- 1Department of Veterinary Tropical Diseases, Faculty of Veterinary Science, University of Pretoria, Onderstepoort, South Africa
- 2Department of Life and Consumer Sciences, College of Agriculture and Environmental Sciences, University of South Africa (UNISA), Roodepoort, South Africa
- 3Department of Agriculture and Animal Health, University of South Africa, Florida, South Africa
- 4Department of Agriculture, Rural Development, Land and Environmental Affairs, Mbombela, South Africa
- 5Department of Water and Environmental Technology, Steffen Robertson Kirten House, Johannesburg, South Africa
- 6Department of Biodiversity, Faculty of Science and Agriculture, University of Limpopo, Sovenga, South Africa
Introduction: Ticks are obligate ectoparasites recognized worldwide as major vectors of several disease-causing pathogens and are good indicators of disease distribution and epidemiology. Recent years have seen a growing concern regarding emerging and re-emerging of economically important tick-borne pathogens of livestock and humans worldwide. The overall objective of the study was to give an insight into current tick distribution and associated bacterial pathogens that may pose a threat to cattle in the sampled study sites.
Methods: A total of 150 cattle were randomly selected from three study sites, Harrismith and Phuthaditjhaba in Free State Province and Bergville in KwaZulu Natal Province, South Africa. Blood samples were collected from the cattle and DNA was subjected to the 16S rRNA gene microbiome sequencing on the circular consensus PacBio sequencing platform. Ticks were also collected from various predilection sites of the sampled animals.
Results: A total of eight tick species were identified and Rhipicephalus evertsi evertsi (79.4%) was the most abundant followed by R. appendiculatus (11.7%), R. afranicus (2.6%), R. simus (2.6%), Hyalomma rufipes (1.2%), R. decoloratus (1.0%), H. truncatum (0.7%) and R. microplus (0.7%). The bacterial microbiome sequence analysis revealed up to 16 phyla and 30 classes in the three study sites. Proteobacteria was the most dominant bacterial phyla with a relative abundance of 67.2% (Bergville), 73.8% (Harrismith) and 84.8% (Phuthaditjhaba), followed by Firmicutes at 9.6% (Phuthaditjhaba), 18.9% (Bergville) and 19.6% (Harrismith).
Conclusion and perspectives: The Chao 1 index estimator revealed significant differences in the α-diversity of microbial communities among three study sites. This study expands the knowledge on tick fauna and microbial communities in the three study sites.
1 Introduction
Ticks are good indicators of disease distribution and epidemiology (1) and are widely distributed particularly in tropical and subtropical areas. Their distribution is dependent on the environment and host for growth and survival (2). Various studies have shown that cattle and other animal species are mostly infested by ticks, transmitting several pathogens (3–10). Ticks transmit obligate intracellular Gram-negative bacterial species of animal and human importance in the order Rickettsiales such as Ehrlichia chaffeensis, E. ruminantium the causative agent of heartwater in ruminants (11), Anaplasma phagocytophilum that causes human granulocytic anaplasmosis, and A. marginale and A. centrale that cause anaplasmosis (gallsickness) in ruminants (12). The other pathogens are Borrelia duttonii which causes tick-borne relapsing fever and Rickettsia species responsible of causing rickettsioses (13), while Coxiella burnetii causes Query fever (Q-fever) in humans or coxiellosis in domestic animals (14). Apart from pathogen transmission, ticks can damage hides, cause blood loss, severe allergic reactions, immune-depression, irritation, and chronic stress (15).
There is a growing concern of emerging and re-emerging tick-borne pathogens of public health concern and of economic significance amongst livestock, but data from African ecosystems is still scanty. The most affected areas are resource-poor, where factors such as poor veterinary services, inability of the farmers to purchase veterinary medicines, uncontrolled translocation of livestock, the wildlife-livestock interface, and burning of grazing land promote emergence and re-emergence of tick-borne pathogens in an environment where climate change is notable (16). Among the few microbiome studies conducted in South Africa, Kolo et al. (17) investigated possible causes of acute febrile illness and found A. phagocytophilum, A. platys and Anaplasma sp. SA/ZAM dog DNA in blood of rodents, dogs, cattle, and humans. In another study, Makgabo et al. (18) revealed the presence of nine novel Anaplasma 16S rRNA genotypes in 11 wild animal species, highlighting the importance of emerging tick-borne pathogens among communities in South Africa.
Global warming has effected the change in the climate and disrupted the ecosystems in agriculture. Consequently, tick distribution and the pathogens they transmit. The other factor is the competence of invasive tick species. The invasive ticks, such as the one-host tick, R. microplus and the three-host tick, A. variegatum have high adaptability and capacity to transmit several pathogens. In South Africa, R. microplus, is the vector of Babesia bigemina, B. bovis and A. marginale. This tick has been reported to displace the indigenous tick, R. decoloratus, in South Africa (7, 19). Amblyomma variegatum is prevalent in various regions within Africa and is considered as the second most invasive tick species after R. microplus (20). It is the vector of E. ruminantium.
Tick distribution has been used as an indication of tick-borne disease distribution, thus demonstrating the presence of various pathogens. However, the aforementioned factors such as climate change, the adaptation of invasive tick species and translocation of animals indicate that the tick distribution might not be a concise marker of the distribution of disease. For instance, the vector may be present, but the pathogen might either not have been reported or the vector might have been displaced by invasive tick species, while other tick-borne pathogens might be concurrently transmitted by other vectors such as biting flies or through contaminated veterinary instruments (21, 22).
In this study, three neighboring small towns: Bergville in KwaZulu Natal Province, Harrismith and Phuthaditjhaba in the Free State Province, South Africa were targeted for the surveillance of ticks and tick-borne pathogens. These towns are separated by the Drakensberg Mountains that serve as the grazing areas for the livestock. Between the three towns, there is a constant movement of livestock for cultural practices, breeding for beef production and barter trade. Tick distribution studies on cattle have previously been carried out to catalog ticks and tick-borne pathogens in Phuthaditjhaba and Harrismith (9, 14, 23, 24). In Bergville, however, only one study has been carried out thus far (25). The studies were conducted eight years ago in the Free State and five years ago in KwaZulu-Natal.
In view of the above, this study sought to determine whether there have been alterations in tick distribution due to factors such as climate change, livestock translocation, and vector displacement and to further determine associated bacterial pathogens and conduct a comparative analysis of cattle blood microbiome present at the three study sites.
2 Materials and methods
2.1 Ethics approval
The study was approved by the Animal Ethics Committee of the University of Pretoria, South Africa (REC029–21), and collection of cattle samples was approved by the Department of Agriculture, Land Reform and Rural Development under section 20 of the Animal Disease Act 35 of 1984 with reference 12/11/1/1/MG.
2.2 Study area
The study was conducted in three neighboring sites in South Africa, one site in KwaZulu-Natal Province (Bergville) and two in the Free State Province (Harrismith, Phuthaditjhaba), from 29 October through 01 November 2021 (Figure 1). The three sites are rural and demarcated by Drakensberg mountains.
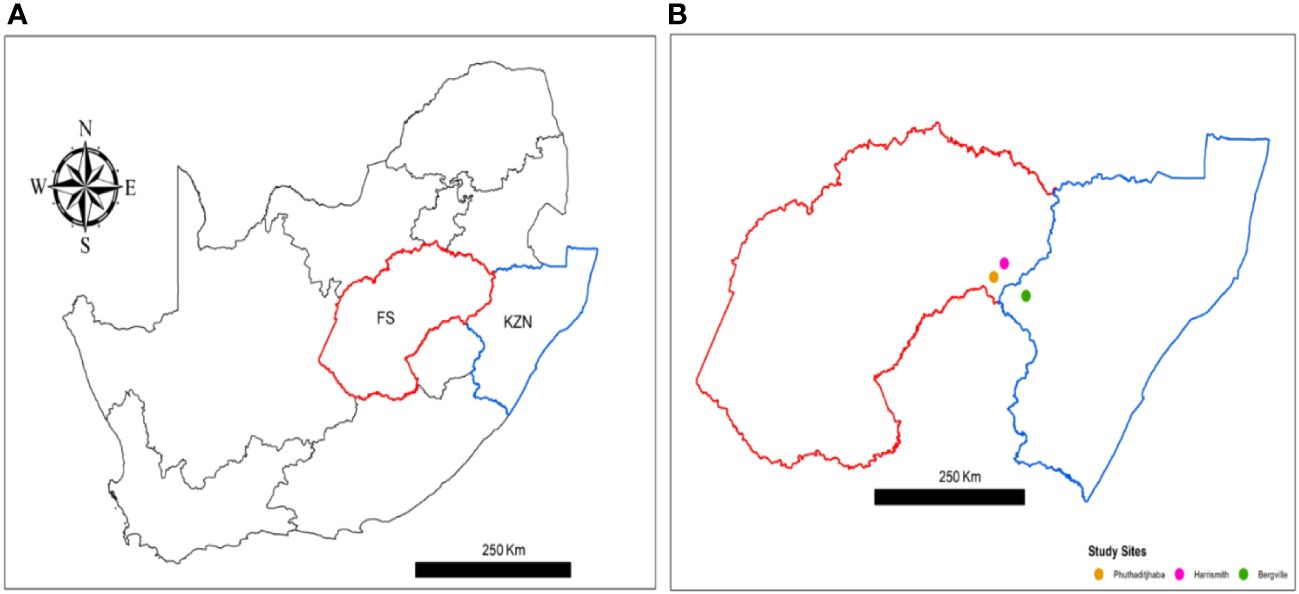
Figure 1 Map of South Africa showing two neighboring provinces where study sites are located; (A) in red = Free State (FS), where Phuthadijthaba and Harrismith are located, blue = KwaZulu Natal (KZN) where Bergville is located. (B) The study sites Harrismith (pink dot), Phuthaditjhaba (yellow dot) and Bergville (green dot). Map were constructed using ArcGIS Desktop 10.8.2 (Esri ArcMap).
