Abstract
Background:
The phenomenon of hybridisation between Schistosoma species has gained a greater degree of significance since the WHO declared that schistosomiasis is to be eliminated, as a public health problem, by 2030. The role hybridisation plays in the transmission of disease is poorly understood and has the potential to complicate this elimination effort. A primary reason for this incomplete understanding of schistosome hybridisation is the lack of suitable, high-throughput and easily accessible methods capable of identifying the species-parentage of individual schistosomes. To address this resource gap, we present the development of a two-tube HRM assay capable of differentiating the species-parentage of schistosomes from a possible range of six species, namely: S. mattheei, S. curassoni, S. bovis, S. haematobium, S. mansoni and S. margrebowiei.
Methods:
The assay was designed using aligned reference sequences for the six target species, with primers designed to amplify PCR products with species-specific melt temperatures for both the nuclear and mitochondrial genomes. The sensitivity and specificity of these novel primer sets were tested against a DNA library comprising representatives of: S. mattheei, S. curassoni, S. bovis, S. haematobium, S. mansoni and S. margrebowiei. The optimal annealing temperature for the real-time PCR (rtPCR) assays was established alongside the efficiency for the different primer pairs. The novel HRM assay was trialled against field samples comprising pooled urine from school-age children collected from 13 schools and miracidial samples preserved on FTA cards. Throughout the optimisation and testing of the novel HRM rtPCR primers targeting nDNA and mtDNA markers comparison against a pre-published S. mansoni and S. haematobium probe-based rtPCR was carried out.
Results:
The assay has a comparable sensitivity to current, probe-based species-specific assays and can detect target DNA at concentrations of 1pg/µL-0.1pg/µL for all six species, with the exception for S. bovis which has a slightly lower sensitivity range of 0.1ng/µL-0.1pg/µL. The analysis of the field samples resulted in all pooled urine samples testing positive for S. haematobium and a further three positive for S. mansoni using the probe-based rtPCR. The HRM rtPCR identified four S. mansoni positive samples in addition to six samples identified as being positive for S. mattheei. Despite identifying non-S. haematobium markers in the urine filter samples analysis of the miracidial samples stored on the FTA cards only identified pure S. haematobium.
Conclusion:
Although no hybrids were detected in this manuscript the novel-two tube assay described, offers the potential to radically increase the number of samples screened for the presence of hybrids in a range of sample types, including biopsy material for FGS screening. This will result in a decrease in cost and time in identifying putative hybrid cases.
Background
Schistosomiasis is an important public health problem with a total of 700 million people at risk in endemic areas, with the greatest burden of disease found in sub-Saharan Africa, where 240 million people require preventive chemotherapy (1) annually as there is currently no effective preventive vaccine available (2, 3). Schistosomiasis predominantly manifests as either urinary or gastrointestinal forms, however, a third manifestation of the disease, genital schistosomiasis, can be found in males (MGS) and females (FGS). Genital schistosomiasis is often misdiagnosed and confused with sexually transmitted diseases due to knowledge and diagnostic limitations in resource poor communities (4). It is estimated that 56 million girls and women in sub-Saharan Africa are affected by FGS (5), with it being the most significant cause of gynaecological morbidity in highly endemic communities (6).
Looking towards the WHO 2030 Roadmap targets, there is an ambition to eliminate schistosomiasis as a public health problem (7) by the end of the decade. With this switch from morbidity control towards elimination, as a public health problem, a broader range of considerations emerge. One of which is the phenomenon of hybridisation between anthroponotic and zoonotic schistosome species, the importance of which is not fully understood. Within the literature there is a growing list of suspected and actual hybrids which are appearing in human transmission cycles (8). With the advent and application of DNA genotyping, particularly of schistosome larvae, a greater number of studies have evidenced ancestral or ongoing introgression between schistosomes of medical and veterinary importance (9–13). In West Africa, introgression between S. haematobium and S. bovis is particularly significant, as this variant has expanded its known range northward to Corsica (14) as well as southward to Malawi (15).
The ability to detect hybrid infections is limited by the availability of suitable methods with traditional microscopy approaches being limited in their ability to detect cases of hybrid schistosomes due to their low sensitivity and specificity (16, 17) in addition to egg morphology alone being insufficient to identify putative hybrids and their parent species (14). The availability of molecular based tools to identify schistosomiasis has expanded over recent years with the development rtPCR and isothermal assays (18–21), although these are limited to the identification of generic or species-specific infections and cannot identify hybrids. To date, most schistosome hybrids have been identified through sequence analysis of the ITS1 and COX1 genes (22, 23) but more recently a new amplification refractory mutation system PCR (ARMS-PCR) has been developed (24) that allows affordable and high-through-put genotyping of potential hybrids. However, this new ARMS-PCR assay is limited to the detection of only three species typically found in West Africa: S. haematobium, S. bovis and S. curassoni. Due to this limited number of targets the ARMS-PCR assay will have limited applicability in other regions of Africa where it will fail to detect non-target, endemic species. This argument is given further relevance as the inter-species interactions in Malawi bring to light an additional species of schistosome, Schistosoma mattheei, which is of particular interest in being able to infect wild and domestic ungulates as well as humans (8). Further, an additional dimension, found in Malawi, is the occurrence of ‘triple’ infections of S. haematobium, S. mansoni and S. mattheei which increases the need for a diagnostic method that targets a broader range of species. Failure to detect these triple infections will make the prospect for elimination, as a public health problem, challenging (25). The unclear role that schistosome hybrids may play in the transmission of the disease is currently being addressed by the Hybridisation in Urogenital Schistosomiasis (HUGS) study. This is a multidisciplinary longitudinal population study seeking to reveal the transmission biology, epidemiological impact and clinical importance of Schistosoma haematobium-hybrids in the Mangochi and Nsanje Districts of Southern Malawi (13). Of particular interest within the HUGS study is the role hybrids may play in both male and female genital schistosomiasis.
