
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 23 December 2016
Sec. Veterinary Epidemiology and Economics
Volume 3 - 2016 | https://doi.org/10.3389/fvets.2016.00116
This article is part of the Research Topic Big Data – The Language and Future of One Medicine, One Science View all 6 articles
A correction has been applied to this article in:
Corrigendum: Evidence in Practice – a Pilot Study Leveraging Companion Animal and Equine Health Data From Primary Care Veterinary Clinics in New Zealand
 Petra Muellner1*
Petra Muellner1* Ulrich Muellner1
Ulrich Muellner1 M. Carolyn Gates2
M. Carolyn Gates2 Trish Pearce3
Trish Pearce3 Christina Ahlstrom1
Christina Ahlstrom1 Dan O’Neill4
Dan O’Neill4 Dave Brodbelt4
Dave Brodbelt4 Nick John Cave2
Nick John Cave2
Veterinary practitioners have extensive knowledge of animal health from their day-to-day observations of clinical patients. There have been several recent initiatives to capture these data from electronic medical records for use in national surveillance systems and clinical research. In response, an approach to surveillance has been evolving that leverages existing computerized veterinary practice management systems to capture animal health data recorded by veterinarians. Work in the United Kingdom within the VetCompass program utilizes routinely recorded clinical data with the addition of further standardized fields. The current study describes a prototype system that was developed based on this approach. In a 4-week pilot study in New Zealand, clinical data on presentation reasons and diagnoses from a total of 344 patient consults were extracted from two veterinary clinics into a dedicated database and analyzed at the population level. New Zealand companion animal and equine veterinary practitioners were engaged to test the feasibility of this national practice-based health information and data system. Strategies to ensure continued engagement and submission of quality data by participating veterinarians were identified, as were important considerations for transitioning the pilot program to a sustainable large-scale and multi-species surveillance system that has the capacity to securely manage big data. The results further emphasized the need for a high degree of usability and smart interface design to make such a system work effectively in practice. The geospatial integration of data from multiple clinical practices into a common operating picture can be used to establish the baseline incidence of disease in New Zealand companion animal and equine populations, detect unusual trends that may indicate an emerging disease threat or welfare issue, improve the management of endemic and exotic infectious diseases, and support research activities. This pilot project is an important step toward developing a national surveillance system for companion animals and equines that moves beyond emerging infectious disease detection to provide important animal health information that can be used by a wide range of stakeholder groups, including participating veterinary practices.
An approach to surveillance has been evolving that utilizes computerized veterinary practice management systems to capture data from individual primary-care veterinary practices and generate population-level information on animal health and welfare (1). While the concept of using veterinary clinical data and electronic medical records for animal health surveillance is not new (2), considerable advances have recently been made to improve compliance, for example, through the development of web-based data collection systems (3). An automated approach to collecting, sharing, and analyzing veterinary primary care data to understand disorders and improve the welfare of animals was initially pioneered for companion animals by the Royal Veterinary College in the UK (VetCompass1) and similar systems or adaptations are now emerging in different institutions and countries (1, 4, 5). The advantage of this approach is that it can be fully integrated in the veterinary workflow and captures information directly from the practice management software, thus minimizing time-consuming additional data entry and the use of linked software or websites. However, only recently resources have become available to process such large volumes of data in real-time (6, 7).
The high value of primary care data for companion animal health research has already been demonstrated. Results from several studies investigating the prevalence of common disorders and associated risk factors in dogs have filled previous knowledge gaps and identified strategies for improving canine health (8–10). Ongoing data collection will provide sufficient big data to identify additional associations and risk factors; indeed, the UK VetCompass database has collected data on more than 6 million unique animals in over 470 participating practices (11). Clinical decision-making and identification of research priorities can also be supported by the system, such as ranking of differential diagnoses, vaccine recommendations, directing veterinary education and training, and investigating changes in disease prevalence (12, 13).
However, the use of such data is by no means restricted to research and clinical decision-making and, instead, potential applications are manifold. For example, systematically collected temporal and spatial data can also be used for surveillance activities (14) and to aid decision-makers in having a sound understanding of disease prevalence, risk factors, and outcomes. A formal assessment recently provided proof-of-concept for the use of electronic medical record data to detect aberrant animal health events in space and time (15). Furthermore, both VetCompass and a similar system also developed in the UK called small animal veterinary surveillance network (SAVSNET) have been identified as mechanisms to collect antimicrobial usage and resistance data (16–18).