There is regular movement of cattle among the three areas. The climate in Bergville is warm and temperate, with temperature varying from 3°C to 28°C (average 15°C). January is the warmest month of the year, with an average temperature of 20.1°C. The summer months (December to March) are wetter than the winters and the annual rainfall is 1657 mm. The driest month is June with 19 mm and the greatest amount of precipitation occurs in January, with an average of 312 mm. Average temperatures in October and November are 17.1°C and 18.5°C, while average rainfall is 148 mm and 196 mm, respectively (https://en.climate-data.org/africa/south-africa/kwazulu-natal/bergville-14872/).
In Harrismith, the temperature typically varies from -1°C to 26°C. The warm season is from October through March and the cold season is from May through August. The rainy period is from August through May, with a sliding 31-day rainfall of at least 12.5 mm. Average temperatures in October and November are 16.3°C and 17.3°C, while average rainfall is 94 mm and 128 mm, respectively (https://en.climate-data.org/africa/south-africa/free-state/harrismith-12770/).
The climate in Phuthaditjhaba is mild, and generally warm and temperate. The temperature averages 13.4°C (monthly average range 7°C to 17.8°C). Precipitation is about 1020 mm (monthly average range 12 mm to 184 mm). The summer months are December, January, February and March. Average temperatures in October and November are 15.2°C and 16.4°C, while average rainfall is 109 mm and 135 mm, respectively (https://en.climate-data.org/africa/south-africa/free-state/phuthaditjhaba-55825/).
2.3 Collection and identification of adult ticks
Representative adult tick samples were collected from the predilection sites: ears, dewlap, tail switch, back, groin, udder, around anus, lower abdomen of the 150 cattle from the three study sites. The ticks were kept in airtight containers with moist cotton wool prior to identification using a stereoscopic microscope at the Ectoparasitology laboratory in the Department of Veterinary Tropical Diseases, University of Pretoria, South Africa, and using morphological keys described by Walker (26).
2.4 Blood sample collection
A total of 150 cattle from the three sites were enrolled for the study. A herd of fifty cattle were randomly selected from each of the three sampling sites, at their respective communal sites or dip tanks. Cattle were apparently healthy but infested with ticks, predominantly adults of both sexes and of Nguni breed (Bergville and Harrismith) or Brahman breed (Phuthaditjhaba). In Harrismith, communal dipping programs are lacking, with the last dipping conducted four months preceding this study (June 2021). However, some farmers, despite limited resources, resorted to alternative methods such as cattle sprays and injectable solutions for tick control. In Phuthaditjhaba, pour-on dipping is the method used every 3–4 months. In Bergville, dipping takes place twice a month at the Woodford dip tank.
Blood samples were collected from the coccygeal vein of individual animals into 10-ml sterile EDTA vacutainer tubes. The blood was stored at 4°C until further analysis.
2.5 DNA extraction, 16S rRNA gene amplification, and PacBio sequencing
Genomic DNA was extracted from 200 µl of blood using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The presence of bacterial communities in the blood samples was analyzed using single molecule real-time PacBio sequencing technology (Pacific Biosciences, Menlo Park, CA, USA). The full-length 16S rRNA gene was amplified from genomic DNA using bacterial-specific primer 27F 5’-AGA GTT TGA TCM TGG CTC AG-3’ and 1492R 5’-TAC GGY TAC CTT GTT ACG ACT T-3’. For multiplexing of amplicons, the 5`-ends of the 16S rRNA forward and reverse primers were tagged with the universal M13F (TGTAAAACGACGGCCAGT) and M13R (GGAAACAGCTATGACCATG) sequences respectively. Two amplification reactions contained water as a template were used as negative controls. SMRTbell libraries were created using SMRTbell™ Template Prep Kit 1.0 (Pacific Biosciences, CA, USA) following instructions in the protocol “Procedure & Checklist – Amplicon Template Preparation and Sequencing” (part number 100–801-600–04). Sequencing was done using the Sequel® Sequencing Kit 2.1 (Pacific Biosciences, CA, USA) with on-plate loading concentration of 4 pM.
2.6 Sequence analysis using divisive amplicon denoising algorithm 2 workflow
The DADA2 package (version 1.12.1) analysis workflow implemented in R statistical software (v3.6.1) was used to analyze raw amplicon sequencing data generated using the PacBio Sequel System (Pacific Biosciences, CA, USA). To infer amplicon sequence variants, error-model learning, and chimera removal were performed on the filtered reads using default DADA2 parameters. Taxonomic assignments were made based on the curated SILVA 16S rRNA database (27). Taxa and abundance tables generated by DADA2 were imported into the phyloseq package v1.28.0 (28) for downstream analysis and visualization, including estimation of richness and visualization of the alpha-diversity, as well as visualization of differences in taxa abundance between the three study sites.
2.7 Statistical analysis
Effect sizes of the differences among the three study sites were calculated using the Cohen’s D measure using the effsize package in R (https://github.com/mtorchiano/effsize), based on Shannon diversity indices. Ordinations for β-diversity between study sites were estimated using Principal Coordinate Analysis (PCoA) based on weighted-UniFrac distance and Non-Metric Multidimensional Scale (NMDS) using Bray distance metric as implemented in the plot_ordination and amp_ordinate functions in PhyloSeq and the ampvis2 package (https://madsalbertsen.github.io/ampvis2/articles/ampvis2.html), respectively. Permutational Multivariate Analysis of Variance (PERMANOVA) using permutation test with pseudo-F ratios as implemented in the Adonis function of the Vegan R package (https://github.com/vegandevs/vegan) was used to determine significance for sample clustering on ordination plots.
We assessed the difference in relative abundance by family, genus, order and species among the three localities using the Kruskal-Wallis test. Analyses were performed in R statistical software (29) at a significant level of 0.05.
3 Results
3.1 Distribution of tick species among cattle in the three study sites
A total of 418 ticks were collected from 150 cattle in the three study sites (Harrismith, Phuthaditjhaba, Bergville). Eight tick species were identified, and the most abundant species in all study sites was R. evertsi evertsi (79.4%), followed by R. appendiculatus (11.7%), R. simus (2.6%) and R. afranicus n. sp. (formerly R. turanicus) (2.6%) (Table 1). Rhipicephalus simus and R. afranicus were found in Phuthaditjhaba and not in other locations. Of the 49 R. appendiculatus ticks collected, 48 were found in Phuthaditjhaba and only one tick found in Bergville (Table 1). Rhipicephalus microplus and R. decoloratus were not found in Bergville (Table 1).
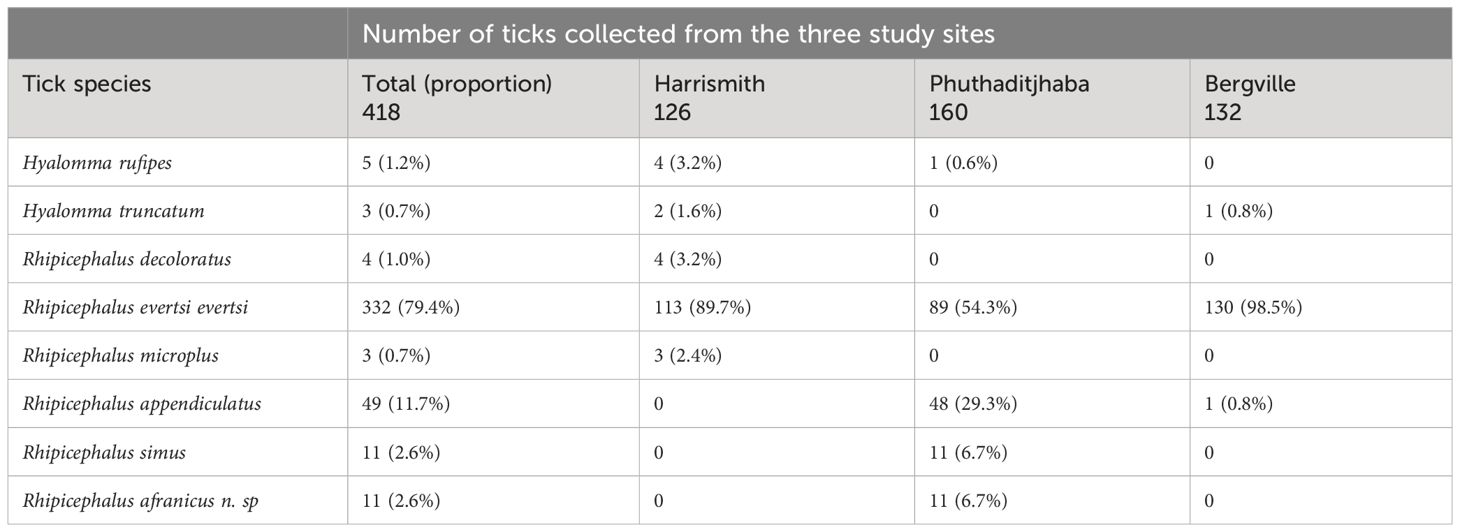
Table 1 Identification and distribution of ticks collected from cattle in three study sites of South Africa.
