- 1Animal Disease Research Unit, United States Department of Agriculture, Agricultural Research Service, Pullman, WA, United States
- 2Department of Veterinary Microbiology and Pathology, Washington State University, Pullman, WA, United States
Bovine anaplasmosis, caused by Anaplasma marginale, is one of the most common tick-borne diseases of cattle and has a worldwide distribution. The high costs of bovine anaplasmosis are due to treatment, decreased production, and outbreaks resulting in high mortality. The impact of bovine anaplasmosis is greatest in tropical and subtropical regions where tick vectors are abundant year around. Prevention generally relies on the use of tetracyclines to prevent disease and acaricides to reduce tick burdens. Thus, additional methods to prevent disease while reducing the use of antibiotics are needed. Protection can be reliably achieved with immunization using outer membrane proteins, thus allowing for the possibility for development of a recombinant vaccine. However, prioritizing the selection and testing of antigens from the protective outer membrane extract remains a challenge. Because A. marginale is an obligate intracellular pathogen, surface proteins that mediate adhesion to host cells, primarily red blood cells (RBCs), are functionally relevant vaccine candidates. With some exceptions, the proteins that bind RBCs remain unknown. To address this gap, a phage display library expressing 66 A. marginale proteins was screened to identify adhesins for bovine RBCs. Of the screened proteins, 73% were eliminated due to poor binding to RBCs. Several potential adhesins were identified, including Msp1b and OmpA, which are known adhesions for bovine RBCs and tick cells, respectively. Additionally, Mlp3, Am779, Msp3, and Omp13 met the criteria as RBCs adhesins and may serve as high priority vaccine candidates for future testing.
1 Introduction
Bovine anaplasmosis, caused by the obligate, intracellular bacterium Anaplasma marginale, is one of the most common tick-borne diseases of cattle worldwide. A. marginale infects red blood cells (RBCs), which are then removed by the spleen, resulting in anemia (1, 2). The significant economic burden of bovine anaplasmosis is due to death, weight loss, costs of treatment and tick prevention (3). This burden is more pronounced in tropical and subtropical regions due to the abundance of tick vectors year around (4). The available methods to control bovine anaplasmosis are currently insufficient (2, 5). In some regions, cattle are intentionally infected with A. centrale, which provides variable protection from bovine anaplasmosis, requires maintenance of a cold chain, and is expensive to produce (6). Additionally, acaricides can be used to control ticks. Though, in regions where ticks are abundant, repeated treatments are necessary, which is expensive. Finaly, long acting tetracyclines can be administered to cattle to prevent disease when the risk of tick transmission is high. This approach is discouraged due to global efforts to reduce antibiotic use. Thus, an affordable subunit vaccine to prevent bovine anaplasmosis would improve cattle health and productivity and thus advance efforts toward improving food security globally.
Protective immunity against bovine anaplasmosis can be achieved through vaccination with outer membrane proteins from A. marginale (7–10). However, the process of isolating outer membranes is laborious, difficult to standardize, and the preparation includes over 20 antigens (11). Thus, a major limitation in development of a subunit vaccine is rational antigen selection for immunization and challenge trials. Because A. marginale is an obligate intracellular pathogen, antibody directed against surface proteins that mediate adhesion to RBCs may serve as an important component of protective immunity achieved through vaccination (12).
Bacterial adhesins are outer membrane proteins that selectively bind specific proteins or glycoproteins on the host cell surface, thus define tissue tropism, and may in the case of intracellular pathogens, trigger pathogen entry (13–15). With a few exceptions, little is known about the molecules and mechanisms employed by A. marginale to enter bovine RBCs. Msp1a and Msp1b when expressed on the surface of E. coli, confer the ability of E. coli to adhere to bovine RBCs (16, 17). Additionally, OmpA is an adhesin for tick cells, including the natural tick vector, Dermacentor andersoni (18, 19). To ensure adequate functional redundancy, it is likely there are additional A. marginale surface proteins that serve as adhesins for bovine RBCs.
Thus, the goal of this work is to identify a broad array of A. marginale surface proteins that potentially mediate adhesion to bovine RBCs. Because we currently lack the ability to generate gene knockouts and culture A. marginale in bovine RBCs, we used bacteriophage (phage) display as a functional screen to test the adhesive capacity of 66 A. marginale proteins. We identified 11 adhesins, including Msp1b and OmpA. These findings help advance our understanding of the molecular interactions between A. marginale and bovine RBCs and will help prioritize vaccine candidates for efficacy testing.
2 Materials and methods
2.1 Ethics statement
This study was carried out in accordance with the Animal Welfare Act, Guide for the care and use of Agricultural Animals in Agricultural Research and Training, and the Public Health Service Policy on Humane Care and Use of Laboratory Animals. The protocol (ASAF #2019–37) involving the use of animals was approved by the University of Idaho Institutional Animal Care and Use Committee.
2.2 Generation of the phage display library
The phage display library was generated using T7 bacteriophages as previously described in Noh et al., 2024 (19). Because these phages can express up to 1200 aa on their capsid, the genes encoding the largest proteins (am072, am366, am387, am712, am811, am1051) were divided into 2 to four segments for cloning. Thus, the phage display library is composed of 74 genes or gene segments encoding 66 A. marginale proteins (19).
Briefly, the selected genes were PCR amplified from genomic DNA isolated from the Saint Maries strain of A. marginale using Green GoTaq MasterMix (Promega, Madison WI, USA) and the following parameters: denaturation 95°C for 2 min, then 35 cycles of 95°C for 30 sec, 58°C for 30 sec, 72°C for 3.5 min with a final extension of 72°C for 5 min.) (20). An EcoR1 and a HindIII restriction site were added to the 5’ and 3’ ends of all amplicons, respectively, to ensure directional cloning.