Importantly, the success of such an approach to animal data collection hinges on veterinary practitioners providing high quality, standardized data and, in the case of VetCompass, this has been supported by the availability of standardized diagnostic terms through the VeNom project (19). The central database dictionary developed by the VeNom Coding Group includes a standard set of clinical veterinary terms and VeNom codes for species, breeds, diagnoses, presenting complaints, procedures, and administrative tasks associated with canine, feline, and other small companion animal veterinary work. A VeNom code list for equines is also currently under development, which provides an exciting opportunity to extend the approach beyond small companion animals. A formal study is currently underway to collect critical baseline information regarding the clinic demographics. Details of veterinary practices employing registered veterinarians are formally published by the New Zealand Veterinary Council,2 and most are believed to work with an electronic veterinary recording system.
Despite the high utility of a VetCompass-like system, no such system currently exists on a large scale in New Zealand. However, New Zealand’s geographic isolation and unique ecosystem (20) and absence of many diseases endemic in other countries underscore the importance of gathering New Zealand-specific data on companion animals and equines to preserve the high health status of its animal population and the continued thriving of its veterinary services. In this pilot study, we explored the feasibility of a multi-species New Zealand veterinary practice-based health information and data system modeled upon the existing VetCompass system. Our main objectives were to gage end-user support, prototype the approach in the New Zealand veterinary landscape, and create preliminary system outputs prior to seeking funding for a nationwide roll-out. A special focus of the project was to work closely with participating veterinarians to gain an understanding of how the system could best be embedded in their clinical workflow.
The project team developed a software add-on in collaboration with Provet Animal Health Practice Solutions (the vendor of Vision VPM), which enabled veterinarians to enter diagnoses and/or reasons for the visit directly into the practice management system. The new code-entry fields (Figure 1) were added in the workflow at a stage where the veterinarian enters and saves a visit or case, with an option to opt-out if they did not want to enter a code at this stage. Up to five diagnoses and five reasons for visit could be entered for each episode of clinical care.
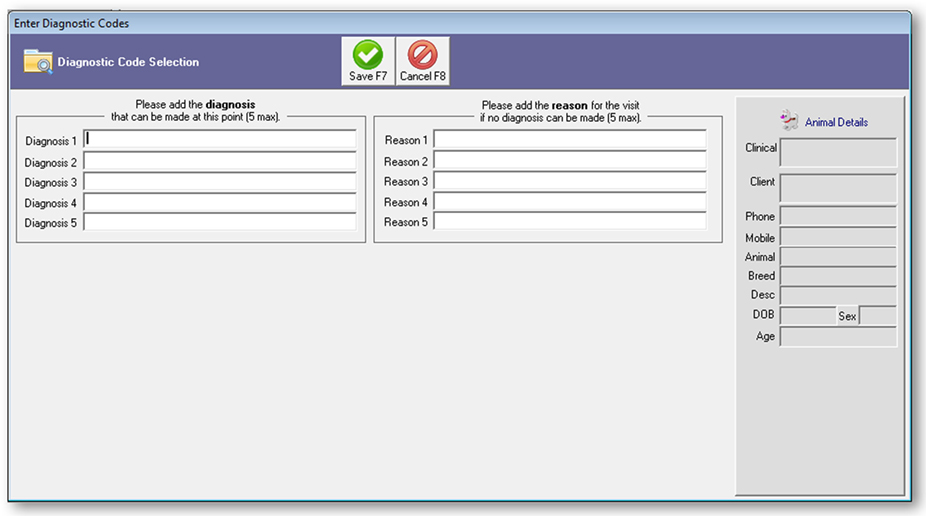
Figure 1. Screenshot of code-entry for diagnosis and reason for the visit installed in the practice management software Vision PM to support the 2016 New Zealand surveillance pilot study.
The software patch was populated with VeNom codes, a suggested standard of language and terms modeled into parent-child hierarchies, that are used internationally in veterinary diagnosis and reports (19) compiled for companion animals (v20 04DEC2012) and equines (pre-release draft version). The number of diagnostic and reason for visit codes in the companion animal list was 2,322 and 1,670, while the number of codes in the equine list was 1,948 and 203, respectively. All terms were available for both series of five boxes. Companion animals were differentiated from equines in this study, as New Zealand has a large population of racehorses. In New Zealand, small animal species kept as pets (e.g., cats and dogs) are commonly referred to as companion animals, whereas horses are referred to as equines.
As the VeNom code list contained more than 6,000 single entries to choose from, a key aspect was the development of an easy-to-use user interface to search and find relevant codes. A “Google-type” autosuggest search (21) allowed users to type keywords or word fragments that filtered the list to the relevant term. Other relevant data fields to be extracted, such as species, breed, age or sex, were already captured by the standard functionality of the existing practice management system.
In addition to the VeNom term entry field, further data were collected and extraction was automated (Figure 2). This utility was password-protected and included the following features:
Opt-in for the coding.
Assignment of Clinic ID (a unique code for a veterinary practice).