3.2 Microbial diversity in cattle samples collected from Harrismith, Phuthaditjhaba, and Bergville
The mean species diversity of bacterial populations detected in the bovine samples plotted using the rarefaction curves showed that the sequencing depth was adequate to capture the existing microbial diversity (Figure 2).
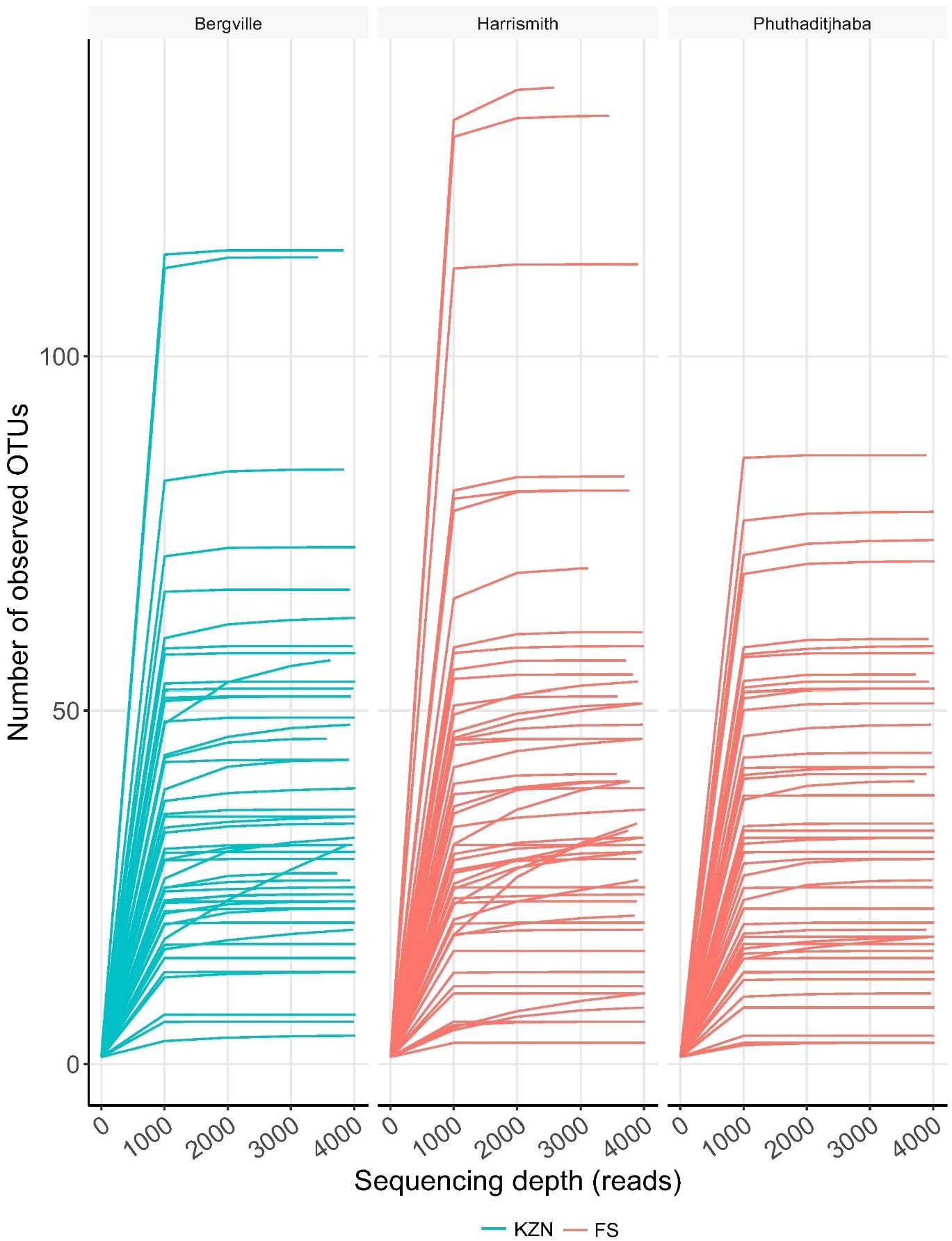
Figure 2 Bovine blood microbiome diversity. Rarefaction curves indicate the effect of sequencing depth (read numbers per sample, X axis) on species richness (Y axis) in bovine blood samples.
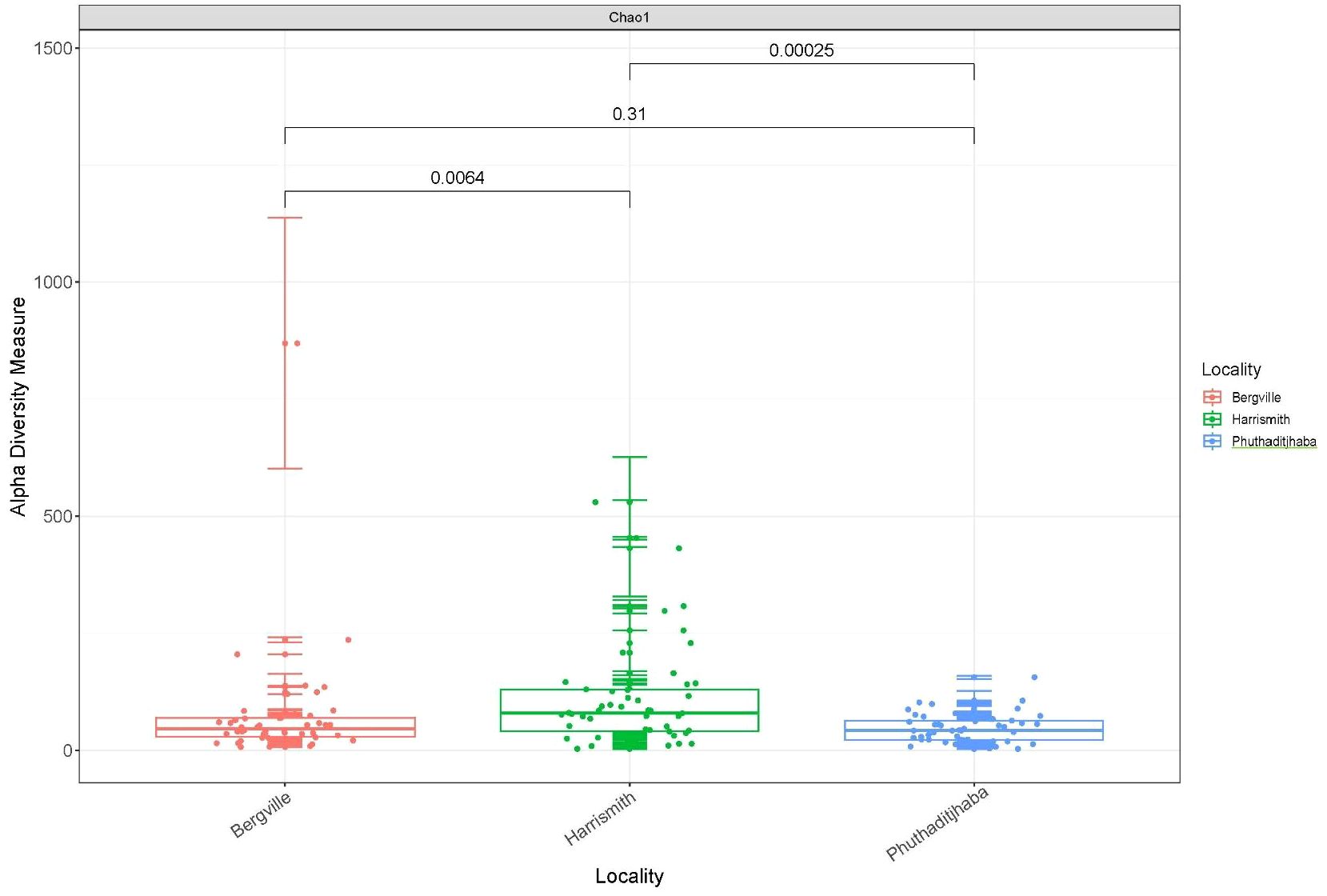
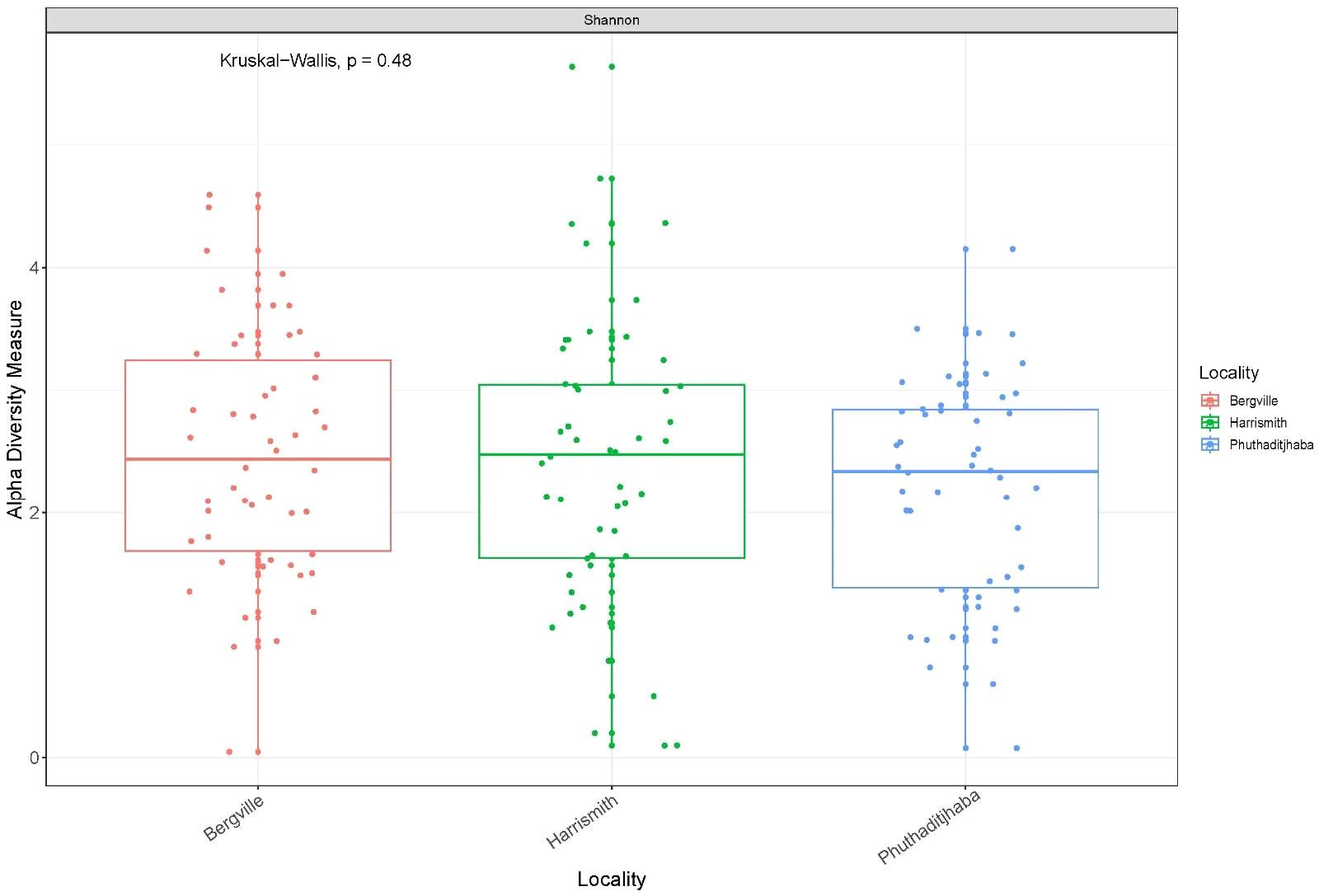
The alpha diversity boxplots reflected the minimal, median, degree of dispersion, maxima and outliers of microbial diversity within study sites. The alpha diversities were estimated through Chao1 index, which measures species richness and the Shannon index, that measures both species richness and evenness (30). The microbial diversity of samples from Harrismith indicated significantly higher species richness than Bergville (p = 0.0064) and Phuthaditjhaba (p = 0.00025) samples as determined using the Chao1 index estimator (Kruskal-Wallis: p = 0.00065) (Figure 3). However, the differences in species diversity between the three-study site was insignificant as per the Shannon estimator (Kruskal-Wallis: p = 0,48) (Figure 4). The effect size measurements (Table 