With a focus on Malawi but mindful of the broader hybridisation potential within the S. haematobium group, we present the development of a novel, two-tube, high-resolution melt (HRM) rtPCR, capable of characterising mitochondrial and nuclear genomic markers to determine the hybrid status of a sample. The following six medically and veterinary important species of Schistosoma will be the focus of this assay: S. haematobium, S. mansoni, S. mattheei, S. bovis, S. curassoni and S. margrebowiei. As an HRM assay, the key diagnostic feature will be the melt temperature of the PCR product generated for the different target species for both the mitochondrial and nuclear rtPCR reactions. As a methodology, HRM relies in the drop in fluorescence caused by the melting of double-stranded DNA in the presence of an intercalating dye, typically EvaGreen® (Biotium). The precise temperature at which half of the DNA is in a single-strand state is referred to as the melting temperature (Tm). The determinants of the Tm are both the length of the DNA molecule and its composition. As the melt process involves breaking the hydrogen bonds of the complimentary DNA strands, the greater the length of the DNA molecule and the higher the percentage of guanine and cytosine bases the higher the Tm. HRM assays have been developed capable of characterising a range of targets from drug-resistance markers in bacteria (26), bloodmeals in hematophagous insects (27), trypanosomes (28) and soil-transmitted helminths (29). It presents a highly flexible and applicable methodology for the development of a high-throughput schistosome-hybrid screen capable of targeting multiple markers of interest.
Methods
Primer design and optimisation
The assay was designed as a two-tube HRM rtPCR, with primers in tube one targeting conserved sites of the ITS2 nuclear gene (nDNA HRM rtPCR) flanking an un-conserved region. Tube two consists of a multiplex assay targeting species-specific regions of the mitochondrial genome (mtDNA HRM rtPCR), specifically the ND6 region of S. mattheei, the ND4 region of S. margrebowiei and the tRNA-Lys regions of S. curassoni, S. mansoni, S. bovis and S. haematobium. In order to design the ITS primers the following reference sequences from the NCBI database were aligned using MEGA7 software (30): Z21718 (S. mattheei), OX104032 (S. margrebowiei), MT580946 (S. curassoni), AY446082 (S. mansoni), MW027650 (S. bovis) and KJ622337 (S. haematobium). Design of the species-specific mitochondrial primers relied on the alignment of the ND1 to ND4 region using the following reference sequences: AP017709 (S. margrebowiei), AP017708 (S. curassoni), NC 002545 (S. mansoni), OX104101 (S. bovis) and MK253578 (S. haematobium). Due to the lack of suitable sequences available for S. mattheei covering the ND1 to ND4 mtDNA, a separate alignment targeting the ND6 region was created for S. mattheei primer design, utilising the prior references with the additional reference sequence: AJ416897 (S. mattheei).
The NCBI Primer 3 design tool (31) was utilised to help improve the specificity of the species-specific primers in addition to the free online melt-curve prediction software U-melt (32), in order to design primers that will amplify products with species-specific melt temperatures (Table 1). The specificity of novel primers were tested using a library of positive controls, (see methods sub-heading: Schistosome species DNA Library). Initially primers were tested in single-plex and later as a multiplex, against the full DNA library, to determine primer specificity. Comparison of Ct values across three annealing temperatures (58°C, 59°C and 60°C) was carried out to determine optimum annealing temperature for the two assays. Further the efficiency of the novel primer sets was tested using a ten-fold dilution series for each species ranging from 1ng/µL – 0.1pg/µL ran as a standard curve. Our novel assay was compared to current published probe-based assays for generic schistosome detection (18), and species-specific identification for S. mansoni and S. haematobium (21) using the standard-curve DNA dilutions.
