
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Urol., 08 March 2024
Sec. Urologic Oncology
Volume 4 - 2024 | https://doi.org/10.3389/fruro.2024.1367720
This article is part of the Research TopicRising Stars in Urologic Oncology: 2023View all 5 articles
 Gianmarco Randazzo
Gianmarco Randazzo Eleonora Bovolenta
Eleonora Bovolenta Tommaso Ceccato
Tommaso Ceccato Giuseppe Reitano
Giuseppe Reitano Giovanni Betto
Giovanni Betto Giacomo Novara
Giacomo Novara Massimo Iafrate
Massimo Iafrate Alessandro Morlacco
Alessandro Morlacco Fabrizio Dal Moro
Fabrizio Dal Moro Fabio Zattoni*
Fabio Zattoni*Introduction: The urinary microbiome (UMB) includes living bacteria, their genomes, and their products from interactions with the host environment. A “core” UMB could potentially exist, with variations between age and sex groups. Changes in UMB composition have been associated with benign urological disorders, but also with urologic cancers. Mechanisms through which UMB can trigger and maintain cancer can be local inflammation and interaction with immune system.
Aim of the study: To describe the association between UMB and development of urologic cancers.
Methods: A non-systematic literature review identified recently published studies (last 5 years), involving human patients, dealing with UMB. The database used for this review was PubMed, and the identified studies served as the base for a narrative analysis of the literature that explored the potential associations between UMB and urological cancers.
Results: In bladder cancer (BC), UMB may play a role in epithelial-mesenchymal transition (and thus to progression to metastasis), as well as in effectiveness of BCG response rate. BC is also associated with changes in UMB, with bacterial richness indices increased in cancer groups compared to non-neoplastic groups and being different between NMIBC vs MIBC patients. In prostate cancer (PCa), there is an abundance in proinflammatory bacteria and uropathogens. In regard to renal cell carcinoma (RCC), penile cancer and testicular cancer there are still too few studies to draw significant conclusions about its relationship with the UMB.
Conclusions: Gaining a deeper understanding of UMB role in urologic tumors could aid in the development of new therapies and improve classification of patients’ risk.
The urinary microbiome (UMB) is a comprehensive concept that includes living bacteria, genes, genomes (identified through 16S ribosomal RNA sequencing), and their products resulting from interactions with the host environment (1, 2). UMB variability is assessed using alpha diversity (diversity of microbial populations within a single sample) and beta diversity (across multiple samples) (2–4). Lewis et al. (5) suggest the existence of a “core” UMB but other studies note variations based on age and greater heterogeneity in bacterial genera among females, with Actinobacteria and Bacteroidetes being more prevalent (5–7). UMB in females shares species and features with the vaginal microbiome, forming a connected system distinct from the gut microbiome (8).
Dysbiosis of the UMB has been associated to various urological disorders, including benign conditions such as interstitial cystitis (9), urgency urinary incontinence (10) and overactive bladder (11), but it also has been associated to prostate cancer, especially in a recurrent antibiotic exposure-setting (12). Moreover, a case-control study showed that regular probiotic intake reduced the risk of bladder cancer in the healthy population (13, 14), suggesting a possible association between UB and bladder cancer. In fact, in the last few years, several studies of UMB have shown potential associations between dysbiosis of UMB and the development and persistence of urological cancers, similarly to what happens for gut microbiome (15–17).
The UMB can influence the host tissues in different ways. Bacteria that are present in the urine can reduce ingested nitrates into nitrites; the formation of endogenous N-nitrosamines in the bladder leads to the initiation of neoplastic events in the cells. The carcinogenic effects of these compounds are related to the ability of the reactive chemical species alkylating microscopic constituents of organs (18–20). Epithelial–mesenchymal transition (EMT) is a series of molecular mechanisms that promotes metastasis in several cancers by detachment from basement membrane, increasing cell mobility and decreasing cell–cell adhesion capabilities (21). EMT is vital in MIBC progression, as indicated by the upregulation of mesenchymal cell markers (N-cadherin and P-cadherin) and the downregulation of epithelial cell markers (E-cadherin) in MIBC tumors (22). The relationship between the local immune response and the microbiome is exemplified by chronic bacterial infections in the prostate, which are linked to reduced expression of the tumor suppressor protein NKX3.1. NKX3.1 regulates prostatic cell growth and DNA repair. Inflammatory cytokines TNF and IL-1β downregulate NKX3.1, increasing susceptibility to oxidative DNA damage. Loss of NKX3.1 (in mice) can lead to prostatic intraepithelial neoplasia and, in combination with Pten loss, prostate cancer. The cause of prostatic inflammation isn’t attributed to a specific organism but likely involves various species over time (23–26). Inizio modulo To date, the specific mechanism is not known, but there is no singular pathway for carcinogenesis. Instead, each mechanism contributes mutations and abnormalities to the cells, thereby promoting cancer progression.