Selection of the species and records to be included for the term entry and data extraction.
Ability to manually bulk export data. This function was used at the start of the pilot to extract historic data.
Diagnostic code import. This function was used to upload the VeNom coding lists and could be used to update the list at a future date.
Details of the external location where the data extract is sent to; this was set up with the details of the FTP server at Massey University.
Fields to select time of the day and frequency of the automatic data extract.
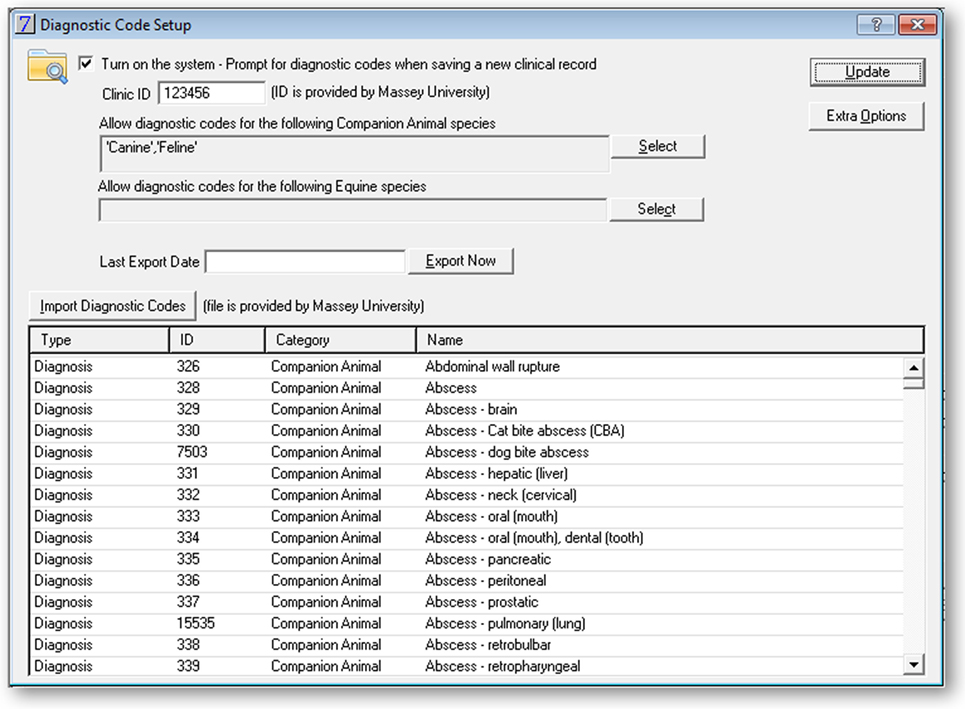
Figure 2. Screenshot of the administration interface in the practice management software Vision PM to support the 2016 New Zealand surveillance pilot study.
Once the data export was set up, the case information was extracted at the specified time of the day into an XML file that was sent via FTP to the secure servers at Massey University. A safe-guard was implemented such that the timestamp of the last successful extract was logged and data were extracted back to the time when the last extract ran successfully. This ensured that data were sent even if an export failed on a particular day.
Two veterinary practices were recruited to participate in the 4-week pilot study, based on convenience sampling. Veterinary Clinic A was a mixed animal practice with seven veterinarians that saw both companion animal and equine patients. Veterinary Clinic B was an exclusively companion animal practice that employed two veterinarians.
Each practice was visited by a project team member to install the software patch, which was developed in collaboration with the practice management system provider, and to introduce the participating veterinarians to the system. At the time of installation, all historical records were extracted from the clinic database to provide baseline demographics and statistics on participating clinic patients. Veterinarians were briefed in VeNom codes prior to the trial and were asked to add at least one diagnosis and/or reason for visit for every patient visit. On completion of the first two weeks of the trial, preliminary results were presented to the veterinarians and their feedback was discussed.
An IT server system hosted by Massey University was set up to collect and store data from the veterinary practices. The system consisted of an FTP server to receive the XML files, an application server, as well as a shared database server (Figure 3). For development purposes, a dedicated development server was set up for testing applications and processes before implementing them in production. Data were anonymized by not extracting owner names and reducing the owner addresses to the post code only.
To process the XML files, a structured query language (SQL) Server Integration Services job was set up that migrated the data from the XML files into the “Staging” SQL server database. This database was used for data collection. Although not yet functional at the time of writing the current paper, a second database for data screening and a third database that uses the schema of the VetCompass system, to allow for international data comparison, were created to enable further development and expansion of the system at a later date. Nightly automated data extractions, which used a scheduled data push process from the practice to the system server, were set up for both veterinary clinics. The following fields were extracted:
Date
Postal code (of owner)
Practice ID
Animal unique ID
Species
Breed
Date of birth
Sex
Neuter status
Body weight (kg)
Body condition score (BCS) (1–9)
Messages and clinical notes added by the veterinarian in free text
VeNom diagnoses
VeNom presentation reason
The collection and use of the data during the pilot complied with the New Zealand Privacy Act 1993 and the Veterinary Council of New Zealand Code of Ethics under the Veterinarian Act 2005.