2) were small to evaluate differences between microbial communities present in the three study sites. Using the pairwise analysis, the α-diversity varied significantly in microbial communities between Bergville and Harrismith and between Harrismith and Phuthadijthaba based on Chao 1 index estimator (p = 0.0064 and p = 0,00025, respectively). In contrast, there was no significant difference between Bergville and Phuthaditjhaba microbial communities based on Chao 1 index estimator (p = 0.31).
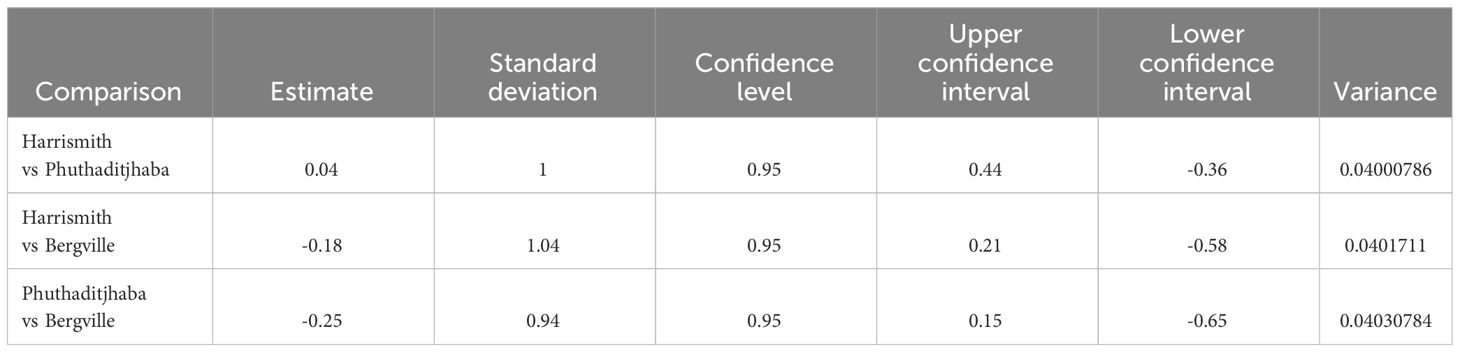
Table 2 Effect sizes of the differences between the three study sites measured using the Cohen’s D with a confidence interval of 95%.
Furthermore, microbial composition dissimilarities among the three study sites were analyzed through PCoA plots. The plots showed one cluster of microbial communities using the weighted UniFrac distance metric on PCoA, which considers abundance and the phylogenetic distance between ASVs. Three clusters indicative of the study sites could be observed (PCoA persanova). The Bergville sample composition was divergent from the Harrismith and Phuthaditjhaba groups: however, the Phuthaditjhaba and Harrismith groups clustered quite closely (Figure 5).
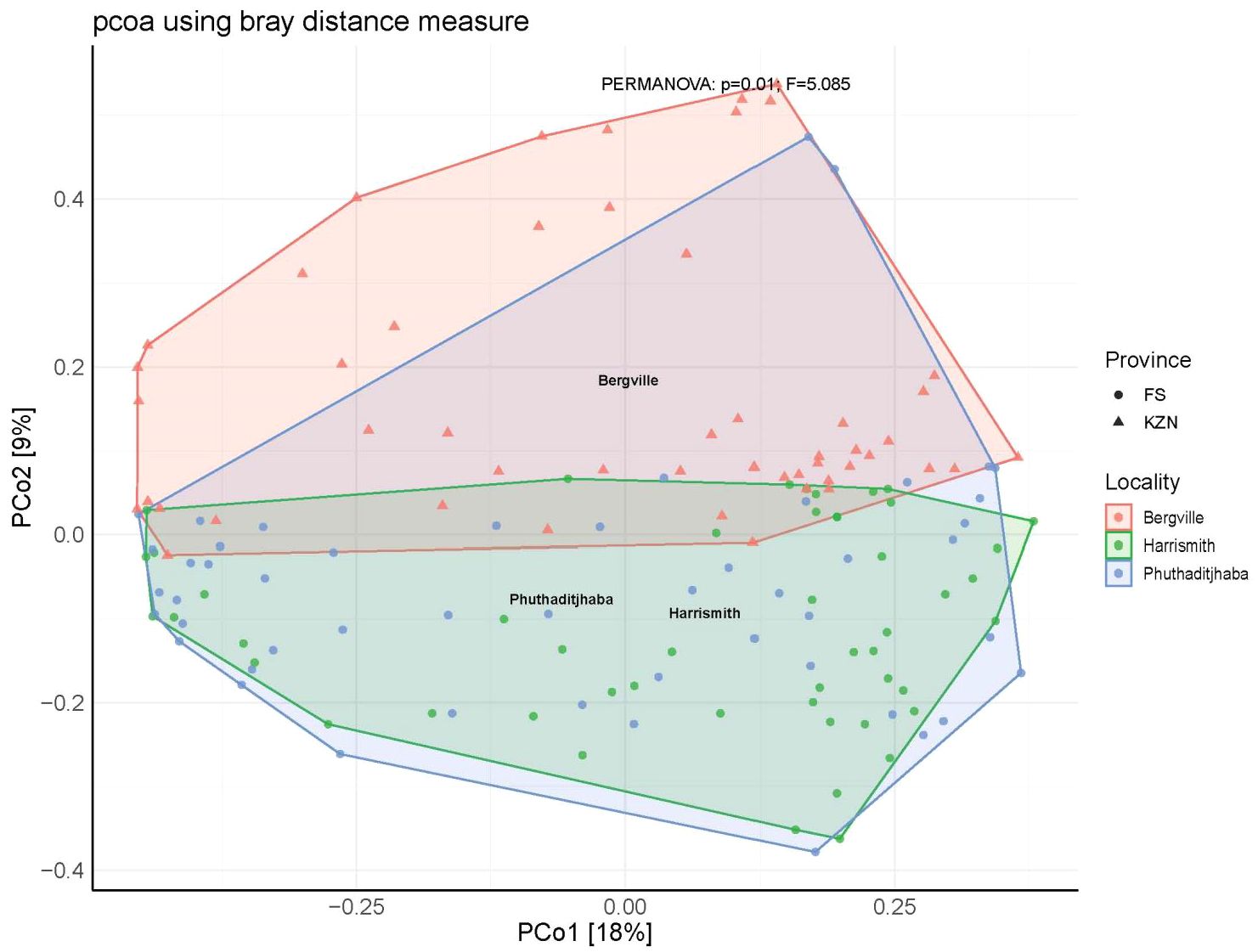
Figure 5 Ordination Plots showing clustering of bacterial diversity at the three study sites estimated using PCoA through Bray distance matrix.
