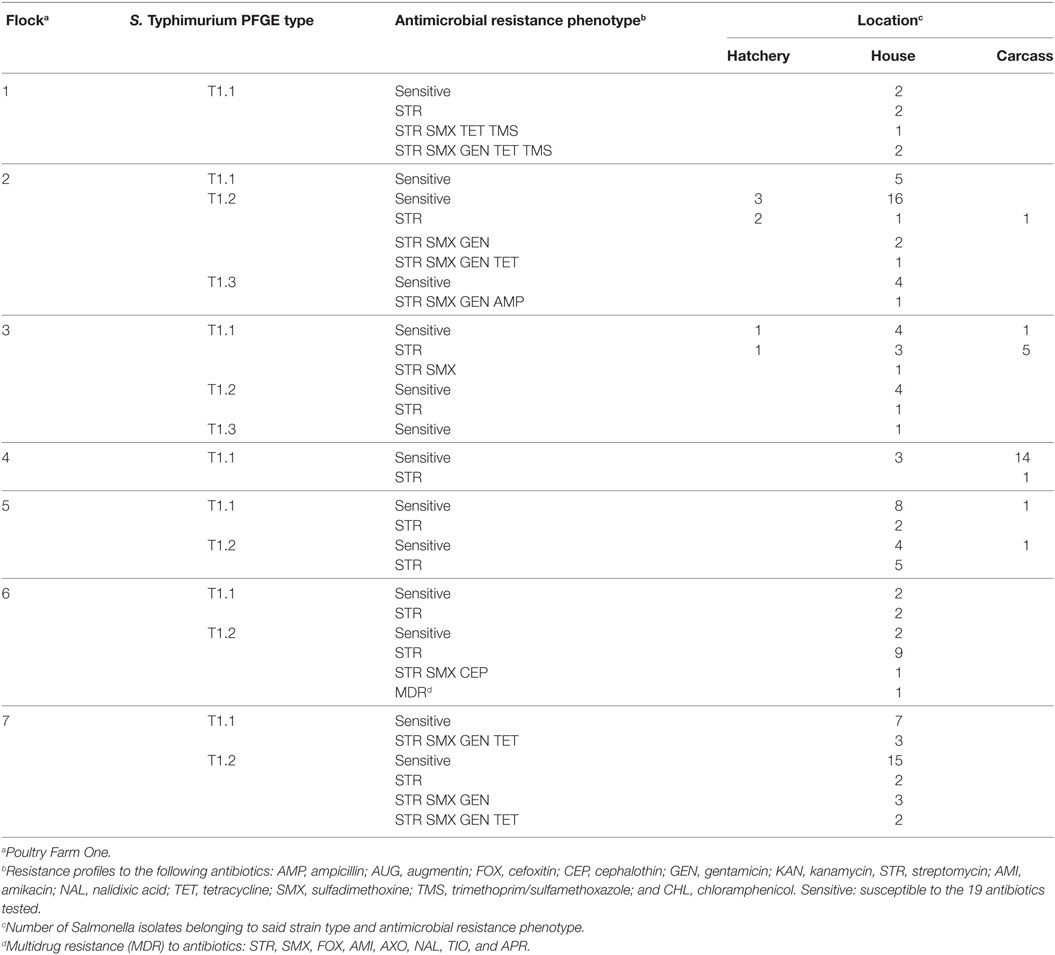
Table 4. Temporal and spatial distribution of resident antibiotic susceptible and resistant S. Typhimurium strain types during the production of seven consecutive commercial broiler flocks.
