
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Urol., 26 July 2023
Sec. Female Urology
Volume 3 - 2023 | https://doi.org/10.3389/fruro.2023.1212590
This article is part of the Research TopicAn Outlook on Urobiome: Advances in Understanding the Role of Urobiome in Urological Health and Disease and its Potential in BiotherapeuticsView all 5 articles
 Robert B. Moreland1
Robert B. Moreland1 Brian I. Choi1
Brian I. Choi1 Wilson Geaman1
Wilson Geaman1 Caroline Gonzalez1
Caroline Gonzalez1 Baylie R. Hochstedler-Kramer1
Baylie R. Hochstedler-Kramer1 Jerrin John1
Jerrin John1 Jacob Kaindl1
Jacob Kaindl1 Nikita Kesav1
Nikita Kesav1 Jyoti Lamichhane1
Jyoti Lamichhane1 Luke Lucio1
Luke Lucio1 Malika Saxena1
Malika Saxena1 Aditi Sharma1
Aditi Sharma1 Lana Tinawi1
Lana Tinawi1 Michael E. Vanek1
Michael E. Vanek1 Catherine Putonti2,3
Catherine Putonti2,3 Linda Brubaker4
Linda Brubaker4 Alan J. Wolfe1*
Alan J. Wolfe1*The advent of sensitive enhanced culture (metaculturomic) and culture-independent DNA-based (metagenomic) methods has revealed a rich collection of microbial species that inhabit the human urinary tract. Known as the urinary microbiome, this community of microbes consists of hundreds of distinct species that range across the entire phylogenetic spectrum. This new knowledge clashes with standard clinical microbiology laboratory methods, established more than 60 years ago, that focus attention on a relatively small subset of universally acknowledged uropathogens. Increasing reports support the hypothesis that this focus is too narrow. Single uropathogen reports are common in women with recurrent urinary tract infection (UTI), although wider disruption of their urinary microbiome is likely. Typical “UTI” symptoms occur in patients with “no growth” reported from standard culture and sometimes antibiotics improve these symptoms. Metaculturomic and metagenomic methods have repeatedly detected fastidious, slow growing, and/or anaerobic microbes that are not detected by the standard test in urine samples of patients with lower urinary tract symptoms. Many of these microbes are also detected in serious non-urinary tract infections, providing evidence that they can be opportunistic pathogens. In this review, we present a set of poorly understood, emerging, and suspected uropathogens. The goal is to stimulate research into the biology of these microbes with a focus on their life as commensals and their transition into pathogens
More than a decade ago, reports began surfacing that challenged the prevailing dogma that urine was typically sterile in the absence of infection (1–10). These studies used high-throughput DNA sequencing (metagenomics) and/or enhanced culture methods (metaculturomics) coupled with matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectroscopy (MS) to detect and identify bacteria in urine samples obtained from diverse sets of study participants. These modern sensitive detection methods documented the presence of microbes in urines deemed “no growth” by the traditional or standard urine culture methodologies used by most clinical microbiological laboratories and highlighted the presence of microbes not typically acknowledged as uropathogens (11, 12). These studies and others have resulted in a list of hundreds of taxa. A few taxa are prevalent in individuals without lower urinary tract symptoms. Many more taxa are present in asymptomatic individuals but are more prevalent in those with symptoms (Table 1, Appendix 1), including those typically associated with urinary tract infection (UTI) and urgency urinary incontinence (UUI), among others (9, 10, 13–21). For a recent review, see (22).
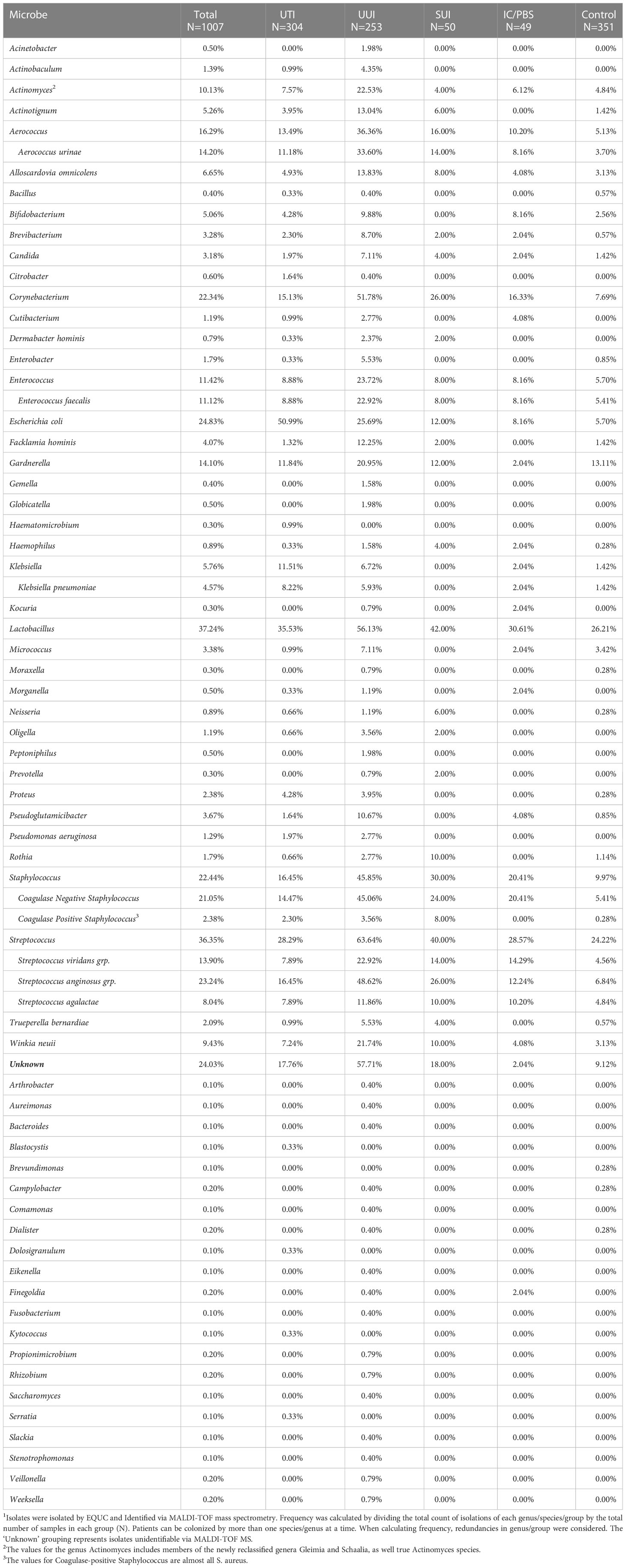
Table 1 Frequency of Microbe Identification via Metaculturomics in Patients with and without LUTS1.
The purpose of this review is to highlight a set of poorly understood, emerging, and suspected uropathogens. The intent is to generate momentum for prospective and retrospective studies to identify risk factors and improve antibiotic surveillance, especially for those species that have no Clinical and Laboratory Standards Institute (CLSI) standards. We also wish to encourage investigations into the pathophysiology of these microbes. Thus, with a few exceptions, this review will focus on these lesser-known microbes, including members of the families Aerococcaceae, Actinomycetaceae, and Bifidobacteriaceae. Also discussed will be members of the Streptococcus anginosus group (SAG) and Enterococcus faecalis. While SAG members have long been considered to be commensals, increasing evidence supports the conclusion that they are more likely opportunistic pathogens (23). Although long accepted as a pathogen, the comparatively well-studied E. faecalis has become increasingly implicated in urinary tract disorders and its pathophysiology within the urinary tract remains understudied (24).
Other species we will review are anaerobes, specifically members of the orders Eubacteriales and Bacteroidales. Traditionally, anaerobes have not been considered to be uropathogenic (25, 26). However, in this age of metagenomics, metaculturomics, and MALDI-TOF identification, this dogma is being reexamined (27). The concept that oxygen is toxic to obligate anaerobes (28) does not account for the strategies these microbes use to survive and flourish in human organ niches (29, 30), including the urinary tract.
Of organisms reviewed here, some are aerobes, some are facultative anaerobes, and some are strict anaerobes. Many are fastidious. As such, classical clinical laboratory diagnosis using standard urine culture (SUC) methods would not detect most of these potential uropathogens in the time frame or atmospheric conditions of the assay (12, 31). In contrast, all taxa reviewed herein have been detected by metagenomic approaches, including 16S rRNA gene sequencing and shotgun metagenomic sequencing (9, 10, 14, 15, 18, 19, 32–34), and/or metaculturomic methods, such as Expanded Quantitative Urine Culture (8, 12, 21) (Table 1). Despite the discovery of some of these microbes as much as a century ago, little is known about their biology.
Since much has been written about the most commonly accepted and best studied uropathogens, including members of the family Enterobacteriaceae (e.g., the genera Escherichia, Klebsiella, Enterobacter, and Proteus), and Gram-negative saprophytes (Pseudomonas aeruginosa and Acinetobacter baumannii), we will discuss them only briefly (35). The same is true for the better-known member(s) of the streptococci (S. agalactiae), the coagulase-positive staphylococi (S. aureus), and the coagulase-negative staphyococci (S. saprophyticus, S. epidermidis, and S. haemolyticus), as well as the yeast genus Candida (C. albicans). We will not review the best-known anaerobes, including but not limited to the genera Porphyromonas, Sneathia, and Peptoniphilus. Finally, some urinary microbes have no cell wall, most notably Mycoplasma and Ureaplasma (36). While specialized techniques exist for the culture of these microbes (27, 37, 38) and rapid molecular diagnoses have been reported for both (39), we will not review these organisms here. For recent reviews on the taxa mentioned above, see (24, 40–46).
The standard approach to treating UTI is based on Koch’s postulates, which assumes a single organism is responsible for pathogenicity, that this organism can be isolated from the diseased tissue/fluid, be able to reproduce the disease state in a healthy experimental system and be recovered afterward in pure culture (47). This highly successful approach was responsible for the elucidation of the bacterial pathogens of nineteen different diseases from 1877 to 1906 including anthrax, bubonic plague, cholera, diphtheria, pediatric diarrhea, bacterial pneumonia, gonorrhea, syphilis, tuberculosis, typhoid fever, and whooping cough (47). However, the discovery of the human microbiome and existence of eubiotic states within human tissues such as the dermis, and the respiratory, gastrointestinal, and urogenital tracts caused a rethinking of the roles played by bacteria in health and disease (48, 49).
An initial cataloging of human urinary bladder isolates revealed 149 distinct species ranging from aerobes and microaerobes to facultative anaerobes and anaerobes (50). While all these microbes can be identified by DNA-dependent methods, most are not culturable or grow poorly under standard urine culture conditions, while others overgrow because they possess adaptive advantages (12). This is particularly true for facultative anaerobes and is consistent with the genera Escherichia, Pseudomonas, Klebsiella, Proteus, Staphylococcus, and Enterococcus being among “The Usual Suspects” and common to standard urine culture diagnoses (29, 51). Microbes within the microbiome can be grouped into six different classes: non-pathogen (not causing disease), a pathogen (causing disease), a commensal (tissue resident, benefiting the host) a symbiont (tissue resident, benefiting the host and is benefited from the host), a colonizer (tissue resident and may or may not be disease causing) and a pathobiont (tissue resident, generally beneficial but can cause disease under special conditions) (52).
To understand microbial communities, one must first isolate and characterize each of the individual species. Establishing the commensal status of a species is much more difficult than reporting pathogen case reports in tissues. Consequently, many of the reports in the literature regarding the species reviewed here are pathological reports from abscesses, blood cultures, or other disease states. The science of understanding the interactions in a microbial community between the six types of microbes mentioned above is in its infancy (52, 53). While reports of infections in tissues other than the urinary tract provide only a worst-case capability of the capacity of these species to cause or contribute to disease, it emphasizes the major thrust of this review - to study these species in context and to begin to understand their interaction within communities.
Approximately 150 million people worldwide are diagnosed each year with UTIs (54). These infections are thought to be caused by uropathogenic bacteria, including but not limited to members of the genera Escherichia, Pseudomonas, Klebsiella, Proteus, Staphylococcus, and Enterococcus (51). Note that many of these species are in the World Health Organization’s ESKAPE list of critical pathogens (55). Escherichia coli is considered to be the most common cause of UTIs. Other bacterial species that are commonly associated with UTI-like symptoms include Pseudomonas aeruginosa, Klebsiella pneumoniae, K. oxytoca, Enterococcus faecalis, E. faecium, Proteus mirabilis, Proteus vulgaris, Staphylococcus aureus, and S. saprophyticus. The yeast species Candida albicans also can cause UTI-like symptoms (56, 57). For example, a study of 727 hospitalized urological patients diagnosed with nosocomial acquired UTI reported the most commonly pathogens detected by SUC to be E. coli (31%), followed by species of the genera Pseudomonas (13%), Enterococcus (10%), Klebsiella (10%), Enterobacter (6%) and Proteus (6%) (58). These taxa are all fast growing, non-fastidious, and able to thrive in the presence of ambient oxygen (PO2 150mmHg, 20kPa) (29, 59), characteristics that facilitate detection by SUC (60). Other microbes that are easily detected by SUC include additional members of the Gram-negative family Enterobacteriaceae, such as the genera Serratia, Citrobacter, Morganella, Providencia, and Pantoea. All have the capacity to be pathogenic, but these genera are detected quite rarely. For example, in a re-examination of several of our previous studies (21), they were each detected in the catheterized (bladder) urine of less than 0.1% of adult females (n=1007) and were rare even in those with UTI-like symptoms (Table 1).
Saprophytes are organisms that obtain their nutrients from decaying organic material. As such, they tend not to be obligate infectious agents of humans. However, they can be opportunistic pathogens, causing wound and nosocomial infections, primarily in immunocompromised individuals. A recent systematic review found saprophytic bacteria to be implicated in hundreds of infections in dozens of countries (42); 5% were UTIs. Most affected individuals had comorbidities and the most common species detected were Pantoea aglomerans, Klebsiella (formerly Enterobacter) aerogenes, and Pseudomonas putida. The authors warn that saprophytes such as these may become more common in healthcare settings like other opportunistic environmental Gram-negative bacteria, especially Acinetobacter baumannii and P. aeruginosa (42).
A. baumannii and P. aeruginosa may cause nosocomial infections, including nosocomial-acquired UTIs, especially in frail or immunocompromised individuals (61). The World Health Organization considers both priority-1 (critical) pathogens because of their tendency to be resistant to carbapenems and third generation cephalosporins, which are considered to be last resort antibiotics (55, 62). Multi-drug resistance and their biofilm-forming capacity makes these infections difficult to treat with antibiotic therapy (63). Whereas efforts to understand P. aeruginosa and A. baumannii pathophysiology have been extensive, uropathogenic strains remain understudied (42, 63–67).
Fungal UTIs are generally caused by members of the genus Candida (68). Of these, the best known and most common UTI-associated species is C. albicans. Other species include C. glabrata, C. parapsilosis, and C. auris. The latter is an emerging pathogen associated with UTIs that the CDC has added to its surveillance list because it tends to be multidrug resistant, is difficult to detect using standard clinical laboratory methodology, and has caused multiple outbreaks in healthcare settings (69–71). Diabetes, catheterization, hospitalization, and broad-spectrum antibiotics are risk factors for Candida infections (72). Azole antifungals are the most common treatment for symptomatic infections; however, increasing resistance has been observed in clinical isolates. Wider surveillance studies are severely needed (73).
The diagnostic criteria for detecting Candida in urine samples are not standardized with continuing debate about reporting thresholds (74, 75). More problematically, typical clinical laboratory methods of detection have poor sensitivity for Candida species, even C. albicans. Several prospective studies that cultured urine on the standard fungal medium, Sabouraud dextrose agar, have reported greater numbers of non-C. albicans species than standard urine culture methods (75–77). Thus, Candida species are often not detected by standard clinical laboratory testing and consequently are underreported.
The genus Staphylococcus is comprised of more than 40 species of Gram-positive, facultative anaerobic cocci (78–80). From a clinical microbiological diagnostic point, the genus can be divided by coagulase activity (conversion of fibrinogen to fibrin). The most common coagulase-positive Staphylococcus is S. aureus, a commensal skin and upper respiratory tract coccus known to be a potent, antibiotic-resistant opportunistic pathogen that can cause diverse infections, especially skin and soft tissue infections and toxic shock syndrome (80, 81). As such, surveillance for this uropathogen is high. In our re-examination of isolates obtained from catheterized bladder urine samples of ~1000 adult females (21), S. aureus, the most common coagulase-positive Staphylococcus, was detected in urine but it was not prevalent (Table 1).
In contrast, coagulase-negative staphylococci (CoNS) are often dismissed as contaminants (80, 82–84). As opportunistic pathogens in the urinary tract, CoNS are associated with UTIs, uncomplicated, catheter-associated, and nosocomial (82, 85–88). The ability for this genus to acquire antibiotic resistance makes this group of microbes an increasing threat to infectious disease control (81). We found them to be quite prevalent, especially in adult females diagnosed with UUI (Table 1). Of the 11 CoNS species detected, here we review the 2 most prevalent species and 1 species commonly associated with UTI (S. epidermidis, S. haemolyticus and S. saprophyticus, respectively).
S. saprophyticus was first recognized as a causative microbe for UTI in young females (82, 86), and does appear to be associated with young females of reproductive age. In contrast, it appears to be very rare in older females; we have never detected it in this population. However, the susceptibility of the host by age and reproductive status remains unclear. Virulence factors, including urease activity, have been described (82, 83, 89).