Overall, 16 phyla, 30 classes, 71 order, 114 families, 159 genera and 71 species were identified from the three sites (Supplementary Table S1). At the phylum level, the composition of bovine blood microbiomes was dominated by Proteobacteria with a relative abundance of 67.2% (Bergville), 73.8% (Harrismith) and 84.8% (Phuthaditjhaba), followed by Firmicutes at 9.6% (Phuthaditjhaba), 18.9% (Bergville) and 19.6% (Harrismith). Bacteroidota was the third most abundant at 4% (Phuthaditjhaba), 5.3% (Harrismith) and 10.2% (Bergville) (Figure 6; Supplementary Table S2). The relative abundance of the Proteobacteria, Firmicutes and Bacteroidota remained higher in Phuthaditjhaba, Harrismith and Bergville, respectively (Supplementary Table S2). The phyla Fusobacteriota and Spirochaetota were only found in Bergville, while Deinococcota was only found in Phuthaditjhaba (Supplementary Table S2). About half of the detected phyla had a relative abundance of ≥0.1% in the various microbiomes (Bergville=7/13, Harrismith=5/11, Phuthaditjhaba=8/13).
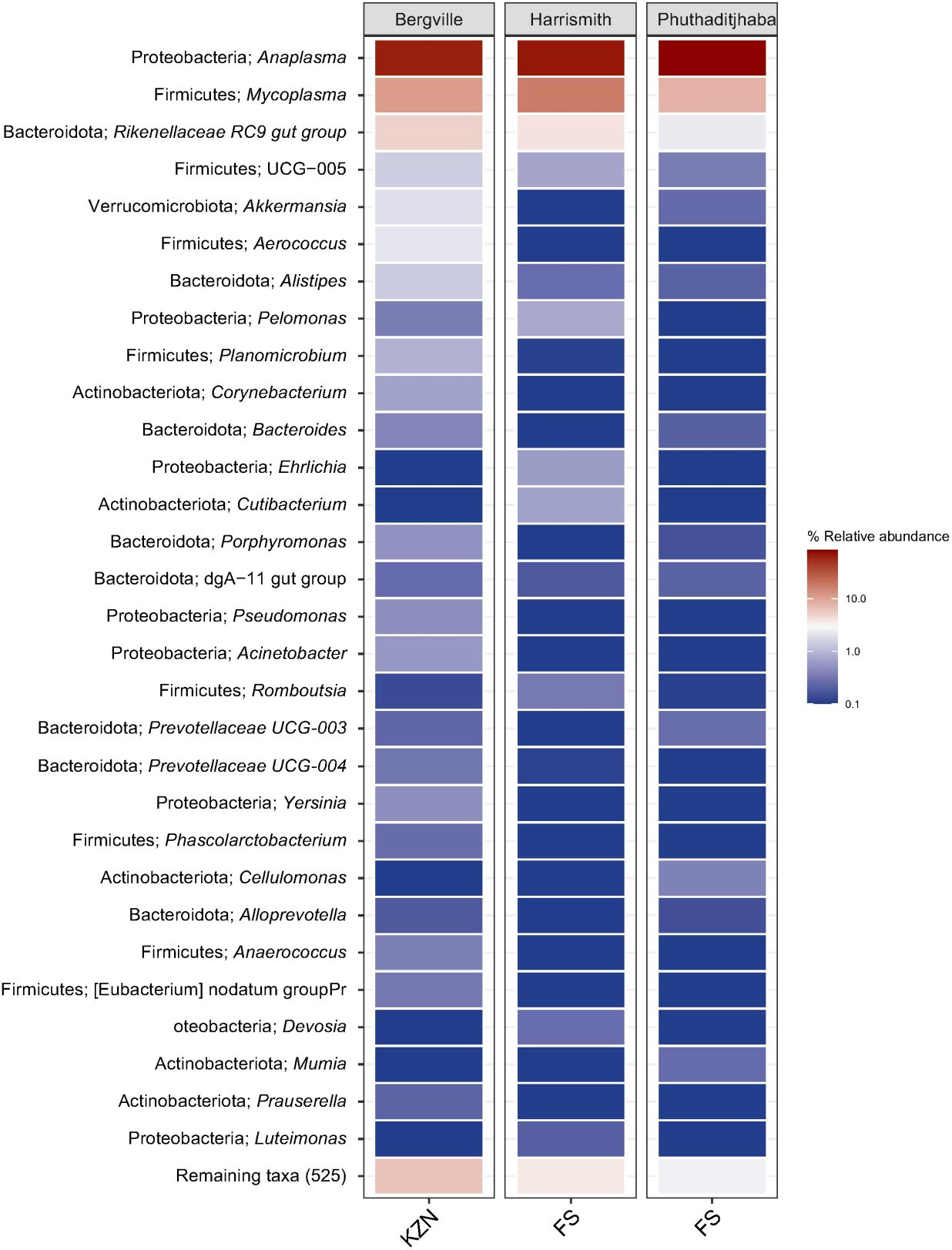
Figure 6 Heat map visualization. The phylum and genus microbiome composition, diversity and associated relative abundance in bovine blood samples collected from Bergville, Harrismith and Phuthaditjhaba in South Africa. The plots show the relative abundance and distribution of 30 most abundant genera (shown on the Y-axis), with the remaining genera grouped as “Remaining taxa”. The distribution and relative abundance of various taxonomic levels in the three study sites are also available in Data supplement Supplementary Table S1.
The most dominant classes in the Bergville, Harrismith and Phuthaditjhaba microbiomes were Alphaproteobacteria (64.8, 72.4%, 84.4%), Bacilli (14.3%, 17.5%, 8.2%) and Bacteroidia (10.2%, 5.3%, 4%) (Supplementary Table S2). Class Spirochaetia was unique to the Bergville microbiome, while classes Blastocatellia, Cynobacteria, Deinococei and Phycisphaerae were unique to the Phuthaditjhaba microbiome (Supplementary Table S2). About half of the detected classes had relative abundance of ≥0.1% in Bergville (11/20), Harrismith (8/20) and Phuthaditjhaba (12/23) (Supplementary Table S2).
The composition of bovine blood microbiomes at the order level in Bergville, Harrismith and Phuthaditjhaba was numerically dominated by Rickettsiales (64.7%, 72%, 84.3%), respectively, followed by Mycoplasmatales (11%, 17.3%, 8.1%) and Bacteroidales (10.2%, 5%, 4%) (Supplementary Table S2). There was a significant difference in the relative abundance among the three study sites (Kruskal Wallis test, p=0.0281). At least 46 orders of bacteria were identified in each of the three microbiomes, of which 13.0% (6/46), 16.0% (8/50) and 19.6% (10/51) were exclusively unique in the Bergville, Harrismith and Phuthaditjhaba microbiome, respectively (Supplementary Table S2). There were more orders with relative percent abundance ≥0.5% in the Bergville microbiome (12/46) than in the Harrismith (6/50) and Phuthaditjhaba (5/51) (Supplementary Table S2).
The unique and shared distribution of bacterial families, genera and species found in the Phuthaditjhaba, Harrismith and Bergville samples are shown by comprehensive Venn diagrams (Figures 7A–C). We detected 114 bacterial families, of which 69, 77 and 81 were found in Phuthaditjhaba, Bergville and Harrismith microbiome, respectively (Figures 7A–C; Supplementary Table S2). Moreover, 41 bacterial families were shared across the three study sites (Figure 7A). The Phuthaditjhaba microbiome had an exclusively unique association with 13 bacterial families, while 12 and 17 bacterial families were uniquely present in Bergville and Harrismith bovine blood samples (Figure 7; Supplementary Table S2). There was significant variation in the relative percent abundance (Kruskal-Walli’s test, p =0.0045) of the bacteria at the family level among the three study sites. More than 80% of the bacterial microbiome in Bergville, Harrismith and Phuthaditjhaba was largely dominated by members of three families: Anaplasmataceae (65.0%, 72.1%, 84.7%, respectively), Mycoplasmataceae (11.1%, 17.4%, 8.1%) and Rikenellaceae (6.6%, 4.3%, 2.7%). There were more families with relative percent abundance ≥0.5% in the Bergville microbiome (14/77) than in the Harrismith (7/81) and Phuthaditjhaba (4/69).