The antibiotic susceptibility and profiles of Salmonella isolated from two poultry farms mirrored antimicrobial resistance data reported in other studies. The majority (51.6%) of Salmonella isolates, from a 2001 NARMS survey, were also pan-susceptible. The most commonly identified resistances were to the antibiotics tetracycline (26.7%), streptomycin (23.7%), sulfadimethoxine (9.1%), gentamicin (6.3%), and ampicillin (15.1%) (26). A 2002 NARMS retail survey also reported that Salmonella isolated from chicken meat were largely pan-susceptible (66.6%), with the most prevalent resistance observed for sulfadimethoxine (18.7%), streptomycin (32.3%), gentamicin (3.4%), ampicillin (5.1%), trimethoprim/sulfamethoxazole (1.7%), and tetracycline (34.3%) (4). The 2003 NARMS retail meats survey, contemporary with the sampling times of this study, reported 47% of Salmonella isolates as pan-susceptible; with resistances observed for tetracycline (27.4%), streptomycin (26.2%), sulfadimethoxine (14.3%), gentamicin (6.0%), and ampicillin (33.3%). In the most recent NARMS retail meats survey (2015), half of the poultry Salmonella isolates were pan-susceptible to a panel of 12 antibiotics. Salmonella isolated from retail meats, in this survey, were resistant to tetracycline (37.3%), streptomycin (37.3%), sulfadimethoxine (8.5%), gentamicin (5.1%), and ampicillin (8.5%) (27).
There was a diversity of antimicrobial resistance phenotypes identified among our poultry Salmonella isolates. Despite this diversity, the antimicrobial resistance phenotype: streptomycin and sulfadimethoxine resistance alone or with other antibiotics was commonly encountered in Salmonella isolated from the commercial poultry farms. The genes conferring resistance to these antimicrobials are frequently found residing on mobile genetic elements which are responsible for the wide-spread dissemination of antimicrobial resistance in nature. The transposon Tn21 contains the mercury resistance gene merA; streptomycin resistance gene aadA1; and sulfadimethoxine resistance gene sul1 (28). This transposon is often responsible for dissemination of mercury and antimicrobial resistance in nature (28) and is prevalent in poultry Salmonella and E. coli (23). While we observed linkage between the resistance genes merA and aadA and streptomycin/sulfadimethoxine resistance, only 17.72% of streptomycin-resistant Salmonella had aadA1, indicating that other antimicrobial resistance gene(s) are responsible for streptomycin resistance and further illustrates the diversity underlying antimicrobial resistance phenotypes observed in these isolates.
Despite the high prevalence of Tn21 in these poultry isolates, antimicrobial resistance phenotypes were not uniformly distributed among Salmonella serovars within as well as between the two commercial broiler chicken farms. Certain Salmonella serovars differed in their antibiotic susceptibility patterns. Salmonella Enteritidis tended to be pan-susceptible while S. Typhimurium exhibited a diversity of antimicrobial resistance phenotypes. Similar trends have been observed for these Salmonella serovars reported in NARMS retail meats (2003, 2015) and HACCP (2003, 2014) surveys (27). Even within S. Typhimurium, there were differences in antibiotic susceptibilities among strain types. The S. Typhimurium PFGE subtype T1.1 from Farm One (21) was identified as phage type (PT) 193, a PT commonly associated with illnesses in humans (29–39). This Salmonella PT has also been isolated from cattle (38, 40, 41), poultry (31, 42), pigs (31, 40), and dogs (40). Like S. Typhimurium DT104, PT 193 isolates generally exhibit resistance to three or more antibiotics, but resistance phenotypes reported have been variable (40, 43). The majority (68.0%) of our S. Typhimurium PT DT193 isolates from Farm One were pan-susceptible, with 32% possessing the following resistance phenotypes to: streptomycin alone; streptomycin, sulfadimethoxine, tetracycline, and trimethoprim/sulfamethoxazole; streptomycin, sulfadimethoxine, gentamicin, tetracycline, and trimethoprim/sulfamethoxazole. The other S. Typhimurium PFGE types, T1.2 and T1.3, were identified, respectively, as PTs DT107 and U302 (21). The S. Typhimurium PTs DT107 and DT193 from this study appear to be genetically related as determined by PFGE (44). Close genetic-relatedness as determined by PFGE among different S. Typhimurium and S. Enteritidis PTs has been reported by others (45, 46). The S. Typhimurium DT107 isolates were similar to the S. Typhimurium DT193 isolates, in that the majority were pan-susceptible (59.6%), with the most prevalent antimicrobial resistance phenotype being resistance to streptomycin only (25.4%).