Predictive tests based on UMB compositions have been proposed, especially involving 16S rRNA sequencing (11, 13), although they have limitations like the inability to detect bacteria at the species level or nonbacterial microorganisms such as viruses and fungi (17). Chipollini et al. (27) found enrichment in unique bacterial communities in invasive bladder cancer patients, suggesting potential for a microbiological signature in high-risk disease. Predictive tests could also help identify non-muscle invasive bladder cancer patients who could benefit from BCG immunotherapy (2). However, UMB signature studies require caution due to issues related to sample collection, biological sample management, and factors like age, menopausal status, sex hormones, and body mass index.
A literature review identified relevant studies on urinary microbiome and its association with urologic cancers, mainly bladder cancer, prostate cancer and renal cell carcinoma. PubMed was used as the database, and the collected studies formed the basis for a narrative analysis of the literature published in the last 5 years. We used the following keywords: urinary microbiome, prostate cancer, bladder cancer, renal cell carcinoma, penile cancer, testicular cancer.
Even though it is not fully demonstrated and understood, the main mechanisms by which microbiome could promote prostate cancer seem to be chronic systemic inflammation and immune modulation (28). Cancerous prostate tissue contains bacterial DNA, unlike healthy tissue. Microbial infection weakens the prostate’s natural defenses, causing epithelial disruption, loss of barrier function and persistent inflammation. Although no specific organism is identified as the main cause of prostate inflammation, the urinary tract is a potential source of microorganisms that may enter the prostate. In the last years some studies have tried to investigate which pathogens could be involved in the pathogenesis of prostate cancer. The studies published so far, differentiate for the type of tissue/fluid analyzed (Table 1). In one study conducted by Cavarretta et al. (33), the microbiome profile of 16 radical prostatectomy specimens was analyzed. Additionally, two separate studies by Shrestha (34) and Alanee (35) concentrated on differences in urine samples from patients with BPH and PCa. Another study conducted by Yu (36) examined the microbiome in urine samples and in samples of expressed prostatic fluid and seminal fluid obtained through prostatic massage, comparing men with BPH and prostate cancer.

Table 1 Summary of analyzed evidence about urinary microbiome and its association with prostate cancer.
In Cavarretta’s study (33), Staphylococcus Spp. were found in higher representation in pathological specimens. Conversely, in Shretsha’s work (34), the clustered group of bacteria species that proportionally had more cancer samples included Streptococcus anginosus, Anaerococcus lactolyticus, Anaerococcus obesiensis, Actinobaculum schaalii, Varibaculum cambriense, and Propionimicrobium lymphophilum. In Alanee’s study (35), the species Veillonella, Streptococcus, and Bacteroides were found to be more abundant. On the other hand, in Yu’s research, an increased presence of Bacteroidetes bacteria, Alphaproteobacteria, Firmicutes bacteria, Lachnospiraceae, Propionicimonas, Sphingomonas, and Ochrobactrum was observed in the PCa group compared to the BPH group (36).
When it comes to the most frequently identified pathogens, comparing these studies can be challenging, even though all of them have discovered significant differences between the group of individuals with prostate cancer (PCa) and the control group. This complexity arises because, as previously explained, the fluids and tissues analyzed in these studies vary. Despite the inherent limitations associated with different sample types (prostate specimens, urine, prostatic secretions), some bacteria, such as Streptococcus spp and Propionimicrobium, appear to recur in two of these research papers. Conducting additional studies that involve comparing samples from urine, prostate tissue, and secretions obtained from the same patient could provide further clarity regarding which uropathogens might be implicated in the pathogenesis of prostate cancer.
Table 2; Figure 1 summarize evidence about urinary microbiome and its association with prostate cancer.