The most common CoNS in the urinary tract is S. epidermidis (41, 87). Whereas S. epidermidis infections are rarely life-threatening, increasing antibiotic resistance and biofilm-forming ability make them difficult to treat with antibiotics. Investigations into the underlying molecular mechanisms have been performed (87). Numerous case reports implicate S. epidermidis in UTIs, especially in children (90–92), but the pathophysiology of urinary isolates has yet to be explored.
S. haemolyticus is the second-most isolated CoNS from urine. It is also common in blood cultures, especially from immunocompromised patients. As such, it is considered an emerging multidrug-resistant nosocomial pathogen (83, 84). Of particular concern is the ability of S. haemolyticus to acquire multiple antibiotic resistance genes, making antibiotic stewardship in the global treatment of UTIs an urgent public health issue (84, 93). The incidence of S. haemolyticus UTIs are increasingly reported (84, 88).
In contrast to several of the universally acknowledged uropathogens, including but not limited to Serratia, Morganella, Citrobacter, and E. faecium, many emerging or suspected uropathogens are considerably more prevalent (Table 1). They have been underappreciated for 2 major reasons. First, as mentioned above, many simply do not grow or grow poorly under SUC conditions (60); however, they can be grown (Appendix 2). Even E. faecalis tends to be underreported, in part due to overgrowth of faster growing species (31). Second, before the advent of MALDI-TOF MS, accurate identification of many species was difficult (94) and many would have been dismissed as contaminants (95–97). This dismissal has its consequences as was suggested by investigators studying polymicrobial infections in urinary sepsis where contamination could be ruled out (98, 99). The growth of microbes at less than 105 colony forming units per milliliter (cfu/mL) has been noted and its significance debated since the initial report of this standard for “infection” (100, 101). On the opposite end of the spectrum and demonstrating that some microbes that do not grow on SUC, negative standard urine cultures was reported in women with lower urinary tract symptoms; with treatment, negative cultures and the symptoms persisted (102), implying that some other causal factor and/or uncultivated microbe was present.
Because its role in lower urinary tract health has been underappreciated, we will review E. faecalis first and then a set of emerging and suspected uropathogens.
Less than 30 years after being recognized as a distinct taxon, the clinical outlook on Enterococcus transitioned from harmless gut commensal to a major public health concern. E. faecalis, the most common clinical enterococcal species, is ubiquitously present in the human gut microbiome where it plays a crucial role in nutrient metabolism and maintenance of a heathy gut environment (103, 104). However, these microbes are also adept at adapting to novel environments and transferring DNA to members of its own genus, as well as other taxa. This latter characteristic has greatly contributed to the worldwide spread of antibiotic resistance, the most notable being the cassette of genes responsible for vancomycin resistance, which is attributed to significantly increased mortality rates (105).
With or without antibiotic resistance genes, enterococcal infections at many body sites exhibit increased risk of persistence and recurrence in comparison to other common pathogens (106). Mechanisms underlying these chronic infection phenotypes are largely unknown, as previous comparative phylogenomic studies have been unable to differentiate between clinical isolates from diverse infection types (107), most likely due to insufficient isolate numbers and metadata. Despite this, E. faecalis epidemiology and pathogenesis are most often studied in the context of nosocomial infections. These investigations have elucidated the presence and putative function of various virulence factors, including proteins that facilitate colonization, aggregation, and toxin production (107). The most severe enterococcal nosocomial infection is bacteremia, which can lead to sepsis and endocarditis. Even with appropriate treatment, this infection is fatal in nearly 30% of cases (108). Enterococcal bacteremia has previously been thought to originate via fecal contamination of venous catheters or other medical devices; however, recent studies have identified ascending bladder infections as a frequent prelude to bacteremia (103, 109).
Patients with long-term indwelling urinary catheters have increased risk for enterococcal bacteremia and sepsis. Therefore, catheter-associated UTI (CAUTI), the most common enterococcal nosocomial infection, is a main model system used to assess E. faecalis behavior in the bladder. Studies have shown that E. faecalis acts as a “founder species” in catheter colonization and that E. faecalis presence in polymicrobial infections increases virulence of other uropathogenic microbes, including P. mirabilis and E. coli (110, 111). CAUTI is thought to result from fecal contamination of indwelling urinary catheters (103); however, the discovery of the bladder microbiome raises the possibility that the bladder and urethra could serve as endogenous reservoirs for E. faecalis, making it possible that community-acquired UTI and subsequent persistent bladder colonization could precede these chronic/recurrent infection phenotypes.
Although E. faecalis is a recognized uropathogen underlying community-acquired UTI, SUC has a detection rate of only 50% relative to EQUC (31). This is because E. faecalis is often cultured alongside other uropathogens or commensals, meaning it is either (1) outcompeted during culture by hardier organisms, such as E. coli, or (2) dismissed as “mixed morphologies” and reported as contamination. Missed detection and empiric treatment of E. faecalis-UTI imparts considerable risk, as the efficacy of many antibiotics commonly used to treat UTI is currently being debated for this species. The adaptability of this species and its ability to acquire antibiotic resistance even to the newest antibiotics has correlated with an increased number of cases reported and represents a substantial health risk (24, 35, 111). These include resistance to aminoglycosides (including gentamycin and kanamycin), β-lactams, chloramphenicol, clindamycin, daptomycin, erythromycin, flouroquinolones, oxazolidinones, rifampin, streptomycin, tetracyclines, and tigecycline (24).
This is extremely problematic, as this species is known to have a tropism for kidneys and once ascended is difficult to eradicate (103, 112). Recently, E. faecalis has also been associated with populations experiencing recurrent UTI (31, 113, 114), defined as 3+ UTI in a year or 2 within 6 months (115). These data suggest that E. faecalis behavior in the bladder mimics that of common nosocomial infections, strengthening the concern that this species could be responsible for more severe infection phenotypes.
Thus far, no studies have reported how E. faecalis alters the host bladder environment to promote its own persistent colonization. Additionally, no studies have identified the virulence genes necessary for persistent bladder colonization or urothelial cell invasion. Understanding enterococcal behavior, especially in connection to recurrent UTI, is crucial for developing more efficacious treatment and prevention of severe infections, such as bacteremia and sepsis.
Understudied and under-detected, members of the family Aerococcaceae are easily mistaken for other Gram-positive cocci with similar morphologies and strict growth requirements. Their taxonomy and identification have been fraught with inconsistency and their relationship with human disease is frustratingly mysterious (116). The increasing isolation of these organisms from the urine of sick humans has earned them the title of emerging uropathogens (116). Indeed, their ability to cause serious disease, such as infectious endocarditis, makes them a clear threat, and yet their ability to cause disease is still uncharacterized. Below, we consider the genera Aerococcus, Facklamia, and Globicatella.
The genus Aerococcus consists of several species, the majority of which are associated with the urogenital tracts of livestock and humans. The most prevalent and threatening species, however, is Aerococcus urinae. Table 1 shows enrichment in adult females diagnosed with UTI or UUI relative to asymptomatic controls (detected in 11%, 34%, and 4%, respectively). Thus, A. urinae is implicated with urine, especially in adult females with LUTS. However, the circumstances and implications of how it ends up there remains a mystery. The natural reservoir of the bacterium is poorly described and the circumstances in which it becomes pathogenic are uncharacterized. Currently, there is a demonstrative need for greater investigation into the involvement of A. urinae in urinary tract disorders such as UTI and UUI, as well as invasive tissue infections. With increasing antibiotic resistance observed in clinical isolates, A. urinae poses a growing threat to the undiagnosed (and misdiagnosed) patient.
The first isolates of A. urinae came from the urine of patients diagnosed with UTI (117). Originally thought of as a rare cause of human infection, the bacterium has since seen a clear rise in diagnoses and case reports alongside improvements in culture techniques and identification technologies (118–121). While lethal cases are rare, A. urinae has been identified in a variety of severe disease complications, such as soft tissue infections and bacteremia, all traced to a urological origin (120, 122–124). Non-invasive infections are associated with UTI and UUI in women (Table 1) (9, 21). However, enhanced culturing of urine from asymptomatic participants also detects this species, complicating characterization of A. urinae’s status, and suggesting that it is an opportunistic pathogen (125).
Monoculture of A. urinae from urine is uncommon; instead, it is often identified alongside several other species, contributing to its dismissal as a contaminant. In cases of bacteremia, however, the majority of infections are monomicrobial with significant risk for endocarditis and septic embolization (126, 127). Thus, it remains unclear whether this bacterium works in concert with others or on its own.
Risk factors for invasive infections include older age and comorbid genitourinary diseases (121, 124, 128). In pediatric settings, A. urinae has been reported as a cause for extraordinary malodorous urine in boys with comorbid urogenital disorders as a risk factor (129, 130). Malodorous urine has been documented in adult patients as well, having been described as ammoniacal and “socially disabling” (120, 131).
In all severe cases of infection, misidentification and lack of resistance testing can lead to fatality (132, 133). Currently, the criterion standard for rapid identification in the clinical setting is via MALDI-TOF MS (125). However, A. urinae is easily missed on routine urine culture and other bacteriological tests and, when isolated, is often misidentified as streptococci, staphylococci, or enterococci because they share many characteristics.
Whole genome sequencing and phenotypic characterization of the organism has revealed substantial diversity within the A. urinae species designation such that subdivision has been suggested (134–136), although the clinical relevance of such divisions remains unknown. Like other invasive uropathogens, A. urinae demonstrates the ability to form biofilms on catheters and heart tissue, as well as the ability to aggregate platelets (137–139). The first UTI mouse model for A. urinae demonstrated a tropism for the kidney, indicating a route for ascending infection despite the bacterium being non-motile (140). Analysis for virulence factors revealed genes predicted to be associated with adhesion and anti-phagocytosis (135). Proteomic studies have supported this finding, revealing an abundance of adhesive surface proteins expressed on the bacterium’s surface (138, 141). Unfortunately, no genetic model currently exists to allow mechanistic studies into these virulence factors.
With proper identification and susceptibility testing, antibiotic therapy is generally effective for A. urinae infection. Isolates from several studies have demonstrated susceptibilities to most antibiotics used against Gram-positive organisms; however, resistances have been indicated to fluoroquinolones, cephalosporins, trimethoprim-sulfamethoxazole, and tetracycline (142–146). There is concern that antibiotic resistance may be increasing; rising resistance has been detected in wastewater samples (147). As with the related streptococci, staphylococci, and enterococci, the possibility of horizontal gene transfer of resistance genes may pose a significant future risk. Another member of this genus, A. urinaeequi, has been found to harbor a plasmid with tetracycline resistance and a transposable element with vancomycin resistance (148, 149). As such, prudent stewardship based on careful microbial identification is foundational for the diagnosis and treatment of A. urinae infections.
Facklamia species are challenging to accurately identify with current microbiologic systems; they are often confused with hemolytic streptococci (150). Thus, F. hominis is an underrecognized pathogen that has been isolated from a variety of clinical specimens, including bacteremia associated with brain and soft tissue abscesses, endocarditis, necrotizing gangrene, and ischemic stroke symptoms (151). It also has been associated with pediatric pyelonephritis (152), acute cystitis and urosepsis (152), as well as bacteremia associated with transurethral resection of the prostate (153). Despite isolation from vaginal specimens and urine, especially in adult females with UUI (Table 1), the role of F. hominis as a commensal and the transition to opportunistic pathogen has yet to be explored (151). Antibiotic resistances have been demonstrated towards cephalosporins, erythromycin, clindamycin, and trimethoprim-sulfamethoxazole (150, 151). More studies are needed to investigate mechanisms of virulence, predisposing risk factors, and rates of infection.
G. sanguinis was first isolated from human blood in 1978; more recently it was proposed to be its own novel genus (154). Globicatella infections have been associated with bacteremia, septicemia, meningitis, infective endocarditis, wound infections, and UTIs in humans on a sporadic basis (154–156). Isolates have been detected in catheter-associated biofilms along with A. urinae (138). As such, G. sanguinis is now considered to be an emerging pathogen with an expanding disease spectrum, recently identified from patients with endophthalmitis and osteomyelitis (157, 158). Since this species also has been considered to be a commensal bacterium (159), it likely should be considered an opportunistic uropathogen. Because of its close resemblance to streptococci and aerococci under microscopic examination and morphologically on blood agar, G. sanguinis is often misidentified. As a result, it can be easily underestimated in clinical settings (160).
The Gram-positive coccus Streptococcus anginosus was originally described in 1906 (161). Early on, S. anginosus was thought to cause strep throat, as it was observed to induce inflammation of the fauces, the arched opening at the back of the mouth that leads to the pharynx (161–163). The high degree of heterogeneity in phenotypic characteristics between strains of S. anginosus (161, 164, 165) led to conflicting taxonomic characterizations during early studies before Whiley and Beighton disambiguated S. anginosus into three separate species: S. intermedius, S. constellatus, and S. anginosus. Together, these species comprise the Streptococcus anginosus Group (SAG), also known as the Streptococcus milleri group, which is one group within the larger set of viridans streptococci (162). Today, it is well known that all three members of SAG are part of the normal human flora, having been isolated from the oropharynx, gastrointestinal tract, and vagina of healthy individuals (23, 162–164, 166). As such, they are generally not considered pathogens. However, in immunocompromised individuals, opportunistic infections leading to bacteremia, pharyngitis, and purulent infections have been reported (23). SAG also may contribute to pulmonary exacerbations in cystic fibrosis patients (167, 168). They also contribute to cases of infective endocarditis (169, 170), and have been reported as complications of otitis media and sinusitis and intracranial infections in children (171, 172).
While SAG is primarily isolated from the upper respiratory tract, an increasing number of studies have detected S. anginosus in the urinary tract (21, 173–176). Isolates of S. anginosus have been identified in urine samples from individuals experiencing various lower urinary tract symptoms; SAG members, particularly S. anginosus are especially enriched in the bladder urine of adult females diagnosed with UUI relative to asymptomatic controls (49% versus 7%) (Table 1) (21).
Genetic sequencing of urinary isolates suggests that they belong to a niche-specific clade that may have implications in disease (176). An extensive list of virulence factors has been annotated in these species; however, their role in establishing opportunistic infections is still unclear (177). Antibiotic resistance towards macrolides, aminoglycosides, sulfonamides, and tetracyclines has been observed (178–181) but the resistance profile of urinary isolates has yet to be reported.
Members of the family Actinomycetaceae are a phylogenetically diverse group of Gram-positive, facultatively anaerobic or micro-aerophilic, branching rod-shaped bacteria. They are part of the flora of the oropharyngeal, gastrointestinal, and genitourinary tracts of humans and many animals (182). First identified in 1896 with A. israelii, these rod-shaped bacilli form colonies with fungus-like branched networks of hyphae, a characteristic that led to the initially incorrect assumption that they were fungi. In most healthy individuals, these organisms are commensal in the mucosal epithelia of hollow organs. However, upon trauma or disruption of the epithelial barrier, access to underlying tissues can lead to actinomycoses characterized by a chronic, granulomatous infectious disease (183–185).
Members of the genus Actinomyces are Gram-positive, pleomorphic, facultative anaerobic rods that exhibit some branching (182, 186). Actinomyces species have been identified in catheterized bladder urine of asymptomatic adult females) but are considerably more prevalent in those with UUI (5% and 23%, respectively) (Table 1). Thus, under certain conditions, these organisms could be opportunistic pathogens (187). For example, initially isolated from urine and vaginal secretions, A. urogenitalis has been reported in infections associated with long-term use of an intrauterine device (183, 185, 188), in a case of bacteremia following in vitro fertilization (189), and in an instance of bacteremia associated with prolonged urinary retention (190). With case reports making up the majority of recorded instances of these organisms, it is clear that studies are needed to determine disease risk factors and antibiotic resistances of infections.
The phylogenetic diversity of this family in combination with new modern diagnostic techniques such as 16S rRNA gene sequencing and MALDI-TOF have led to multiple taxonomic revisions and the introduction of many novel species termed Actinomyces-like organisms (ALOs). These include Actinotignum, Gleimia, Schaallia, Trueperella, Varibaculum, and Winkia (182, 191).
This genus of facultatively anaerobic Gram-positive rods consists of 4 species: Actinotignum schaalii, Actinotignum urinale, Actinotignum sanguinis, and Actinotignum timonense (192–195). A. schaalii and A. urinale were first described under the basonyms Actinobaculum schaalii and Actinobaculum urinale, respectively (192, 193). However, based on the 16S rRNA gene sequence, a remote relationship with the Actinobaculum suis type strain was found (Soltys 50052), resulting in reclassification to the Actinotignum genus (194, 196).
Although first isolated from blood, both A. schaalii, A. sanguinis, and A. urinale have since been isolated from urine. A. schaalii has most often been reported in the context of UTI (197). For example, it is reported to be an emerging uropathogen of elderly people suffering from UTI with comorbidities (198, 199). It also has been detected in children with urinary tract disorders (200, 201). The genus as a whole has been reported to be significantly more common in adult women with UUI than in unaffected controls (9, 21) (Table 1), but the species A. schaalii specifically has been found at significantly higher mean abundances in adult women with UUI compared to unaffected controls (202).
Other species have been associated with infections. A. urinale was first isolated from human urine of patients with UTI (194); however, it also has been isolated from human blood cultures (203). A. sanguinis was first isolated from a human blood culture of a patient with septicemia and has been co-isolated with Trueperella bernardiae from breast abscesses in women (204). The first report of A. timonense concerned an isolate from the urine of a 59-year-old man with end-stage renal disease (195).