The nightly data extracts were aggregated and imported into R statistical software for analysis (22). Each variable was checked for coding errors, and a data dictionary was developed to map variations in terminology to describe the patient sex, breed, and neuter status within and between practices into a single unified coding system. Detailed summaries were prepared for and emailed to each participating practice and included descriptive statistics on the daily caseloads, patient demographics, reasons for presentation, and diagnoses.
From 18 September 2015 to 15 October 2015, participating veterinarians from Veterinary Clinic A recorded 120 visits for 86 unique equine patients and 241 visits for 210 unique companion animal patients, whereas participating veterinarians from Veterinary Clinic B recorded 224 visits for 168 companion animal patients. The average VeNom coding rates (i.e., visits with at least one diagnosis or reason for visit coded) were 83% for Veterinary Clinic A equine visits, 87% for Veterinary Clinic A companion animal visits, and 74% for Veterinary Clinic B companion animal visits. The average number of daily records totaled across the clinics for the 27-day pilot trial was approximately 21.
Data from the remaining fields required extensive cleaning in preparation for analysis to correct spelling errors and ensure consistent use of terminology. Equine visits were more likely to be missing information in the core data fields than companion animal visits (Table 1). The placement of the coding field was very well received by the participating veterinarians, and only minor changes were requested, for example, functionality to close the window by a single button action.
Across both veterinary clinics, the most common reasons for cats to present were for vaccination (19%), gastrointestinal problems (11%), and non-specific clinical signs (anorexia, lethargy, and weight loss) (9%). For dogs, the most common reasons were vaccination (20%), skin disease (11%), and lameness (5%). Most Veterinary Clinic A equine cases were seen for wellness visits (17%), lameness (8%), or therapeutic interventions (3%). Over 100 unique diagnostic codes were utilized during the pilot trial.
The practice-specific summaries prepared for and discussed with the participating veterinarians were enthusiastically received by the veterinarians. Care was taken to provide metrics not only of value to population health analyses but also to assist the veterinarians in improving their practice management. These included maps showing the distribution of clients by postal code, caseload statistics by weekday, and detailed analyses of common reasons for presentation and diagnoses. It was particularly interesting to note the number of animals that presented for routine wellness visits, but were diagnosed with other medical conditions, which highlights the importance of preventative veterinary care. Participating veterinary practices continued to code beyond the pilot period even though the data collection process was not actively monitored during this time. Coding rates remained high and informal feedback to the practices continued (data not shown).
The data migration worked seamlessly over the 4-week window, and there were no issues with transferring the nightly uploads to the server. Feedback from the participating veterinarians indicated that the VeNom codes could be improved to remove redundancies. Though they could readily work with the VeNom codes, in some circumstances they expressed uncertainty as to the best suited code(s) and noted some omissions, particularly some that are parochial or idiosyncratic to New Zealand, such as trace element deficiencies and toxicities. Otherwise, they found the search format to be user friendly and easy to operate.
This study provided proof-of-concept for the feasibility of establishing a VetCompass-like system for collecting veterinary clinical data on small animals and equines in New Zealand. The observed high percentage of coded cases demonstrates that veterinarians are not just willing to code clinical cases according to a set of standardized terms, but if properly engaged they show considerable enthusiasm to improve the quality and usability of the data they collect. In this study, this enthusiasm was driven by a sustainable information feedback loop providing veterinarians with easily accessible “practice health” metrics that could potentially help them to improve their clinical decision-making and business strategy (e.g., by understanding daily caseloads, patient demographics, temporal trends, and frequency of specific disorders/procedures). This, in addition to seamless integration into the existing management system and workflow, resulted in the veterinarians continuing to participate and code visits well beyond the conclusion of the pilot trial.
In human health, classification of diseases and other health problems is already standardized (23) with the International Classification of Disease published by the World Health Organization. This provides a set of agreed upon diagnostic codes (24), enabling national, regional, and global comparisons. Further development of the VeNom codes can equally enable such international collaboration in the field of veterinary medicine. As such, additional refinement and adoption of the VeNom codes to New Zealand and other countries will establish an analogous comprehensive and robust international language system for global comparisons. Furthermore, long-term national disease baseline establishment would support local comparisons, such as benchmarking to gage individual practice disease rates against a national standard.