Poultry litter contains a large reservoir of antimicrobial resistance genes. We had shown in a previous study that many of these antimicrobial resistance genes are shared among diverse bacterial species in poultry litter (ex. aadA1 in Corynebacterium and Salmonella) (20). Therefore, the potential exists for environmental transfer of antimicrobial resistance to Salmonella and subsequent horizontal transmission of emergent resistant Salmonella strains to poultry in this environment. Of the eight antibiotic resistant phenotypes solely present in S. Typhimurium isolated from the farm environment, none were identified in S. Typhimurium isolated from processed chicken carcasses. This finding suggests that despite the diversity of antibiotic-resistant S. Typhimurium resident in the broiler house environment, none of these antibiotic resistant strains were being transmitted through the processing plant to the poultry carcass. Only those antibiotic-resistant strain types present with the chicks at placement remained on birds at processing. Therefore, our data support the importance of vertical transmission routes in the dissemination of antibiotic-resistant Salmonella through the food chain.
Therapeutic tetracycline antibiotic usage was not a significant predictor of emergent antimicrobial resistance in Salmonella. This result is not surprising, considering that the all-in, all-out production method used in the commercial poultry industry is designed to break disease cycles and should minimize antimicrobial resistance development, as long as pathogen persistence from flock-to-flock is prevented (47). However, the reservoir for antimicrobial resistance remains within the farm environment. Additional measures involving litter management and pest control may be needed to prevent future emergence of antimicrobial resistance zoonotic bacteria on treated farms. In addition, the prevalence of streptomycin resistance in poultry Salmonella was surprisingly high considering that streptomycin is rarely used in poultry production medicine and to our knowledge had not been used at these farms. This is most likely due to linkage of streptomycin resistance gene(s) with other resistance genes, or competitively advantageous genes (bacteriocins, siderophores, etc.); or its integration into the chromosome that has maintained streptomycin resistance in Salmonella, even in the absence of antibiotic selection (19). However, gentamicin is commonly used with in ovo poultry vaccines as a prophylaxis against peritonitis in chicks and therefore may explain, in part, the level of resistance to this antibiotic in Salmonella. The physical linkage of resistance genes associated with gentamicin with streptomycin resistance may also explain the persistence of streptomycin resistance in the absence of usage (19). As gentamicin was used by both poultry companies, it is uncertain whether gentamicin resistance in Salmonella will persist with time. The other antibiotics used by the poultry farms in this study, bacitracin and virginiamycin, are used to control C. perfringens infections in poultry. While these antibiotics do not affect Salmonella or other Gram-negative enterics, they do have an impact in the Gram positive, intestinal microbiota of chickens (48). It is currently not known how changes to the chicken intestinal microbiota, in response to bacitracin and virginiamycin, affect Salmonella prevalence, abundance, or antibiotic resistance patterns.
Vertical transmission from the breeder flock, rather than horizontal transmission from the environment, appears to play a significant role in carcass contamination with antibiotic-resistant Salmonella. If antibiotic usage is involved in the emergence and spread of antibiotic-resistant Salmonella to chicken meat, it may exist at the breeder, not broiler level of poultry production. One way to block transmission of antimicrobial-resistant Salmonella would be to apply an intervention such as competitive exclusion or vaccination at the breeder level (49, 50). The poultry integrator for Poultry Farm One has recently instituted a company-wide Salmonella vaccination program at the broiler-breeder level. It will be interesting to see if this mitigation strategy has significantly changed antimicrobial resistance profiles of Salmonella isolated from broiler chicken farms and poultry charges, especially on Poultry Farm One.
JM, CH, DW, and ML contributed to the conception and design of this study. KL and SA were responsible for the acquisition of data analyzed in this study. JM, KL, and DW were involved in the analysis and interpretation associated with this work. KL was responsible for writing the first draft. All the authors were involved in manuscript revisions and final approval of the version to be published.
None of the authors received any financial gain or applied for patents associated with the work described in this article.
The reviewer, TP, and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
We wish to thank the United States Department of Agriculture and the College of Veterinary Medicine at the University of Georgia for support of this work and the poultry companies that participated in this study. This work, presented here, was part of KL PhD dissertation at the University of Georgia (51).
This work was supported by a grant from the United States Department of Agriculture (99-35212-8680). The State of Georgia Veterinary Medical Experimental Station supported KL.
1. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States – major pathogens. Emerg Infect Dis (2011) 17(1):7–15. doi:10.3201/eid1707.110572
2. Guo C, Hoekstra RM, Schroeder CM, Pires SM, Ong KL, Hartnett E, et al. Application of Bayesian techniques to model the burden of human salmonellosis attributable to U.S. food commodities at the point of processing: adaptation of a Danish model. Foodborne Pathog Dis (2011) 8(4):509–16. doi:10.1089/fpd.2010.0714
3. Williams MS, Ebel ED. Estimating changes in public health following implementation of hazard analysis and critical control point in the United States broiler slaughter industry. Foodborne Pathog Dis (2012) 9(1):59–67. doi:10.1089/fpd.2011.0951
4. White DG, Zhao S, Sudleer R, Ayers S, Friedman S, Chen S, et al. The isolation of antibiotic-resistant Salmonella from retail ground meats. N Engl J Med (2001) 345(16):1147–54. doi:10.1056/NEJMoa010315
5. Schlosser W, Hogue A, Ebel E, Rose B, Umholtz R, Ferris K, et al. Analysis of Salmonella serotypes from selected carcasses and raw ground products sampled prior to implementation of the pathogen reduction; hazard analysis and critical control point final rule in the US. Int J Food Microbiol (2000) 58:107–11. doi:10.1016/S0168-1605(00)00293-2
6. Cohen M, Tauxe RV. Drug-resistant Salmonella in the United States: an epidemiological perspective. Science (1986) 234:964–9. doi:10.1126/science.3535069
7. Martin JL, Fyfe M, Dore K, Buxton JA, Pollari F, Henry B, et al. Increased burden of illness associated with antimicrobial resistant Salmonella enterica serotype Typhimurium infections. J Infect Dis (2004) 189:377–84. doi:10.1086/381270
8. Helms M, Vastrup P, Gerner-Smidt P, Molbak K. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium DT104. Emerg Infect Dis (2002) 8:490–5. doi:10.3201/eid0805.010267
9. Barza M. Potential mechanisms of increased disease in humans from antimicrobial resistance in food animals. Clin Infect Dis (1987) 34(S3):123–5. doi:10.1086/340249
10. Smith KE, Besser JM, Hedberg CW, Leano FT, Bender JB, Wicklund JH, et al. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. N Engl J Med (1999) 340:1525–31. doi:10.1056/NEJM199905203402001
11. Glynn MK, Bopp C, Dewitt W, Dabney P, Mokhta M, Angulo FJ. Emergence of multi-drug resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med (1998) 338(19):1333–8. doi:10.1056/NEJM199805073381901
12. Hollinger K, Wray C, Evans D, Pascoe S, Chappell S, Jones Y. Salmonella Typhimurium DT104 in cattle in Great Britain. J Am Vet Med Assoc (1998) 213(12):1732–3.
13. Rankin SC, Aceto H, Cassidy J, Holt J, Young S, Love B, et al. Molecular characterization of cephalosporin-resistant Salmonella enterica serotype Newport isolates from animals in Pennsylvania. J Clin Microbiol (2002) 40:4679–84. doi:10.1128/JCM.40.12.4679-4684.2002
14. Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, Iwen PC, et al. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N Engl J Med (2000) 342:1242–9. doi:10.1056/NEJM200004273421703
15. Folster JP, Pecic G, Bolcen S, Theobald L, Hise K, Carattoli A, et al. Characterization of extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg isolated from humans in the United States. Foodborne Pathog Dis (2010) 7(2):181–7. doi:10.1089/fpd.2009.0376
16. Nield BS, Holmes AJ, Gillings MR, Recchia GD, Mabbutt BC, Nevalainen KM, et al. Recovery of new integron classes from environmental DNA. FEMS Microbiol Lett (2001) 195:59–65. doi:10.1111/j.1574-6968.2001.tb10498.x
17. Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Register B, et al. Incidence of class 1 and class 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob Agents Chemother (2001) 45(3):723–6. doi:10.1128/AAC.45.3.723-726.2001
18. Marshall B, Petrowski D, Levy SB. Inter- and intraspecies spread of Escherichia coli in a farm environment in the absence of antibiotic usage. Proc Natl Acad Sci U S A (1990) 87:6609–13. doi:10.1073/pnas.87.17.6609