Like many of the species reviewed here, Actinotignum species grow slowly under ambient atmospheric conditions typically used by clinical microbiology laboratories; thus, they are often overgrown by faster-growing bacteria. Furthermore, because these species resemble commensal skin and mucosal species, they are often mistakenly identified as contaminants (205). Also, until recently, Actinotignum species were difficult to identify after cultivation. However, the advent of molecular techniques has resulted in increasing reports of A. schaalii in the context of human infection (198, 199). Further research is essential to determine whether these Actinotignum species are uropathogens.
Formerly belonging to the Actinomyces genus, the new Gleimia genus consists of three members: G. europea, G. hominis, and G. coleocanis. The first two have been isolated in humans and the latter in dogs. Human isolates have been implicated in UTIs (182, 206) and have been suggested as a bladder cancer marker (207). They can present clinically with persistent ear infections and recurrent soft tissue infections (208–211), as well as abscesses of the neck, back, feet, brain, and genital area in both men and women of various ages (182, 185, 187, 206, 208, 212–214). Recent cases have linked G. europaea with necrotizing fasciitis (210, 211) with a recent case report of rapid infection progression and Fournier’s Gangrene (215). Due to ineffective identification techniques, taxon reclassification, and inadequate research, Gleimia species remain misunderstood with few reports concerning their pathophysiology.
Like Gleimia, a former member of the Actinomyces genus, S. turicensis and S. radingae are Gram-positive, catalase- and urease-negative, anaerobic, filamentous bacilli (182, 216) that have both been isolated from catheterized urine. Originally isolated from a perianal abscess, they were originally classed as CDC Coryneform Group E, from which A. radingae and A. turicensis were purified and characterized (216). After reassessment of phylogenetic positioning and chemotaxonomic characteristics, these species were reassigned to Schaalia along with eleven other species (191), including S. meyeri and S. odontolytica, all part of the human commensal urinary bladder microbiome.
S. turicensis is a commensal of the skin, gut, oral cavity, and female urogenital tract (182) but is also an opportunistic pathogen. Clinical isolates have been reported in bacteremia (217), purent mastoiditis and meningitis (218), infection following rotator cuff repair (219), gonococcal urethritis (220), and a perianal abscess (216). The circumstances and conditions that transform S. turicensis from commensal to pathogen remain to be elucidated.
T. bernardiae is an emerging opportunistic pathogen in both humans (204, 221–229) and animals (230, 231). In the 1980s, isolates recovered from blood cultures, wounds, abscesses, and skin infections were found to be similar by biochemical testing and described using the provisional name CDC fermentative Coryneform group 2 (CDC group 2) (230). CDC group 2 was formally assigned to the species Actinomyces bernardiae based on 16S rRNA gene sequencing and other features for strains recovered from infections in the United States, Canada, and Switzerland (226). In 1997, following reanalysis of the 16S rRNA gene sequences and after comparison with species in the genus Actinomyces, A. bernardiae was assigned to the genus Arcanobacterium (232). After reassessment of phylogenetic positioning and chemotaxonomic characteristics, this species was reassigned to Trueperella, along with A. abortisuis, A. bialowiezensis, A. bonasi and A. pyogenes (233). Of these five organisms, only T. bernardiae and T. pyrogenes have been reported in humans, all associated with mild to severe infections and abscesses. As all 5 species have been reported as pathogens in animals, it has yet to be established if T. bernardiae and T. pyrogenes are commensals in skin, oropharynx, and urinary tract or are opportunistic zoonotic pathogens of humans (225, 231, 233). The occurrence of T. bernardiae in polymicrobial infections may reflect dependence of this organism on nutrients provided by other species. Immunosuppressed patients appear to be more at risk for infection by T. bernardiae (94).
The first member of this anaerobic, diphtheroid, Gram-positive genus was initially characterized as a distinct species with resemblance to the genus Actinomyces in 2003 (234). Case reports have associated V. cambriensis in polymicrobial, anaerobic human abscess infections (235). The source of these infections remains unknown, and it is unclear if this microbe depends on one or more partner species for survival or infection. Before the advent of MALDI-TOF MS, accurate identification of V. cambriense in routine clinical microbiology laboratories was difficult (233). Thus, in the past, this species may have been dismissed as contamination (97). Indeed, the use of more modern detection methods have identified members of the genus Varibaculum in human urine, as well as prostate and bladder cancer (236–238). Whereas metaculturomic methods rarely detect this anaerobe, it is frequently detected by metagenomic approaches. How Varibaculum species cause disease remains poorly understood.
Actinomyces neuii was discovered in 1994 (239). Recently, it was given its own genus Winkia (191). This catalase-positive coccobacillus has been found in asymptomatic women (240) but may be an opportunistic emerging pathogen in humans. Infections include abscesses and infected atheromas (241), cellulitis (242), endophthalmitis, and UTIs (185), as well as bacteremia, including endocarditis (243). Isolates have also been implicated in neonatal sepsis (244–246) and bacterial vaginosis (247). In all cases, how W. neuii is mechanistically involved in these diseases is poorly described. Antibiotic resistance to fluoroquinolones has been observed (248), but wider studies into urinary isolate resistances are needed. Like other Gram-positive rods, it is often dismissed as a contaminant (249). We have found it to be highly enriched in adult females with UUI relative to asymptomatic controls (28% and 3%, respectively) (21) (Table 1).
The Gram-positive genus Corynebacterium includes approximately 80 recognized species. These are catalase-positive rods with occasional swelling or club-like ends. The envelopes of most but not all contain mycolic acid. Nine species are lipophilic (able to metabolize lipids), asaccharolytic (unable to metabolize carbohydrates), and strictly aerobic. The rest are non-lipophilic and saccharolytic; some of these are fermentative facultative anaerobes, while others are non-fermentative aerobes (250).
Although Corynebacterium species are typically commensals of the mucous membranes of hollow organs and skin, some are opportunistic pathogens. Many species have been isolated from the bladder urine of asymptomatic adult females (50) but the genus is particularly enriched in those diagnosed with UUI (8% and 52%, respectively (21), (Table 1). Seven species that occur often and either are or could be urinary tract opportunists are summarized in Table 2 (250). Here, we review 2 of them: C. amycolatum and C. urealyticum.
is a facultative anaerobic fermenter that is non-lipophilic (250, 251). It is unusual, as it lacks mycolic acid, which is common in other coryneforms. Although a commensal of skin and mucous membranes, C. amycolatum can be an opportunistic pathogen, especially in immunosuppressed patents and nosocomial environments. It has been isolated from blood cultures, cellulitis, wounds, endocarditis, and peritonitis (250, 253). A recent pan-genomic study of drug resistant and commensal isolates of C. amycolatum gave insight into the core genome and the transition from commensal to pathogenic phenotype (262).
is an asaccharolytic, lipophilic coryneform that expresses lipase and strong urease activity (250, 261, 263). C. urealyticum is an opportunistic nosocomial pathogen that can cause acute cystitis, pyelonephritis, alkaline-encrusted cystitis, and encrusted pyelitis (250). It has been associated with bacteremia, mainly in patients with chronic urological diseases and its strong urease activity is a major factor in urinary stone formation (260). Painful and persistent pathologies occur associated with encrustations in the kidney, ureters, and urethra due to alkalinization of urine from metabolism of urea (261). It tends to be multi-drug resistant. In a study of C. urealyticum infections in kidney transplant recipients, between 40 to 85% of the isolates tested were resistant to azithromycin, cefotaxime, chloramphenicol, ciprofloxacin, clindamycin, erythromycin, gentamycin, norfloxacin, penicillin G, or tetracycline (261) One non-antibiotic treatment relies on oral L-methionine, which when metabolized acidifies urine (264, 265). It is unclear if this treatment is bactericidal or bacteriostatic, as in the original report removing methionine resulted in return of the uropathogens (264). C. urealyticum is also a zoonotic pathogen associated with UTIs in dogs, cats and other animals (250).
The order Micrococcales is comprised of eighteen families including Dermabacteraceae and Micrococcaceae (191), comprised of 3 and 19 genera, respectively. Several of these genera are detected in human bladder urine including Dermabacter, Kokura, Pseudoglutamicibacter, and Rothia (Table 1). Here, we review D. hominis and both Pseudoglutamicibacter albus and P. cumminsii.
The Dermabacter genus contains three species: D. hominis, D. jinjuensis, and D. vaginalis, of which only D. hominis has been found in human catheterized urine (Table 1). First described in 1988 (266), this Gram-positive, non-spore forming, non-acid fast, facultative anaerobic short rod is considered to be a commensal of human skin (267). However, D. hominis has been reported in diverse clinically relevant scenarios, almost always as part of polymicrobial communities in patients that are immunocompromised or suffering with significant comorbidities, most often cardiovascular disease, diabetes mellitus, and chronic kidney disease (267). Other reports exist of its isolation from biopsies of bone and joint infections and swabs of soft tissue infection (267), a case of trichobacteriosis axillaris (268), a neck sebaceous cyst (269), blood cultures of patients with bacteremia (270), peritoneal fluid from a patient with end stage renal disease (271), recurrent abscesses (272), bone deposits from a patient with chronic osteomyelitis (273), breast implant infections (274), and cerebral abscess of a renal transplant patient (275). In one report, D. hominis isolated from human semen was found to be capable of forming a strong biofilm, which could potentially be a cause of prostatitis (276). Thus, in immunocompromised patients or those with comorbidities, D. hominis may be pathogenic. Although we have detected it in the bladder urine of adult females with UTI and UUI (Table 1), the relationship between this microbe and the urinary tract remains unclear.
Originally assigned to Centers for Disease Control and Prevention coryneform group B-1 and B-3 (277) and later the genus Arthrobacter (278), the recently established genus Pseudoglutamicibacter contains two species: P. cumminsii and P. albus (279). It is unknown whether these species are commensal microbes or opportunistic pathogens, and we have detected both species in the bladder urine of both asymptomatic and affected adult females (1% and 11%, respectively) (Table 1). However, most samples collected from humans have been associated with severe infections and abscesses, including infected amniotic fluid, chronic cervicitis, chronic otorrhea, external otitis, calcaneus osteomyelitis, sepsis, and UTI (277, 280). Isolation sites have included blood, bone, amniotic fluid, leg wounds, and urine (277, 280).
Both P. cumminsii and P. albus are Gram-positive, mesophilic, catalase-positive, obligate aerobe coccobacilli (278, 279). P. cumminisii is the most frequently encountered member of the genus in human clinical specimens (280). A recent case study identified P. cumminsii in the urine culture of a woman with UTI (281). The first clinical specimen of P. albus was isolated from a blood culture of a surgical patient with severe phlebitis (278). The 16S rRNA genes of P. cumminsii and P. albus share a high degree of homology. This makes distinguishing these two organisms difficult (279). Better differentiation will require whole genome sequencing of isolates and better defined MALDI-TOF profiles.
The family Bifidobacteriales consist of 5 genera: Bifidobacterium, Gardnerella, Alloscardovia, Scardovia and Parascardovia (191). Of these, Bifidobacterium, Gardnerella and Alloscardovia have been detected in human bladder urine (Table 1). While the role of Bifidobacterium in colonizing the gastrointestinal tract is well known (282), its role in the urinary tract remains undefined. Gardnerella species are prevalent and abundant in the urinary tracts of asymptomatic adult females but are somewhat more prevalent in women with UUI (13% and 21%, respectively (9, 10) (Table 1). The role of G. vaginalis in bacterial vaginosis and a link to UTI have been observed, but the mechanisms remain elusive (82). Below, we review a lesser-known member of this family Alloscardovia omnicolens.
Several species belong to the genus Alloscardovia, but only A. omnicolens has been described in human urine, especially from UUI patients (Table 1). It is a Gram-positive, oxidase and catalase-negative, non-spore-forming, anaerobic rod (283, 284). While originally considered to be a commensal of the gastrointestinal tract and oral cavity, there is evidence that this microbe is clinically significant and should not be ignored if found in clinical specimens, especially if isolated from the urinary tract (285). The Alloscardovia genus was first described in 2007 by Huys and co-authors after sampling various clinical sites, including the urethra, urine, blood, abscesses of the lung and aorta, and tonsils (286). A. omnicolens has been identified in urine cultures of bladder cancer patients with concurrent UTI (285). It also has been considered the probable cause of infection for at least one UTI (283) and a case of bacteremia due to a UTI in a 70-year-old woman with advanced uterine cancer (287). Therefore, A. omnicolens appears to be an opportunistic pathogen. Antibiotic resistance to metronidazole and moxifloxacin has been described (288, 289).
The order Eubacteriales is comprised of at least 25 families, all anaerobes. Of these families, members of Clostridiaceae (290, 291) and Peptostreptococcacae (40, 292) are reviewed here. Anaerobes are not detected by SUC (27). Oxygen toxicity complicates the collection and culturing these microbes (28, 30), making it difficult to obtain sufficient material for characterization (27, 293). However, with the advent of molecular diagnostic techniques and enhanced culture methods, the role of anaerobes in both the commensal flora and as opportunistic pathogens is becoming recognized (27).
Is a Gram-positive obligate anaerobic bacillus with the ability to hydrolyze esculin (290, 293–297). As such, it is rarely cultured, even by EQUC, but it is observed by metagenomic approaches.
Discovered in 1898, it was named Bacillus ramosum then renamed Ramibacterium ramosum (295, 297). The demonstration of sporulation led to its reclassification as Clostridium ramosum (294, 297). Further dissections of the genus Clostridium, using a combination of genetic markers, led to another name change, this time to Erysipelatoclostridium ramosum and most recently to Thomasclavelia ramosum (290, 293, 296).
While found as part of the commensal flora in the gastrointestinal and urinary tracts, this organism has been documented in infections, such as appendicitis, blood, brain abscess, bacteremia, joint infections, and pulmonary gangrene (295, 297, 298). It also is one of the few sporulating bacteria detected in the urinary microbiome. Further study is warranted to understand how this commensal becomes an opportunistic and potent pathogen.
Originally described in 1936, the genus Peptostreptococcus consists of four species, P. anaerobius, P. canis, P. russellii, and P. stomatis (40, 292, 299). These are Gram-positive anaerobic cocci; thus, they are rarely cultured but are frequently detected by metagenomic approaches. They have weak fermentative and proteolytic metabolisms. Consequently, it may be symbiotic with other organisms from which it derives nutrients (300). Indeed, P. anaerobius has been isolated most often from polymicrobial infections of soft tissue, bone, brain, implant-related and respiratory tract infections (40, 292, 299). P. anaerobius is also one of the most common Gram-positive, anaerobic cocci isolated from infections of the female urogenital tract and the abdominal cavity (292). Thus, while P. anaerobius is probably a commensal of the gastrointestinal, vaginal, and urinary tracts, it can be an opportunistic pathogen, particularly within polymicrobial infections.
The order Bacteroidales is comprised of 17 families. The best known are Bacteroidaceae and Prevotellaceae. The latter family consists of Gram-negative anaerobes split into four genera: Hallella, Paraprevotella, Prevotella, and Alloprevotella (301). Below, we will review a few species.
The genus Prevotella consists of 55 distinct species of Gram-negative coccobacilli anaerobes that are commonly found in the oral, gastrointestinal, and urogenital tracts of humans and animals. It was originally proposed to characterize the moderately saccharolytic, oral Bacteroides species (302). Until recently, the lack of characteristic phenotypic and biochemical traits had hampered identification at the species level among this group of obligatory anaerobes. They are rarely cultured, but the availability of 16S rRNA sequence analysis has improved detection, and thus the number of recognized Prevotella species has increased over the last few years (303). Recently, a genomic and functional analysis of the 55 phenotypically, ecologically and functionally diverse species comprising Prevotella identified 7 distinct clades and thus reassignment across 7 genera, with 4 of them being new genera: Segatella, Hoylesella, Leyella and Palleniella (304). Below, we review Prevotella bivia and Hoylesella timonensis.
Previously classified as Bacteroides bivius, P. bivia is commonly found in the human vaginal microbiome (305). Though normally a commensal, P. bivia has pathogenic potential to trigger severe infection and induce tissue destruction, especially when there is excess estrogen and with the synergistic cooperation of other species (306). P. bivia is one of the most frequently isolated anaerobic bacteria in cases of bacterial vaginosis. The presence of P. bivia creates an environment that facilitates growth of Peptostreptococcus anerobius (300) and Gardnerella vaginalis (307). P. anaerobius growth is enhanced with production of certain amino acids by P. bivia. Likewise, G. vaginalis growth is enhanced with production of ammonia by P. bivia.
P. bivia is one of the most frequently isolated bacteria in women with pelvic inflammatory disease, as noted in a retrospective, cross-sectional study (308), It has also been implicated in cases of recurrent UTIs, osteomyelitis (309), osteitis (310), endocarditis (311), necrobacillosis (312), sinusitis (313), wound infections from animal bites (314, 315), intracranial abscesses (316), periodontal and tubo-ovarian abscesses (317), and adverse pregnancy outcomes such as preterm labor (318). The virulence factors of P. bivia are not fully understood, but research suggests that they may include adhesins that allow the bacteria to attach to host cells, enzymes that degrade host tissue, and toxins that damage host cells and stimulate inflammation. As with other members of the genus Prevotella, antibiotic resistance is becoming an increasing concern; the most common are amoxicillin-clavinate, clindamycin, and moxifloxacin (319). Thus, further research into P. bivia is warranted.