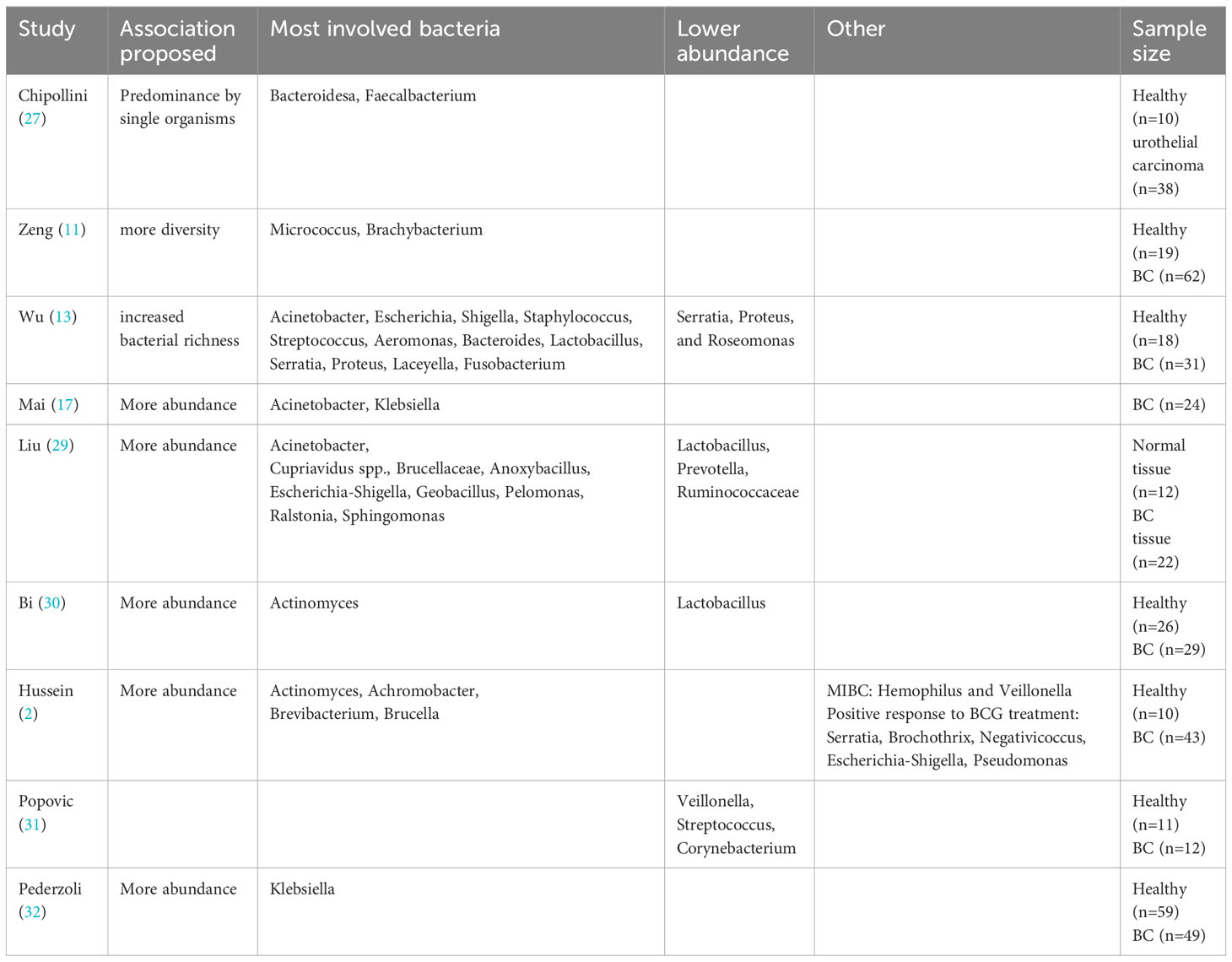
Table 2 Summary of analyzed evidence about urinary microbiome and its association with bladder cancer.
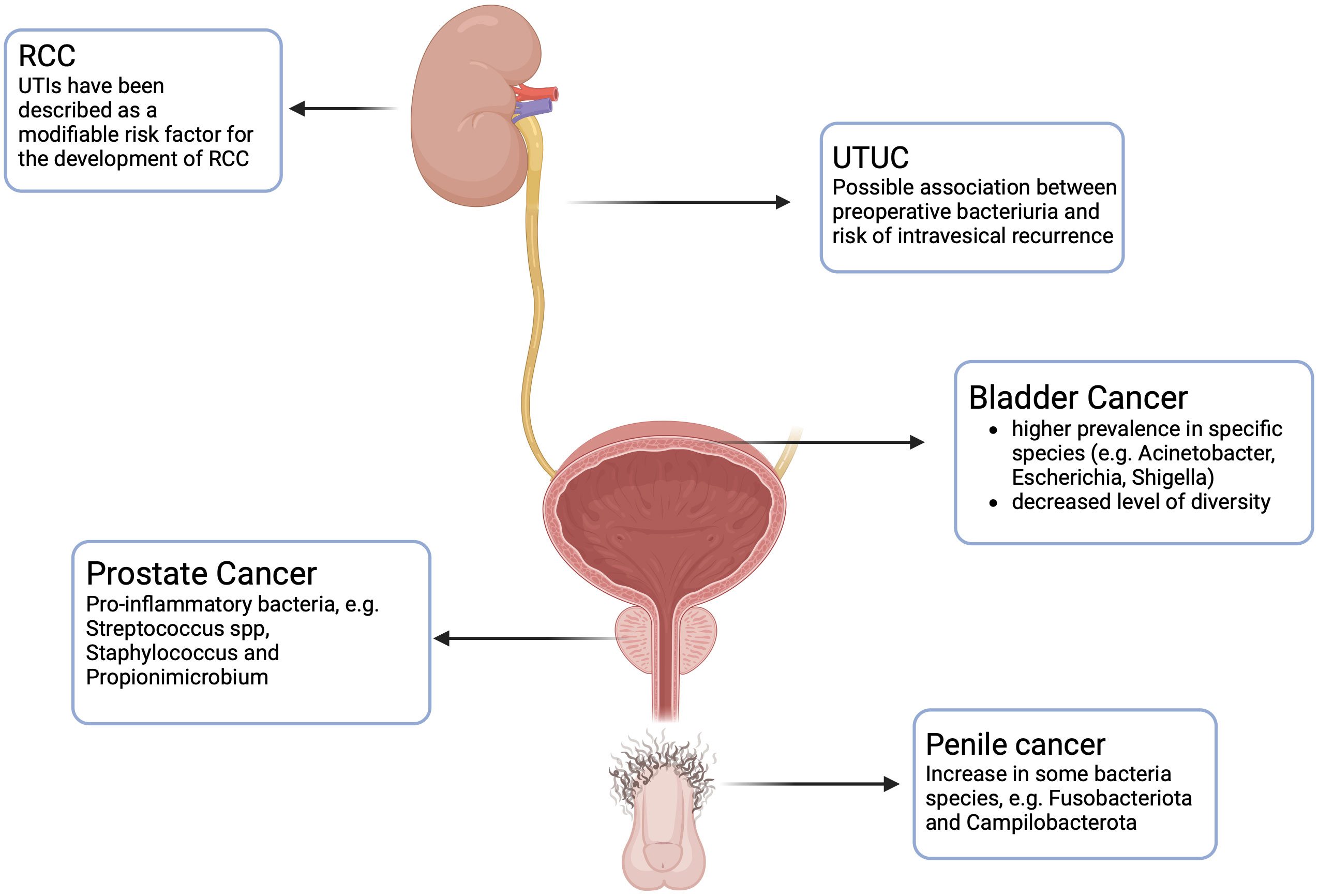
Figure 1 Urinary microbiome and its association with urologic cancers. RCC: Renal Cell Carcinoma. UTUC: Upper Urinary Tract Urothelial Cell Carcinoma. Created with BioRender.com.
The studies examining bladder cancer and its relationship with the UMB primarily suggest two main types of associations: either a higher prevalence in specific species or an elevated level of diversity.
Chipollini et al. (27) provide evidence of a reduction in both species’ richness and evenness in the urine of bladder cancer Patients, suggesting a higher probability of a dominant presence of an individual organism.
Acinetobacter is found to be more prevalent in patients with bladder cancer, as described in a study by Mai et al. (17). In a similar manner, Liu et al. (29) revealed higher relative abundances of Acinetobacter in cancerous compared to normal tissues, also with Cupriavidus spp., Brucellaceae, Anoxybacillus, Escherichia-Shigella, Geobacillus, Pelomonas, Ralstonia, and Sphingomonas. Lower relative abundances of the microbial genera Lactobacillus, Prevotella, as well as Ruminococcaceae was observed. Hussein et al. (2) found significant differences in beta-diversity, with Actinomyces, Achromobacter, Brevibacterium, and Brucella being significantly more abundant in urine samples from bladder cancer patients. These findings are partially consistent with those reported by Bi et