The T7Select10–3b vector (New England Biolabs, Ipswich MA, USA) was restriction digested, cleaned, concentrated, and ligated to the amplicons using T4 ligase and ligase buffer (New England Biolabs, Ipswich MA, USA). The recombinant DNA was then packaged in vitro using the T7Select Phage Display System according to the manufacturer’s instructions (Novagen Biotech, Great Neck NY, USA). Plaque assays were done to enumerate the phages in each input pool following the manual of the T7Select Phage Display System.
2.3 Selection for phages expressing A. marginale proteins that bind bovine RBCs
Biopanning, the enrichment for adherent phages, was accomplished through multiple rounds of selection using bovine RBCs or empty wells (Figure 1). The bovine RBCs were kindly provided by Massaro Ueti, at the United States Department of Agriculture-Agricultural Research Service, Animal Disease Research Unit in Pullman, WA. Following collection, RBCs were defibrinated with glass beads and centrifuged at 1,000 RCF for 20 min. The plasma and buffy coat were removed by aspiration and the RBCs were washed three times in Puck’s G Balanced Salt solution with an additional 20 mg/ml glucose, 100 μg/ml streptomycin, and 100 units/ml penicillin at a pH of 7.2. RBCs were then resuspended as a 50% solution using the medium described above and stored at 4°C until use (less than two weeks) (21).
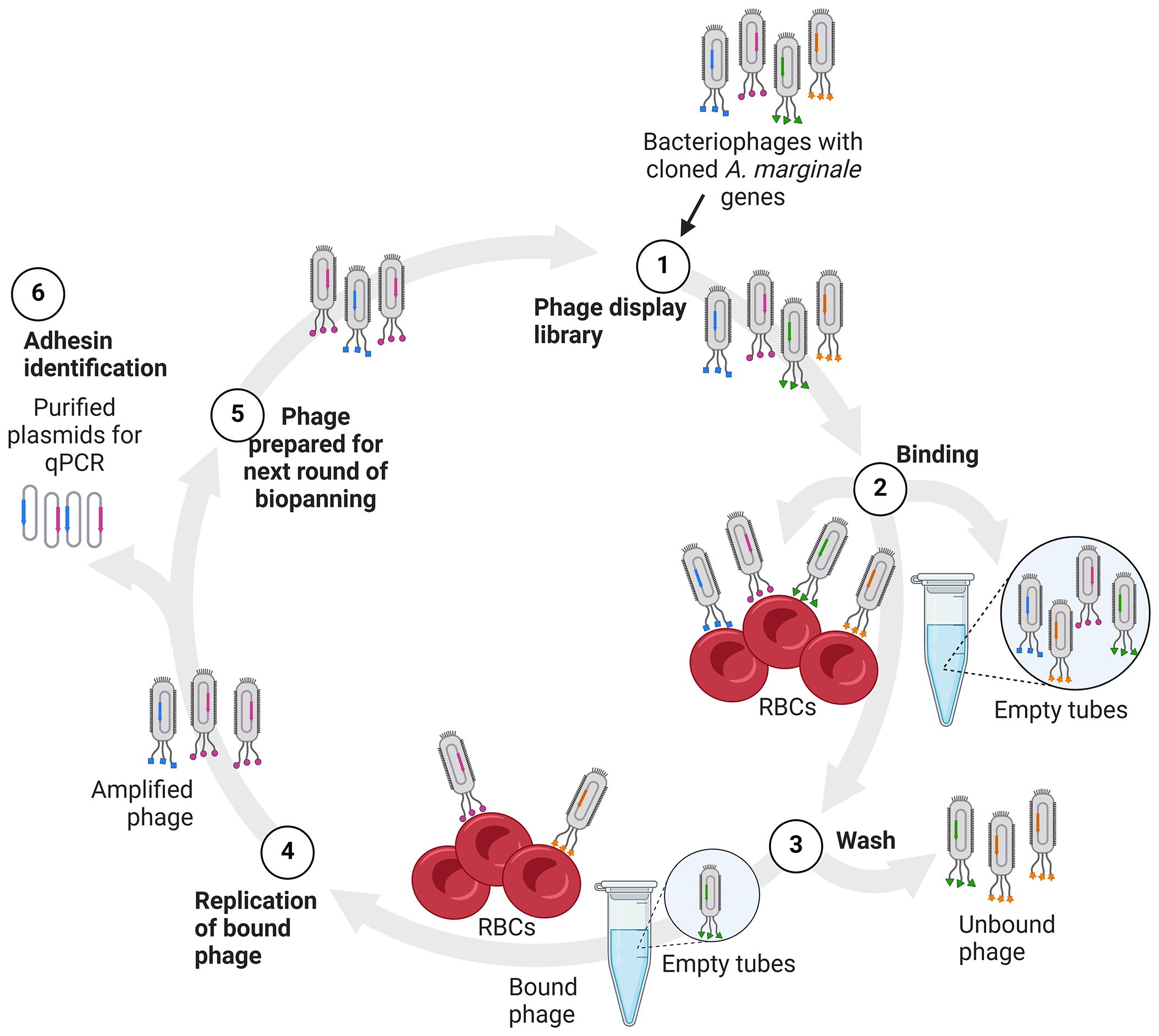
Figure 1 Diagram of biopanning and identification of potential adhesins. (1) The phages displaying A. marginale proteins (1), depicted as colored shapes, are added to bovine RBCs or empty tubes for binding (2). The RBCs and empty tubes are washed to remove unbound phages (3). The bound phages are recovered and replicated in E. coli (4). Recovered phages are then quantified and used in the next round of biopanning (5). Alternatively, plasmid DNA is isolated from the recovered phages and used as template in real time PCR to detect the A. marginale genes in the bound phages (6).