19. Salyers AA, Amabile-Cuevas CF. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother (1997) 41(11):2321–5.
20. Nandi S, Maurer JJ, Hofacre C, Summers AO. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc Natl Acad Sci U S A (2004) 101(18):7118–22. doi:10.1073/pnas.0306466101
21. Liljebjelke KA, Hofacre CL, Liu T, White DG, Ayers S, Young S, et al. Vertical and horizontal transmission of Salmonella within integrated broiler production system. Foodborne Pathog Dis (2005) 2(1):90–102. doi:10.1089/fpd.2005.2.90
22. Pope MJ, Cherry TE. An evaluation of the presence of pathogens on broilers raised on poultry litter treatment-treated litter. Poult Sci (2000) 79(9):1351–5. doi:10.1093/ps/79.9.1351
23. Bass L, Liebert CA, Lee MD, Summers AO, White DG, Thayer SG, et al. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob Agents Chemother (1999) 43(12):2925–9.
24. Anonymous. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Villanova, PA: National Committee for Clinical Laboratory Standards (2000).
25. Anonymous. Performance Standard for Antimicrobial Disk Susceptibility Tests. 7th ed. Villanova, PA: National Committee for Clinical Laboratory Standards (2000).
26. Anonymous. National Antimicrobial Susceptibility Monitoring Program for Veterinary and Human Isolates. Laurel, MD: United States Food and Drug Administration (2001).
27. Anonymous. NARMS Now: Interactive Data Displays. United States Food and Drug Administration (2016). Available from: https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm416741.htm
28. Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev (1999) 63(3):507–22.
29. Ang-Kucuker M, Tolun V, Helmuth R, Rabsch W, Buyukbaba-Boral O, Torumkuney-Akbulut D, et al. Phage types, antibiotic susceptibilities and plasmid profiles of Salmonella Typhimurium and Salmonella enteritidis strains isolated in Istanbul, Turkey. Clin Microbiol Infect (2000) 6:593–9. doi:10.1046/j.1469-0691.2000.00132.x
30. Asensi MD, Costa AP, Moura E, dos Reis EM, Hofer E. Lysotypes and plasmidial profile of Salmonella serovar Typhimurium isolated from children with enteric processes in the cities of Rio de Janeiro, RJ, and Salvador, BA-Brazil. Rev Inst Med Trop Sao Paulo (1995) 37:297–302. doi:10.1590/S0036-46651995000400003
31. Baggesen DL, Aarestrup FM. Characterization of recently emerged multiple antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 and other multiresistant phage types from Danish pig herds. Vet Rec (1998) 143:95–7. doi:10.1136/vr.143.4.95
32. Cruchaga S, Echeita A, Aladuena A, Garcia-Pena J, Frias N, Usera MA. Antimicrobial resistance in salmonellae from humans, food and animals in Spain in 1998. J Antimicrob Chemother (2001) 47:315–21. doi:10.1093/jac/47.3.315
33. Hannu T, Mattila L, Siitonen A, Lerisalo-Repo M. Reactive arthritis following an outbreak of Salmonella Typhimurium phage type 193 infection. Ann Rheum Dis (2002) 61:264–6. doi:10.1136/ard.61.3.264
34. Nastasi A, Mammina C. Surveillance of multidrug-resistant strains of Salmonella enterica serotype Typhimurium in southern Italy in the years 1992–1997. Eur J Epidemiol (2000) 16:135–9. doi:10.1023/A:1007680500678
35. Pontello M, Sodano L, Nastasi A, Mammina C, Astuti M, Domenichini M, et al. A community-based outbreak of Salmonella enterica serotype Typhimurium associated with salami consumption in Northern Italy. Epidemiol Infect (1998) 120:209–14. doi:10.1017/S095026889800870X
36. Robins-Browne RM, Rowe B, Ramsaroop R, Naran AD, Threlfall EJ, Ward LR, et al. A hospital outbreak of multiresistant Salmonella Typhimurium belonging to phage type 193. J Infect Dis (1983) 147:210–6. doi:10.1093/infdis/147.2.210