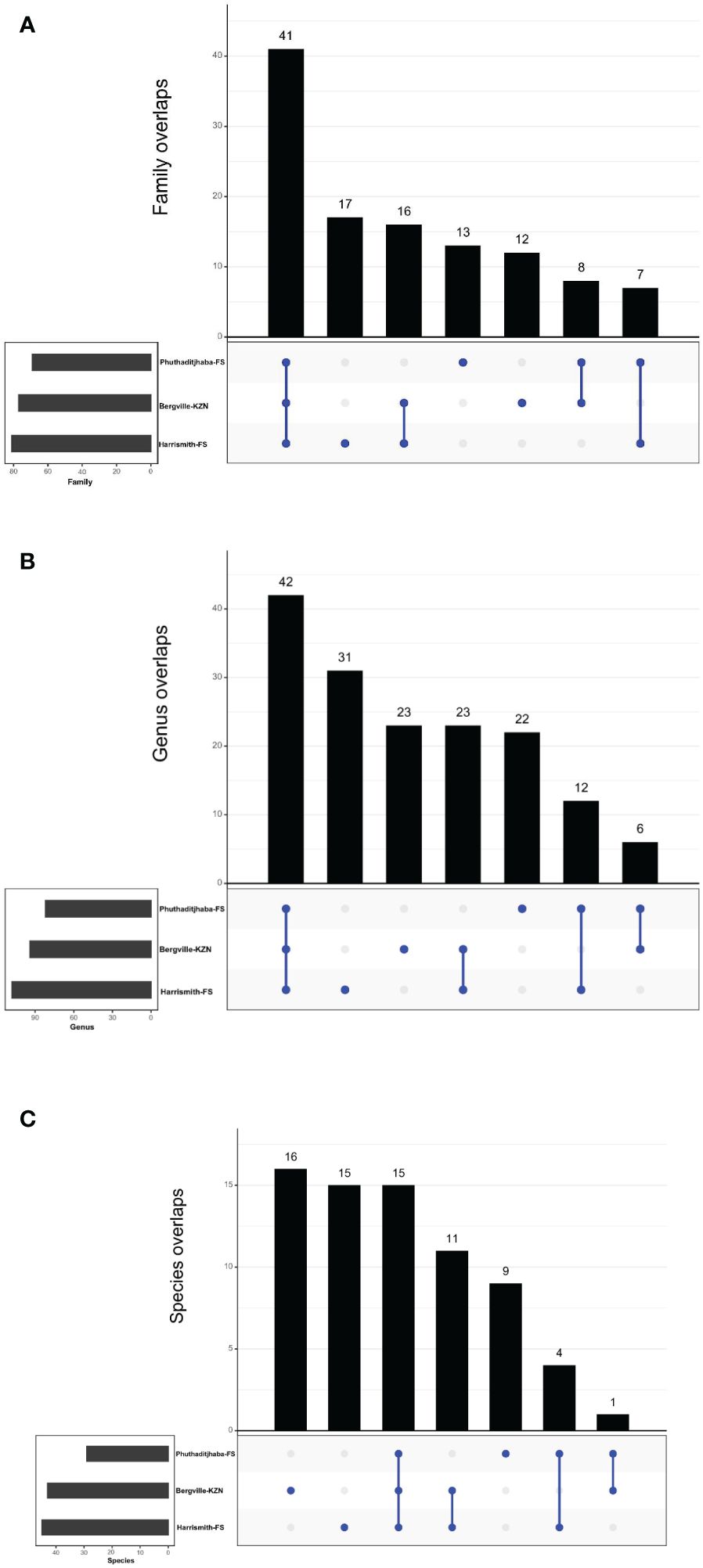
Figure 7 (A–C) Taxonomic composition of microbiomes from three locations in South Africa. Venn diagrams representing the unique and shared microbiomes in the Phuthaditjhaba, Bergville and Harrismith bovine blood samples. (A) Venn diagram representing unique and shared bacterial families, (B) Venn diagram comparison of bacteria at genus level by MR analysis, (C) Venn diagram showing unique and shared bacterial species in blood samples as determined using the MR pipeline. Microbiome uniqueness or sharing among the locations is symbolized by blue dots or lines, respectively.
Overall, we detected 159 microbiome genera, of which 94, 108 and 82 were found in Bergville, Harrismith and Phuthaditjhaba, respectively (Figure 7B). Forty-two genera were shared across the three study sites (Figure 7B), while 23, 31 and 22 exclusively associated microbiome genera were found in the Bergville, Harrismith and Phuthaditjhaba, respectively (Figure 7B; Supplementary Table S2). There were significant differences in the relative percent abundance (Kruskal-Wallis test, P=0.00042) of the bacteria at the genus level in the locality microbiome. Anaplasma was the most abundant bacterial genus in the Phuthaditjhaba, Harrismith and Bergville samples, with a relative abundance of 85.4%, 72.4% and 67.1%, respectively. This was followed by Mycoplasma (8.2%, 17.6% and 11.4%, respectively) (Supplementary Table S2). The Phuthaditjhaba microbiome was also predominated with Rikenellaceae RC9 gut group (2.3%). Other abundant genera in the Bergville blood microbiome were Rikenellaceae RC9 gut group (5.2%), Aerococcus (2.1%), Akkermansia (1.9%), Alistipes (1.4%), UCG-005 (1.4%), Planomicrobium (0.9%), Corynebacterium (0.7%), Acinetobacter (0.6%), Porphyromonas (0.5%) and Pseudomonas (0.5%). Other top abundant genera in the Harrismith microbiome were Pelomonas (0.7%), Cutibacterium (0.7%), UCG-005 (0.6%) and Ehrlichia (0.6%) (Supplementary Table S2). The rest of the genera (82, 102, 79) had <0.5% relative abundance in the three study sites (Bergville, Harrismith, Phuthaditjhaba, respectively) (Supplementary Table S2).
We investigated the species-level differences of microbial communities across the three study sites, which showed no significant differences (Kruskal Wallis test, p=0.081) in the microbiome relevant abundance (Supplementary Table S2). Of the 71 detected species (159 genera), 12.67%, 22.54% and 21.13% had a sole association with Phuthaditjhaba, Bergville and Harrismith study sites respectively (Figure 7C). Anaplasma marginale (relative abundance 56.2%, 43.5%, 54.2%, respectively) was the most abundant bacterial pathogen in the Bergville, Harrismith and Phuthaditjhaba samples, followed by A. platys (22.6%, 31.5%, 32.9%) and Mycoplasma wenyonii (14%, 19.6%, 7.8%) (Supplementary Table S2). Other predominant species in Bergville bovine blood microbiome were Aerococcus vaginalis (2.6%), Acinetobacter lwoffii (0.6%), Pseudomonas poae (0.5%) and Yersinia pestis (0.5%). In contrast, Anaplasma bovis (1.3%), Anaplasma centrale (0.9%), Anaplasma ovis (0.8%), Cutibacterium namnetense (0.8%) and Ehrlichia canis (0.6%) were the other relatively predominant species in Harrismith. Moreover, Anaplasma bovis (3.0%) and Mycoplasma haemobos (1.0%) were the other predominant bacterial species in Phuthaditjhaba (Supplementary Table S2). The rest of the species identified in these samples had relative abundance lower than 0.5% (Supplementary Table S2). The presence of few predominating bacterial genera in the three study sites could have suggested that these differences might also occur in composition at the species level, but instead most of the genera identified in each microbiome were represented on average by two species.
4 Discussion
The most abundant tick species collected in the three study sites was R. evertsi evertsi (78.7%). The tick species is the most widespread rhipicephalid in Africa (26), and transmits the bacterium A. marginale (31), the main cause of anaplasmosis in cattle, as well as protozoan parasites Theileria equi and Babesia caballi (32), which cause piroplasmosis in horses, and induces tick paralysis in sheep (33). This was not surprising as previous studies have shown that the tick is widely distributed (26) and is the most prevalent tick species present in different parts of South Africa (6, 14, 24). Cattle sampled in Bergville belonged to different farmers under one systematic dipping regimen. This could mean that R. evertisi evertsi is resistant to the dipping chemicals used at these sites. In reference to a pilot study conducted in Bergville in March 2015 at the same dip tanks (25), the most prevalent tick species was also R. evertsi evertsi (55%). In the Free State, R. evertsi evertsi was identified to be 44.7% and was prevalent in cattle over a period of two years (34). In the present study, Phuthadijthaba had the most diverse tick species, namely R. evertsi evertsi, H. rufipes, R. decoloratus, R. appendiculatus, R. simus and R. africanus, followed by Harrismith which had five tick species, namely H. rufipes, H. truncatum, R. decolaratus and R. microplus, in addition to R. eversti evesti, while Bergville had the least tick species diversity, with H. truncatum and R. appendiculatus as well as R. evertsi evertsi. Other relatively abundant tick species were R. appendiculatus, R. simus and R. afranicus. Rhipicephalus appendiculatus was the second most collected tick and was present in Phuthaditjhaba and Bergville. Rhipicephalus appendiculatus transmits Theileria parva, the main causative agent of Corridor disease in cattle, T. taurotragi that causes benign bovine theileriosis and A. bovis and R. conorii. The latter causes tick typhus in humans. The presence of this tick was not recorded in the eastern Free State during a 3-year long tick survey study (23) and during the following years (9, 35). Whereas in Bergville it was identified in 2017 (25). Rhipicephalus simus transmits A. centrale, the cause of subacute bovine anaplasmosis.
Rhipicephalus afranicus n sp., is a recently described species in South Africa (36) and was subsequently identified in Uganda (37). The genetic divergence between the Ugandan and Southern African sequences is 2.4%, demonstrating that two different populations of this species may be occurring between Southern and East Africa (37). A study conducted in Israel speculate that this tick species could be the source of transmission for A. bovis causing bovine anaplasmosis in cattle (36). In our knowledge, this study is the first to report the presence of Rhipicephalus afranicus n sp. in Phuthaditjhaba.
Only three R. microplus ticks were collected, and this was in Harrismith. The tick is an invasive species of Asian origin and considered one of the most widespread ectoparasites of livestock. This tick has been recovered in the Free State Province from very few sites including wild animals (9, 19, 35). This is an interesting finding from this study, and it has been found in one locality (Harrismith) because there are very few reports of this tick species in the Free State province. Therefore, considering its high dispersal rate as reported in South-East Africa (19, 38), the occurrence of R. microplus as determined by this study warrants further investigation about its distribution in the Free State.
The tick species identified in this study are associated with transmission of bacterial pathogens: Anaplasma, Rickettsia and Erhlirchia. The bacterial microbiome in Bergville, Harrismith and Phuthaditjhaba was largely dominated by members of three families: Anaplasmataceae (65.0%, 72.1%, 84.7%, respectively), Mycoplasmataceae (11.1%, 17.4%, 8.1%) and Rikenellaceae (6.6%, 4.3%, 2.7%). The high proportion of Anaplasmataceae is in correlation with the abundance of biological vectors present at the three study sites. The latter two families are associated with respiratory and gastrointestinal tract microbiomes in various animals.