Rapid and intuitive selection of the desired diagnosis and/or reason for visit codes by the vets within the practice management system is essential if high compliance for clinical data is to be achieved. To support this, participating veterinarians were consulted regarding the positioning of the coding in the system (i.e., between the clinical record and billing fields) to optimize workflow. Previous studies relied to varying extents on assigning codes retrospectively by processing the clinical notes added by the veterinarian either manually (25, 26) or via natural language processing (27). Given the remarkably high coding compliance in this study, data mining of the clinical notes could be implemented to compare and potentially validate these two different approaches and assess the accuracy of retrospectively coded cases based on clinical notes versus real-time coding at the time of the consult. The autosuggest search method for diagnostic codes and presentation reason incorporated into the New Zealand pilot system reduced the effort required by veterinarians; rather than having to scroll through long lists or decision-trees of diagnostic codes and presentation reasons, they could simply use a Google-like autosuggest function. This was well-received by all participating veterinarians and was easy to implement during development of the software patch. Additionally, the automatic daily upload of clinical data eliminated a further obstacle to compliance, as veterinarians were not tasked with manually extracting and sending data. Still, the adoption of new informatics tools depends on their usability and integration into routine workflow (28, 29), and challenges remain in convincing some veterinarians to modify existing routines and systematically record data in a usable way.
Primary care animal health data have successfully been used to estimate disorder prevalence in dogs (10), and recent work has clearly demonstrated the ability of such systems to produce reliable disease outbreak alerts (15). At the same time, this information can also provide easy and immediate access to epidemiological information relevant to a full investigation and assessment, as effective management or eradication of a new disease is often only technically feasible and affordable if emerging threats are identified early. Veterinary practitioners are often the first trained professionals to observe clinical signs in affected animals and therefore play an important role in early disease detection (30). The development of standardized and easy-to-use systems for recording clinical observation data in the field would remove a significant barrier to implementing veterinary practitioner-based disease surveillance in real-time (31–33). This would be an improvement over current systems that rely on manual curation, periodic reporting, and retrospective recording of diagnoses based on clinical notes. With the increasing sophistication of IT systems, such big data can now be securely collected, analyzed, and presented automatically.
The primary care data collection described here can easily form part of national animal health surveillance activities by collecting data on specific disorders diagnosed (4, 34–36). As such, we believe this new approach to surveillance should not be classified as being “syndromic” in nature (37) but rather be perceived as access to a new type of (big) data. To date, such big data on companion animal health have not been immediately available and had to be, with considerable effort, collected outside of the primary veterinary care environment (e.g., through data entry on dedicated websites) (33, 38). However, the overall system design and the nature of the data extracted will ultimately define the characteristics of the surveillance conducted (39).
This pilot demonstrated high feasibility for primary care data collection and surveillance in different species groups of animals under veterinary care (i.e., companion animals and equines), and the system could in theory be extended to every species for which electronic primary care data exist. It extends previous companion animal focused work to also include equine, which includes not only horses held for recreational use but also for racing and other sporting purposes. As a result, the project was supported by the New Zealand equine industry that has a strong international reputation for producing high-quality performance horses and breeding stock (40). A similar extension to livestock is possible with appropriately developed VeNom-like codes and a customized data entry interface. This could expand early warning capacity and support biosecurity preparedness and government-industry collaboration in the livestock sector. It could also facilitate trade by providing assurance to trade partners using baseline information on the number of animals seen by a veterinarian and thereby demonstrating evidence for sufficient coverage of care by veterinary services. Furthermore, such a system could be adapted to monitor antimicrobial usage (16) and resistance (AMR) in companion animals and livestock; hence, the value of the piloted system to AMR monitoring should be further explored along with necessary requirements.
As this was a pilot study, there were some limitations. Available funding was only sufficient to enroll two practices and to work with one software provider to build the modifications required to extract the data and enter the coding field software. However, many additional software providers as well as veterinary clinics expressed interest in participating, which has supported the further development of this pilot system. An additional limitation was that the equine VeNom codes were only a draft version compiled by the RVC and were not yet publically released at the time of data collection. Furthermore, we observed that the BCS coding field was coded at a lower rate than other variables in Clinic A. Going forward, the epidemiological value and validity of this (and other coding fields) will have to be formally assessed to define in more detail how the information extracted can be best utilized. Furthermore, such disparities in coding rates between clinics may encourage improved coding if these data are communicated back to the clinics. As with all analyses based on big data, inherent biases stemming from multi-source data collection must be considered (7).