For each round of biopanning, 5x105 RBCs were added to a microfuge tube and 10 PFU of phage/erythrocyte were added to the tubes. The RBCs were incubated with gentle rotation at 38.5°C for 2 h. Following incubation, RBCs were pelleted and washed using Tris-buffered saline with 0.1% Tween 20 (MilliporeSigma Burlington, MA, USA) ten times. RBCs were resuspended in 1ml lysogeny (LB), added to 4 ml of log-phase BLT5403 E. coli, and incubated shaking until lysis of E. coli occurred, approximately 4 hr following inoculation.
Plastic binding of phages displaying foreign proteins and peptides, particularly those rich in tyrosine and tryptophan, is common and can lead to false positive selection of binding candidates (22, 23). Thus, to control for plastic binding, biopanning was done using empty microfuge tubes and equivalent numbers of phage as used with RBCs, described above.
The phages recovered from RBCs or empty tubes were enumerated using plaque assays, normalized and used in the next round of biopanning, as previously described (19). This experiment was done two independent times in triplicate.Three rounds of biopanning were done in experiment one and four rounds of biopanning were done in experiment two.
2.4 Real-time PCR to detect A. marginale genes in recovered phages
Following the final round of biopanning, plasmid DNA was extracted from clarified phage lysates using a phage DNA isolation kit (Norgen Biotek Corp, Thorold ON, Canada) according to the manufacturer’s instructions. Real-time PCR using Biorad iQ SYBR Green Supermix (Thermo Fisher Scientific, Waltham, MA, USA), a CFX96 Touch Real-Time PCR Detection System Thermocycler (Bio-Rad, Hercules, CA, USA) and A. marginale gene-specific primers were used to detect A. marginale genes in each output pool, using previously published primers and amplification conditions (19). If no template was detected after 40 rounds of PCR, the gene was considered absent and given a cycle threshold (Ct) of 40 for computational purposes.
2.5 Data analysis and criteria for designation of an A. marginale protein as a potential adhesin
Following PCR, the average Ct for each A. marginale gene in each replicate and the percentage of replicates from which a gene was detected were calculated using Excel.
To determine the criteria for a screened A. marginale gene expressed in the phage display library to be considered a potential adhesin, we calculated a frequency distribution of the percentage recovery using GraphPad Prism v.10.0.3 (Dotmatics, Boston, MA, USA) (Supplementary Figure S1). The screened proteins recovered from empty tubes in 100% of the replicates were considered high plastic binders and were excluded from this frequency distribution. A. marginale genes or gene segments in the 75th percentile for percentage recovery (≥ 33%) were selected as potential adhesins. Additionally, the percentage recovery from RBCs had to be twice that of the percentage recovery from the empty tubes to be considered as a potential adhesin.
For analysis and data visualization, the screened proteins were divided into functional categories including the Major Surface Protein 1 superfamily, the Major Surface Protein 2 superfamily, outer membrane proteins not in superfamilies, type 4 secretion system proteins (T4SS), proteins with conserved domains, and proteins of unknown function.
3 Results
3.1 Validation of phage display
There are several potential biases of phage display that can affect the outcome of biopanning, including the size of an expressed gene and abundance of a phage expressing a particular gene within the library. Thus, we first calculated the correlation between the size of each A. marginale gene and its percentage recovery in the output pool. The Pearson’s correlation coefficient (r) was - 0.44 and the R2 = 0.2, indicating 20% of the variance was shared between insert size and percent recovery (p < 0.0001) (Figure 2A). Next, we determined that the correlation between the quantity of each A. marginale gene in the inoculum and its percentage recovery was r = - 0.51 with a R2 of 0.26 (Figure 2B). Thus, there is a moderate negative correlation between the size and amount of each A. marginale gene in the input pool and the percentage recovery of that gene following biopanning. Finally, we determined that there is a strong negative correlation between the quantity of an A. marginale gene in the output pool and its percentage recovery (r = -0.88, R2 = 0.69) (Figure 2C). Thus, the percentage recovery was used to evaluate recovery of phages displaying A. marginale proteins following biopanning.