Anaplasma species are widespread in South Africa, with a seemingly wide host range. Anaplasma marginale was the most abundant species in the three study sites, confirming that the ticks present are vectors of this pathogen. The second most abundant pathogen was A. platys, which has previously been detected in ruminants’ blood (39, 40). A recent 16S microbiome analysis revealed the presence of both species and nine novel species in nine different free-roaming wildlife species (African buffalo, impala, kudu, zebra, warthog, hyena, leopard, lion, and elephant) in the Kruger National Park and surroundings game reserves (18). Anaplasma platys is an emergent zoonotic pathogen (41, 42) that causes canine cyclic thrombocytopenia (43) and the presence of this species in cattle may pose a threat to human health. The pathogen has a worldwide distribution, and the suspected tick vector is R. sanguineus sensu lato ticks (44). The pathogen was detected in 12 African tick species in seven African countries in all regions (45), which may imply that various tick species are involved in its epidemiology. Detection of A. platys in cattle can be attributed to the close association of dogs with cattle in the study areas. Given the widespread occurrence of R. sanguineus s.l, this is likely to increase the risk of infections in humans. Further studies on the occurrence of platys in other mammalian and tick and evaluation of the vectorial capacity of tick species can assist in elucidating the epidemiology of A. platys. The other species found in the present study was M. wenyonii, whose mode of transmission is unknown, but presumably through mechanical transmission by blood-sucking arthropods (46). Many cattle infected with M. wenyonii do not exhibit clinical illness, unless is rare when concurrent illness results in immunosuppression (46, 47).
High abundance of A. marginale, the main cause of bovine anaplamsosis, can be attributed to the various tick species involved in transmission (Rhipicephalus, Dermacentor, Ixodes, Hyalomma) and other means of transmission, namely mechanical transmission by contaminated fomites and biting flies and the transplacental route (48, 49). A recent study involving African ticks showed that the pathogen was detected in 17 species of the genera Amblyomma, Rhipicephalus and Hyalomma (45). High A. marginale abundance in the study area may increase selection for genetically distinct strains and thus further complicating the efforts towards the development of a safe and effective vaccine. Rikenellaceae RC9 gut group, belonging to the Rikenellaceae family, plays an important role in the digestion of crude fiber (50). Therefore, the presence of the Rikenellaceae RC9 gut group indicates the roughage diet consumed by cattle in the three study sites. The presence of this bacteria could suggest that while drawing blood from the caudal vein there were remnants of feaces that got into the vacutainer.
5 Conclusions
We determined tick species distribution and abundance in three selected study sites in two provinces of South Africa, and established diversity of bacterial tick-borne pathogens from blood of cattle using the 16S rRNA PacBio sequencing method. Rhipicephalus evertsi evertsi, followed by R. simus and R. appendiculatus were the most abundant tick species. High abundance of R. evertsi evertsi has important implications for the transmission and occurrence of bovine anaplasmosis and equine piroplasmosis. Effective control of the ticks will minimize severe disease cases, taking into consideration the development and spread of acaricide resistance and maintenance of endemic stability. There was a high diversity of tick-borne bacterial pathogens in the blood of cattle, that can be attributed to emergence and change in distribution of pathogens as well close interactions among domestic and peri-domestic animals and humans. Anaplasma marginale and A. platys were the most abundance tick-borne bacterial pathogens, highlighting the risk of anaplasmosis in cattle, dogs, and humans. Control of anaplasmosis requires consideration of various factors such as climate change, acaricide resistance and animal movements.
6 Limitations of the study
Few cattle were sampled from a limited number of locations; however, given the common practice of communal grazing, it is probable that other cattle in and around the sampled locations harbor similar pathogens, as observed in this study.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1031221.
Ethics statement
The animal studies were approved by Animal Ethics Committee of the University of Pretoria, South Africa. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
BK: Formal analysis, Writing – original draft, Investigation. CB: Formal analysis, Writing – original draft, Methodology, Writing – review & editing. SM: Formal analysis, Writing – review & editing. NN: Writing – review & editing, Methodology. TM: Writing – review & editing. SN: Writing – review & editing, Methodology. MO: Writing – review & editing, Conceptualization. ZM: Conceptualization, Writing – review & editing, Formal analysis, Funding acquisition, Methodology, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Research Foundation of South Africa (Grant no: 2392782 awarded to ZM).
Acknowledgments
We thank the technicians from the Department of Agriculture who assisted with blood sample collection in Bergville, Harrismith, and Phuthaditjhaba.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2024.1399364/full#supplementary-material
Supplementary Table 1 | 16S combined rank totals.
Supplementary Table 2 | Locality abundances.
References
1. Perry BD, Young AS. The past and future roles of epidemiology and economics in the control of tick-borne diseases of livestock in africa: the case of theileriosis. Prev Vet Med (1995) 25:107–20.
2. Norval RAI, Walker JB, Colborne J. The ecology of rhipicephalus zambeziensis and rhipicephalus appendiculatus (Acarina, ixodidae) with particular reference to zimbabwe. Onderstepoort J Vet Res (1982) 49:181–90.
3. Rikhotso BO, Stoltsz WH, Bryson NR, Sommerville JEM. The impact of 2 dipping systems on endemic stability to bovine babesiosis and anaplasmosis in cattle in 4 communally grazed areas in Limpopo Province, South Africa. J South Afr Vet Assoc. (2005) 76:217–23. doi: 10.4102/jsava.v76i4.430
4. Mtshali MS, de la Fuente J, Ruybal P, Kocan KM, Vicente J, Mbati PA, et al. Prevalence and genetic diversity of Anaplasma marginale strains in cattle in South Africa. Zoonoses Public Health. (2007) 54:23–30. doi: 10.1111/j.1863-2378.2007.00998.x
5. Andreotti R, Pérez de León AA, Dowd SE, Guerrero FD, Bendele KG, Scoles GA. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. (2011) 11:1–11. doi: 10.1186/1471-2180-11-6
6. Spickett AM, Heyne IH, Williams R. Survey of the livestock ticks of the North West province, South Africa. Onderstepoort J Vet Res. (2011) 78:1–12. doi: 10.4102/ojvr.v78i1.305
7. Nyangiwe N, Harrison A, Horak IG. Displacement of Rhipicephalus decoloratus by Rhipicephalus microplus (Acari: ixodidae) in the eastern cape province, South Africa. Exp Appl Acarol. (2013) 61:371–82. doi: 10.1007/s10493-013-9705-7
8. Mutshembele AM, Cabezas-Cruz A, Mtshali MS, Thekisoe OMM, Galindo RC, de la Fuente J. Epidemiology and evolution of the genetic variability of Anaplasma marginale in South Africa. Ticks tick-borne Dis. (2014) 5:624–31. doi: 10.1016/j.ttbdis.2014.04.011
9. Horak IG, Jordaan AJ, Van Dalen EM, Heyne H. Distribution of endemic and introduced tick species in Free State Province, South Africa. J South Afr Vet Assoc. (2015) 86:1–9. doi: 10.4102/jsava.v86i1.1255
10. Khumalo ZTH, Brayton KA, Collins NE, Chaisi ME, Quan M, Oosthuizen MC. Evidence confirming the phylogenetic position of Anaplasma centrale (ex Theiler 1911) Ristic and Kreier 1984. Int J system evolution Microbiol. (2018) 68:2682–91. doi: 10.1099/ijsem.0.002832
11. Allsopp BA. Natural history of Ehrlichia ruminantium. Vet Parasitol. (2010) 167:123–35. doi: 10.1016/j.vetpar.2009.09.014
12. Khumalo ZTH, Catanese HN, Liesching N, Hove P, Collins NE, Chaisi ME, et al. Characterization of Anaplasma marginale subsp. centrale strains by use of msp1aS genotyping reveals a wildlife reservoir. J Clin Microbiol. (2016) 54:2503–12. doi: 10.1128/JCM.01029-16
13. De Boni L. Occurrence of Tick-Borne Haemoparasites in Selected South African Wild Rodent Species in the Mnisi Communal Area. South Africa: University of Pretoria (2017).
14. Mtshali K, Khumalo ZTH, Nakao R, Grab DJ, Sugimoto C, Thekisoe OMM. Molecular detection of zoonotic tick-borne pathogens from ticks collected from ruminants in four South African provinces. J Vet Med Sci. (2015) 77:1573–9. doi: 10.1292/jvms.15-0170
15. Abbas RZ, Zaman MA, Colwell DD, Gilleard J, Iqbal Z. Acaricide resistance in cattle ticks and approaches to its management: the state of play. Vet Parasitol. (2014) 203:6–20. doi: 10.1016/j.vetpar.2014.03.006
16. Trollope WSW, Trollope LA, Hartnett DC. Fire behaviour a key factor in the fire ecology of African grasslands and savannas. In Viegas DX (ed)., Forest Fire Research & Wildland Fire Safety. Proceedings of the IV International Conference on Forest Fire Research. Rotterdam, the Netherlands: Millpress (2002), pp. 1–15.