Table 1
| mtDNA primers | |||||
|---|---|---|---|---|---|
| Target sp. | Primer name | Primer sequence | Primer Tm (°C) | Product size (bp) | molecular target |
| S. mattheei | SchMattF | GTTGGTTTCGTATTTTTTTATGTTAAGG | 61.3 | 74 | ND6 |
| SchMattR | CTAACTTAGCGCTTCACAAAATGC | 62 | |||
| S. curassoni | SchCurrF | GTCGTGCTTTTGGTGATTAGC | 59 | 120 | tRNA-Lys |
| SchCurrR | CCTACGCCCGATAAACTAAAC | 59 | |||
| S. bovis | SchBovF | CAACATAAGATGATTGTAGTTAGC | 58 | 156 | tRNA-Lys |
| SchBovR | CTTTATTACTCGGCCACGATATG | 60.9 | |||
| S. mansoni | SchMnF | GGTTGAAGAGGAGGTTCGTG | 60 | 121 | tRNA-Lys |
| SchMnR | GGTCGCAATATACTCGACACC | 61.2 | |||
| S. haematobium | SchHmF2 | GCTGTAAAGGTGGCTGATAGTAGC | 65 | 126 | tRNA-Lys |
| SchHmR2 | TATCAACTTAACTATGCACCTAGTG | 60 | |||
| S. margrebowiei | SchMrgF2 | GGATCACGAAGTTGGGCTATAC | 62 | 78 | ND4 |
| SchMrgR2 | GAATATCAGCACAGACAATACTTGAAC | 63 | |||
| nDNA primers | |||||
| Target sp. | Primer name | Primer sequence | Primer Tm (°C) | Product size (bp) | molecular target |
| Schistosoma sp. | NUC ITS F | GCTCGAGTCGTGGCTTAATGAC | 64 | 163 | ITS2 |
| NUC ITS R | CTGATCCGAGGTCRGGGTCAATTA | 65.2-66.9 | |||
The primer list for the species-specific mtDNA HRM rtPCR assay and nDNA HRM rtPCR assay.
Field samples
Schistosome positive urine samples were collected from representative children of 13 schools during a spot-check epidemiological survey within the Lower Shire River region. The samples were processed as follows, from each school 10mL of urine from boys and girls (n= ~20) underwent urine filtration and microscopy examination to determine the presence of Schistosoma eggs. The remaining urine was pooled by sex, and the resultant filters were preserved in 100% ethanol for later DNA extraction and molecular analysis resulted in a total of 26 urine samples. Two urine pools from schools 5 and 10 were observed to have non-S. haematobium eggs present, notably zoonotic-shaped eggs were observed in samples from school 5 and S. mansoni eggs were observed in school 10 samples. To better characterise the non-S. haematobium eggs observed, the microscopy filters from schools 5 and 10 were placed in water to induce hatching and ~100 individual miracidia from each school were then stored on Whatman Flinders Technology Associates (FTA) cards. Upon arrival at the Liverpool School of Tropical Medicine (LSTM), the urine filters and FTA cards were processed, using the methods described below. A total of 26 urine filters and 184 FTA spots underwent DNA extraction and rtPCR screening. The processing of the field samples is explained via flow-diagram in the Supplementary Materials (Supplementary Figure 1).
Schistosome species DNA library
To properly validate and test the novel oligos designed in this study, a DNA library of the six target species was created. Whole worms for species S. margrebowiei, S. curassoni, and S. bovis were sourced from the schistosomiasis collection at the Natural History Museum (SCAN) supplemented by six S. mattheei worms collected from Malawian cattle, with the DNA extraction process described below. Schistosoma haematobium and S. mansoni DNA was sourced from the collections at LSTM.
DNA extraction
The worm and urine filter samples underwent DNA extraction using the DNAmini blood and tissue kit (Qiagen) following the manufacturer’s instructions, with the addition of a bead-beating treatment using ~0.5g of 1.4mm ceramic beads, per-sample, prior to the incubation of the sample with the ATL/Proteinase K buffer. FTA card material underwent alkaline extraction following protocol by B. Webster et al. (2019) (15). For both the Qiagen and FTA DNA extraction Phocine herpes virus (PhHV) was added alongside the addition of the first buffers, to act as an internal extraction control; negative extraction controls were similarly included.
rtPCR
The HRM ITS2 assay and the mtDNA species-specific HRM assay were ran in 12µL reactions comprised of 6µL of Type-it HRM supermix (Qiagen), 400nM of each primer and 2µL of template, with the remaining volume being made up of nuclease free water up to 12 µL.
The probe-based reactions comprised similar volumes and concentrations of appPROBE No ROX supermix (Appleton Woods), primer and DNA template with the addition of 100nM of reaction-specific probes. The two probe-based reactions comprised a duplex reaction utilising the ITS1 generic schistosome primer/probe set and the primer-probes for the detection of the internal PhHV control. The second probe-based reaction was a triplex reaction consisting of the 16S species-specific primer-probe sets for S. haematobium and S. mansoni plus the PhHV primer-probe set. All rtPCR assays were run for 40 cycles with the exception of the HRM analysis of the urine filters and FTA spots which ran for 35 cycles. Both the urine filters and miracidia FTA spots were screened using the triplex probe-based rtPCR, whilst the ITS1 generic probe assay was used to screen the urine filters only. The mtDNA HRM assay was used to screen the filters and the FTA spots, with the modification of excluding the S. haematobium specific primers from the urine filter assay, so as to better identify low numbers of non-S. haematobium eggs. Both the nDNA and mtDNA HRM assays were used to screen the FTA spots to characterise the species and hybrid status of individual miracidia.
Ethics statement
Parasitological surveys took place as part of the “Hybridisation in UroGenital Schistosomiasis (HUGS)” study which was approved in the UK by the Research Ethics Committee of the Liverpool School of Tropical Medicine (LSTM), study protocol (22-028), and in Malawi by the College of Medicine Research and Ethics Committee (COMREC), study protocol P.08/21/3381. All human participants who provided microscopy positive urine samples were treated on site with praziquantel (40mg/kg).