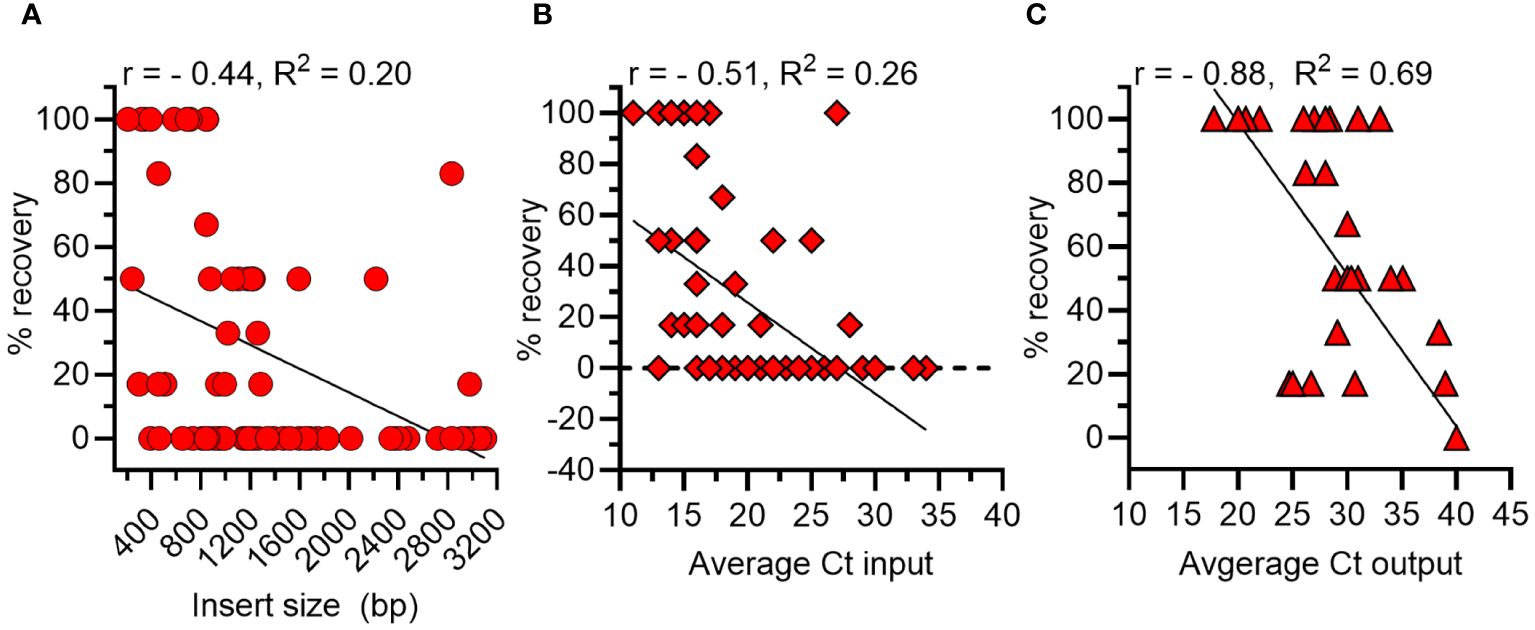
Figure 2 Correlations between features of A. marginale genes in the phage display library and outcome of biopanning using bovine RBCs. (A) There is a moderate negative correlation between the size in base pairs (bp) of a gene and its percentage recovery (% recovery). (B) There is moderate negative correlation between the amount of a gene in the input pool (Ct) prior to biopanning and its percentage recovery from RBCs after biopanning. (C) There is a strong negative correlation between the amount of a gene in the output pool (Ct) and its percentage recovery following biopanning. r = Pearson’s Correlation Coefficient.
3.2 Results of biopanning
Importantly, 73% (48/66) of A. marginale genes encoding phage displayed proteins were eliminated due to poor binding to RBCs (Supplementary Tables S1–S6). Additionally, 11% (7/66) of the A. marginale genes were recovered from 100% of the empty tubes. Thus, because these phages bound plastic, their capacity to bind RBCs could not be assessed. Overall, 17% (11/66) of the A. marginale phage displayed proteins met the criteria for selection as a potential adhesin for RBCs (Supplementary Table S7).
Of the four proteins from the Msp1 superfamily tested, 50% met the criteria as potential adhesins, including Msp1b and Mlp3. Mlp2 bound plastic and Mlp4 was eliminated as a potential adhesin due to poor binding to RBCs (Figures 3, 4A). Thirty-six percent of proteins with predicted functions were deemed potential adhesins including Aaap1, Alp2, Am573, Am560, and SucC (Figures 3, 4B). Among the outer membrane proteins not in a superfamily, 22% met the criteria as potential adhesins including OmpA and Am779 (Figures 3, 4C). Of the Msp2 superfamily proteins, 10%, including Msp3 and Omp13, met the criteria as potential adhesins (Figures 3, 4D). Interestingly, no potential adhesins were identified among the T4SS proteins (Figure 4E) or the proteins with unknown function (Figure 4F), though 8% and 14% of proteins in these groups bound plastic, respectively (Figure 3).