Prevotella timonensis was first isolated from a human breast abscess (320). After reassessment of phylogenetic positioning and chemotaxonomic characteristics, this species was reassigned to Hoylesella (304). Like the previously described anaerobes, H. timonensis is rarely cultured but detected often by metagenomics. The breast abscess isolate mentioned above ferments glucose, maltose, and lactose. It also hydrolyzes esculin but is urease and catalase negative. Although the species has most often been isolated from cutaneous/soft tissue abscesses and bone infections, it is also prevalent in human genitourinary samples, including from urine (304, 321). While H. timonensis may be a commensal organism in the genitourinary tract, it has been associated with bacterial vaginosis (322). Thus, some evidence supports its role as an emerging opportunistic pathogen (321, 322).
This review provides a robust description of lesser-known microbes to support our recommendation that our understanding of uropathogens should go beyond the “usual suspects.” Current clinical diagnosis is hampered by limitations in what clinical microbiologists detect in culture in as little incubation time as possible, most often under atmospheric oxygen conditions (323). This current approach underreports fastidious, slow growing, and anaerobic bacteria, many of which are generally excluded as uropathogens despite evidence to the contrary (12, 27, 292). The clinical consequences include the common scenario of repeated negative standard urine culture results despite relevant, persistent symptoms (102). Common sense should lead one to suspect that some undetected agent(s) play(s) a role in these persistent symptoms and the most likely candidates are fastidious, slow growing and/or anaerobic bacteria. The standard method dismisses, underreports, or does not detect these microbes but they are repeatedly detected by more sensitive metaculturomic (enhanced culture-dependent) or metagenomic (culture-independent, DNA-based methods) (324).
None of the reviewed species were discovered recently. For example, Thomasaclava (formerly Clostridium) ramnosum dates back more than a century, while others (e.g., A. urinae) were discovered at least 10 years ago. Yet, little is known of their pathophysiology. Even their phylogeny is poorly understood, as highlighted by the plethora of name changes (191, 293, 304). As microbial detection technologies have improved, especially with the advent of MALDI-TOF MS, so too have the detection and reporting of these microbes. However, description of these microbes in relation to disease should not be confined to sporadic case reports as has been the case so far.
This review is a call to action to fill this knowledge gap, to begin studies designed to determine first their functioning as commensals and then their transition to opportunistic pathogens. Both retrospective and prospective studies are greatly needed to determine risk factors for symptomatic infections, especially for microbes that have the ability to cause severe complications that might be entirely preventable with proper and early diagnosis. The global rise in antibiotic resistance is well established but only for microbes under active surveillance. Until the resistance profiles of urinary isolates are better reported, patients will continue to experience therapy failures. Thus, as long as we remain blind to the activities and capabilities of these emerging uropathogens, preventable damage will continue to afflict patients and will most definitely worsen as these microbes continue to evolve.
AW conceived of the manuscript. All authors contributed to the article and approved the submitted version.
This work is funded by internal funds to AW. No funding source had a role in the process of writing this manuscript.
We acknowledge the efforts of the clinical and scientific team members of the Loyola Urinary Education and Research Collaborative (LUEREC).
LB discloses editorial stipends from JAMA, Female Pelvic Medicine & Reconstructive Surgery, and UpToDate. AW discloses advisory board membership for Pathnostics and Urobiome Therapeutics, funding from Pathnostics, the Craig Neilsen Foundation, NIH, and an anonymous donor.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author AW declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fruro.2023.1212590/full#supplementary-material
1. Nelson DE, van der Pol B, Dong Q, Revanna KV, Fan B, Easwaran S, et al. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PloS One (2010) 5:e14116. doi: 10.1371/journal.pone.0014116
2. Dong Q, Nelson DE, Toh E, Diao L, Gao X, Fortenberry JD, et al. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PloS One (2011) 6:e19709. doi: 10.1371/journal.pone.0019709
3. Siddiqui H, Nederbragt AJ, Lagesen K, Jeansson SL, Jakobsen KS. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol (2011) 11:244. doi: 10.1186/1471-2180-11-244
4. Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med (2012) 10:174. doi: 10.1186/1479-5876-10-174
5. Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, Fitzgerald M, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol (2012) 50:1376–83. doi: 10.1128/JCM.05852-11
6. Khasriya R, Sathiananthamoorthy S, Ismail S, Kelsey M, Wilson M, Rohn JL, et al. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clin Microbiol (2013) 51:2054–62. doi: 10.1128/JCM.03314-12
7. Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi JR, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol (2013) 3:41. doi: 10.3389/fcimb.2013.00041
8. Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol (2014) 52:871–6. doi: 10.1128/JCM.02876-13
9. Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio (2014) 5:e01283–14. doi: 10.1128/mBio.01283-14
10. Pearce MM, Zilliox MJ, Rosenfeld AB, Thomas-White KJ, Richter HE, Nager CW, et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol (2015) 213:347 e1–11. doi: 10.1016/j.ajog.2015.07.009
11. Brubaker L, Wolfe AJ. The new world of the urinary microbiota in women. Am J Obstet Gynecol (2015) 213:644–9. doi: 10.1016/j.ajog.2015.05.032
12. Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, Brincat C, et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol (2016) 54:1216–22. doi: 10.1128/JCM.00044-16
13. Barr-Beare E, Saxena V, Hilt EE, Thomas-White K, Schober M, Li B, et al. The interaction between enterobacteriaceae and calcium oxalate deposits. PloS One (2015) 10:e0139575. doi: 10.1371/journal.pone.0139575
14. Karstens L, Asquith M, Davin S, Stauffer P, Fair D, Gregory WT, et al. Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front Cell Infect Microbiol (2016) 6:78. doi: 10.3389/fcimb.2016.00078
15. Thomas-White KJ, Hilt EE, Fok C, Pearce MM, Mueller ER, Kliethermes S, et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J (2016) 27:723–33. doi: 10.1007/s00192-015-2847-x
16. Dune TJ, Price TK, Hilt EE, Thomas-White KJ, Kliethermes S, Brincat C, et al. Urinary symptoms and their associations with urinary tract infections in urogynecologic patients. Obstet Gynecol (2017) 130:718–25. doi: 10.1097/AOG.0000000000002239
17. Fok CS, Gao X, Lin H, Thomas-White KJ, Mueller ER, Wolfe AJ, et al. Urinary symptoms are associated with certain urinary microbes in urogynecologic surgical patients. Int Urogynecol J (2018) 29:1765–71. doi: 10.1007/s00192-018-3732-1
18. Komesu YM, Richter HE, Carper B, Dinwiddie DL, Lukacz ES, Siddiqui NY, et al. The urinary microbiome in women with mixed urinary incontinence compared to similarly aged controls. Int Urogynecol J (2018) 29:1785–95. doi: 10.1007/s00192-018-3683-6
19. Thomas-White KJ, Gao X, Lin H, Fok CS, Ghanayem K, Mueller ER, et al. Urinary microbes and postoperative urinary tract infection risk in urogynecologic surgical patients. Int Urogynecol J (2018) 29:1797–805. doi: 10.1007/s00192-018-3767-3
20. Dornbier RA, Bajic P, Van Kuiken M, Jardaneh A, Lin H, Gao X, et al. The microbiome of calcium-based urinary stones. Urolithiasis (2019) 48(3):191–9. doi: 10.1007/s00240-019-01146-w
21. Joyce C, Halverson T, Gonzalez C, Brubaker L, Wolfe AJ. The urobiomes of adult women with various lower urinary tract symptoms status differ: a re-analysis. Front Cell Infect Microbiol (2022) 12:860408. doi: 10.3389/fcimb.2022.860408
22. Brubaker L, Gourdine JF, Siddiqui NY, Holland A, Halverson T, Limeria R, et al. Forming consensus to advance urobiome research. mSystems (2021) 6:e0137120. doi: 10.1128/mSystems.01371-20
23. Pilarczyk-Zurek M, Sitkiewicz I, Koziel J. The clinical view on Streptococcus anginosus group - opportunistic pathogens coming out of hiding. Front Microbiol (2022) 13:956677. doi: 10.3389/fmicb.2022.956677
24. Garcia-Solache M, Rice LB. The Enterococcus: a model of adaptability to its environment. Clin Microbiol Rev (2019) 32(2):e00058–18. doi: 10.1128/CMR.00058-18
25. Aarnoudse JG, Meijer-Severs GJ, Dankert J. Do anaerobes cause urinary tract infection? Lancet (1980) 1:368–9. doi: 10.1016/S0140-6736(80)90917-4
26. Ribot S, Gal K, Goldblat MV, Eslami HH. The role of anaerobic bacteria in the pathogenesis of urinary tract infections. J Urol (1981) 126:852–3. doi: 10.1016/S0022-5347(17)54781-3
27. Legaria MC, Barberis C, Famiglietti A, De Gregorio S, Stecher D, Rodriguez CH, et al. Urinary tract infections caused by anaerobic bacteria. utility of anaerobic urine culture. Anaerobe (2022) 78:102636. doi: 10.1016/j.anaerobe.2022.102636
28. Imlay JA. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv Microb Physiol (2002) 46:111–53. doi: 10.1016/S0065-2911(02)46003-1
29. Andre AC, Debande L, Marteyn BS. The selective advantage of facultative anaerobes relies on their unique ability to cope with changing oxygen levels during infection. Cell Microbiol (2021) 23:e13338. doi: 10.1111/cmi.13338
30. Lu Z, Imlay JA. When anaerobes encounter oxygen: mechanisms of oxygen toxicity, tolerance and defence. Nat Rev Microbiol (2021) 19:774–85. doi: 10.1038/s41579-021-00583-y
31. Hochstedler BR, Burnett L, Price TK, Jung C, Wolfe AJ, Brubaker L. Urinary microbiota of women with recurrent urinary tract infection: collection and culture methods. Int Urogynecol J (2022) 33:563–70. doi: 10.1007/s00192-021-04780-4
32. Jacobs KM, Thomas-White KJ, Hilt EE, Wolfe AJ, Waters TP. Microorganisms identified in the maternal bladder: discovery of the maternal bladder microbiota. AJP Rep (2017) 7:e188–e96. doi: 10.1055/s-0037-1606860
33. Adebayo AS, Ackermann G, Bowyer RCE, Wells PM, Humphrey G, Knight R, et al. The urinary tract microbiome in older women exhibits host genetics and environmental influences. Cell Host Microbe (2020) 28(2):298–305.e3. doi: 10.1016/j.chom.2020.06.022
34. Pohl HG, Groah SL, Perez-Losada M, Ljungberg I, Sprague BM, Chandal N, et al. The urine microbiome of healthy men and women differs by urine collection method. Int Neurourol J (2020) 24:41–51. doi: 10.5213/inj.1938244.122
35. Mancuso G, Midiri A, Gerace E, Marra M, Zummo S, Biondo C. Urinary tract infections: the current scenario and future prospects. Pathogens (2023) 12:623. doi: 10.3390/pathogens12040623
36. Horner P, Donders G, Cusini M, Gomberg M, Jensen JS, Unemo M. Should we be testing for urogenital Mycoplasma hominis, ureaplasma parvum and Ureaplasma urealyticum in men and women? - a position statement from the european STI guidelines editorial board. J Eur Acad Dermatol Venereol (2018) 32(11):1845–51. doi: 10.1111/jdv.15146
37. La Scola B, Fournier PE, Raoult D. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe. (2011) 17(3):106–12. doi: 10.1016/j.anaerobe.2011.05.010
38. Dubourg G, Morand A, Mekhalif F, Godefroy R, Corthier A, Yacouba A, et al. Deciphering the urinary microbiota repertoire by culturomics reveals mostly anaerobic bacteria from the gut. Front Microbiol (2020) 11:513305. doi: 10.3389/fmicb.2020.513305
39. Fathizadeh Z, Ghasemi Tehrani H, Kazemi M, Karbasizade V. Rapid detection and simultaneous identification of the Mycoplasma and Ureaplasma species by real-time PCR and melt curve analysis among fertile and infertile females. Iran J Basic Med Sci (2023) 26(6):628–34. doi: 10.22038/IJBMS.2023.66170.14545
40. Murphy EC, Frick IM. Gram-positive anaerobic cocci–commensals and opportunistic pathogens. FEMS Microbiol Rev (2013) 37:520–53. doi: 10.1111/1574-6976.12005
41. Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev (2014) 27:870–926. doi: 10.1128/CMR.00109-13
42. Raphael E, Riley LW. Infections caused by antimicrobial drug-resistant saprophytic gram-negative bacteria in the environment. Front Med (Lausanne) (2017) 4:183. doi: 10.3389/fmed.2017.00183
43. Raabe VN, Shane AL. Group B Streptococcus (Streptococcus agalatiae). Microbiol Spectr (2019) 7(2):10.1128/microbiolspec.GPP3-0007-2018. doi: 10.1128/microbiolspec.GPP3-0007-2018
44. Du H, Bing J, Hu T, Ennis CL, Nobile CJ, Huang G. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PloS Pathog (2020) 16:e1008921. doi: 10.1371/journal.ppat.1008921
45. Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence (2021) 12:547–69. doi: 10.1080/21505594.2021.1878688
46. Theis KR, Florova V, Romero R, Borisov AB, Winters AD, Galaz J, et al. Sneathia: an emerging pathogen in female reproductive disease and adverse perinatal outcomes. Crit Rev Microbiol (2021) 47:517–42. doi: 10.1080/1040841X.2021.1905606
47. Blevins SM, Bronze MS. Robert Koch And the golden age’ of bacteriology. Int J Infect Dis (2010) 14:e744–51. doi: 10.1016/j.ijid.2009.12.003
48. Iebba V, Totino V, Gagliardi A, Santangelo F, Cacciotti F, Trancassini M, et al. Eubiosis and dysbiosis: the two sides of the microbiota. New Microbiol (2016) 39:1–12.
49. Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med (2016) 8:51. doi: 10.1186/s13073-016-0307-y
50. Thomas-White KJ, Forster SC, Kumar N, Van Kuiken M, Putonti C, Stares MD, et al. Culturing of female bladder bacteria reveals an interconnected urogenital microbiota. Nat Commun (2018) 9:1557. doi: 10.1038/s41467-018-03968-5
51. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol (2015) 13:269–84. doi: 10.1038/nrmicro3432
52. Dey P, Ray Chaudhuri S. The opportunistic nature of gut commensal microbiota. Crit Rev Microbiol (2022), 1–25. doi: 10.1080/1040841X.2022.2133987
53. Gaston JR, Johnson AO, Bair KL, White AN, Armbruster CE. Polymicrobial interactions in the urinary tract: is the enemy of my enemy my friend? Infect Immun (2021) 89:e00652–20. doi: 10.1128/IAI.00652-20
54. Ozturk R, Murt A. Epidemiology of urological infections: a global burden. World J Urol (2020) 38:2669–79. doi: 10.1007/s00345-019-03071-4
55. Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial antibiotic resistance: the most critical pathogens. Pathogens (2021) 10(10):1310. doi: 10.3390/pathogens10101310
56. Ronald A. The etiology of urinary tract infection: traditional and emerging pathogens. Am J Med (2002) 113 Suppl 1A:14S–9S. doi: 10.1016/S0002-9343(02)01055-0
57. Zhang L, Foxman B, Marrs C. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J Clin Microbiol (2002) 40:3951–5. doi: 10.1128/JCM.40.11.3951-3955.2002
58. Johansen TE, Cek M, Naber KG, Stratchounski L, Svendsen MV, Tenke P, et al. Hospital acquired urinary tract infections in urology departments: pathogens, susceptibility and use of antibiotics. data from the PEP and PEAP-studies. Int J Antimicrob Agents (2006) 28 Suppl 1:S91–107. doi: 10.1016/j.ijantimicag.2006.05.005
59. Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C, Kieda C. Why is the partial oxygen pressure of human tissues a crucial parameter? small molecules and hypoxia. J Cell Mol Med (2011) 15:1239–53. doi: 10.1111/j.1582-4934.2011.01258.x
60. Price TK, Hilt EE, Dune TJ, Mueller ER, Wolfe AJ, Brubaker L. Urine trouble: should we think differently about UTI? Int Urogynecol J (2017) 29:205–10. doi: 10.1007/s00192-017-3528-8
61. Gootz TD, Marra A. Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert Rev Anti Infect Ther (2008) 6:309–25. doi: 10.1586/14787210.6.3.309
62. Organization, World Health. WHO publishes list of bacteria for which new antibiotics are urgently needed. In: Media centre. news release. New York City, NY: World Health Organization News Release. (2017). Available at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
63. Baginska N, Cieslik M, Gorski A, Jonczyk-Matysiak E. The role of antibiotic resistant A. baumannii in the pathogenesis of urinary tract infection and the potential of its treatment with the use of bacteriophage therapy. Antibiotics (Basel) (2021) 10(3):281. doi: 10.3390/antibiotics10030281
64. Braun G, Vidotto MC. Evaluation of adherence, hemagglutination, and presence of genes codifying for virulence factors of Acinetobacter baumannii causing urinary tract infection. Mem Inst Oswaldo Cruz (2004) 99:839–44. doi: 10.1590/S0074-02762004000800010
65. Mittal R, Aggarwal S, Sharma S, Chhibber S, Harjai K. Urinary tract infections caused by Pseudomonas aeruginosa: a minireview. J Infect Public Health (2009) 2:101–11. doi: 10.1016/j.jiph.2009.08.003
66. Subramanian P, Shanmugam N, Sivaraman U, Kumar S, Selvaraj S. Antiobiotic resistance pattern of biofilm-forming uropathogens isolated from catheterised patients in Pondicherry, India. Australas Med J (2012) 5:344–8. doi: 10.4066/AMJ.2012.1193
67. He Y, Wu L, Liao P, Shen L, Yang H. Phenotypic and genotypic characterization of multi-drug resistance Pseudomonas aeruginosa isolated from urinary tract infections of non-catheterized and catheterized Chinese patients: a descriptive study over 3 years. Med (Baltimore) (2022) 101:e31373. doi: 10.1097/MD.0000000000031373
68. Gharanfoli A, Mahmoudi E, Torabizadeh R, Katiraee F, Faraji S. Isolation, characterization, and molecular identification of Candida species from urinary tract infections. Curr Med Mycol (2019) 5:33–6. doi: 10.18502/cmm.5.2.1159
69. Garcia Rivera MV, Heyl JJ, Oh MC. Candida auris urinary tract infection in a nursing home patient with multicomorbidities. Cureus (2020) 12:e12322. doi: 10.7759/cureus.12322
70. Griffith N, Danziger L. Candida auris urinary tract infections and possible treatment. Antibiotics (Basel) (2020) 9(12):898. doi: 10.3390/antibiotics9120898
71. Poloni JAT, Rotta LN. Urine sediment findings and the immune response to pathologies in fungal urinary tract infections caused by candida spp. J Fungi (Basel) (2020) 6(4):245. doi: 10.3390/jof6040245
72. Odabasi Z, Mert A. Candida urinary tract infections in adults. World J Urol (2020) 38:2699–707. doi: 10.1007/s00345-019-02991-5
73. Zaidi KU, Mani A, Thawani V, Mehra A. Total protein profile and drug resistance in Candida albicans isolated from clinical samples. Mol Biol Int (2016) 2016:4982131. doi: 10.1155/2016/4982131
74. Colodner R, Nuri Y, Chazan B, Raz R. Community-acquired and hospital-acquired candiduria: comparison of prevalence and clinical characteristics. Eur J Clin Microbiol Infect Dis (2008) 27:301–5. doi: 10.1007/s10096-007-0438-6
75. Achkar JM, Fries BC. Candida infections of the genitourinary tract. Clin Microbiol Rev (2010) 23:253–73. doi: 10.1128/CMR.00076-09
76. Jain N, Kohli R, Cook E, Gialanella P, Chang T, Fries BC. Biofilm formation by and antifungal susceptibility of Candida isolates from urine. Appl Environ Microbiol (2007) 73:1697–703. doi: 10.1128/AEM.02439-06
77. Okulicz JF, Rivard RG, Conger NG, Nguyen MX, Hospenthal DR. Primary isolation of Candida species from urine specimens using chromogenic medium. Mycoses (2008) 51:141–6. doi: 10.1111/j.1439-0507.2007.01456.x
78. Lyell A. Alexander Ogston, micrococci, and Joseph Lister. J Am Acad Dermatol (1989) 20:302–10. doi: 10.1016/S0190-9622(89)70035-9
79. Takahashi T, Satoh I, Kikuchi N. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int J Syst Bacteriol (1999) 49 Pt 2:725–8. doi: 10.1099/00207713-49-2-725
80. Becker K, Bierbaum G, von Eiff C, Engelmann S, Gotz F, Hacker J, et al. Understanding the physiology and adaptation of staphylococci: a post-genomic approach. Int J Med Microbiol (2007) 297:483–501. doi: 10.1016/j.ijmm.2007.04.004
81. Shariati A, Dadashi M, Chegini Z, van Belkum A, Mirzaii M, Khoramrooz SS, et al. The global prevalence of daptomycin, tigecycline, Quinupristin/Dalfopristin, and linezolid-resistant Staphylococcus aureus and coagulase-negative staphylococci strains: a systematic review and meta-analysis. Antimicrob Resist Infect Control (2020) 9:56. doi: 10.1186/s13756-020-00714-9
82. Kline KA, Lewis AL. Gram-positive uropathogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol Spectr (2016) 4(2):UTI–0012–2012. doi: 10.1128/microbiolspec.UTI-0012-2012
83. Argemi X, Hansmann Y, Prola K, Prevost G. Coagulase-negative staphylococci pathogenomics. Int J Mol Sci (2019) 20(5):1215. doi: 10.3390/ijms20051215
84. Eltwisy HO, Twisy HO, Hafez MH, Sayed IM, El-Mokhtar MA. Clinical infections, antibiotic resistance, and pathogenesis of Staphylococcus haemolyticus. Microorganisms (2022) 10(6):1130. doi: 10.3390/microorganisms10061130
85. Pead L, Crump J, Maskell R. Staphylococci as urinary pathogens. J Clin Pathol (1977) 30:427–31. doi: 10.1136/jcp.30.5.427
86. Pead L, Maskell R, Morris J. Staphylococcus saprophyticus as a urinary pathogen: a six year prospective survey. Br Med J (Clin Res Ed) (1985) 291:1157–9. doi: 10.1136/bmj.291.6503.1157
87. Otto M. Staphylococcus epidermidis–the a’ccidental’ pathogen. Nat Rev Microbiol (2009) 7:555–67. doi: 10.1038/nrmicro2182
88. Phillip S, Mushi MF, Decano AG, Seni J, Mmbaga BT, Kumburu H, et al. Molecular characterizations of the coagulase-negative staphylococci species causing urinary tract infection in Tanzania: a laboratory-based cross-sectional study. Pathogens (2023) 2(2):180. doi: 10.3390/pathogens12020180
89. de Paiva-Santos W, de Sousa VS, Giambiagi-deMarval M. Occurrence of virulence-associated genes among Staphylococcus saprophyticus isolated from different sources. Microb Pathog (2018) 119:9–11. doi: 10.1016/j.micpath.2018.03.054
90. Hall DE, Snitzer 3JA. Staphylococcus epidermidis as a cause of urinary tract infections in children. J Pediatr (1994) 124:437–8. doi: 10.1016/S0022-3476(94)70370-1
91. Lozano V, Fernandez G, Spencer PL, Taylor SL, Hatch R. Staphylococcus epidermidis in urine is not always benign: a case report of pyelonephritis in a child. J Am Board Fam Med (2015) 28:151–3. doi: 10.3122/jabfm.2015.01.140118
92. Parsons E, Albert C, Forouhar M, Kunz A, Sainato R. Recurrent severe Staphylococcus epidermidis urinary tract infections in a 7-Year-Old boy. Clin Pediatr (Phila) (2021) 60:346–49. doi: 10.1177/00099228211021277
93. Li L, Li Y, Yang J, Xie X, Chen H. The immune responses to different uropathogens call individual interventions for bladder infection. Front Immunol (2022) 13:953354. doi: 10.3389/fimmu.2022.953354
94. Hijazin M, Ulbegi-Mohyla H, Alber J, Lammler C, Hassan AA, Timke M, et al. Identification of Arcanobacterium (Trueperella) abortisuis, a novel species of veterinary importance, by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Berl Munch Tierarztl Wochenschr (2012) 125:32–7. doi: 10.1016/j.vetmic.2011.12.022
95. Valenstein P, Meier F. Urine culture contamination: a College of American Pathologists Q-Probes study of contaminated urine cultures in 906 institutions. Arch Pathol Lab Med (1998) 122:123–9.
96. Bekeris LG, Jones BA, Walsh MK, Wagar EA. Urine culture contamination: a College of American Pathologists Q-Probes study of 127 laboratories. Arch Pathol Lab Med (2008) 132:913–7. doi: 10.5858/2008-132-913-UCCACO
97. Clarke TM, Citron DM, Towfigh S. The conundrum of the gram-positive rod: are we missing important pathogens in complicated skin and soft-tissue infections? a case report and review of the literature. Surg Infect (Larchmt) (2010) 11:65–72. doi: 10.1089/sur.2008.085
98. Siegman-Igra Y, Kulka T, Schwartz D, Konforti N. The significance of polymicrobial growth in urine: contamination or true infection. Scand J Infect Dis (1993) 25:85–91. doi: 10.1080/00365549309169675
99. Siegman-Igra Y. The significance of urine culture with mixed flora. Curr Opin Nephrol Hypertens (1994) 3:656–9. doi: 10.1097/00041552-199411000-00017
100. Kass EH. Asymptomatic infections of the urinary tract. 1956. J Urol (2002) 167:1016–9; discussion 19-2. doi: 10.1016/s0022-5347(02)80328-7
101. Brubaker L, Wolfe AJ. The female urinary Microbiota/Microbiome: clinical and research implications. Rambam Maimonides Med J (2017) 28:8(2):e0015. doi: 10.5041/RMMJ.10292
102. Cohen JE, Yura EM, Chen L, Schaeffer AJ. Predictive utility of prior negative urine cultures in women with suspected recurrent uncomplicated urinary tract infections. J Urol (2019) 202:979–85. doi: 10.1097/JU.0000000000000325
103. Lebreton F, Willems RJL, Gilmore MS. Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Boston: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection (2014).
104. Krawczyk B, Wityk P, Galecka M, Michalik M. The many faces of Enterococcus spp.-commensal, probiotic and opportunistic pathogen. Microorganisms (2021) 9(9):1900. doi: 10.3390/microorganisms9091900
105. Johnson CN, Sheriff EK, Duerkop BA, Chatterjee A. Let me upgrade you: impact of mobile genetic elements on enterococcal adaptation and evolution. J Bacteriol (2021) 203:e0017721. doi: 10.1128/JB.00177-21
106. Kao PHN, Kline KA. Dr. Jekyll and Mr. Hide: How Enterococcus faecalis subverts the host immune response to cause infection. J Mol Biol (2019) 431:2932–45. doi: 10.1016/j.jmb.2019.05.030
107. He Q, Hou Q, Wang Y, Li J, Li W, Kwok LY, et al. Comparative genomic analysis of Enterococcus faecalis: insights into their environmental adaptations. BMC Genomics (2018) 19:527. doi: 10.1186/s12864-018-4887-3
108. Suppli M, Aabenhus R, Harboe ZB, Andersen LP, Tvede M, Jensen JU. Mortality in enterococcal bloodstream infections increases with inappropriate antimicrobial therapy. Clin Microbiol Infect (2011) 17:1078–83. doi: 10.1111/j.1469-0691.2010.03394.x
109. Fernandez Guerrero ML, Goyenechea A, Verdejo C, Roblas RF, de Gorgolas M. Enterococcal endocarditis on native and prosthetic valves: a review of clinical and prognostic factors with emphasis on hospital-acquired infections as a major determinant of outcome. Med (Baltimore) (2007) 86:363–77. doi: 10.1097/MD.0b013e31815d5386
110. Goh HMS, Yong MHA, Chong KKL, Kline KA. Model systems for the study of enterococcal colonization and infection. Virulence (2017) 8:1525–62. doi: 10.1080/21505594.2017.1279766
111. Gaston JR, Andersen MJ, Johnson AO, Bair KL, Sullivan CM, Guterman LB, et al. Enterococcus faecalis polymicrobial interactions facilitate biofilm formation, antibiotic recalcitrance, and persistent colonization of the catheterized urinary tract. Pathogens (2020) 9(10):835. doi: 10.3390/pathogens9100835
112. Kau AL, Martin SM, Lyon W, Hayes E, Caparon MG, Hultgren SJ. Enterococcus faecalis tropism for the kidneys in the urinary tract of C57BL/6J mice. Infect Immun (2005) 73:2461–8. doi: 10.1128/IAI.73.4.2461-2468.2005
113. Horsley H, Malone-Lee J, Holland D, Tuz M, Hibbert A, Kelsey M, et al. Enterococcus faecalis subverts and invades the host urothelium in patients with chronic urinary tract infection. PloS One (2013) 8:e83637. doi: 10.1371/journal.pone.0083637
114. Whiteside SA, Dave S, Seney SL, Wang P, Reid G, Burton JP. Enterococcus faecalis persistence in pediatric patients treated with antibiotic prophylaxis for recurrent urinary tract infections. Future Microbiol (2018) 13:1095–115. doi: 10.2217/fmb-2018-0048
115. Anger JT, Bixler BR, Holmes RS, Lee UJ, Santiago-Lastra Y, Selph SS. Updates to recurrent uncomplicated urinary tract infections in women: AUA/CUA/SUFU guideline. J Urol (2022) 208:536–41. doi: 10.1097/JU.0000000000002860
116. Ruoff KL. Miscellaneous catalase-negative, gram-positive cocci: emerging opportunists. J Clin Microbiol (2002) 40:1129–33. doi: 10.1128/JCM.40.4.1129-1133.2002
117. Aguirre M, Collins MD. Phylogenetic analysis of some Aerococcus-like organisms from urinary tract infections: description of Aerococcus urinae sp. nov. J Gen Microbiol (1992) 138:401–5. doi: 10.1099/00221287-138-2-401
118. Senneby E, Petersson AC, Rasmussen M. Epidemiology and antibiotic susceptibility of aerococci in urinary cultures. Diagn Microbiol Infect Dis (2015) 81:149–51. doi: 10.1016/j.diagmicrobio.2014.11.009
119. Narayanasamy S, King K, Dennison A, Spelman DW, Aung AK. Clinical characteristics and laboratory identification of Aerococcus infections: an Australian tertiary centre perspective. Int J Microbiol (2017) 2017:5684614. doi: 10.1155/2017/5684614
120. Greco M, Rua-Figueroa I, Ghiglione S, Santana Medina E, Quevedo Abeledo JC, Rodriguez-Lozano C. Musculoskeletal infections caused by Aerococcus urinae: a case-based review. Clin Rheumatol (2018) 37:2587–94. doi: 10.1007/s10067-018-4072-8
121. Sihvonen R, Turunen M, Lehtola L, Pakarinen L, Gronroos JO, Rantakokko-Jalava K, et al. Clinical and microbiological characterization of Aerococcus urinae bacteraemias at Helsinki metropolitan area, Finland. Eur J Clin Microbiol Infect Dis (2022) 41:751–60. doi: 10.1007/s10096-022-04415-6
122. Forsvall A, Wagenius M, Rasmussen M. Perigenital necrotizing soft tissue infection caused by Aerococcus urinae. IDCases (2019) 18:e00590. doi: 10.1016/j.idcr.2019.e00590
123. Lyagoubi A, Souffi C, Baroiller V, Vallee E. Aerococcus urinae spondylodiscitis: an increasingly described localization. EJIFCC (2020) 31:169–73.
124. Tai DBG, Go JR, Fida M, Saleh OA. Management and treatment of Aerococcus bacteremia and endocarditis. Int J Infect Dis (2021) 102:584–89. doi: 10.1016/j.ijid.2020.10.096
125. Rasmussen M. Aerococcus: an increasingly acknowledged human pathogen. Clin Microbiol Infect (2016) 22:22–7. doi: 10.1016/j.cmi.2015.09.026
126. Senneby E, Goransson L, Weiber S, Rasmussen M. A population-based study of aerococcal bacteraemia in the MALDI-TOF MS-era. Eur J Clin Microbiol Infect Dis (2016) 35:755–62. doi: 10.1007/s10096-016-2594-z
127. Sunnerhagen T, Nilson B, Olaison L, Rasmussen M. Clinical and microbiological features of infective endocarditis caused by aerococci. Infection (2016) 44:167–73. doi: 10.1007/s15010-015-0812-8
128. Sahu KK, Lal A, Mishra AK, Abraham GM. Aerococcus-related infections and their significance: a 9-year retrospective study. J Microsc Ultrastruct (2021) 9:18–25. doi: 10.4103/JMAU.JMAU_61_19
129. Lenherr N, Berndt A, Ritz N, Rudin C. Aerococcus urinae: a possible reason for malodorous urine in otherwise healthy children. Eur J Pediatr (2014) 173:1115–7. doi: 10.1007/s00431-014-2348-9
130. Rast D, Evers KS, Egli A, Rudin C, Goischke A, Ritz N. Aerococcus urinae - significance of detection in the paediatric urinary tract: a case series. Eur J Pediatr (2023) 182:749–56. doi: 10.1007/s00431-022-04730-2
131. Geeraedts F, Stoffers C, Smidt H, Schijffelen M. Foul smelling urine in an adult caused by Aerococcus urinae. IDCases (2023) 31:e01657. doi: 10.1016/j.idcr.2022.e01657
132. Higgins A, Garg T. Aerococcus urinae: an emerging cause of urinary tract infection in older adults with multimorbidity and urologic cancer. Urol Case Rep (2017) 13:24–5. doi: 10.1016/j.eucr.2017.03.022
133. Akinboboye O, Babalola TO, Onoriode K, Appiah-Pippim J. Fatal Aerococcus urinae aortic valve endocarditis with severe regurgitation. Am J Case Rep (2022) 23:e937596. doi: 10.12659/AJCR.937596
134. Christensen JJ, Whitney AM, Teixeira LM, Steigerwalt AG, Facklam RR, Korner B, et al. Aerococcus urinae: intraspecies genetic and phenotypic relatedness. Int J Syst Bacteriol (1997) 47:28–32. doi: 10.1099/00207713-47-1-28
135. Carkaci D, Hojholt K, Nielsen XC, Dargis R, Rasmussen S, Skovgaard O, et al. Genomic characterization, phylogenetic analysis, and identification of virulence factors in Aerococcus sanguinicola and Aerococcus urinae strains isolated from infection episodes. Microb Pathog (2017) 112:327–40. doi: 10.1016/j.micpath.2017.09.042
136. Hilt EE, Putonti C, Thomas-White K, Lewis AL, Visick KL, Gilbert NM, et al. Aerococcus urinae isolated from women with lower urinary tract symptoms: In vitro aggregation and genome analysis. J Bacteriol (2020) 202(13):e00170–20. doi: 10.1128/JB.00170-20
137. Shannon O, Morgelin M, Rasmussen M. Platelet activation and biofilm formation by Aerococcus urinae, an endocarditis-causing pathogen. Infect Immun (2010) 78:4268–75. doi: 10.1128/IAI.00469-10
138. Yu Y, Tsitrin T, Bekele S, Thovarai V, Torralba MG, Singh H, et al. Aerococcus urinae and Globicatella sanguinis persist in polymicrobial urethral catheter biofilms examined in longitudinal profiles at the proteomic level. Biochem Insights (2019) 12:1178626419875089. doi: 10.1177/1178626419875089
139. Yaban B, Kikhney J, Musci M, Petrich A, Schmidt J, Hajduczenia M, et al. Aerococcus urinae - a potent biofilm builder in endocarditis. PloS One (2020) 15:e0231827. doi: 10.1371/journal.pone.0231827
140. Gilbert NM, Choi B, Du J, Collins C, Lewis AL, Putonti C, et al. A mouse model displays host and bacterial strain differences in Aerococcus urinae urinary tract infection. Biol Open (2021) 10(8):bio058931. doi: 10.1242/bio.058931
141. Senneby E, Sunnerhagen T, Hallstrom B, Lood R, Malmstrom J, Karlsson C, et al. Identification of two abundant Aerococcus urinae cell wall-anchored proteins. Int J Med Microbiol (2019) 309:151325. doi: 10.1016/j.ijmm.2019.06.005
142. Skov R, Christensen JJ, Korner B, Frimodt-Moller N, Espersen F. In vitro antimicrobial susceptibility of Aerococcus urinae to 14 antibiotics, and time-kill curves for penicillin, gentamicin and vancomycin. J Antimicrob Chemother (2001) 48:653–8. doi: 10.1093/jac/48.5.653
143. Humphries RM, Hindler JA. In vitro antimicrobial susceptibility of Aerococcus urinae. J Clin Microbiol (2014) 52:2177–80. doi: 10.1128/JCM.00418-14
144. Lupo A, Guilarte YN, Droz S, Hirzel C, Furrer H, Endimiani A. In vitro activity of clinically implemented beta-lactams against Aerococcus urinae: presence of non-susceptible isolates in Switzerland. New Microbiol (2014) 37:563–6.