Results
Design and optimisation
The specificity tests for the mtDNA HRM rtPCR found that, in-multiplex, there were no instances of cross-reaction of between target species from the DNA library. The optimum annealing temperature was found to be 58°C for the mtDNA HRM rtPCR and 60°C for the nDNA HRM rtPCR. It was also found that at 40 cycles the mtDNA HRM rtPCR produced non-specific products, likely primer dimers, in negative controls. Reducing the number of cycles to 35 reduced this background fluorescence. It should be noted that little to no none-specific amplification was observed in the the Alkaline extracted, negative controls, at 40 cycles. The melt peaks for both the mitochondria rtPCR (mtDNA HRM rtPCR) and nuclear rtPCR (nDNA HRM rtPCR) are shown in Figure 1 and can be cross-plotted against each other allowing for more accurate characterisation of target species and the detection of samples with mixed-species parentage as demonstrated by Figure 2. It was noted that the different DNA extraction methods used (Qiagen or alkaline) effected the species-specific melt temperatures (Tm) for both the nDNA and mtDNA HRM rtPCRs, although the relative temperature differences remained the same for the different schistosome species (Supplementary Table 1, Supplementary Figure 2).
Figure 1

Melt peak profiles for the nDNA HRM rtPCR (A) and mtDNA HRM rtPCR (B). Profile A depicts S. mansoni (i), S. margrebowiei (ii), S. mattheei (iii), S. bovis (iv), S. curassoni (v) and S. haematobium (vi). Profile B depicts S. mattheei (i), S. curassoni (ii), S. bovis (iii), S. haematobium (iv), S. mansoni (v) and S. margrebowiei (vi).
Figure 2
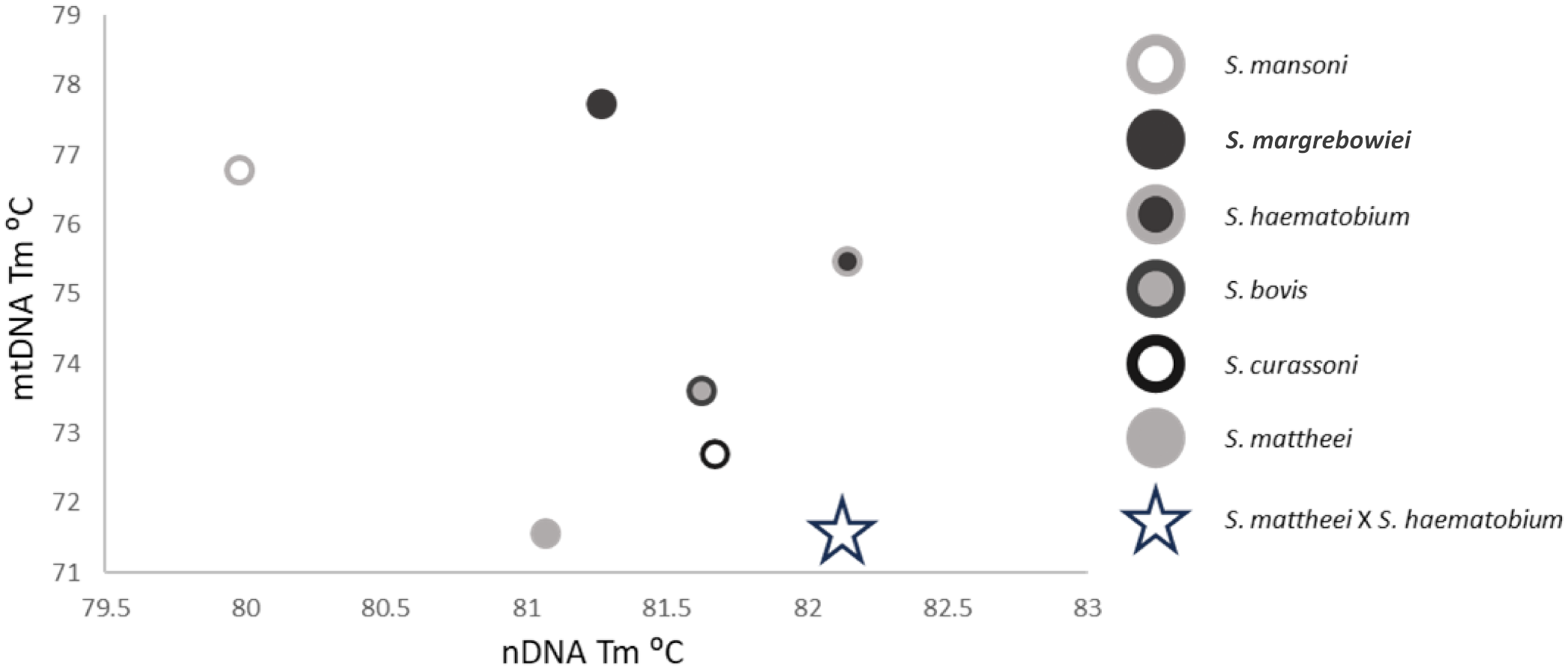
Cross-plot showing the nDNA HRM rtPCR and mtDNA HRM rtPCR Tm positions of the six target species plus a hypothetical S. mattheei x S. haematobium hybrid denoted by the open star. This hybrid has a similar mtDNA Tm to S. mattheei but also an nDNA Tm like that of S. haematobium.