al. (30), in which Actinomyces had a higher abundance in bladder cancer patients, being other four genera of bacteria more prevalent in healthy controls (Streptococcus, Bifidobacterium, Lactobacillus, Veillonella). Particularly notable was the higher prevalence of Lactobacillus in healthy individuals, a bacterium that has been shown to be a component of the microbiome and to confer protective effects against tumors in various organ systems, including gastrointestinal tumors (37) and gynecological tumors (38). Pederzoli et al. (32) found Klebsiella enrichment in urine of females with bladder cancer, similarly to a previous UMB study (17). Notably, Klebsiella’s colibactin toxin may cause direct DNA-strand damage, leading to genomic instability (39).
Zeng et al. (11) describe an increased bacterial richness index (Observed Species index, Chao1 index, Ace index; all P < 0.01) in cancer group compared to non-neoplastic group. Furthermore, in patients with NMIBC following TURBT, it was observed that the recurrence group displayed significantly greater alpha diversity when compared to the non-recurrence group. Additionally, higher alpha diversity was associated with a shorter time to recurrence.
Similarly, Wu et al. (13) noted elevated bacterial richness levels (including the Observed Species, Chao1, and Ace indexes) along with concurrent enrichment in certain bacterial genera (such as Acinetobacter, Anaerococcus, and Sphingobacterium), and a reduction in others (like Serratia, Proteus, and Roseomonas) when comparing the cancer group to the non-cancer group.