Ideally, any practitioner-based disease surveillance system would be supported by a multi-tiered user interface where veterinary clinics could access their individual practice information and benchmark their metrics to national/regional levels. It is believed that tangible and even monetizable benefits (such as practice health summaries, evidence for clinical decision-support, or performance indicators) are required for the participating practitioners to comply fully and that without these benefits larger national or international research objectives are insufficient to motivate participation. A customized suite of data analysis and visualization tools are currently being developed that allows users to easily explore animal health trends while maintaining the confidentiality of animal owners and their veterinarians. Indeed, sophisticated web-based applications are increasingly being used for surveillance and complex epidemiological data analysis/visualization (28). For example, a prototype of an animal health and slaughterhouse surveillance data visualization system was recently developed that demonstrated the value of an interactive data hub for the early detection of outbreaks and health trends for equine surveillance in Switzerland (3, 33). The SAVSNET in the United Kingdom has recently started publishing quarterly reports (4) on national practice- and laboratory-based surveillance, though the presentation reasons and diagnoses collected are not based on standardized terms. By integrating knowledge and experience of multiple clinical practices into a common operating picture, it would be possible to observe high-level trends that are valuable for not only veterinarians but also researchers and decision-makers as well. This, supplemented with coding at the time of consult, would facilitate timely communication, as data would be available near real-time and analyzed automatically.
Further work should consider an enrollment strategy for a nationwide surveillance system, such as whether all New Zealand veterinary clinics should be included or only selected “sentinel” practices. Given the number of registered veterinary practices in New Zealand and that the average number of daily records recorded during the pilot was 21, we could conservatively expect at least 3,000 records per day coming into the system. Insights from the analysis of surveillance reports and participant feedback highlight important considerations for transitioning the pilot program to a sustainable large-scale and multi-species surveillance system. Other veterinary practitioner-based surveillance systems have used financial incentives such as laboratory testing credits (32) and direct financial compensation (41) to encourage user participation. Although this has been highlighted as an important incentive for some veterinarians (31), it may not be sustainable depending on long-term funding availability. The strong support from veterinary clinics in the current study suggests alternative engagement strategies may be more effective, as intrinsic motivation (e.g., to understand the clinic’s metrics and how it compares to regional statistics) may be a stronger driver for increased participation. As the system develops, it will be important to investigate additional practice management software programs, their market share in New Zealand and how they differ to Vision VPM used in this pilot.
In conclusion, recent advances in technology have made it possible to integrate large volumes of clinical data from practicing veterinarians into population health data in near real-time. This provides new opportunities in many areas including detection of unusual health events (36), provision of benchmark information, and provision of information on breed health (42). The high usability demonstrated in this New Zealand pilot study has provided a first step in implementing sustainable surveillance based around primary care data. Without adding undue burdens on participating veterinarians and veterinary clinics, this system captured information on presentation reason and subsequent diagnoses. By embedding into the existing workflow, we have created an interface that allows extraction of information from veterinarians that was previously poorly accessible. The set-up creates a win-win for veterinary practices, population health, and individual animal evidence-based medicine. The insights gained from these data will be used to refine the surveillance system design for implementation on a national scale, and a strategy for rollout to practices across the country is currently being developed.
PM, CA, and UM drafted the manuscript and made a lead contribution to the conception, design, analysis, and interpretation of the work. NC and TP critically reviewed the paper for important intellectual content and made a lead contribution to the conception, design, analysis, and interpretation of the work. MG contributed to drafting and reviewed the manuscript and made contributions to analysis and interpretation of the work. DO and DB contributed to the conception and design of the work and reviewed the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge the contributions from participating veterinary clinics. This project would not have been possible without their enthusiasm and dedication. Furthermore, they would like to thank their Royal Veterinary colleagues (RVC; in particular David Church and Noel Kennedy) for openly sharing VetCompass methods and experiences and supporting international collaboration and harmonization. They also acknowledge the contribution made by Mark Stevenson (University of Melbourne) to initial discussions on the system design and the VeNom Coding group (http://www.venomcoding.org) for sharing the diagnostic codes used in this project.
This project was supported by funds from the Companion Animal Society (CAS) of the New Zealand Veterinary Association (NZVA) and the Equine Health Association of New Zealand (EHA).
1. O’Neill DG, Church DB, McGreevy PD, Thomson PC, Brodbelt DC. Approaches to canine health surveillance. Canine Genet Epidemiol (2014) 1(1):1. doi:10.1186/2052-6687-1-2
2. Lund EM, Armstrong PJ, Kirk CA, Kolar LM, Klausnor JS. Health status and population characteristics of dogs and cats examined at private veterinary practices in the United States. J Am Vet Med Assoc (1999) 214:1336–41.