37. Thorton L, Gray S, Gingham P, Salmon RL, Hutchinson DN, Rowe B, et al. The problems of tracing a geographically widespread outbreak of salmonellosis from a commonly eaten food: Salmonella Typhimurium DT193 in Northwest England and North Wales in 1991. Epidemiol Infect (1993) 111:465–71. doi:10.1017/S0950268800057198
38. Threlfall EJ, Ward LR, Skinner JA, Graham A. Antimicrobial drug resistance in non-typhoidal Salmonellas from humans in England and Wales in 1999: decrease in multiple resistance in Salmonella enterica serotypes Typhimurium, Virchow, and Hadar. Microb Drug Resist (2000) 6:319–25. doi:10.1089/mdr.2000.6.319
39. van Leeuwen WJ, Voogd CE, Guinee PA, Manten A. Incidence of resistance to ampicillin, chloramphenicol, kanamycin, tetracycline and trimethoprim of Salmonella strains isolated in the Netherlands during 1975–1980. Antonie Van Leeuwenhoek (1982) 48:85–96. doi:10.1007/BF00399490
40. Daly M, Buckley J, Power E, O’Hare C, Cormican M, Cryan B, et al. Molecular characterization of Irish Salmonella enterica serotype Typhimurium: detection of class I integrons and assessment of genetic relationships by DNA amplification fingerprinting. Appl Environ Microbiol (2000) 66:614–9. doi:10.1128/AEM.66.2.614-619.2000
41. Hinton M, Ali EA, Allen V, Linton AH. The excretion of Salmonella Typhimurium in the faeces of calves fed milk substitute. J Hyg (1983) 91:33–45. doi:10.1017/S0022172400060009
42. Rajashekara G, Haverly E, Halvorson DA, Ferris KE, Lauer DC, Nagaraja KV. Multidrug-resistant Salmonella Typhimurium DT104 in poultry. J Food Prot (2000) 63:155–61.
43. Gebreyes WA, Altier C. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J Clin Microbiol (2002) 40:2813–22. doi:10.1128/JCM.40.8.2813-2822.2002
44. Tenover FC, Arbeit RD, Goering RV, Mickelson PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol (1995) 33(9):2233–9.
45. Olsen SJ, Bishop R, Brenner FW, Roels TH, Bean N, Tauxe RV, et al. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987–1997. J Infect Dis (2001) 183(5):753–61. doi:10.1086/318832
46. Hudson CR, Charlotte Q, Lee MD, Keyes K, Dodson SV, Morales C, et al. The genetic-relatedness of Salmonella from non-domestic birds in the Southeastern United States. J Clin Microbiol (2000) 38:1860–5.
47. Hofacre C. The health and management of poultry production. Int J Infect Dis (2002) 6:353–7. doi:10.1016/S1201-9712(02)90177-3
48. Lu J, Hofacre C, Smith F, Lee MD. Effects of feed additives on the development on the ileal bacterial community of the broiler chicken. Animal (2008) 2(5):669–76. doi:10.1017/S1751731108001894
49. Dórea FC, Cole DJ, Hofacre C, Zamperini K, Mathis D, Doyle MP, et al. Effect of Salmonella vaccination of breeder chickens on contamination of broiler chicken carcasses in integrated poultry operations. Appl Environ Microbiol (2010) 76(23):7820–5. doi:10.1128/AEM.01320-10
50. Berghaus RD, Thayer SG, Maurer JJ, Hofacre CL. Effect of vaccinating breeder chickens with a killed Salmonella vaccine on Salmonella prevalences and loads in breeder and broiler chicken flocks. J Food Prot (2011) 74(5):727–34. doi:10.4315/0362-028X.JFP-10-542
Keywords: Salmonella, strain type, antimicrobial resistance, poultry, vertical transmission
Citation: Liljebjelke KA, Hofacre CL, White DG, Ayers S, Lee MD and Maurer JJ (2017) Diversity of Antimicrobial Resistance Phenotypes in Salmonella Isolated from Commercial Poultry Farms. Front. Vet. Sci. 4:96. doi: 10.3389/fvets.2017.00096
Received: 20 March 2017; Accepted: 08 June 2017;
Published: 23 June 2017
Edited by:
Michael Kogut, United States Department of Agriculture, Agricultural Research Service, United StatesReviewed by:
Sherry Layton, Vetanco, ArgentinaCopyright: © 2017 Liljebjelke, Hofacre, White, Ayers, Lee and Maurer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John J. Maurer, am1hdXJlckB1Z2EuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.