Pederzoli et al. showed that the UMB shares over 80% of the bacterial families found in the paired bladder tissue, indicating that the UMB can serve as a reliable representation of the tissue bacterial environment (32).
On the other hand, Mansour et al. (40) demonstrated a higher presence of certain species (Akkermansia, Bacteroides, Clostridium, Enterobacter and Klebsiella) in bladder tissues compared to the urine.
When comparing NMIBC and MIBC patients, Hussein et al. (2) discovered higher Hemophilus and Veillonella levels in MIBC patients’ urine, while Cupriavidus predominated in NMIBC patients. This aligns with Oresta et al. (41) study, where high-grade NMIBC and T2 tumor patients had more Veillonella in their urine samples and reduced Bifidobacterium and Ruminococcus 1, both of which have anti-inflammatory roles in mucosal homeostasis (42, 43). In contrast, Popovic et al.’s study (31) found Veillonella, along with Streptococcus and Corynebacterium, as the most common bacteria in healthy individuals.
The concept that certain bacteria may offer protection against cancer is notably apparent in urinary bladder cancer. This is unique as it is the only cancer treated with live microorganisms, specifically Mycobacterium bovis - Bacillus Calmette-Guérin (BCG) (44). BCG is believed to function by binding to fibronectin and integrin α5β1, subsequently triggering an immune response (45, 46). It is conceivable that, similarly to BCG, specific commensal bacteria naturally inhabiting a healthy bladder may serve in tumor surveillance or confer different beneficial effects (31). Additionally, the microbiome was proposed to influence responses to adjuvant BCG therapy and systemic immunochemotherapy in individuals with high-risk or advanced bladder cancer cases (2, 47).
Table 1; Figure 1 summarize evidence about urinary microbiome and its association with bladder cancer.
The evidence regarding the association between UTUC and urinary microbiome is still very limited, and there are few studies on the topic. Fukushima et al. (48) in their study investigated the effect of perioperative bacteriuria and pyuria on intravesical recurrences in patients with UTUC undergoing radical nephroureterectomy and they found that bacteriuria and pyuria independently predicted a decreased risk of intravesical recurrences (Figure 1). Since serial cystoscopies for follow-up are costly and create discomfort for the patient, being able to stratify patients based on preoperative parameters can be useful in understanding which patients are at low risk of recurrence, thus avoiding such stringent follow-up. A different result was obtained by Liang et al., who instead demonstrated that preoperative pyuria among UTUC patients undergoing radical nephroureterectomy was significantly associated with advanced pathological tumor stage and worse survival (49).
The association between local and systemic inflammation and cancer is still controversial. While certain inflammatory and immune responses exhibit anti-tumor activity, inflammation itself can also promote cancer progression. A heightened preoperative CRP level is indicative of a reduced survival and worst prognosis for patients with UTUC (50). Unlike the studies of bladder urothelial cancer, there is no literature regarding the abundance or specific differences of urinary microbiome in patients with upper tract urothelial cancer compared to controls.
The role of UB in the development of renal cell carcinoma (RCC) is still a debate. An association between prior UTIs and RCC is still unclear, even if UTIs have been described as a modifiable risk factor for the development of RCC (51) (Figure 1). Further studies are necessary to clarify the presence of UB in the kidney tissue and its role in the development of RCC.
A recent study conducted by De Deus et al. (52) endeavored to delineate the presence of a microbiome in penile carcinoma tissue. As in other studies previously mentioned, the 16S rRNA was analyzed in both tumor and non-tumor adjacent tissues to assess the presence of different pathogens. They found that Fusobacteriota and Campilobacterota were the two species significantly increased in tumors compared to non-tumor tissues (Figure 1).
Furthermore, how penile microbiome can modulate immune response is already well known in other circumstances, as reported by Onywera et al. (53). In fact, changes in the microbiota after circumcision can lead to altered susceptibility to HIV and HPV infection.
These studies represent a starting point to explore the role of microbiome in penile cancer. However, further works are required to elucidate the potential role of microbiome in the pathogenesis of this condition and its implications for developing prevention strategies and treatment modalities.
The concept of the urinary microbiome is a recent development with potential applications in urologic tumor diagnosis, risk assessment, and treatment. While our analysis of several studies has yielded conflicting outcomes in some instances and inconclusive findings in others, available evidence indicates that certain bacteria may actively contribute to the initiation and progression of tumors. Moreover, these bacteria may also have a role in influencing the response to therapy through immunomodulation. Additional research is required to comprehensively define the characteristics of a healthy urinary microbiome, identify dysbiosis, and understand its potential impact on tumorigenesis and the host’s response.