Comparison of the sensitivity of the novel HRM assay against both the ITS and species-specific (S. mansoni/S. haematobium) probe-based rtPCR assays revealed a comparable sensitivity between the two. The average Ct difference between the probe assay and mtDNA rtPCR, across all template concentrations (1ng/µL-0.1pg/µL) was 1.55 for S. haematobium and 2.25 for S. mansoni, which equates to the HRM assay having, on average, a 3x and 5x greater degree of sensitivity than the probe-based assay respectively (Supplementary Table 2). The difference between the generic ITS1 probe assay and the nDNA rtPCR provided similar results for S. mattheei, S. bovis, S. haematobium and S. mansoni with the average Ct difference between the probe-based assay and HRM assay being 0.53, 0.89, 1.10 and 0.96 respectively. This shows that the HRM assay is moderately more sensitive than the probe-based assay. However, for S. curassoni and S. margrebowiei the inverse was true, with the probe-based assay showing a far higher degree of sensitivity with S. curassoni results being on average 3.55 Ct’s lower than the HRM Ct values and S. margrebowiei being 5.91 Ct points lower than the average HRM Ct values for the same concentration ranges (1ng/µL-0.1pg/µL). These Ct differences roughly equating to a difference of 11x and 60x greater degree of sensitivity of the ITS1 probe-based assay as compared to the HRM nDNA assay (Supplementary Table 3). The limit of detection LOD for the mtDNA and nDNA are given in Table 2.
Table 2
| Species | LOD mtDNA | LOD nDNA |
|---|---|---|
| S.mattheei | 0.1pg/µL | 0.1pg/µL |
| S.curassoni | 1pg/µL | 1pg/µL |
| S.bovis | 0.1ng/µL | 0.1pg/µL |
| S.haematobium | 0.1pg/µL | 0.1pg/µL |
| S.mansoni | 0.1pg/µL | 0.1pg/µL |
| S.margrebowiei | 1pg/µL | 1pg/µL |
LOD for each target species for the mtDNA HRM assay and nDNA HRM assay.
Analysis of field samples
All urine filters produced a positive PhHV result indicating successful isolation of DNA, similarly all urine filters were positive for schistosome DNA with the ITS1 generic rtPCR. Both male and female filter samples produced an average Ct value of 17.20 and 18.88 respectively. The 16S probe based species-specific assays successfully identified S. haematobium in all 26 urine samples, of which three also produced positive results for S. mansoni. The mtDNA HRM assay identified four S. mansoni positives (two males and two females, of which three were S. mansoni positive by the probe-based 16S rtPCR. The mtDNA rtPCR also identified six S. mattheei positives from four males and two females (Table 3).
Table 3
| Probe rtPCR | mtDNA HRM rtPCR | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS1 | S. haematobium | S. mansoni | S. mattheei | S. currassoni | S. bovis | S. mansoni | S. margrebowei | |||||||||
| Sample type | n (%) | Ct IQR | n (%) | Ct IQR | n (%) | Ct IQR | n (%) | Ct IQR | n (%) | Ct IQR | n (%) | Ct IQR | n (%) | Ct IQR | n (%) | Ct IQR |
| Male (n=13) | 13 (100) | 1.55 | 13 (100) | 1.71 | 1 (7.7) | Not applicable | 4 (30.8) | 2.65 | 0 | Not applicable | 0 | Not applicable | 2 (15.4) | 0.7 | 0 | 0 |
| Female (n=13) | 13 (100) | 3.52 | 13 (100) | 3.94 | 2 (15.4) | 3.8 | 2 (15.4) | 2.07 | 0 | Not applicable | 0 | Not applicable | 2 (15.4) | 1.26 | 0 | 0 |
| Total | 26 (100) | 3.07 | 26 (100) | 3.47 | 3 (11.5) | 3.8 | 6 (23.1) | 2.1 | 0 | Not applicable | 0 | Not applicable | 4 (15.4) | 1 | 0 | 0 |
Results of the urine filters screened using the probe and mtDNA HRM rtPCRs, with the Ct interquartile ranges given (Ct IQR).
Of the 184 FTA spots that underwent DNA extraction 183 produced a positive PhHV result, indicating one sample failed to undergo reliable DNA isolation, this sample was excluded from all subsequent analysis. From the remaining 183 successfully extracted FTA spots 132 were positive for S. haematobium using the probe-based 16S rtPCR and none were positive for S. mansoni. The nDNA and mtDNA HRM assays successfully amplified schistosome DNA from 142 samples and 150 samples respectively of which 139 were positive for both. The melt temperatures of the nDNA and mtDNA rtPCRs averaged 82.41°C and 75.64°C which closely matches the expected temperatures of pure S. haematobium genotypes. When plotted against each other the FTA samples cluster closely with S. haematobium, Figure 3.
Figure 3

The nDNA and mtDNA melt temperatures for the FTA spot results, cross-plotted against each other. Open circles denote the relative positions of the six target species, S. mansoni (i), S. margrebowiei (ii), S. haematobium (iii), S. bovis (iv), S. curassoni (v) and S. mattheei (vi). The open triangles are the 139 FTA results that produced a product in both HRM assays.