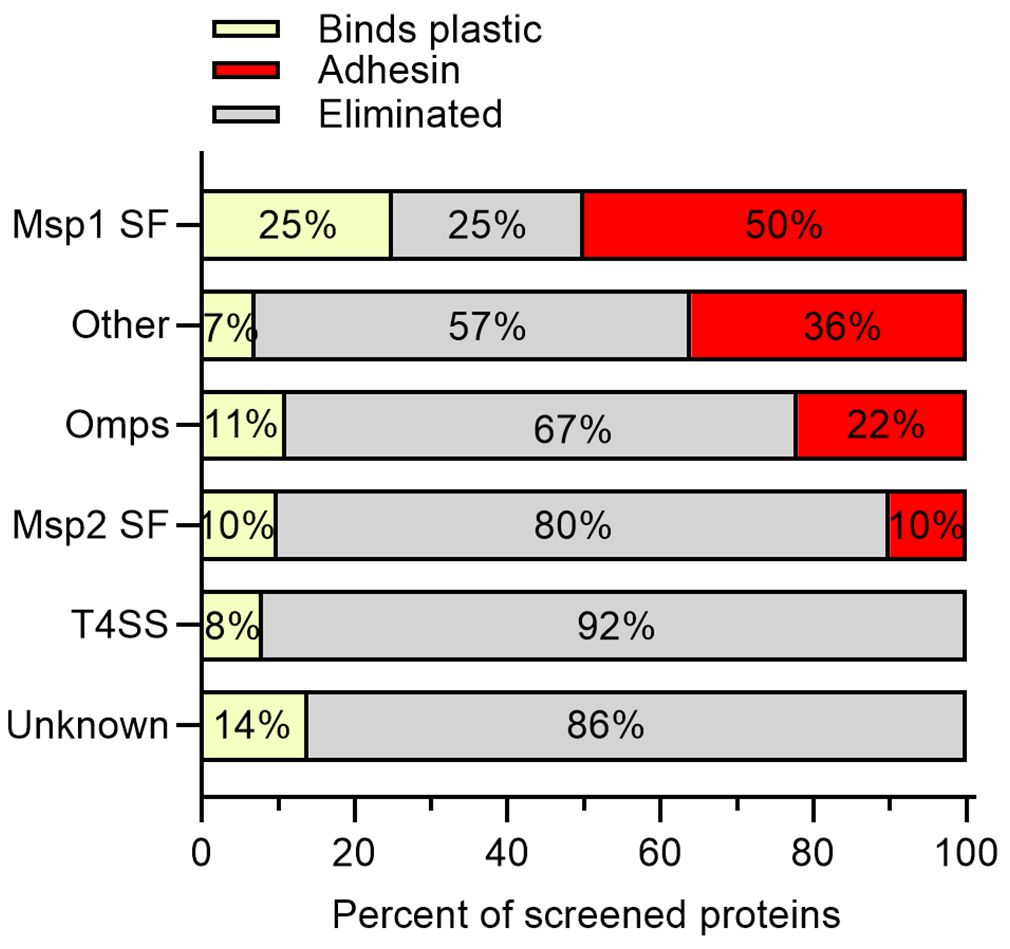
Figure 3 Outcome of biopanning summarized by functional protein groups. The screened proteins were grouped into the Msp1 superfamily (Msp1 SF), proteins with predicted functions or conserved domains (Other), outer membrane proteins not included in superfamilies (Omps), the Msp2 superfamily (Msp2 SF), type IV secretion system proteins (T4SS), and proteins with unknown function and lacking conserved domains (Unknown). Except for the Msp1a SF, the majority of proteins in each category were eliminated due to poor binding to RBCs (gray bars). Potential adhesins were identified in the Msp1 SF, Other category, Omps, and the Msp2 SF, but not the T4SS or proteins of unknown function. A smaller proportion of proteins in each category were recovered from empty tubes in 100% of replicates (yellow bars), and thus bound plastic.
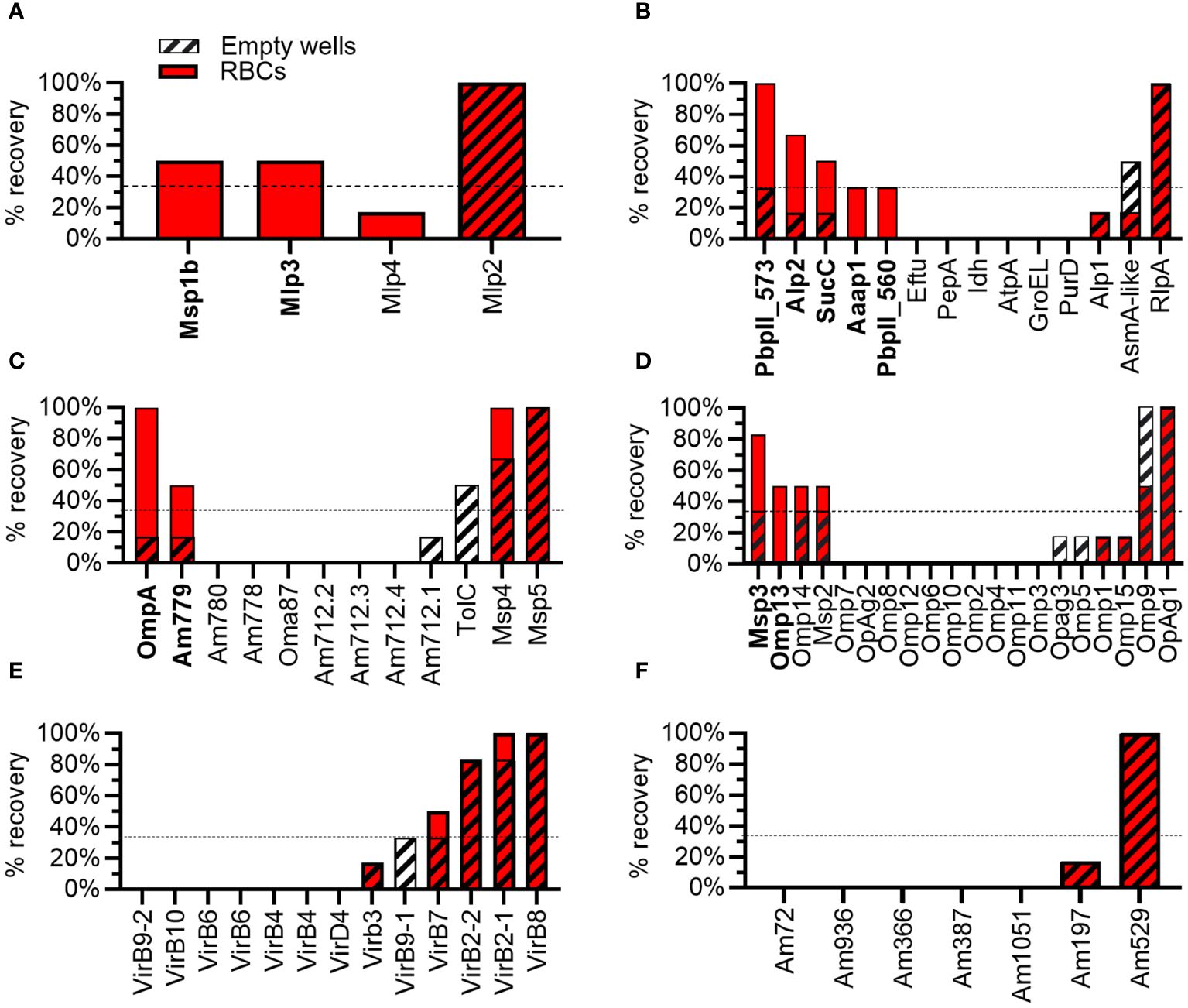
Figure 4 Results from biopanning for individual A. marginale proteins. Percentage recovery (% recovery) is the proportion of replicates from which the A. marginale gene was recovered following biopanning using RBCs (solid red bars) or empty tubes (striped bars). To meet the criteria as a potential adhesin (bold type) the percentage recovery from RBCs must be ≥ 33% (dotted line) and greater than twice the percentage recovery from empty tubes. Each graph represents a group of proteins including the (A) Msp1 superfamily, (B) proteins with predicted functions or conserved domains, (C) outer membrane proteins not included in superfamilies, (D) Msp2 superfamily, (E) T4SS proteins, and (F) proteins of unknown function and lacking conserved domains. Data represent two independent experiments.