GRa: Writing – original draft, Writing – review & editing. EB: Writing – original draft, Writing – review & editing. TC: Writing – original draft. GRe: Writing – original draft. GB: Writing – review & editing. GN: Writing – review & editing. MI: Writing – review & editing. AM: Writing – review & editing. FM: Writing – review & editing. FZ: Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Whiteside SA, Razvi H, Dave S, Reid G, Burton JP. The microbiome of the urinary tract–a role beyond infection. Nat Rev Urol. (2015) 12:81–90. doi: 10.1038/nrurol.2014.361
2. Hussein AA, Elsayed AS, Durrani M, Jing Z, Iqbal U, Gomez EC, et al. Investigating the association between the urinary microbiome and bladder cancer: An exploratory study. Urologic Oncol. (2021) 39:370.e9–370.e19. doi: 10.1016/J.UROLONC.2020.12.011
3. Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, et al. Conducting a microbiome study. Cell. (2014) 158:250–62. doi: 10.1016/J.CELL.2014.06.037
4. Markowski MC, Boorjian SA, Burton JP, Hahn NM, Ingersoll MA, Vareki SM, et al. The microbiome and genitourinary cancer: A collaborative review. Eur Urol. (2019) 75:637–46. doi: 10.1016/J.EURURO.2018.12.043
5. Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi JR, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol. (2013) 4:41/BIBTEX. doi: 10.3389/FCIMB.2013.00041/BIBTEX
6. Dong Q, Nelson DE, Toh E, Diao L, Gao X, Fortenberry JD, et al. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PloS One. (2011) 6:e19709. doi: 10.1371/JOURNAL.PONE.0019709
7. Nelson DE, van der Pol B, Dong Q, Revanna KV, Fan B, Easwaran S, et al. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PloS One. (2010) 5:e14116. doi: 10.1371/JOURNAL.PONE.0014116
8. Thomas-White K, Forster SC, Kumar N, Van Kuiken M, Putonti C, Stares MD, et al. Culturing of female bladder bacteria reveals an interconnected urogenital microbiota. Nat Commun. (2018) 9:1557. doi: 10.1038/s41467-018-03968-5
9. Siddiqui H, Lagesen K, Nederbragt AJ, Jeansson SL, Jakobsen KS. Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol. (2012) 12. doi: 10.1186/1471-2180-12-205
10. Pearce MM, Zilliox MJ, Rosenfeld AB, Thomas-White KJ, Richter HE, Nager CW, et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol. (2015) 213:347.e1–347.e11. doi: 10.1016/J.AJOG.2015.07.009
11. Zeng J, Zhang G, Chen C, Li K, Wen Y, Zhao J, et al. Alterations in urobiome in patients with bladder cancer and implications for clinical outcome: A single-institution study. Front Cell Infect Microbiol. (2020) 10:555508. doi: 10.3389/FCIMB.2020.555508
12. Boursi B, Mamtani R, Haynes K, Yang YX. Recurrent antibiotic exposure may promote cancer formation–another step in understanding the role of the human microbiota? Eur J Cancer. (2015) 51:2655–64. doi: 10.1016/J.EJCA.2015.08.015
13. Wu P, Zhang G, Zhao J, Chen J, Chen Y, Huang W, et al. Profiling the urinary microbiota in male patients with bladder cancer in china. Front Cell Infect Microbiol. (2018) 8:167/BIBTEX. doi: 10.3389/FCIMB.2018.00167/BIBTEX
14. Ohashi Y, Nakai S, Tsukamoto T, Masumori N, Akaza H, Miyanaga N, et al. Habitual intake of lactic acid bacteria and risk reduction of bladder cancer. Urol Int. (2002) 68:273–80. doi: 10.1159/000058450
15. Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reida G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. (2016) 82:5039–48. doi: 10.1128/AEM.01235-16
16. Irrazábal T, Belcheva A, Girardin SE, Martin A, Philpott DJ. The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell. (2014) 54:309–20. doi: 10.1016/J.MOLCEL.2014.03.039
17. Mai G, Chen L, Li R, Liu Q, Zhang H, Ma Y. Common core bacterial biomarkers of bladder cancer based on multiple datasets. BioMed Res Int. (2019) 2019. doi: 10.1155/2019/4824909
18. Tricker AR, Stickler DJ, Chawla JC, Preussmann R. Increased urinary nitrosamine excretion in paraplegic patients. Carcinogenesis. (1991) 12:943–6. doi: 10.1093/carcin/12.5.943