Discussion
The novel two-tube HRM rtPCR developed in this paper has comparable sensitivity to current, probe-based schistosome rtPCRs, but has the added advantage of also being able to plot both nDNA identity against mtDNA to determine if individual worms, miracidia or cercariae have mixed-species parentage. Such samples would present with either a double ITS2 peak, or a mtDNA peak that correlates with a separate species to that identified from the nDNA result. When the Tm of the nDNA and mtDNA HRM rtPCRs are cross plotted unusual results will fall out of the expected species cluster (Figure 2). Further, the range of species capable of being detected is broad, with the mtDNA HRM rtPCR detecting six schistosome species of medical and veterinary importance and the nDNA HRM rtPCR detecting a broader range of species, as the primer sites are conserved across multiple species.
Whilst the development of the assay in this paper was to predominantly screen individual schistosomes at different life-cycle stages, it is also capable of screening pooled samples, in this case, by removing the S. haematobium primers from the multiplex mtDNA HRM rtPCR it was possible to not only readily detect cases of urinary S. mansoni but also cases of S. mattheei. The presence of a zoonotic species in the human samples increases the likelihood that hybridisation can occur, due to the presence of two closely related species co-existing within the same host. Due to the nature of these samples being pooled eggs it was not possible to confirm if the S. mattheei positives were pure or hybrids.
Despite the presence of both S. mansoni and S. mattheei genetic material in the urine filter samples the analysis of the FTA card material was unable to detect non-S. haematobium species. This is likely due to a high ratio of S. haematobium eggs against non-S. haematobium eggs meaning it likely requires a greater number of samples to be screened to detect low numbers of non-S. haematobium individuals. Although no hybrids were detected in this manuscript the novel two-tube assay described here has been successfully used in the detection of S. mattheei x S. haematobium hybrids as well as pure S. haematobium found in cattle in a sister publication (33). The results of the Tm for the mtDNA and nDNA amplicons from the cattle survey can be seen in Supplementary Materials (Supplementary Figure 3). Our assay described here was also successfully used to identify S. mattheei in FGS samples collected as part of the HUGS study, these results are published in Kayuni et al. (34).
Whilst not exclusively mentioned in the main body of the text it should be noted that the cost of the two-tube HRM assay is approximately £0.82 in total for the two 12µL reactions per sample, which is considerably cheaper than Sanger-sequencing which can cost ~£3.50, per-read, from a commercial company. Therefore, for a single read from both the mitochondrial and nuclear DNA markers, the cost per sample would be ~£7, and that is not taking into account the additional costs of PCR reagents and consumables.
Study limitations
The nDNA and mtDNA HRM rtPCR were not able to detect all six schistosome species with a similar level of sensitivity, notably the nDNA HRM rtPCR had a lower sensitivity in the detection of S. margrebowiei and to a lesser extent S. curassoni (Supplementary Table 3). Similarly, the mtDNA rtPCR had a reduced sensitivity in the detection of S. bovis in comparison to the five other target species, as determined by the efficiency calculations. Another limitation in the design of the novel HRM assays described in this paper is the challenge of screening a pooled sample for the presence of unexpected species, as this is hindered by the likelihood that the dominant species present would outcompete other species. With the mtDNA HRM this can be mitigated with the removal of specific primer-pairs from the reaction, allowing for the detection of low prevalence schistosome species. However, such an approach cannot be taken with the nDNA rtPCR assay as this is single-plex, targeting conserved primer sites. This means that it is only possible to detect markers of interest from the maternal line of the schistosomes present and not the paternal line. This allows for the detection of female zoonotic schistosomes, however, male zoonotic schistosomes would be harder to detect in a pooled sample comprising multiple eggs. As mentioned above, a final limitation of this study is the lack of confirmed hybrid samples detected in this study. A larger sample size would have likely yielded the identification of a hybrid sample, going forward efforts will need to be made to increase the number of miracidia collected and screened.
Conclusions
The two-tube HRM rtPCR described in this paper allows for the rapid screening of individual eggs, worms and larvae of schistosomes in order to identify the species and determine the hybrid status of the worm in question, improving upon the capacity of previously described methods that required the sequencing of the mtDNA and nDNA of individual worms. This methodology will greatly improve the detection and sample processing time for the HUGS study and future studies as exemplified by the recent One Health paper (33) and FGS investigation (34).