3. Wohlfender-Remy F, Struchen R, Graubner C, Balmer S, Hadorn D. Re-launch of Equinella: a web-based equine disease reporting and information platform. J Equine Vet Sci (2016) 39:S17. doi:10.1016/j.jevs.2016.02.035
4. Sánchez-Vizcaíno F, Jones PH, Menacere T, Heayns B, Wardeh M, Newman J, et al. Small animal disease surveillance. Vet Rec (2015) 177(23):591–4. doi:10.1136/vr.h6174
5. Jones PH, Radford AD, Noble P-JM, Sánchez-Vizcaíno F, Menacere T, Heayns B, et al. SAVSNET: collating veterinary electronic health records for research and surveillance. Online J Public Health Inform (2016) 8(1). doi:10.5210/ojphi.v8i1.6543
6. Kao RR, Haydon DT, Lycett SJ, Murcia PR. Supersize me: how whole-genome sequencing and big data are transforming epidemiology. Trends Microbiol (2014) 22(5):282–91. doi:10.1016/j.tim.2014.02.011
7. Pfeiffer DU, Stevens KB. Spatial and temporal epidemiological analysis in the big data era. Prev Vet Med (2015) 122(1):213–20. doi:10.1016/j.prevetmed.2015.05.012
8. Kearsley-Fleet L, O’neill DG, Volk HA, Church DB, Brodbelt DC. Prevalence and risk factors for canine epilepsy of unknown origin in the UK. Vet Rec (2013) 172(13):338. doi:10.1136/vr.101133
9. Mattin M, O’Neill D, Church D, McGreevy PD, Thomson PC, Brodbelt D. An epidemiological study of diabetes mellitus in dogs attending first opinion practice in the UK. Vet Rec (2014) 174(14):349. doi:10.1136/vr.101950
10. O’Neill DG, Church DB, McGreevy PD, Thomson PC, Brodbelt DC. Prevalence of disorders recorded in dogs attending primary care veterinary practices in England. PLoS One (2014) 9(3):e90501. doi:10.1371/journal.pone.0090501
11. O’Neill D. Building practice-based research in the UK. Vet Rec (2015) 176(18):461–2. doi:10.1136/vr.h2067
12. Lanyon L. Collecting the evidence for EBVM: who pays? Vet Rec (2016) 178(5):120–1. doi:10.1136/vr.i254
13. Robinson NJ, Dean RS, Cobb M, Brennan ML. Paper: investigating common clinical presentations in first opinion small animal consultations using direct observation. Vet Rec (2015) 176(18):463. doi:10.1136/vr.102751
14. Klompas M, Murphy M, Lankiewicz J, McVetta J, Lazarus R, Eggleston E, et al. Harnessing electronic health records for public health surveillance. Online J Public Health Inform (2011) 3(3). doi:10.5210/ojphi.v3i3.3794
15. Kass PH, Weng H-Y, Gaona MAL, Hille A, Sydow MH, Lund EM, et al. Syndromic surveillance in companion animals utilizing electronic medical records data: development and proof of concept. PeerJ (2016) 4:e1940. doi:10.7717/peerj.1940
16. Mateus A, Brodbelt DC, Barber N, Stärk KDC. Antimicrobial usage in dogs and cats in first opinion veterinary practices in the UK. J Small Anim Pract (2011) 52(10):515–21. doi:10.1111/j.1748-5827.2011.01098.x
17. Radford AD, Noble PJ, Coyne KP, Gaskell RM, Jones PH, Bryan JGE, et al. Antibacterial prescribing patterns in small animal veterinary practice identified via SAVSNET: the small animal veterinary surveillance network. Vet Rec (2011) 169(12):310. doi:10.1136/vr.d5062
18. Buckland EL, O’Neill D, Summers J, Mateus A, Church D, Redmond L, et al. Characterisation of antimicrobial usage in cats and dogs attending UK primary care companion animal veterinary practices. Vet Rec (2016) 179(19):489. doi:10.1136/vr.103830
19. Venom Coding Group. VeNom Veterinary Nomenclature. (2015). Available from: http://www.venomcoding.org/VeNom/Welcome.html
20. Crump JA, Murdoch DR, Baker MG. Emerging infectious diseases in an island ecosystem: the New Zealand perspective. Emerg Infect Dis (2001) 7(5):767. doi:10.3201/eid0705.017501
21. Li G, Ji S, Li C, Feng J. Efficient fuzzy full-text type-ahead search. VLDB J (2011) 20(4):617–40. doi:10.1007/s00778-011-0218-x
22. Core Team R. R: A Language and Environment for Statistical Computing. Vienna Austria: R Foundation for Statistical Computing (2016).
23. Spackman KA, Campbell KE, Côté RA. SNOMED RT: a reference terminology for health care. Proceedings of the AMIA Annual Fall Symposium. Nashville, TN: American Medical Informatics Association (1997). 640 p.