19. Calmels S, Ohshima H, Bartsch H. Nitrosamine formation by denitrifying and non-denitrifying bacteria: Implication of nitrite reductase and nitrate reductase in nitrosation catalysis. Microbiol (NY). (1988) 134:221–6. doi: 10.1099/00221287-134-1-221
20. Hicks RM, Ismail MM, Walters CL, Beecham PT, Rabie MF, El Alamy MA. Association of bacteriuria and urinary nitrosamine formation with schistosoma haematobium infection in the qalyub area of egypt. Trans R Soc Trop Med Hyg. (1982) 76:519–27. doi: 10.1016/0035-9203(82)90153-5
21. Ogawa K, Shimizu Y, Uketa S, Utsunomiya N, Kanamaru S. Prognosis of patients with muscle invasive bladder cancer who are intolerable to receive any anti-cancer treatment. Cancer Treat Res Commun. (2020) 24. doi: 10.1016/J.CTARC.2020.100195
22. Franzen CA, Blackwell RH, Todorovic V, Greco KA, Foreman KE, Flanigan RC, et al. Urothelial cells undergo epithelial-to-mesenchymal transition after exposure to muscle invasive bladder cancer exosomes. Oncogenesis. (2015) 4. doi: 10.1038/ONCSIS.2015.21
23. Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-Function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res. (2005) 65:6773–9. doi: 10.1158/0008-5472.CAN-05-1948
24. Markowski MC, Bowen C, Gelmann EP. Inflammatory cytokines induce phosphorylation and ubiquitination of prostate suppressor protein NKX3.1. Cancer Res. (2008) 68:6896–901. doi: 10.1158/0008-5472.CAN-08-0578
25. Bowen C, Gelmann EP. NKX3.1 activates cellular response to DNA damage. Cancer Res. (2010) 70:3089–97. doi: 10.1158/0008-5472.CAN-09-3138
26. Khalili M, Mutton LN, Gurel B, Hicks JL, De Marzo AM, Bieberich CJ. Loss of Nkx3.1 expression in bacterial prostatitis. Am J Pathol. (2010) 176:2259–68. doi: 10.2353/ajpath.2010.080747
27. Chipollini J, Wright JR, Nwanosike H, Kepler CY, Batai K, Lee BR, et al. Characterization of urinary microbiome in patients with bladder cancer: Results from a single-institution, feasibility study. Urologic Oncology: Semin Original Investigations. (2020) 38:615–21. doi: 10.1016/J.UROLONC.2020.04.014
28. Katongole P, Sande OJ, Joloba M, Reynolds SJ, Niyonzima N. The human microbiome and its link in prostate cancer risk and pathogenesis. Infect Agent Cancer. (2020) 15:53. doi: 10.1186/s13027-020-00319-2
29. Cavarretta I, Ferrarese R, Cazzaniga W, Saita D, Lucianò R, Ceresola ER, et al. The microbiome of the prostate tumor microenvironment. Eur Urol. (2017) 72:625–31. doi: 10.1016/j.eururo.2017.03.029
30. Shrestha E, White JR, Yu SH, Kulac I, Ertunc O, De Marzo AM, et al. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J Urol. (2018) 199:161–71. doi: 10.1016/J.JURO.2017.08.001
31. Alanee S, El-Zawahry A, Dynda D, Dabaja A, McVary K, Karr M, et al. A prospective study to examine the association of the urinary and fecal microbiota with prostate cancer diagnosis after transrectal biopsy of the prostate using 16sRNA gene analysis. Prostate. (2019) 79:81–7. doi: 10.1002/PROS.23713
32. Yu H, Meng H, Zhou F, Ni X, Shen S, Das UN. Urinary microbiota in patients with prostate cancer and benign prostatic hyperplasia. Arch Med Sci. (2015) 11:385–94. doi: 10.5114/AOMS.2015.50970
33. Liu F, Liu A, Lu X, Zhang Z, Xue Y, Xu J, et al. Dysbiosis signatures of the microbial profile in tissue from bladder cancer. Cancer Med. (2019) 8:6904–14. doi: 10.1002/cam4.2419
34. Bi H, Tian Y, Song C, Li J, Liu T, Chen Z, et al. Urinary microbiota - a potential biomarker and therapeutic target for bladder cancer. J Med Microbiol. (2021) 68:1471–8. doi: 10.1099/JMM.0.001058/CITE/REFWORKS
35. Eslami M, Yousefi B, Kokhaei P, Hemati M, Nejad ZR, Arabkari V, et al. Importance of probiotics in the prevention and treatment of colorectal cancer. J Cell Physiol. (2019) 234:17127–43. doi: 10.1002/jcp.28473
36. Łaniewski P, Ilhan ZE, Herbst-Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol. (2020) 17:232–50. doi: 10.1038/s41585-020-0286-z