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Liverpool School of Tropical Medicine Review Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
LC: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Writing – original draft. SK: Writing – review & editing. AJ: Writing – review & editing. PM: Writing – review & editing. DL: Writing – review & editing. GN: Writing – review & editing. DK: Writing – review & editing. PC: Writing – review & editing. BM: Writing – review & editing. SJ: Writing – review & editing. JA: Writing – review & editing. EL: Writing – review & editing. JM: Funding acquisition, Writing – review & editing. JS: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research is funded by National Institute for Health Research (NIHR) and Wellcome Trust through a Joint Investigator Award, grant number WT 220818/Z/20/Z.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2024.1350680/full#supplementary-material
References
1
Mutapi F Maizels R Fenwick A Woolhouse M . Human schistosomiasis in the post mass drug administration era. Lancet Infect Diseases. (2017) 17:E42–E8. doi: 10.1016/S1473-3099(16)30475-3
2
Panzner U Boissier J . Natural intra- and interclade human hybrid schistosomes in Africa with considerations on prevention through vaccination. Microorganisms. (2021) 9. doi: 10.3390/microorganisms9071465
3
Driciru E Koopman JPR Cose S Siddiqui AA Yazdanbakhsh M Elliott AM et al . Immunological considerations for schistosoma vaccine development: transitioning to endemic settings. Front Immunol. (2021) 12:635985. doi: 10.3389/fimmu.2021.635985
4
Shukla JD Kleppa E Holmen S Ndhlovu PD Mtshali A Sebitloane M et al . The association between female genital schistosomiasis and other infections of the lower genital tract in adolescent girls and young women: A cross-sectional study in South Africa. J Low Genit Tract Dis. (2023) 27:291–6. doi: 10.1097/LGT.0000000000000756
5
Sturt AS Webb EL Francis SC Hayes RJ Bustinduy AL . Beyond the barrier: Female Genital Schistosomiasis as a potential risk factor for HIV-1 acquisition. Acta Trop. (2020) 209:105524. doi: 10.1016/j.actatropica.2020.105524
6
Christinet V Lazdins-Helds JK Stothard JR Reinhard-Rupp J . Female genital schistosomiasis (FGS): from case reports to a call for concerted action against this neglected gynaecological disease. Int J Parasitology. (2016) 46:395–404. doi: 10.1016/j.ijpara.2016.02.006
7
WHO . Ending the neglect to attain the Sustainable Development Goals: a road map for neglected tropical diseases 2021–2030. Geneva: World Health Institution (2020).
8
Leger E Webster JP . Hybridizations within the Genus: implications for evolution, epidemiology and control. Parasitology. (2017) 144:65–80. doi: 10.1017/S0031182016001190
9
Morgan JAT DeJong RJ Lwambo NJS Mungai BN Mkoji GM Loker ES . First report of a natural hybrid between Schistosoma mansoni and S. rodhaini. J Parasitology. (2003) 89:416–8. doi: 10.1645/0022-3395(2003)089[0416:FROANH]2.0.CO;2
10
Wright CA Ross GC . Hybrids between Schistosoma haematobium and S. mattheei and their identification by isoelectric-focusing of enzymes. Trans R Soc Trop Med Hygiene. (1980) 74:326–32. doi: 10.1016/0035-9203(80)90091-7
11
Rey O Toulza E Chaparro C Allienne JF Kincaid-Smith J Mathieu-Begné E et al . Diverging patterns of introgression from across. African lineages. PloS Pathogens. (2021) 17.
12
Huyse T Van den Broeck F Hellemans B Volckaert FAM Polman K . Hybridisation between the two major African schistosome species of humans. Int J Parasitology. (2013) 43:687–9. doi: 10.1016/j.ijpara.2013.04.001
13
Huyse T Webster BL Geldof S Stothard JR Diaw OT Polman K et al . Bidirectional introgressive hybridization between a cattle and human schistosome species. PloS Pathog. (2009) 5. doi: 10.1371/journal.ppat.1000571
14
Kincaid-Smith J Tracey A Augusto RD Bulla I Holroyd N Rognon A et al . Morphological and genomic characterisation of the hybrid infecting humans in Europe reveals admixture between and. PloS Negl Trop Diseases. (2021) 15.
15
Webster BL Alharbi MH Kayuni S Makaula P Halstead F Christiansen R et al . Schistosome interactions within the Schistosoma haematobium Group, Malawi. Emerging Infect Diseases. (2019) 25:1245–7. doi: 10.3201/eid2506.190020
16
Fuss A Mazigo HD Tappe D Kasang C Mueller A . Comparison of sensitivity and specificity of three diagnostic tests to detect Schistosoma mansoni infections in school children in Mwanza region, Tanzania. PloS One. (2018) 13:e0202499. doi: 10.1371/journal.pone.0202499
17
Knopp S Ame SM Hattendorf J Ali SM Khamis IS Bakar F et al . Urogenital schistosomiasis elimination in Zanzibar: accuracy of urine filtration and haematuria reagent strips for diagnosing light intensity Schistosoma haematobium infections. Parasit Vectors. (2018) 11:552. doi: 10.1186/s13071-018-3136-6
18
Obeng BB Aryeetey YA de Dood CJ Amoah AS Larbi IA Deelder AM et al . Application of a circulating-cathodic-antigen (CCA) strip test and real-time PCR, in comparison with microscopy, for the detection of Schistosoma haematobium in urine samples from Ghana. Ann Trop Med Parasitology. (2008) 102:625–33. doi: 10.1179/136485908X337490
19
Sady H Al-Mekhlafi HM Ngui R Atroosh WM Al-Delaimy AK Nasr NA et al . Detection of Schistosoma mansoni and Schistosoma haematobium by Real-Time PCR with High Resolution Melting Analysis. Int J Mol Sci. (2015) 16:16085–103. doi: 10.3390/ijms160716085
20
Mesquita SG Lugli EB Matera G Fonseca CT Caldeira RL Webster B . Development of real-time and lateral flow recombinase polymerase amplification assays for rapid detection of Schistosoma mansoni. Front Microbiol. (2022) 13:1043596. doi: 10.3389/fmicb.2022.1043596
21
Alzaylaee H Collins RA Rinaldi G Shechonge A Ngatunga B Morgan ER et al . Schistosoma species detection by environmental DNA assays in African freshwaters. PloS Negl Trop Dis. (2020) 14:e0008129.