24. WHO. International Classification of Disease (ICD). (2016). Available from: http://www.who.int/classifications/icd/en/
25. O’Neill DG, Scudder C, Faire JM, Church DB, McGreevy PD, Thomson PC, et al. Epidemiology of hyperadrenocorticism among 210,824 dogs attending primary-care veterinary practices in the UK from 2009 to 2014. J Small Anim Pract (2016). doi:10.1111/jsap.12523
26. O’Neill DG, Meeson RL, Sheridan A, Church DB, Brodbelt DC. The epidemiology of patellar luxation in dogs attending primary-care veterinary practices in England. Canine Genet Epidemiol (2016) 3(1):1. doi:10.1186/s40575-016-0034-0
27. Wang X, Chase H, Markatou M, Hripcsak G, Friedman C. Selecting information in electronic health records for knowledge acquisition. J Biomed Inform (2010) 43(4):595–601. doi:10.1016/j.jbi.2010.03.011
28. Carroll LN, Au AP, Detwiler LT, Fu T, Painter IS, Abernethy NF. Visualization and analytics tools for infectious disease epidemiology: a systematic review. J Biomed Inform (2014) 51:287–98. doi:10.1016/j.jbi.2014.04.006
29. Reeder B, Revere D, Hills RA, Baseman JG, Lober WB. Public health practice within a health information exchange: information needs and barriers to disease surveillance. Online J Public Health Inform (2012) 4(3). doi:10.5210/ojphi.v4i3.4277
30. Vourc’h G, Bridges VE, Gibbens J, De Groot BD, McIntyre L, Poland R, et al. Detecting emerging diseases in farm animals through clinical observations. Emerg Infect Dis (2006) 12(2):204–10. doi:10.3201/eid1202.050498
31. Amezcua MDR, Pearl DL, Friendship RM, McNab WB. Evaluation of a veterinary-based syndromic surveillance system implemented for swine. Can J Vet Res (2010) 74(4):241–51.
32. Zurbrigg KJ, Van den Borre NM. Factors associated with good compliance and long-term sustainability in a practitioner-based livestock disease surveillance system. Can Vet J (2013) 54(3):243.
33. Muellner UJ, Vial F, Wohlfender F, Hadorn D, Reist M, Muellner P. Timely reporting and interactive visualization of animal health and slaughterhouse surveillance data in Switzerland. Front Vet Sci (2015) 2:47. doi:10.3389/fvets.2015.00047
35. Chaudhry B, Wang J, Wu S, Maglione M, Mojica W, Roth E, et al. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Ann Intern Med (2006) 144(10):742–52. doi:10.7326/0003-4819-144-10-200605160-00125
36. Moore GE, Lund E. Disease reporting and surveillance: where do companion animal diseases fit in? Vet Clin North Am Small Anim Pract (2009) 39(2):225–40. doi:10.1016/j.cvsm.2008.10.009
37. Dórea FC, Sanchez J, Revie CW. Veterinary syndromic surveillance: current initiatives and potential for development. Prev Vet Med (2011) 101(1):1–17. doi:10.1016/j.prevetmed.2011.05.004
38. Ward MP, Kelman M. Companion animal disease surveillance: a new solution to an old problem? Spat Spatiotemporal Epidemiol (2011) 2(3):147–57. doi:10.1016/j.sste.2011.07.009
39. Robertson C, Nelson TA, MacNab YC, Lawson AB. Review of methods for space–time disease surveillance. Spat Spatiotemporal Epidemiol (2010) 1(2):105–16. doi:10.1016/j.sste.2009.12.001
40. Rogers CW, Cogger N. A cross-sectional survey of biosecurity practices on thoroughbred stud farms in New Zealand. N Z Vet J (2010) 58(2):64–8. doi:10.1080/00480169.2010.65087
41. Sawford K, Vollman AR, Stephen C. A focused ethnographic study of Alberta cattle veterinarians’ decision making about diagnostic laboratory submissions and perceptions of surveillance programs. PLoS One (2013) 8(5):e64811. doi:10.1371/journal.pone.0064811
Keywords: surveillance, veterinary, primary care, early warning, interface design, IT, companion animal, equine
Citation: Muellner P, Muellner U, Gates MC, Pearce T, Ahlstrom C, O’Neill D, Brodbelt D and Cave NJ (2016) Evidence in Practice – A Pilot Study Leveraging Companion Animal and Equine Health Data from Primary Care Veterinary Clinics in New Zealand. Front. Vet. Sci. 3:116. doi: 10.3389/fvets.2016.00116
Received: 29 September 2016; Accepted: 07 December 2016;
Published: 23 December 2016
Edited by:
Andres M. Perez, University of Minnesota, USACopyright: © 2016 Muellner, Muellner, Gates, Pearce, Ahlstrom, O’Neill, Brodbelt and Cave. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Muellner, cGV0cmFAZXBpLWludGVyYWN0aXZlLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.