145. Hirzel C, Guilarte YN, Hirzberger L, Furrer H, Marschall J, Endimiani A. In vitro susceptibility of Aerococcus urinae isolates to antibiotics used for uncomplicated urinary tract infection. J Infect (2015) 71:395–7. doi: 10.1016/j.jinf.2015.04.020
146. Scholtz SL, Faron ML, Buchan BW, Ledeboer NA. Comparison of methods for determining the antibiotic susceptibility of Aerococcus species in a clinical setting. Am J Clin Pathol (2022) 157:781–88. doi: 10.1093/ajcp/aqab195
147. Siew SW, Musa SM, Sabri N, Farida Asras MF, Ahmad HF. Evaluation of pre-treated healthcare wastes during COVID-19 pandemic reveals pathogenic microbiota, antibiotics residues, and antibiotic resistance genes against beta-lactams. Environ Res (2023) 219:115139. doi: 10.1016/j.envres.2022.115139
148. Yang Q, Zhu Y, Schwarz S, Wang L, Liu W, Yang W, et al. A novel plasmid from Aerococcus urinaeequi of porcine origin co-harboring the tetracycline resistance genes tet(58) and tet(61). Vet Microbiol (2021) 257:109065. doi: 10.1016/j.vetmic.2021.109065
149. Zhou W, Gao S, Zheng J, Zhang Y, Zhou H, Zhang Z, et al. Identification of an Aerococcus urinaeequi isolate by whole genome sequencing and average nucleotide identity analysis. J Glob Antimicrob Resist (2022) 29:353–59. doi: 10.1016/j.jgar.2022.04.013
150. LaClaire LL, Facklam RR. Comparison of three commercial rapid identification systems for the unusual gram-positive cocci Dolosigranulum pigrum, Ignavigranum ruoffiae, and Facklamia species. J Clin Microbiol (2000) 38:2037–42. doi: 10.1128/JCM.38.6.2037-2042.2000
151. Rahmati E, Martin V, Wong D, Sattler F, Petterson J, Ward P, et al. Facklamia species as an underrecognized pathogen. Open Forum Infect Dis (2017) 4:ofw272. doi: 10.1093/ofid/ofw272
152. Perez-Cavazos S, Cisneros-Saldana D, Espinosa-Villasenor F, Castillo-Bejarano JI, Vaquera-Aparicio DN, Sanchez-Alanis H, et al. Facklamia hominis pyelonephritis in a pediatric patient: first case report and review of the literature. Ann Clin Microbiol Antimicrob (2022) 21:4. doi: 10.1186/s12941-022-00497-4
153. Gahl M, Stockli T, Fahrner R. Facklamia hominis bacteremia after transurethral resection of the prostate: a case report. BMC Urol (2020) 20:192. doi: 10.1186/s12894-020-00762-8
154. Facklam R, Elliott JA. Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin Microbiol Rev (1995) 8:479–95. doi: 10.1128/CMR.8.4.479
155. Takahashi S, Xu C, Sakai T, Fujii K, Nakamura M. Infective endocarditis following urinary tract infection caused by Globicatella sanguinis. IDCases (2018) 11:18–21. doi: 10.1016/j.idcr.2017.12.001
156. Skali H, Hanchi AL, Laghmari M, Ghannane H, Benali SA, Soraa N. A case report of pediatric bacterial meningitis due to the rare isolate, Globicatella sanguinis. Sch J Med Case Rep (2022) 10:182–84. doi: 10.36347/sjmcr.2022.v10i03.002
157. Miller AO, Buckwalter SP, Henry MW, Wu F, Maloney KF, Abraham BK, et al. Globicatella sanguinis osteomyelitis and bacteremia: review of an emerging human pathogen with an expanding spectrum of disease. Open Forum Infect Dis (2017) 4:ofw277. doi: 10.1093/ofid/ofw277
158. Gupta B, Jain AK, Saini M, Sardana M, Soni R, Angrup A. Globicatella sanguinis corneal abscess with endophthalmitis. J AAPOS (2022) 26:46–8. doi: 10.1016/j.jaapos.2021.08.305
159. Hery-Arnaud G, Doloy A, Ansart S, Le Lay G, Le Fleche-Mateos A, Seizeur R, et al. Globicatella sanguinis meningitis associated with human carriage. J Clin Microbiol (2010) 48:1491–3. doi: 10.1128/JCM.01299-09
160. Lau SK, Woo PC, Li NK, Teng JL, Leung KW, Ng KH, et al. Globicatella bacteraemia identified by 16S ribosomal RNA gene sequencing. J Clin Pathol (2006) 59:303–7. doi: 10.1136/jcp.2005.028878
161. Andrewes FW, Horder TJ. A study of the streptococci pathogenic for man. Lancet (1906) 168:708–13. doi: 10.1016/S0140-6736(01)31538-6
162. Whiley RA, Beighton D. Emended descriptions and recognition of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus anginosus as distinct species. Int J Syst Bacteriol (1991) 41:1–5. doi: 10.1099/00207713-41-1-1
163. Jiang S, Li M, Fu T, Shan F, Jiang L, Shao Z. Clinical characteristics of infections caused by Streptococcus anginosus group. Sci Rep (2020) 10:9032. doi: 10.1038/s41598-020-65977-z
164. Ruoff KL. Streptococcus anginosus (“Streptococcus milleri”): the unrecognized pathogen. Clin Microbiol Rev (1988) 1:102–8. doi: 10.1128/CMR.1.1.102
165. Summanen PH, Rowlinson MC, Wooton J, Finegold SM. Evaluation of genotypic and phenotypic methods for differentiation of the members of the anginosus group streptococci. Eur J Clin Microbiol Infect Dis (2009) 28:1123–8. doi: 10.1007/s10096-009-0758-9
166. Coykendall AL. Classification and identification of the viridans streptococci. Clin Microbiol Rev (1989) 2:315–28. doi: 10.1128/CMR.2.3.315
167. Parkins MD, Sibley CD, Surette MG, Rabin HR. The Streptococcus anginosus group–an unrecognized cause of disease in cystic fibrosis: a case series and literature review. Pediatr Pulmonol (2008) 43:490–7. doi: 10.1002/ppul.20809
168. Navratilova L, Bardon J, Novotny R, Zatloukal J, Jakubec P, Kolek V, et al. The Streptococcus milleri group in chronic obstructive pulmonary disease. BioMed Pap Med Fac Univ Palacky Olomouc Czech Repub (2016) 160:378–84. doi: 10.5507/bp.2016.017
169. Finn T, Schattner A, Dubin I, Cohen R. Streptococcus anginosus endocarditis and multiple liver abscesses in a splenectomised patient. BMJ Case Rep (2018) 2018:bcr2018224266.
170. Chang KM, Hsieh SL, Koshy R. An unusual case of Streptococcus anginosus endocarditis in a healthy host with bicuspid aortic valve. Cureus (2021) 13:e13171. doi: 10.7759/cureus.13171
171. McNeil JC, Dunn JJ, Kaplan SL, Vallejo JG. Complications of otitis media and sinusitis caused by Streptococcus anginosus group organisms in children. Pediatr Infect Dis J (2020) 39:108–13. doi: 10.1097/INF.0000000000002514
172. Dodson DS, Heizer HR, Gaensbauer JT. Sequential intravenous-oral therapy for pediatric Streptococcus anginosus intracranial infections. Open Forum Infect Dis (2022) 9:ofab628. doi: 10.1093/ofid/ofab628
173. Brassil B, Mores CR, Wolfe AJ, Putonti C. Characterization and spontaneous induction of urinary tract Streptococcus anginosus prophages. J Gen Virol (2020) 101:685–91. doi: 10.1099/jgv.0.001407
174. Miller S, Miller-Ensminger T, Voukadinova A, Wolfe AJ, Putonti C. Draft genome sequence of Streptococcus anginosus UMB7768, isolated from a woman with recurrent UTI symptoms. Microbiol Resour Announc (2020) 9(21):e00418–20. doi: 10.1128/MRA.00418-20
175. Wu H, Zheng R. Splenic abscess caused by Streptococcus anginosus bacteremia secondary to urinary tract infection: a case report and literature review. Open Med (Wars) (2020) 15:997–1002. doi: 10.1515/med-2020-0117
176. Prasad A, Ene A, Jablonska S, Du J, Wolfe AJ, Putonti C. Comparative genomic study of Streptococcus anginosus reveals distinct group of urinary strains. mSphere (2023) 8(2):e0068722. doi: 10.1128/msphere.00687-22
177. Kurylek A, Stasiak M, Kern-Zdanowicz I. Virulence factors of Streptococcus anginosus - a molecular perspective. Front Microbiol (2022) 13:1025136. doi: 10.3389/fmicb.2022.1025136
178. Tracy M, Wanahita A, Shuhatovich Y, Goldsmith EA, Clarridge 3JE, Musher DM. Antibiotic susceptibilities of genetically characterized Streptococcus milleri group strains. Antimicrob Agents Chemother (2001) 45:1511–4. doi: 10.1128/AAC.45.5.1511-1514.2001
179. Stelzmueller I, Biebl M, Berger N, Eller M, Mendez J, Fille M, et al. Relevance of group milleri streptococci in thoracic surgery: a clinical update. Am Surg (2007) 73:492–7. doi: 10.1177/000313480707300515
180. Asmah N, Eberspacher B, Regnath T, Arvand M. Prevalence of erythromycin and clindamycin resistance among clinical isolates of the Streptococcus anginosus group in Germany. J Med Microbiol (2009) 58:222–27. doi: 10.1099/jmm.0.001560-0
181. Kaplan NM, Khader YS, Ghabashineh DM. Laboratory diagnosis, antimicrobial susceptibility and genuine clinical spectrum of Streptococcus anginosus group; our experience At a university hospital. Med Arch (2022) 76:252–58. doi: 10.5455/medarh.2022.76.252-258
182. Kononen E, Wade WG. Actinomyces and related organisms in human infections. Clin Microbiol Rev (2015) 28:419–42. doi: 10.1128/CMR.00100-14
183. Boyanova L, Kolarov R, Mateva L, Markovska R, Mitov I. Actinomycosis: a frequently forgotten disease. Future Microbiol (2015) 10:613–28. doi: 10.2217/fmb.14.130
184. Gajdacs M, Urban E, Terhes G. Microbiological and clinical aspects of cervicofacial Actinomyces infections: an overview. Dent J (Basel) (2019) 7(3):85. doi: 10.3390/dj7030085
185. Gajdacs M, Urban E. The pathogenic role of Actinomyces spp. and related organisms in genitourinary infections: discoveries in the new, modern diagnostic era. Antibiotics (Basel) (2020) 9(8):524. doi: 10.3390/antibiotics9080524
186. Nikolaitchouk N, Hoyles L, Falsen E, Grainger JM, Collins MD. Characterization of actinomyces isolates from samples from the human urogenital tract: description of Actinomyces urogenitalis sp. nov. Int J Syst Evol Microbiol (2000) 50 Pt 4:1649–54. doi: 10.1099/00207713-50-4-1649
187. Clarridge JE 3rd, Zhang Q. Genotypic diversity of clinical Actinomyces species: phenotype, source, and disease correlation among genospecies. J Clin Microbiol (2002) 40:3442–8. doi: 10.1128/JCM.40.9.3442-3448.2002
188. Elsayed S, George A, Zhang K. Intrauterine contraceptive device-associated pelvic actinomycosis caused by Actinomyces urogenitalis. Anaerobe (2006) 12:67–70. doi: 10.1016/j.anaerobe.2005.12.004
189. Van Hoecke F, Beuckelaers E, Lissens P, Boudewijns M. Actinomyces urogenitalis bacteremia and tubo-ovarian abscess after an in vitro fertilization (IVF) procedure. J Clin Microbiol (2013) 51:4252–4. doi: 10.1128/JCM.02142-13
190. Holmgaard DB, Marina D, Hansen F, Christensen JJ. Bacteremia and urogenital infection with Actinomyces urogenitalis following prolonged urinary retention. APMIS (2020) 128:20–4. doi: 10.1111/apm.13000
191. Nouioui I, Carro L, Garcia-Lopez M, Meier-Kolthoff JP, Woyke T, Kyrpides NC, et al. Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol (2018) 9:2007. doi: 10.3389/fmicb.2018.02007
192. Lawson PA, Falsen E, Akervall E, Vandamme P, Collins MD. Characterization of some Actinomyces-like isolates from human clinical specimens: reclassification of Actinomyces suis (Soltys and Spratling) as Actinobaculum suis comb. nov. and description of Actinobaculum schaalii sp. nov. Int J Syst Bacteriol (1997) 47:899–903. doi: 10.1099/00207713-47-3-899
193. Hall V, Collins MD, Hutson RA, Falsen E, Inganas E, Duerden BI. Actinobaculum urinale sp. nov., from human urine. Int J Syst Evol Microbiol (2003) 53:679–82. doi: 10.1099/ijs.0.02422-0
194. Yassin AF, Sproer C, Pukall R, Sylvester M, Siering C, Schumann P. Dissection of the genus Actinobaculum: reclassification of Actinobaculum schaalii Lawson et al. 1997 and Actinobaculum urinale hall et al. 2003 as Actinobaculum schaalii gen. nov., comb. nov. and Actinotignum urinale comb. nov., description of Actinotignum sanguinis sp. nov. and emended descriptions of the genus Actinobaculum and Actinobaculum suis; and re-examination of the culture deposited as Actinobaculum massiliense CCUG 47753T (= DSM 19118T), revealing that it does not represent a strain of this species. Int J Syst Evol Microbiol (2015) 65:615–24. doi: 10.1099/ijs.0.069294-0
195. Brahimi S, Cadoret F, Fournier PE, Moal V, Raoult D. Actinotignum timonense’ sp. nov., a new bacterial species isolated from a human urine sample. New Microbes New Infect (2017) 16:47–8. doi: 10.1016/j.nmni.2017.01.002
196. Soltys MA, Spratling FR. Infectious cystitis and pyelonephritis of pigs: a preliminary communication. Veterinary Rec (1951) 69:500–04.
197. Lotte R, Lotte L, Ruimy R. Actinotignum schaalii (formerly Actinobaculum schaalii): a newly recognized pathogen-review of the literature. Clin Microbiol Infect (2016) 22:28–36. doi: 10.1016/j.cmi.2015.10.038
198. Bank S, Jensen A, Hansen TM, Soby KM, Prag J. Actinobaculum schaalii, a common uropathogen in elderly patients, Denmark. Emerg Infect Dis (2010) 16:76–80. doi: 10.3201/eid1601.090761
199. Nielsen HL, Soby KM, Christensen JJ, Prag J. Actinobaculum schaalii: a common cause of urinary tract infection in the elderly population. bacteriological and clinical characteristics. Scand J Infect Dis (2010) 42:43–7. doi: 10.3109/00365540903289662
200. Pajkrt D, Simoons-Smit AM, Savelkoul PH, van den Hoek J, Hack WW, Furth AMv. Pyelonephritis caused by Actinobaculum schaalii in a child with pyeloureteral junction obstruction. Eur J Clin Microbiol Infect Dis (2003) 22:438–40. doi: 10.1007/s10096-003-0933-3
201. Zimmermann P, Berlinger L, Liniger B, Grunt S, Agyeman P, Ritz N. Actinobaculum schaalii an emerging pediatric pathogen? BMC Infect Dis (2012) 12:201. doi: 10.1186/1471-2334-12-201
202. Price TK, Lin H, Gao X, Thomas-White KJ, Hilt EE, Mueller ER, et al. Bladder bacterial diversity differs in continent and incontinent women: a cross-sectional study. Am J Obstet Gynecol (2020) S0002-9378:30512–3. doi: 10.1016/j.ajog.2020.04.033
203. Pedersen H, Senneby E, Rasmussen M. Clinical and microbiological features of Actinotignum bacteremia: a retrospective observational study of 57 cases. Eur J Clin Microbiol Infect Dis (2017) 36:791–96. doi: 10.1007/s10096-016-2862-y
204. Calatrava E, Borrego J, Cobo F. Breast abscess due to Trueperella bernardiae and Actinotignum sanguinis. Rev Esp Quimioter (2019) 32:200–02.