4 Discussion
The 66 proteins in the phage display library include the majority of A. marginale outer membranes proteins (8, 24–27), nearly all A. marginale proteins bound by antibodies in serum from protectively immunized animals (11, 28, 29), and proteins of unknown function shown to be surface exposed through cross-linking and proteomics (8). Of these proteins, 11 bound bovine RBCs when expressed on the surface of T7 phages (Supplementary Tables S1-S7). Two of these proteins, Msp1b and OmpA, are known adhesins. Specifically, Msp1b is an adhesin for bovine RBCs and was more recently demonstrated to bind Dermacentor andersoni cells, derived from a natural vector of A. marginale (16, 17, 19). OmpA serves as an adhesin for endothelial cells, Ixodes scapularis cells, and D. andersoni cells (18, 30). Additionally, the majority (73%) of screened proteins were eliminated. Together these results demonstrate the utility of phage display to identify A. marginale adhesins for RBCs.
The Msp1 superfamily includes Msp1a, Msp1b, and three Msp1a-like proteins named Mlp2–4. Msp1a and Msp1b are covalently expressed on the surface of A. marginale and are both demonstrated adhesins for bovine RBCs (16, 17, 19, 31, 32). The N-terminal portion of Msp1a is composed of variable numbers of tandem repeats that form the host cell binding domain and are commonly used for genotyping (33, 34). The C-terminal portion of Msp1a and Mlp2-Mlp4 have four helical, transmembrane domains, suggesting these proteins may have a similar function despite low amino acid identity (16–34%) (24). Interestingly, Mlp3 met the criteria as a potential adhesin for RBCs in this study. Mlp2 bound plastic and thus could not be assessed. Mlp 4 was eliminated due to poor binding to RBCs. Thus, it is possible Mlp3 may function in concert with Msp1a and Msp1b to bind bovine RBCs. A role for Mlp2 in adhesion to RBCs cannot be determined without further studies.
The outer membrane proteins Am779, Msp3, and Omp13 met the criteria as RBC adhesins. Msp3 (Uniprot Q84H51) plays a prominent role in antigenic variation and is predicted to have an OMP-like beta barrel (35). Omp13 (Uniprot Q5P9G5) is invariant through time, but like Msp3, has an OMP-like beta barrel (36). Am779 (AOA643LT3_ANAMA) has a porin domain. Thus, all three of these proteins may serve as both transporters and adhesins for RBCs, though additional work is required to confirm these functions.
Am573, Am560, Aaap, Alp1, and succinate CoA-ligase (SucC), all have conserved domains or predicted functions and were identified as potential adhesins. It is likely these A. marginale proteins interact with host cell proteins that are not necessarily exposed on the surface of bovine RBCs. For example, Aaap and Alp1 form F-actin appendages in the cytoplasm of the RBC which are closely associated with the A. marginale containing vacuole (37–40). The A. marginale containing vacuole is likely derived from the RBC plasma membrane, thus accounting for the binding of the phage displayed Aaap and Alp1 to RBCs following biopanning. SucC plays a central role in the TCA cycle in the bacterial cytoplasm. While many cytoplasmic proteins moonlight as cell surface exposed adhesins in bacteria, moonlighting functions for SucC have not been identified (41). Finally, Am560 and Am573 have type II periplasmic binding protein domains, and thus likely shuttle substrates across the bacterial periplasm, though it is possible they have additional functions in A. marginale.
Biases in phage display may affect the outcome of biopanning. The moderate negative correlation between the size and amount of each A. marginale gene in the input pool of phages and the percentage recovery of that gene following biopanning indicates smaller and more abundant genes were more likely to be recovered following biopanning. For example, am573 was abundant in the input pool of phages and encodes the smallest protein identified as a potential adhesin, suggesting it may be a false positive. Thus, additional experiments are necessary to confirm the function of these potential adhesins.
Overall, this work supports the previous conclusion that Msp1b is an adhesin for RBCs and identified additional potential A. marginale adhesins for bovine RBCs, including OmpA, Mlp3, Omp13, Msp3, and Am779. Future work, including biopanning using RBCs from non-host species and pull-down assays will help provide additional evidence of the functional relevance of these proteins for adhesion and A. marginale entry into RBCs. The specific binding domains will serve as high priority vaccine candidates for future immunization and challenge studies. Ultimately, a cost-effective vaccine to prevent bovine anaplasmosis would improve cattle health and help improve global food security.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by University of Idaho Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SN: Conceptualization, Data curation, Formal analysis, Funding acquisition, Supervision, Visualization, Writing – original draft, Writing – review & editing. JU: Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. DA: Investigation, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. USDA-ARS CRIS# 2090-32000-038-000D and 2090-32000-043-000D.