22
Webster BL Diaw OT Seye MM Webster JP Rollinson D . Introgressive hybridization of Schistosoma haematobium group species in Senegal: species barrier break down between ruminant and human schistosomes. PloS Negl Trop Dis. (2013) 7:e2110. doi: 10.1371/journal.pntd.0002110
23
Onyekwere AM Rey O Allienne JF Nwanchor MC Alo M Uwa C et al . Population genetic structure and hybridization of Schistosoma haematobium in Nigeria. Pathogens. (2022) 11. doi: 10.3390/pathogens11040425
24
Blin M Dametto S Agniwo P Webster BL Angora E Dabo A et al . A duplex tetra-primer ARMS-PCR assay to discriminate three species of the Schistosoma haematobium group: Schistosoma curassoni, S. bovis, S. haematobium and their hybrids. Parasites Vectors. (2023) 16.
25
Kayuni SA O'Ferrall AM Baxter H Hesketh J Mainga B Lally D et al . An outbreak of intestinal schistosomiasis, alongside increasing urogenital schistosomiasis prevalence, in primary school children on the shoreline of Lake Malawi, Mangochi District, Malawi. Infect Dis Poverty. (2020) 9. doi: 10.1186/s40249-020-00736-w
26
Edwards T Williams C Teethaisong Y Sealey J Sasaki S Hobbs G et al . A highly multiplexed melt-curve assay for detecting the most prevalent carbapenemase, ESBL, and AmpC genes. Diagn Micr Infect Dis. (2020) 97. doi: 10.1016/j.diagmicrobio.2020.115076
27
Pena VH Fernandez GJ Gomez-Palacio AM Mejia-Jaramillo AM Cantillo O Triana-Chavez O . High-resolution melting (HRM) of the cytochrome B gene: A powerful approach to identify blood-meal sources in chagas disease vectors. PloS Negl Trop Diseases. (2012) 6. doi: 10.1371/journal.pntd.0001530
28
Garrod G Adams ER Lingley JK Saldanha I Torr SJ Cunningham LJ . A pilot study demonstrating the identification of Trypanosoma brucei gambiense and T. b. rhodesiense in vectors using a multiplexed high-resolution melt qPCR. PloS Negl Trop Diseases. (2020) 14.
29
Cunningham LJ Stothard JR Osei-Atweneboana M Armoo S Verweij JJ Adams ER . Developing a real-time PCR assay based on multiplex high-resolution melt-curve analysis: a pilot study in detection and discrimination of soil-transmitted helminth and schistosome species. Parasitology. (2018) 145:1733–8. doi: 10.1017/S0031182018001361
30
Kumar S Stecher G Tamura K . MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. doi: 10.1093/molbev/msw054
31
NCBI . Primer-BLAST. Available online at: https://www.ncbi.nlm.nih.gov/tools/primer-blast/.
32
Dwight Z Palais R Wittwer CT . uMELT: prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics. (2011) 27:1019–20. doi: 10.1093/bioinformatics/btr065
33
Juhász A Makaula P Cunningham LJ Jones S Archer J Lally D et al . Revealing bovine schistosomiasis in Malawi: Connecting human and hybrid schistosomes within cattle. One Health. (2024). doi: 10.1016/j.onehlt.2024.100761
34
Kayuni SA Cunningham LJ Kumwenda D Lally D Chammudzi P Kapira D et al . Challenges in diagnosis and control of Female Genital Schistosomiasis (FGS) in sub-Saharan Africa: An exemplar case report associated with mixed and putative hybrid schistosome infection in Nsanje District. Front Trop Dis. (2024). doi: 10.3389/fitd.2024.1354119
Summary
Keywords
HRM, schistosomiasis hybrids, female genital schistosomiasis, qPCR, real-time PCR diagnostic, Malawi
Citation
Cunningham LJ, Kayuni S, Juhász A, Makaula P, Lally D, Namacha G, Kapira D, Chammudzi P, Mainga B, Jones S, Archer J, LaCourse EJ, Musaya J and Stothard JR (2024) A rapid DNA screening method using high-resolution melt analysis to detect putative Schistosoma haematobium and Schistosoma mattheei hybrids alongside other introgressing schistosomes. Front. Trop. Dis 5:1350680. doi: 10.3389/fitd.2024.1350680
Received
05 December 2023
Accepted
15 July 2024
Published
04 September 2024
Volume
5 - 2024
Edited by
Thiago Almeida Pereira, Stanford University, United States
Reviewed by
Hope Onohuean, Kampala International University, Uganda
Javier Sotillo, Carlos III Health Institute (ISCIII), Spain
Updates
Copyright
© 2024 Cunningham, Kayuni, Juhász, Makaula, Lally, Namacha, Kapira, Chammudzi, Mainga, Jones, Archer, LaCourse, Musaya and Stothard.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucas J. Cunningham, lucas.cunningham@lstmed.ac.uk
†These authors share senior authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.