205. Reinhard M, Prag J, Kemp M, Andresen K, Klemmensen B, Hojlyng N, et al. Ten cases of Actinobaculum schaalii infection: clinical relevance, bacterial identification, and antibiotic susceptibility. J Clin Microbiol (2005) 43:5305–8. doi: 10.1128/JCM.43.10.5305-5308.2005
206. Sabbe LJ, Van De Merwe D, Schouls L, Bergmans A, Vaneechoutte M, Vandamme P. Clinical spectrum of infections due to the newly described Actinomyces species A. turicensis, A. radingae, and A. europaeus. J Clin Microbiol (1999) 37:8–13. doi: 10.1128/JCM.37.1.8-13.1999
207. Bi H, Tian Y, Song C, Li J, Liu T, Chen Z, et al. Urinary microbiota - a potential biomarker and therapeutic target for bladder cancer. J Med Microbiol (2019) 68:1471–78. doi: 10.1099/jmm.0.001058
208. Nielsen HL. First report of Actinomyces europaeus bacteraemia result from a breast abscess in a 53-year-old man. New Microbes New Infect (2015) 7:21–2. doi: 10.1016/j.nmni.2015.05.001
209. Zarrif-Nabbali H, Bolanos-Rivero M, Navarro-Navarro R, Martin-Sanchez AM. [A sebaceous cyst infection by Actinomyces europaeus]. Enferm Infecc Microbiol Clin (2016) 34:324–5. doi: 10.1016/j.eimc.2015.08.001
210. Kus NJ, Kim BJ, Ross HM. A case report of necrotizing fasciitis with growth of Actinomyces europaeus and Actinotignum schaalii. J Surg Case Rep (2019) 2019:rjz286. doi: 10.1093/jscr/rjz286
211. Allen N, James G, Jain Y. A rare case of abdominal wall necrotizing fasciitis caused by Actinomyces europaeus-a novel pathogen. J Surg Case Rep (2021) 2021:rjab533. doi: 10.1093/jscr/rjab533
212. Funke G, Alvarez N, Pascual C, Falsen E, Akervall E, Sabbe L, et al. Actinomyces europaeus sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol (1997) 47:687–92. doi: 10.1099/00207713-47-3-687
213. Alvarez C, Almuzara M, Tosello C, Stecher D, Vay C, Barberis C. [Actinomyces europaeus (Gleimia europaea) associated with brain abscess: a report of three cases]. Rev Argent Microbiol (2023) 13:S0325–7541(22)00108–0. doi: 10.1016/j.ram.2022.07.003
214. Yamamoto Y, Shiroyama T, Hirata H, Matsumoto K, Kuge T, Yoneda M, et al. Secondary subcutaneous abscess due to mixed infections by Peptoniphilus olsenii and Gleimia europaea after COVID-19. Clin Case Rep (2023) 11:e6844. doi: 10.1002/ccr3.6844
215. Zhang S, Xie Y, Wang Y, Jin G, Cui R, Zou Y. Fournier’s gangrene with growth of Actinomyces europaeus: a case report. Infect Dis Ther (2023) 12:1007–11. doi: 10.1007/s40121-023-00781-6
216. Wust J, Stubbs S, Weiss N, Funke G, Collins MD. Assignment of Actinomyces pyrogenes-like (CDC coryneform group e) bacteria to the genus Actinomyces as Actinomyces radingae sp. nov. and Actinomyces turicensis sp. nov. Lett Appl Microbiol (1995) 20:76–81. doi: 10.1111/j.1472-765X.1995.tb01290.x
217. Kansara T, Majmundar M, Doshi R, Ghosh K, Saeed M. A case of life-threatening Actinomyces turicensis bacteremia. Cureus (2020) 12:e6761. doi: 10.7759/cureus.6761
218. Kocsis B, Tiszlavicz Z, Jakab G, Brassay R, Orban M, Sarkany A, et al. Case report of Actinomyces turicensis meningitis as a complication of purulent mastoiditis. BMC Infect Dis (2018) 18:686. doi: 10.1186/s12879-018-3610-y
219. Cronin JT, Richards BW, Skedros JG. Schaalia (formerly Actinomyces) turicensis infection following open rotator cuff repair. Cureus (2023) 15:e34242. doi: 10.7759/cureus.34242
220. Nokchan N, Wongsurawat T, Jenjaroenpun P, Nitayanon P, Tribuddharat C. Complete genome sequence of Schaalia turicensis strain CT001, isolated from a patient with gonococcal urethritis in Thailand. Microbiol Resour Announc (2021) 10:e0083621. doi: 10.1128/MRA.00836-21
221. Ieven M, Verhoeven J, Gentens P, Goossens H. Severe infection due to Actinomyces bernardiae: case report. Clin Infect Dis (1996) 22:157–8. doi: 10.1093/clinids/22.1.157
222. Adderson EE, Croft A, Leonard R, Carroll K. Septic arthritis due to Arcanobacterium bernardiae in an immunocompromised patient. Clin Infect Dis (1998) 27:211–2. doi: 10.1086/514603
223. Lepargneur JP, Heller R, Soulie R, Riegel P. Urinary tract infection due to Arcanobacterium bernardiae in a patient with a urinary tract diversion. Eur J Clin Microbiol Infect Dis (1998) 17:399–401. doi: 10.1007/s100960050095
224. Bemer P, Eveillard M, Touchais S, Redon H, Corvec S. A case of osteitis due to Staphylococcus aureus and Arcanobacterium bernardiae coinfection. Diagn Microbiol Infect Dis (2009) 63:327–9. doi: 10.1016/j.diagmicrobio.2008.10.016
225. Hijazin M, Metzner M, Erhard M, Nagib S, Alber J, Lammler C, et al. First description of Trueperella (Arcanobacterium) bernardiae of animal origin. Vet Microbiol (2012) 159:515–8. doi: 10.1016/j.vetmic.2012.04.018
226. Bernier AM, Bernard K. Draft genome sequence of Trueperella bernardiae LCDC 89-0504T, isolated from a human blood culture. Genome Announc (2016) 4(1):e01634–15. doi: 10.1128/genomeA.01634-15
227. Lawrence CHD, Waseem S, Newsholme W, Klein JL. Trueperella bernardiae: an unusual cause of septic thrombophlebitis in an injection drug user. New Microbes New Infect (2018) 26:89–91. doi: 10.1016/j.nmni.2018.09.001
228. Roh J, Kim M, Kim D, Yong D, Lee K. First case of Trueperella bernardiae bacteremia in an immunocompromised patient in Korea. Ann Lab Med (2019) 39:593–95. doi: 10.3343/alm.2019.39.6.593
229. Casale R, Bianco G, Cosma S, Micheletti L, Comini S, Iannaccone M, et al. Trueperella bernardiae bloodstream infection following onco-gynaecologic surgery and literature review. Infez Med (2022) 30:124–28. doi: 10.53854/liim-3001-15
230. Na’Was TE, Hollis DG, Moss CW, Weaver RE. Comparison of biochemical, morphologic, and chemical characteristics of centers for disease control fermentative coryneform groups 1, 2, and a-4. J Clin Microbiol (1987) 25:1354–8. doi: 10.1128/jcm.25.8.1354-1358.1987
231. Ahmed MFE, Alssahen M, Lammler C, Kohler B, Metzner M, Plotz M, et al. Identification of Trueperella bernardiae isolated from peking ducks (Anas platyrhynchos domesticus) by phenotypical and genotypical investigations and by a newly developed loop-mediated isothermal amplification (LAMP) assay. Folia Microbiol (Praha) (2022) 67:277–84. doi: 10.1007/s12223-021-00927-4
232. Funke G, Ramos CP, Fernandez-Garayzabal JF, Weiss N, Collins MD. Description of human-derived centers for disease control coryneform group 2 bacteria as Actinomyces bernardiae sp. nov. Int J Syst Bacteriol (1995) 45:57–60. doi: 10.1099/00207713-45-1-57
233. Yassin AF, Hupfer H, Siering C, Schumann P. Comparative chemotaxonomic and phylogenetic studies on the genus Arcanobacterium Collins et al. 1982 emend. lehnen et al. 2006: proposal for Trueperella gen. nov. and emended description of the genus Arcanobacterium. Int J Syst Evol Microbiol (2011) 61:1265–74. doi: 10.1099/ijs.0.020032-0
234. Hall V, Collins MD, Lawson PA, Hutson RA, Falsen E, Inganas E, et al. Characterization of some Actinomyces-like isolates from human clinical sources: description of Varibaculum cambriensis gen nov, sp nov. J Clin Microbiol (2003) 41:640–4. doi: 10.1128/JCM.41.2.640-644.2003
235. Chu YW, Wong CH, Chu MY, Cheung CP, Cheung TK, Tse C, et al. Varibaculum cambriensis infections in Hong Kong, China 2006. Emerg Infect Dis (2009) 15:1137–9. doi: 10.3201/eid1507.081291
236. Niang EHA, Lo CI, Brahimi S, Armstrong N, Raoult D, Fournier PE, et al. Varibaculum massiliense sp. nov., a new bacterium isolated from human urine with culturomics. New Microbes New Infect (2019) 32:100591. doi: 10.1016/j.nmni.2019.100591
237. Hurst R, Meader E, Gihawi A, Rallapalli G, Clark J, Kay GL, et al. Microbiomes of urine and the prostate are linked to human prostate cancer risk groups. Eur Urol Oncol (2022) 5:412–19. doi: 10.1016/j.euo.2022.03.006
238. Hrbacek J, Tlaskal V, Cermak P, Hanacek V, Zachoval R. Bladder cancer is associated with decreased urinary microbiota diversity and alterations in microbial community composition. Urol Oncol (2023) 41:107:e15–07 e22. doi: 10.1016/j.urolonc.2022.09.018
239. Funke G, Stubbs S, von Graevenitz A, Collins MD. Assignment of human-derived CDC group 1 coryneform bacteria and CDC group 1-like coryneform bacteria to the genus Actinomyces as Actinomyces neuii subsp. neuii sp. nov., subsp. nov., and Actinomyces neuii subsp. anitratus subsp nov. Int J Syst Bacteriol (1994) 44:167–71. doi: 10.1099/00207713-44-1-167
240. Ugarcina Perovic S, Ksiezarek M, Rocha J, Cappelli EA, Sousa M, Ribeiro TG, et al. Urinary microbiome of reproductive-age asymptomatic European women. Microbiol Spectr (2022) 10:e0130822. doi: 10.1128/spectrum.01308-22
241. von Graevenitz A. Actinomyces neuii: review of an unusual infectious agent. Infection (2011) 39:97–100. doi: 10.1007/s15010-011-0088-6
242. Gomez-Garces JL, Burillo A, Gil Y, Saez-Nieto JA. Soft tissue infections caused by Actinomyces neuii, a rare pathogen. J Clin Microbiol (2010) 48:1508–9. doi: 10.1128/JCM.02139-09
243. Yang WT, Grant M. Actinomyces neuii: a case report of a rare cause of acute infective endocarditis and literature review. BMC Infect Dis (2019) 19:511. doi: 10.1186/s12879-019-4149-2
244. Mann C, Dertinger S, Hartmann G, Schurz R, Simma B. Actinomyces neuii and neonatal sepsis. Infection (2002) 30:178–80. doi: 10.1007/s15010-002-2165-3
245. Giron de Velasco-Sada P, Peinado H, Romero-Gomez MP. Neonatal sepsis secondary to chorioamnionitis by Actinomyces neuii in a 25 weeks pregnant woman. Med Clin (Barc) (2018) 150:407–08. doi: 10.1016/j.medcli.2017.10.007
246. Alsohime F, Assiri RA, Al-Shahrani F, Bakeet H, Elhazmi M, Somily AM. Premature labor and neonatal sepsis caused by Actinomyces neuii. J Infect Public Health (2019) 12:282–84. doi: 10.1016/j.jiph.2018.04.001
247. Castro J, Machado D, Cerca N. Unveiling the role of Gardnerella vaginalis in polymicrobial bacterial vaginosis biofilms: the impact of other vaginal pathogens living as neighbors. ISME J (2019) 13:1306–17. doi: 10.1038/s41396-018-0337-0
248. Wolff A, Rodloff AC, Vielkind P, Borgmann T, Stingu CS. Antimicrobial susceptibility of clinical oral isolates of actinomyces spp. Microorganisms (2022) 10(1):125. doi: 10.3390/microorganisms10010125
249. Leal SM Jr., Jones M, Gilligan PH. Clinical significance of commensal gram-positive rods routinely isolated from patient samples. J Clin Microbiol (2016) 54:2928–36. doi: 10.1128/JCM.01393-16
250. Bernard K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol (2012) 50:3152–8. doi: 10.1128/JCM.00796-12
251. Collins MD, Burton RA, Jones D. Corynebacterium amycolatum sp. nov. a new mycolic acid-less Corynebacterium species from human skin. FEMS Microbiol Lett (1988) 49:349–52. doi: 10.1111/j.1574-6968.1988.tb02755.x
252. Paviour S, Musaad S, Roberts S, Taylor G, Taylor S, Shore K, et al. Corynebacterium species isolated from patients with mastitis. Clin Infect Dis (2002) 35(11):1434–40. doi: 10.1086/344463
253. Knox KL, Holmes AH. Nosocomial endocarditis caused by Corynebacterium amycolatum and other nondiphtheriae corynebacteria. Emerg Infect Dis (2002) 8:97–9. doi: 10.3201/eid0801.010151
254. Yassin AF, Steiner U, Ludwig W. Corynebacterium aurimucosum sp. nov. and emended description of Corynebacterium minutissimum Collins and Jones, (1983). Int J Syst Evol Microbiol (2002) 52(Pt 3):1001–5. doi: 10.1099/00207713-52-3-1001
255. Devriese LA, Riegel P, Hommez J, Vaneechoutte M, de Baere T, Haesebrouck F. Identification of Corynebacterium glucuronolyticum strains from the urogenital tract of humans and pigs. J Clin Microbiol (2000) 38(12):4657–9. doi: 10.1128/JCM.38.12.4657-4659.2000
256. Ruiz-Pino M, Foronda-García-Hidalgo C, Alarcón-Blanco P, Gutiérrez-Fernández J. Male Genitourinary infections by Corynebacterium glucuronolyticum. a review and clinical experience. Rev Esp Quimioter. (2019) 32(5):479–48.