17. Kolo AO, Collins NE, Brayton KA, Chaisi M, Blumberg L, Frean J, et al. Anaplasma phagocytophilum and other Anaplasma spp. in various hosts in the Mnisi community, Mpumalanga province, South Africa. Microorganisms. (2020) 8:1812. doi: 10.3390/microorganisms8111812
18. Makgabo SM, Brayton KA, Oosthuizen MC, Collins NE. Unravelling the diversity of Anaplasma species circulating in selected African wildlife hosts by targeted 16S microbiome analysis. Curr Res Microbial Sci. (2023) 5:100198. doi: 10.1016/j.crmicr.2023.100198
19. Nyangiwe N, Horak IG, van der Mescht L, Matthee S. Range expansion of the economically important Asiatic blue tick, Rhipicephalus microplus, in South Africa. J South Afr Vet Assoc. (2017) 88:1–7. doi: 10.4102/jsava.v88i0.1482
20. Barré N, Uilenberg G. Spread of parasites transported with their hosts: case study of two species of cattle tick. Rev scientif tech. (2010) 29:149.
21. Kocan KM, de la Fuente J, Blouin EF, Garcia-Garcia JC. Anaplasma marginale (Rickettsiales: Anaplasmataceae): recent advances in defining host–pathogen adaptations of a tick-borne rickettsia. Parasitology. (2004) 129:S285–300. doi: 10.1017/S0031182003004700
22. Estrada-Peña A, Gray JS, Kahl O, Lane RS, Nijhof AM. Research on the ecology of ticks and tick-borne pathogens—methodological principles and caveats. Front Cell infect Microbiol. (2013) 3:29. doi: 10.3389/fcimb.2013.00029
23. Hlatshwayo M, Mbati PA, Dipeolu OO. Seasonal abundance of adult ixodid ticks infesting cattle belonging to resource-limited farmers in the north-eastern Free State Province of South Africa. Onderstepoort Journal of Veterinary Research (2002) 69(1):55–62.
24. Mbati PA, Hlatshwayo M, Mtshali MS, Mogaswane KR, De Waal TD, Dipeolu OO. In: Ticks and Tick-Borne Pathogens: Proceedings of the 4th International Conference on Ticks and Tick-Borne Pathogens The Banff Centre Banff, Alberta, Canada, 21–26 July 2002. Dordrecht: Springer Netherlands (2003), pp. 217–24.
25. Khumalo ZTH. Anaplasma centrale in South Africa: Occurrence Phylogeny and Genetic Diversity. South Africa: University of Pretoria (2017).
26. Walker AR. Ticks of domestic animals in Africa: a guide to identification of species. Edinburgh: University of Edinburgh, Centre for Tropical Veterinary Medicine (2003) 74. Bioscience Reports.
27. Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol. (2017) 261:169–76. doi: 10.1016/j.jbiotec.2017.06.1198
28. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
29. R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2022). Available at: https://www.R-project.org/.
30. Hoque MN, Istiaq A, Rahman MS, Islam MR, Anwar A, Siddiki AMAMZ, et al. Microbiome dynamics and genomic determinants of bovine mastitis. Genomics. (2020) 112:5188–203. doi: 10.1016/j.ygeno.2020.09.039
31. Potgieter F. Tick transmission of anaplasmosis in south africa. In: International conference on tick biology and control. Grahamstown, South Africa (1981). p. 222.
32. De Waal DT, Potgieter FT. The transstadial transmission of babesia caballi by rhipicephalus evertsi evertsi. Onderstepoort J Vet Res (1987) 54:655–66.
33. Hamel HD, Gothe R. Influence of infestation rate of tick-paralysis in sheep induced by rhipicephalus evertsi evertsi neumann, 1897. Vet Parasitol (1978) 4:183–91.
34. Mtshali MS, De Waal DT, Mbati PA. A sero-epidemiological survey of blood parasites in cattle in the north-eastern Free State, South Africa. Onderstepoort J Vet Res. (2004) 71:67–75. doi: 10.4102/ojvr.v71i1.287
35. Tonetti N, Berggoetz M, Rühle C, Pretorius AM, Gern L. Ticks and tick-borne pathogens from wildlife in the Free State Province, South Africa. J Wildl Dis. (2009) 45:437–46. doi: 10.7589/0090-3558-45.2.437
36. Bakkes DK, Chitimia-Dobler L, Matloa D, Oosthuysen M, Mumcuoglu KY, Mans BJ, et al. Integrative taxonomy and species delimitation of rhipicephalus turanicus (Acari: Ixodida: Ixodidae). Int J Parasitol (2020) 50:577–94. doi: 10.1016/j.ijpara.2020.04.005
37. Balinandi S, Chitimia-Dobler L, Grandi G, Nakayiki T, Kabasa W, Bbira J, et al. Morphological and molecular identification of ixodid tick species (Acari: Ixodidae) infesting cattle in uganda. Parasitol Res (2020) 119:2411–20. doi: 10.1007/s00436-020-06742-z
38. Lynen G, Zeman P, Bakuname C, Di Giulio G, Mtui P, Sanka P, et al. Shifts in the distributional ranges of boophilus ticks in tanzania: evidence that a parapatric boundary between boophilus microplus and b. decoloratus follows climate gradients. Exp Appl Acarol. (2008) 44:147–64. doi: 10.1007/s10493-008-9134-1
39. Chien NTH, Nguyen TL, Bui KL, Nguyen TV, Le TH. Anaplasma marginale and A. platys characterized from dairy and indigenous cattle and dogs in northern Vietnam. Korean J Parasitol. (2019) 57:43. doi: 10.3347/kjp.2019.57.1.43
40. Nguyen AHL, Tiawsirisup S, Kaewthamasorn M. Molecular detection and genetic characterization of Anaplasma marginale and Anaplasma platys-like (Rickettsiales: Anaplasmataceae) in water buffalo from eight provinces of Thailand. BMC vet Res. (2020) 16:1–12. doi: 10.1186/s12917-020-02585-z
41. Maggi RG, Mascarelli PE, Havenga LN, Naidoo V, Breitschwerdt EB. Co-infection with Anaplasma platys, Bartonella henselae and Candidatus Mycoplasma haematoparvum in a veterinarian. Parasites Vectors. (2013) 6:1–10. doi: 10.1186/1756-3305-6-103
42. Arraga-Alvarado CM, Qurollo BA, Parra OC, Berrueta MA, Hegarty BC, Breitschwerdt EB. Case report: molecular evidence of Anaplasma platys infection in two women from Venezuela. Am J Trop Med hyg. (2014) 91:1161. doi: 10.4269/ajtmh.14-0372
43. Abarca K, López J, González P, Dabanch J, Torres M. Seroepidemiological evidence of human exposure to Anaplasma sp in Santiago, Chile. Rev Chil Infectol. (2008) 25:358–61.
44. Selim A, Almohammed H, Abdelhady A, Alouffi A, Alshammari FA. Molecular detection and risk factors for Anaplasma platys infection in dogs from Egypt. Parasites Vectors. (2021) 14:429. doi: 10.1186/s13071-021-04943-8
45. Cossu CA, Collins NE, Oosthuizen MC, Menandro ML, Bhoora RV, Vorster I, et al. Distribution and prevalence of anaplasmataceae, rickettsiaceae and coxiellaceae in African ticks: A systematic review and meta-analysis. Microorganisms. (2023) 11:714. doi: 10.3390/microorganisms11030714
46. Persson Waller K, Dahlgren K, Grandi G, Holding ML, Näslund K, Omazic A, et al. A disease outbreak in beef cattle associated with Anaplasma and Mycoplasma infections. Animals. (2023) 13:286. doi: 10.3390/ani13020286
47. Genova SG, Streeter RN, Velguth KE, Snider TA, Kocan KM, Simpson KM. Severe anemia associated with Mycoplasma wenyonii infection in a mature cow. Can Vet J. (2011) 52:1018.
48. Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. (2010) 167:95–107. doi: 10.1016/j.vetpar.2009.09.012
49. Kumar B, Manjunathachar HV, Ghosh S. A review on Hyalomma species infestations on human and animals and progress on management strategies. Heliyon. (2020) 6. doi: 10.1016/j.heliyon.2020.e05675
Keywords: PacBio, Anaplasma, Bergville, Harrismith, Phuthaditjhaba, ticks, 16S rRNA, South Africa
Citation: Khoza BL, Byaruhanga C, Makgabo SM, Nyangiwe N, Mnisi T, Nxumalo S, Oosthuizen MC and Mnisi ZTH (2024) Tick distribution and comparative analysis of bovine blood microbiome in two provinces of South Africa using 16S rRNA PacBio sequencing approach. Front. Trop. Dis 5:1399364. doi: 10.3389/fitd.2024.1399364
Received: 11 March 2024; Accepted: 07 May 2024;
Published: 28 May 2024.
Edited by:
Amira Adel Taha Abdel Aleem AL-Hosary, Assiut University, EgyptReviewed by:
Ahmed Mahmoud Elsify, University of Sadat City, EgyptSammuel Shahzad, United States Department of Agriculture (USDA), United States
Maha Hamed, Assiut University, Egypt
Copyright © 2024 Khoza, Byaruhanga, Makgabo, Nyangiwe, Mnisi, Nxumalo, Oosthuizen and Mnisi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zamantungwa T. H. Mnisi, WmFtYW50dW5nd2EubW5pc2lAdWwuYWMuemE=
 Bongekile L. Khoza
Bongekile L. Khoza Charles Byaruhanga
Charles Byaruhanga S. Marcus Makgabo
S. Marcus Makgabo Nkululeko Nyangiwe
Nkululeko Nyangiwe Themba Mnisi4
Themba Mnisi4 Marinda C. Oosthuizen
Marinda C. Oosthuizen Zamantungwa T. H. Mnisi
Zamantungwa T. H. Mnisi