Acknowledgments
We appreciate the kind help and advice of Holly Wichman at the University of Idaho concerning handling phage and Lowell Kappmeyer of the Animal Disease Research Unit concerning cloning.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2024.1422860/full#supplementary-material
Supplementary Figure 1 | Histogram of the number of A. marginale genes recovered following biopanning binned by percentage recovery from RBCs. The dotted line at 33% recovery is the seventy-fifth percentile for percentage recovery. This value was used as a cut-off for identification of potential adhesins. Phages displaying A. marginale genes recovered from 100% of empty wells were excluded from this analysis.
References
1. Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. (2010) 167:95–107. doi: 10.1016/j.vetpar.2009.09.012
2. Salinas-Estrella E, Amaro-Estrada I, Cobaxin-Cardenas ME, Preciado de la Torre JF, Rodriguez SD. Bovine Anaplasmosis: Will there ever be an almighty effective vaccine? Front Vet Sci. (2022) 9:946545. doi: 10.3389/fvets.2022.946545
3. Railey AF, Marsh TL. Economic benefits of diagnostic testing in livestock: Anaplasmosis in cattle. Front Vet Sci. (2021) 8:626420. doi: 10.3389/fvets.2021.626420
4. Brown DC. Dynamics and impact of tick-borne diseases of cattle. Trop Anim Health Prod. (1997) 29:1S–3S. doi: 10.1007/BF02632905
5. Pipano E. Live vaccines against hemoparasitic diseases in livestock. Vet Parasitol. (1995) 57:213–31. doi: 10.1016/0304-4017(94)03122-D
6. Bock RE, de Vos AJ. Immunity following use of Australian tick fever vaccine: a review of the evidence. Aust Vet J. (2001) 79:832–9. doi: 10.1111/j.1751-0813.2001.tb10931.x
7. Brown WC, Shkap V, Zhu D, McGuire TC, Tuo W, McElwain TF, et al. CD4(+) T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect Immun. (1998) 66:5406–13. doi: 10.1128/IAI.66.11.5406-5413.1998
8. Noh SM, Brayton KA, Brown WC, Norimine J, Munske GR, Davitt CM, et al. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect Immun. (2008) 76:2219–26. doi: 10.1128/IAI.00008-08
9. Noh SM, Turse JE, Brown WC, Norimine J, Palmer GH. Linkage between Anaplasma marginale outer membrane proteins enhances immunogenicity but is not required for protection from challenge. Clin Vaccine Immunol. (2013) 20:651–6. doi: 10.1128/CVI.00600-12
10. Tebele N, McGuire TC, Palmer GH. Induction of protective immunity by using Anaplasma marginale initial body membranes. Infect Immun. (1991) 59:3199–204. doi: 10.1128/iai.59.9.3199-3204.1991
11. Lopez JE, Siems WF, Palmer GH, Brayton KA, McGuire TC, Norimine J, et al. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect Immun. (2005) 73:8109–18. doi: 10.1128/IAI.73.12.8109-8118.2005
12. Wizemann TM, Adamou JE, Langermann S. Adhesins as targets for vaccine development. Emerg Infect Dis. (1999) 5:395–403. doi: 10.3201/eid0503.990310
13. Pizarro-Cerda J, Cossart P. Bacterial adhesion and entry into host cells. Cell. (2006) 124:715–27. doi: 10.1016/j.cell.2006.02.012
14. Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. (2015) 17:173–83. doi: 10.1016/j.micinf.2015.01.004
15. Vimonish R, Capelli-Peixoto J, Johnson WC, Hussein HE, Taus NS, Brayton KA, et al. Anaplasma marginale infection of Dermacentor andersoni primary midgut cell culture is dependent on fucosylated glycans. Front Cell Infect Microbiol. (2022) 12:877525. doi: 10.3389/fcimb.2022.877525
16. McGarey DJ, Barbet AF, Palmer GH, McGuire TC, Allred DR. Putative adhesins of Anaplasma marginale: major surface polypeptides 1a and 1b. Infect Immun. (1994) 62:4594–601. doi: 10.1128/iai.62.10.4594-4601.1994
17. de la Fuente J, Garcia-Garcia JC, Blouin EF, Kocan KM. Differential adhesion of major surface proteins 1a and 1b of the ehrlichial cattle pathogen Anaplasma marginale to bovine erythrocytes and tick cells. Int J Parasitol. (2001) 31:145–53. doi: 10.1016/s0020-7519(00)00162-4
18. Hebert KS, Seidman D, Oki AT, Izac J, Emani S, Oliver LD Jr., et al. Anaplasma marginale Outer Membrane Protein A is an adhesin that recognizes sialylated and fucosylated glycans and functionally depends on an essential binding domain. Infect Immun. (2017) 85:e00968–16(3). doi: 10.1128/IAI.00968-16
19. Noh SM, Ujczo J, Alperin DC, Jarvis SM, Solyman MSM, Koku R, et al. Identification of Anaplasma marginale adhesins for entry into Dermacentor andersoni tick cells using phage display. Infect Immun. (2024) 92:e00540-23. doi: 10.1128/iai.00540-23
20. Eriks IS, Stiller D, Goff WL, Panton M, Parish SM, McElwain TF, et al. Molecular and biological characterization of a newly isolated Anaplasma marginale strain. J Vet Diagn Invest. (1994) 6:435–41. doi: 10.1177/104063879400600406
21. Hussein HE, Johnson WC, Ueti MW. Differential paired stage-specific expression of Babesia bovis cysteine-rich GCC2/GCC3 domain family proteins (BboGDP) during development within Rhipicephalus microplus. Parasit Vectors. (2023) 16:16. doi: 10.1186/s13071-022-05628-6