37. Pederzoli F, Ferrarese R, Amato V, Locatelli I, Alchera E, Lucianò R, et al. Sex-specific alterations in the urinary and tissue microbiome in therapy-naïve urothelial bladder cancer patients. Eur Urol Oncol. (2020) 3:784–8. doi: 10.1016/J.EUO.2020.04.002
38. Kaur CP, Vadivelu J, Chandramathi S. Impact of klebsiella pneumoniae in lower gastrointestinal tract diseases. J Dig Dis. (2018) 19:262–71. doi: 10.1111/1751-2980.12595
39. Mansour B, Monyók Á, Makra N, Gajdács M, Vadnay I, Ligeti B, et al. Bladder cancer-related microbiota: examining differences in urine and tissue samples. Sci Rep. (2020) 10:11042. doi: 10.1038/s41598-020-67443-2
40. Oresta B, Braga D, Lazzeri M, Frego N, Saita A, Faccani C, et al. The microbiome of catheter collected urine in males with bladder cancer according to disease stage. J Urol. (2021) 205:86–93. doi: 10.1097/JU.0000000000001336
41. Burrello C, Garavaglia F, Cribiù FM, Ercoli G, Lopez G, Troisi J, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. (2018) 9. doi: 10.1038/S41467-018-07359-8
42. Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J Immunol. (2005) 174:3237–46. doi: 10.4049/JIMMUNOL.174.6.3237
43. Bučević Popović V, Šitum M, Chow C-ET, Chan LS, Roje B, Terzić J. The urinary microbiome associated with bladder cancer. Sci Rep. (2018) 8:12157. doi: 10.1038/s41598-018-29054-w
45. Bao Y, Al KF, Chanyi RM, Whiteside S, Dewar M, Razvi H, et al. Questions and challenges associated with studying the microbiome of the urinary tract. Ann Transl Med. (2017) 5. doi: 10.21037/ATM.2016.12.14
46. Redelman-Sidi G, Glickman MS, Bochner BH. The mechanism of action of BCG therapy for bladder cancer–a current perspective. Nat Rev Urol. (2014) 11:153–62. doi: 10.1038/NRUROL.2014.15
47. Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science. (2018) 359:1366–70. doi: 10.1126/SCIENCE.AAR6918
48. Fukushima H, Kobayashi M, Kawano K, Morimoto S. Effect of preoperative bacteriuria and pyuria on intravesical recurrence in patients with upper tract urothelial carcinoma undergoing radical nephroureterectomy. In Vivo. (2017) 31:1215–20. doi: 10.21873/INVIVO.11193
49. Liang C, Wang J, Liu H, Huang L, Xu D, Qian S, et al. Preoperative pyuria predicts advanced pathologic tumor stage and worse survival in patients with urothelial carcinoma of the upper urinary tract treated by radical nephroureterectomy. Urol Oncol. (2016) 34:418.e1–7. doi: 10.1016/J.UROLONC.2016.04.011
50. Saito K, Kawakami S, Ohtsuka Y, Fujii Y, Masuda H, Kumagai J, et al. The impact of preoperative serum c-reactive protein on the prognosis of patients with upper urinary tract urothelial carcinoma treated surgically. BJU Int. (2007) 100:269–73. doi: 10.1111/J.1464-410X.2007.06934.X
51. Dhote R, Thiounn N, Debré B, Vidal-Trecan G. Risk factors for adult renal cell carcinoma. Urologic Clinics North America. (2004) 31:237–47. doi: 10.1016/J.UCL.2004.01.004
52. de Deus A, Gonçalves G, da Silva J, de Jesus LC, Azevedo-Santos APS, Dall Agnol H, et al. Microbiome reveals inflammatory-related bacteria and putative functional pathways involved in human papillomavirus-associated penile squamous cell carcinoma. Andrology. (2023). doi: 10.1111/ANDR.13545
Keywords: urinary microbiome, bladder cancer, prostate cancer, renal cell carcinoma, UTUC (renal pelvis and ureter), microbiome and dysbiosis, BCG (Bacille Calmette-Guérin)
Citation: Randazzo G, Bovolenta E, Ceccato T, Reitano G, Betto G, Novara G, Iafrate M, Morlacco A, Dal Moro F and Zattoni F (2024) Urinary microbiome and urological cancers: a mini review. Front. Urol. 4:1367720. doi: 10.3389/fruro.2024.1367720
Received: 09 January 2024; Accepted: 13 February 2024;
Published: 08 March 2024.
Edited by:
Riccardo Campi, Careggi Hospital, ItalyReviewed by:
Alessio Pecoraro, University of Florence, ItalyCopyright © 2024 Randazzo, Bovolenta, Ceccato, Reitano, Betto, Novara, Iafrate, Morlacco, Dal Moro and Zattoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabio Zattoni, ZmFiaW8uemF0dG9uaUB1bmlwZC5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.