257. Funke G, Lawson PA, Collins MD. Corynebacterium riegelii sp. nov., an unusual species isolated from female patients with urinary tract infections. J Clin Microbiol (1998) 36(3):624–7. doi: 10.1128/JCM.36.3.624-627.1998
258. Aygun G, Midilli K, Cilingir H, Yilmaz M, Kutukcu A, Eker E. A fatal case of urosepsis due to. Corynebacterium riegelii. Braz J Microbiol (2013) 44(2):475–6. doi: 10.1590/S1517-83822013000200022
259. Feurer C, Clermont D, Bimet F, Candréa A, Jackson M, Glaser P, et al. Taxonomic characterization of nine strains isolated from clinical and environmental specimens, and proposal of Corynebacterium tuberculostearicum sp. nov. Int J Syst Evol Microbiol (2004) 54(Pt 4):1055–61. doi: 10.1099/ijs.0.02907-0
260. Soriano F, Tauch A. Microbiological and clinical features of Corynebacterium urealyticum: urinary tract stones and genomics as the Rosetta stone. Clin Microbiol Infect (2008) 14:632–43. doi: 10.1111/j.1469-0691.2008.02023.x
261. Salem N, Salem L, Saber S, Ismail G, Bluth MH. Corynebacterium urealyticum: a comprehensive review of an understated organism. Infect Drug Resist (2015) 8:129–45. doi: 10.2147/IDR.S74795
262. Jesus HNR, Djpg Rocha RTJ, Silva A, Brenig B, Goes-Neto A, Costa MM, et al. Pan-genomic analysis of Corynebacterium amycolatum gives insights into molecular mechanisms underpinning the transition to a pathogenic phenotype. Front Microbiol (2022) 13:1011578. doi: 10.3389/fmicb.2022.1011578
263. Pitcher D, Soto A, Soriano F, Valero-Guillen P. Classification of coryneform bacteria associated with human urinary tract infection (group D2) as Corynebacterium urealyticum sp. nov. Int J Syst Bacteriol (1992) 42:178–81. doi: 10.1099/00207713-42-1-178
265. Sabiote L, Emiliani E, Kanashiro AK, Balana J, Mosquera L, Sanchez-Martin FM, et al. Oral acidification with l-methionine as a noninvasive treatment for encrusted uropathy. J Endourol Case Rep (2020) 6:143–46. doi: 10.1089/cren.2019.0164
266. Jones D, Collins MD. Taxonomic studies on some human cutaneous coryneform bacteria: description of Dermabacter hominis gen. nov., sp. nov. FEMS Microbiol Lett (1988) 51:51–5. doi: 10.1111/j.1574-6968.1988.tb02967.x
267. Schaub C, Drager S, Hinic V, Bassetti S, Frei R, Osthoff M. Relevance of Dermabacter hominis isolated from clinical samples 2012-2016: a retrospective case series. Diagn Microbiol Infect Dis (2020) 98:115118. doi: 10.1016/j.diagmicrobio.2020.115118
268. Larrondo J, Porte L, Gosch M, Cabrera R, Weitzel T. Trichobacteriosis axillaris caused by Dermabacter hominis. J Eur Acad Dermatol Venereol (2017) 31:e267–e68. doi: 10.1111/jdv.14082
269. Bertona E, De Paulis AN, Gutierrez MA, Santa Maria V, Vay CA, Predari SC. [Unusually infected sebaceous cyst by Dermabacter hominis]. Rev Argent Microbiol (2016) 48:303–07. doi: 10.1016/j.ram.2016.09.003
270. Gomez-Garces JL, Oteo J, Garcia G, Aracil B, Alos JI, Funke G. Bacteremia by Dermabacter hominis, a rare pathogen. J Clin Microbiol (2001) 39:2356–7. doi: 10.1128/JCM.39.6.2356-2357.2001
271. Radtke A, Bergh K, Oien CM, Bevanger LS. Peritoneal dialysis-associated peritonitis caused by Dermabacter hominis. J Clin Microbiol (2001) 39:3420–1. doi: 10.1128/JCM.39.9.3420-3421.2001
272. Martin J, Bemer P, Touchais S, Asseray N, Corvec S. Recurrent abscesses due to finegoldia magna, Dermabacter hominis and Staphylococcus aureus in an immunocompetent patient. Anaerobe (2009) 15:201–3. doi: 10.1016/j.anaerobe.2009.03.006
273. Van Bosterhaut B, Boucquey P, Janssens M, Wauters G, Delmee M. Chronic osteomyelitis due to Actinomyces neuii subspecies neuii and Dermabacter hominis. Eur J Clin Microbiol Infect Dis (2002) 21:486–7. doi: 10.1007/s10096-002-0747-8
274. Seng P, Bayle S, Alliez A, Romain F, Casanova D, Stein A. The microbial epidemiology of breast implant infections in a regional referral centre for plastic and reconstructive surgery in the south of France. Int J Infect Dis (2015) 35:62–6. doi: 10.1016/j.ijid.2015.04.010
275. Bavbek M, Caner H, Arslan H, Demirhan B, Tuncbilek S, Altinors N. Cerebral Dermabacter hominis abscess. Infection (1998) 26:181–3. doi: 10.1007/BF02771848
276. Turk S, Mazzoli S, Stsepetova J, Kuznetsova J, Mandar R. Coryneform bacteria in human semen: inter-assay variability in species composition detection and biofilm production ability. Microb Ecol Health Dis (2014) 25:22701. doi: 10.3402/mehd.v25.22701
277. Funke G, Hutson RA, Bernard KA, Pfyffer GE, Wauters G, Collins MD. Isolation of Arthrobacter spp. from clinical specimens and description of Arthrobacter cumminsii sp. nov. and Arthrobacter woluwensis sp. nov. J Clin Microbiol (1996) 34:2356–63. doi: 10.1128/jcm.34.10.2356-2363.1996
278. Wauters G, Charlier J, Janssens M, Delmee M. Identification of Arthrobacter oxydans, Arthrobacter luteolus sp. nov., and Arthrobacter albus sp. nov., isolated from human clinical specimens. J Clin Microbiol (2000) 38:2412–5. doi: 10.1128/JCM.38.6.2412-2415.2000
279. Busse HJ. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseu. Int J Syst Evol Microbiol (2016) 66:9–37. doi: 10.1099/ijsem.0.000702
280. Funke G, Pagano-Niederer M, Sjoden B, Falsen E. Characteristics of Arthrobacter cumminsii, the most frequently encountered Arthrobacter species in human clinical specimens. J Clin Microbiol (1998) 36:1539–43. doi: 10.1128/JCM.36.6.1539-1543.1998
281. Chaves Blanco L, Gomez-Camarasa C, Illescas Lopez M, Chueca Porcuna N, Rojas-Garcia LL. Unusual isolation of Pseudoglutamicibacter cumminsii in urine culture. Rev Esp Quimioter (2022) 35:95–6. doi: 10.37201/req/088.2021
282. Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sanchez B, Margolles A. Bifidobacteriaand their health-promoting effects. Microbiol Spectr (2017) 5(3). doi: 10.1128/microbiolspec.BAD-0010-2016
283. Brown MK, Forbes BA, Stitley K, Doern CD. Defining the clinical significance of Alloscardovia omnicolens in the urinary tract. J Clin Microbiol (2016) 54:1552–56. doi: 10.1128/JCM.03084-15
284. Lainhart W, Burnham CA. Enhanced recovery of fastidious organisms from urine culture in the setting of total laboratory automation. J Clin Microbiol (2018) 56(8):e00546–18. doi: 10.1128/JCM.00546-18
285. Mahlen SD, Clarridge 3JE. Site and clinical significance of Alloscardovia omnicolens and Bifidobacterium species isolated in the clinical laboratory. J Clin Microbiol (2009) 47:3289–93. doi: 10.1128/JCM.00555-09
286. Huys G, Vancanneyt M, D’Haene K, Falsen E, Wauters G, Vandamme P. Alloscardovia omnicolens gen. nov., sp. nov., from human clinical samples. Int J Syst Evol Microbiol (2007) 57:1442–46. doi: 10.1099/ijs.0.64812-0
287. Ogawa Y, Koizumi A, Kasahara K, Lee ST, Yamada Y, Nakano R, et al. Bacteremia secondary to Alloscardovia omnicolens urinary tract infection. J Infect Chemother (2016) 22:424–5. doi: 10.1016/j.jiac.2015.12.013
288. Isnard C, Lienhard R, Reissier S, Rodriguez S, Krahenbuhl J, Liassine N, et al. In vitro antimicrobial susceptibility of Alloscardovia omnicolens and molecular mechanisms of acquired resistance. Diagn Microbiol Infect Dis (2016) 84:227–9. doi: 10.1016/j.diagmicrobio.2015.08.009
289. Cardona-Benavides I, Puertas-Prieto A, Pinilla-Martin FJ, Navarro-Mari JM, Gutierrez-Fernandez J. Alloscardovia omnicolens emerging presence in premature rupture of membranes. New Microbiol (2019) 42:237–39.
290. Yutin N, Galperin MY. A genomic update on Clostridial phylogeny: gram-negative spore formers and other misplaced Clostridia. Environ Microbiol (2013) 15:2631–41. doi: 10.1111/1462-2920.12173
291. Moore RJ, Lacey JA. Genomics of the pathogenic Clostridia. Microbiol Spectr (2019) 7:GPP3–0033–2018. doi: 10.1128/microbiolspec.GPP3-0033-2018
292. Murdoch DA. Gram-positive anaerobic cocci. Clin Microbiol Rev (1998) 11:81–120. doi: 10.1128/CMR.11.1.81
293. Lawson PA, Saavedra Perez L, Sankaranarayanan K. Reclassification of Clostridium cocleatum, Clostridium ramosum, Clostridium spiroforme and Clostridium saccharogumia as Thomasclavelia cocleata gen. nov., comb. nov., Thomasclavelia ramosa comb. nov., gen. nov., Thomasclavelia spiroformis comb. nov. and Thomasclavelia saccharogumia comb. nov. Int J Syst Evol Microbiol (2023) 73(1). doi: 10.1099/ijsem.0.005694
294. Holdeman LV, Cato EP, Moore WEC. Clostridium ramosum (Vuillemin) comb. nov.: emended description and proposed neotype strain. Int J Syst Bacteriol (1971) 21:35–9. doi: 10.1099/00207713-21-1-35
295. Tally FP, Armfield AY, Dowell VR Jr., Kwok YY, Sutter VL, Finegold SM. Susceptibility of Clostridium ramosum to antimicrobial agents. Antimicrob Agents Chemother (1974) 5:589–93. doi: 10.1128/AAC.5.6.589
296. Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol (2018) 36:996–1004. doi: 10.1038/nbt.4229
297. Legaria MC, Garcia SD, Tudanca V, Barberis C, Cipolla L, Cornet L, et al. Clostridium ramosum rapidly identified by MALDI-TOF MS. a rare gram-variable agent of bacteraemia. Access Microbiol (2020) 2:acmi000137. doi: 10.1099/acmi.0.000137
298. Milosavljevic MN, Kostic M, Milovanovic J, Zaric RZ, Stojadinovic M, Jankovic SM, et al. Antimicrobial treatment of Erysipelatoclostridium ramosum invasive infections: a systematic review. Rev Inst Med Trop Sao Paulo (2021) 63:e30. doi: 10.1590/s1678-9946202163030
299. Legaria MC, Nastro M, Camporro J, Heger F, Barberis C, Stecher D, et al. Peptostreptococcus anaerobius: pathogenicity, identification, and antimicrobial susceptibility. review of monobacterial infections and addition of a case of urinary tract infection directly identified from a urine sample by MALDI-TOF MS. Anaerobe (2021) 72:102461. doi: 10.1016/j.anaerobe.2021.102461
300. Pybus V, Onderdonk AB. A commensal symbiosis between prevotella bivia and Peptostreptococcus anaerobius involves amino acids: potential significance to the pathogenesis of bacterial vaginosis. FEMS Immunol Med Microbiol (1998) 22:317–27. doi: 10.1111/j.1574-695X.1998.tb01221.x
301. Garcia-Lopez M, Meier-Kolthoff JP, Tindall BJ, Gronow S, Woyke T, Kyrpides NC, et al. Analysis of 1,000 type-strain genomes improves taxonomic classification of Bacteroidetes. Front Microbiol (2019) 10:2083. doi: 10.3389/fmicb.2019.02083
302. Shah HN, Collins DM. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Bacteriol (1990) 40:205–8. doi: 10.1099/00207713-40-2-205
303. Nagy E, Urban E, Soki J, Terhes G, Nagy K. The place of molecular genetic methods in the diagnostics of human pathogenic anaerobic bacteria. a minireview. Acta Microbiol Immunol Hung (2006) 53:183–94. doi: 10.1556/AMicr.53.2006.2.5
304. Hitch TCA, Bisdorf K, Afrizal A, Riedel T, Overmann J, Strowig T, et al. A taxonomic note on the genus Prevotella: description of four novel genera and emended description of the genera Hallella and Xylanibacter. Syst Appl Microbiol (2022) 45:126354. doi: 10.1016/j.syapm.2022.126354
305. Snydman DR, Tally FP, Knuppel R, Landrigan J, Gorbach SL, Bartlett JG. Bacteroides bivius and Bacteroides disiens in obstetrical patients: clinical findings and antimicrobial susceptibilities. J Antimicrob Chemother (1980) 6:519–25. doi: 10.1093/jac/6.4.519
306. Grande-Del-Arco J, Jimenez-Cristino MD, Garcia-de-la-Pena R, Fernandez-Espejo E, Cordoba-Fernandez A. A rare paronychia with superinfection with Prevotella bivia and Staphylococcus haemolyticus: the importance of early microbiological diagnosis. Pathogens (2020) 9(12):999. doi: 10.3390/pathogens9120999
307. Pybus V, Onderdonk AB. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J Infect Dis (1997) 175:406–13. doi: 10.1093/infdis/175.2.406
308. Chen PC, Li PC, Ding DC. Pelvic inflammatory disease and causative pathogens in older women in a medical center in eastern Taiwan: a retrospective cross-sectional study. PloS One (2021) 16:e0257627. doi: 10.1371/journal.pone.0257627
309. Fukuoka M, Aita K, Aoki Y, Hayashi S, Satoh T, Hotokebuchi T, et al. Pyogenic vertebral osteomyelitis caused by Prevotella intermedia. J Infect Chemother (2002) 8:182–4. doi: 10.1007/s101560200032
310. Riesbeck K. Paronychia due to Prevotella bivia that resulted in amputation: fast and correct bacteriological diagnosis is crucial. J Clin Microbiol (2003) 41:4901–3. doi: 10.1128/JCM.41.10.4901-4903.2003
311. Kentos A, Motte S, Nonhoff C, Jacobs F, De Smet JM, Serruys E, et al. Prevotella bivia as an unusual cause of endocarditis. Eur J Clin Microbiol Infect Dis (1994) 13:142–5. doi: 10.1007/BF01982187
312. Huits RM, van Assen S, Wildeboer-Veloo AC, Verschuuren EA, Koeter GH. Prevotella bivia necrobacillosis following infectious mononucleosis. J Infect (2006) 53:e59–63. doi: 10.1016/j.jinf.2005.10.016
313. Busch N, Mertens PR, Schonfelder T, Nguyen H, Marschall HU, Kierdorf H, et al. [Lemierre’s post-tonsillitis sepsis with meningitis and intravascular consumption coagulopathy as complication of infectious mononucleosis with pansinusitis]. Dtsch Med Wochenschr (1996) 121:94–8. doi: 10.1055/s-2008-1042978
314. Alexander CJ, Citron DM, Hunt Gerardo S, Claros MC, Talan D, Goldstein EJ. Characterization of saccharolytic Bacteroides and Prevotella isolates from infected dog and cat bite wounds in humans. J Clin Microbiol (1997) 35:406–11. doi: 10.1128/jcm.35.2.406-411.1997
315. Schindl A, Schon H. Foot infection with Prevotella bivia, P. oralis and P. loescheii after wound licking. J Med Microbiol (1999) 48:109.
316. Hoshino T, Nakamura A. [Clinical and bacteriological features of six cases with intracranial abscess in childhood]. Kansenshogaku Zasshi (2002) 76:83–8. doi: 10.11150/kansenshogakuzasshi1970.76.83
317. Dumitriu S, Bancescu G, Murea A, Skaug N. Isolation and speciation of Prevotella strains from periodontal abscesses. Roum Arch Microbiol Immunol (1998) 57:5–10.
318. Mikamo H, Kawazoe K, Izumi K, Watanabe K, Ueno K, Tamaya T. Studies on the pathogenicity of anaerobes, especially prevotella bivia, in a rat pyometra model. Infect Dis Obstet Gynecol (1998) 6:61–5. doi: 10.1155/S1064744998000155
319. Brook I, Wexler HM, Goldstein EJ. Antianaerobic antimicrobials: spectrum and susceptibility testing. Clin Microbiol Rev (2013) 26:526–46. doi: 10.1128/CMR.00086-12
320. Glazunova OO, Launay T, Raoult D, Roux V. Prevotella timonensis sp. nov., isolated from a human breast abscess. Int J Syst Evol Microbiol (2007) 57:883–86. doi: 10.1099/ijs.0.64609-0
321. Aberkane S, Pradel B, Dumont Y, Veloo ACM, Laurens C, Bonzon L, et al. Clinical sources and antimicrobial susceptibility of Prevotella timonensis at the University Hospital of Montpellier, France. Anaerobe (2018) 50:19–21. doi: 10.1016/j.anaerobe.2018.01.002
322. Lehtoranta L, Hibberd AA, Reimari J, Junnila J, Yeung N, Maukonen J, et al. Recovery of vaginal microbiota after standard treatment for bacterial vaginosis infection: an observational study. Microorganisms (2020) 8(6):875. doi: 10.3390/microorganisms8060875
323. Joho KL, Soliman H, Weinstein MP. Comparison of one-day versus two-day incubation of urine cultures. Diagn Microbiol Infect Dis (1995) 21:55–6. doi: 10.1016/0732-8893(94)00115-D
324. Szlachta-McGinn A, Douglass KM, Chung UYR, Jackson NJ, Nickel JC, Ackerman AL. Molecular diagnostic methods versus conventional urine culture for diagnosis and treatment of urinary tract infection: a systematic review and meta-analysis. Eur Urol Open Sci (2022) 44:113–24. doi: 10.1016/j.euros.2022.08.009
Keywords: 16S rRNA gene sequencing, anaerobe, antibiotic resistance, facultative anaerobe, metaculturomics, urinary microbiome, urinary tract infection, uropathogen
Citation: Moreland RB, Choi BI, Geaman W, Gonzalez C, Hochstedler-Kramer BR, John J, Kaindl J, Kesav N, Lamichhane J, Lucio L, Saxena M, Sharma A, Tinawi L, Vanek ME, Putonti C, Brubaker L and Wolfe AJ (2023) Beyond the usual suspects: emerging uropathogens in the microbiome age. Front. Urol. 3:1212590. doi: 10.3389/fruro.2023.1212590
Received: 26 April 2023; Accepted: 27 June 2023;
Published: 26 July 2023.
Edited by:
Evann Hilt, University of Minnesota Twin Cities, United StatesReviewed by:
Robert Fredrick Potter, Washington University in St. Louis, United StatesCopyright © 2023 Moreland, Choi, Geaman, Gonzalez, Hochstedler-Kramer, John, Kaindl, Kesav, Lamichhane, Lucio, Saxena, Sharma, Tinawi, Vanek, Putonti, Brubaker and Wolfe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan J. Wolfe, YXdvbGZlQGx1Yy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.