22. Adey NB, Mataragnon AH, Rider JE, Carter JM, Kay BK. Characterization of phage that bind plastic from phage-displayed random peptide libraries. Gene. (1995) 156:27–31. doi: 10.1016/0378-1119(95)00058-E
23. Newton-Northup JR, Deutscher SL. Contending with target unrelated peptides from phage display. J Mol Imaging Dynam. (2012) 2:1000e101. doi: 10.4172/2155-9937.1000e101
24. Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci U.S.A. (2005) 102:844–9. doi: 10.1073/pnas.0406656102
25. Brayton KA, Palmer GH, Brown WC. Genomic and proteomic approaches to vaccine candidate identification for Anaplasma marginale. Expert Rev Vaccines. (2006) 5:95–101. doi: 10.1586/14760584.5.1.95
26. Dark MJ, Al-Khedery B, Barbet AF. Multistrain genome analysis identifies candidate vaccine antigens of Anaplasma marginale. Vaccine. (2011) 29:4923–32. doi: 10.1016/j.vaccine.2011.04.131
27. Palmer GH, McGuire TC. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J Immunol. (1984) 133:1010–5. doi: 10.4049/jimmunol.133.2.1010
28. Agnes JT, Brayton KA, LaFollett M, Norimine J, Brown WC, Palmer GH. Identification of Anaplasma marginale outer membrane protein antigens conserved between A. marginale sensu stricto strains and the live A. marginale subsp. centrale vaccine. Infect Immun. (2011) 79:1311–8. doi: 10.1128/IAI.01174-10
29. Lopez JE, Palmer GH, Brayton KA, Dark MJ, Leach SE, Brown WC. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect Immun. (2007) 75:2333–42. doi: 10.1128/IAI.00061-07
30. Ojogun N, Kahlon A, Ragland SA, Troese MJ, Mastronunzio JE, Walker NJ, et al. Anaplasma phagocytophilum outer membrane protein A interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect Immun. (2012) 80:3748–60. doi: 10.1128/IAI.00654-12
31. Vidotto MC, McGuire TC, McElwain TF, Palmer GH, Knowles DP Jr. Intermolecular relationships of major surface proteins of. Anaplasma marginale. Infect Immun. (1994) 62:2940–6. doi: 10.1128/iai.62.7.2940-2946.1994
32. de la Fuente J, Garcia-Garcia JC, Blouin EF, McEwen BR, Clawson D, Kocan KM. Major surface protein 1a effects tick infection and transmission of Anaplasma marginale. Int J Parasitol. (2001) 31:1705–14. doi: 10.1016/s0020-7519(01)00287-9
33. de la Fuente J, Van Den Bussche RA, Prado TM, Kocan KM. Anaplasma marginale msp1alpha genotypes evolved under positive selection pressure but are not markers for geographic isolates. J Clin Microbiol. (2003) 41:1609–16. doi: 10.1128/JCM.41.4.1609-1616.2003
34. Palmer GH, Rurangirwa FR, McElwain TF. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J Clin Microbiol. (2001) 39:631–5. doi: 10.1128/JCM.39.2.631-635.2001
35. Brayton KA, Knowles DP, McGuire TC, Palmer GH. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc Natl Acad Sci U.S.A. (2001) 98:4130–5. doi: 10.1073/pnas.071056298
36. Noh SM, Brayton KA, Knowles DP, Agnes JT, Dark MJ, Brown WC, et al. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect Immun. (2006) 74:3471–9. doi: 10.1128/IAI.01843-05
37. Fallquist HM, Tao J, Cheng X, Pierle SA, Broschat SL, Brayton KA. Dynamics of repeat-associated plasticity in the aaap gene family in Anaplasma marginale. Gene. (2019) 721S:100010. doi: 10.1016/j.gene.2019.100010
38. Kocan KM, Venable JH, Brock WE. Ultrastructure of anaplasmal inclusions (Pawhuska isolate) and their appendages in intact and hemolyzed erythrocytes and in complement-fixation antigen. Am J Vet Res. (1978) 39:1123–30.
39. Stich RW, Kocan KM, Damian RT, Fechheimer M. Inclusion appendages associated with the intraerythrocytic rickettsial parasite Anaplasma marginale are composed of bundled actin filaments. Protoplasm. (1997) 199:93–8. doi: 10.1007/BF02539810
40. Stich RW, Olah GA, Brayton KA, Brown WC, Fechheimer M, Green-Church K, et al. Identification of a novel Anaplasma marginale appendage-associated protein that localizes with actin filaments during intraerythrocytic infection. Infect Immun. (2004) 72:7257–64. doi: 10.1128/IAI.72.12.7257-7264.2004
Keywords: bovine anaplasmosis, adhesins, obligate intracellular pathogen, vaccine, Anaplasma marginale, red blood cells, phage display
Citation: Noh SM, Ujczo J and Alperin DC (2024) Identification of Anaplasma marginale adhesins for bovine erythrocytes using phage display. Front. Trop. Dis 5:1422860. doi: 10.3389/fitd.2024.1422860
Received: 24 April 2024; Accepted: 11 June 2024;
Published: 25 June 2024.
Edited by:
Seham Hendawy, National Research Centre, EgyptReviewed by:
Sergio D. Rodríguez Camarillo, INIFAP, MexicoPablo Andrés Parodi Thexeira, National Institute for Agricultural Research (INIA), Uruguay
Amira Adel Taha Abdel Aleem AL-Hosary, Assiut University, Egypt
Copyright © 2024 Noh, Ujczo and Alperin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susan M. Noh, c3VzYW4ubm9oQHVzZGEuZ292
 Susan M. Noh
Susan M. Noh Jessica Ujczo1
Jessica Ujczo1